Emergence of Neonatal Sepsis Caused by MCR-9- and NDM-1-Co-Producing Enterobacter hormaechei in China
- 1Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, the First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 2Jinan Microecological Biomedicine Shandong Laboratory, Jinan, China
- 3Department of Laboratory Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 4Department of Infectious Diseases, The First Affiliated Hospital, College of Clinical Medicine, Henan University of Science and Technology, Luoyang, China
- 5Sansure Biotech Inc. Medical Affairs Department, National Joint Local Engineering Research Center for Genetic Diagnosis of Infection Diseases and Tumors, Beijing, China
- 6Research Units of Infectious Diseases and Microecology, Chinese Academy of Medical Sciences, Beijing, China
Mobile colistin resistance (mcr) genes represent an emerging threat to public health. Reports on the prevalence, antimicrobial profiles, and clonality of MCR-9-producing Enterobacter cloacae complex (ECC) isolates on a national scale in China are limited. We screened 3,373 samples from humans, animals, and the environment and identified eleven MCR-9-positive ECC isolates. We further investigated their susceptibility, epidemiology, plasmid profiles, genetic features, and virulence potential. Ten strains were isolated from severe bloodstream infection cases, especially three of them were recovered from neonatal sepsis. Enterobacter hormaechei was the most predominant species among the MCR-9-producing ECC population. Moreover, the co-existence of MCR-9, CTX-M, and SHV-12 encoding genes in MCR-9-positive isolates was globally observed. Notably, mcr-9 was mainly carried by IncHI2 plasmids, and we found a novel ~187 kb IncFII plasmid harboring mcr-9, with low similarity with known plasmids. In summary, our study presented genomic insights into genetic characteristics of MCR-9-producing ECC isolates retrieved from human, animal, and environment samples with one health perspective. This study is the first to reveal NDM-1- and MCR-9-co-producing ECC from neonatal sepsis in China. Our data highlights the risk for the hidden spread of the mcr-9 colistin resistance gene.
Introduction
Enterobacter cloacae complex (ECC) are clinically significant Gram-negative, facultatively anaerobic, rod-shaped pathogens, which cause severe nosocomial infections, including bloodstream infections, pneumonia, urinary tract infections, endocarditis, and intra-abdominal infections (Mezzatesta et al., 2012; Davin-Regli and Pages, 2015). It is worthy to note that ECC has now become the third broad spectrum Enterobacteriaceae species involved in hospital-acquired infections after Escherichia coli and Klebsiella pneumoniae (Potron et al., 2013). Currently, E. cloacae is the most frequent cluster of ECC isolated from adult intensive care unit (ICU) patients (Fernandez et al., 2015; Stoesser et al., 2015; Ferry et al., 2020), even in neonatal patients (Yaffee et al., 2016; Rahal et al., 2021).
E. cloacae complex produced an AmpC β-lactamase, which has intrinsic resistance to many front-line treatment options, including penicillins, penicillin-penicillinase inhibitor combinations, first-generation cephalosporins, and cefoxitin (Davin-Regli and Pages, 2015). It exhibits a high frequency of enzymatic resistance to third-generation cephalosporins when AmpC β-lactamases become overproduced (Ellington et al., 2019). Colistin has been the last resort of CRE treatment for several decades, which ensures it serves as an essential global priority for use to manage infections resistant to carbapenems and other β-lactams. Previous work evidenced that colistin show effective in vitro activities against severe infections caused by carbapenem-resistant ECC (Yang et al., 2018).
Worryingly, since the first mobile colistin resistance gene (mcr-1), conferring transmissible colistin resistance, was detected in 2015, ten major classes of mcr genes have been detected so far (Wang et al., 2020; Xu et al., 2021). Despite these reports, most of mcr genes prevalence in ECC has remained relatively lower than that in E. coli and K. pneumoniae, except for mcr-9 (Macesic et al., 2021a). Since MCR-9 was identified in Salmonella enterica serotype Typhimurium in 2019 (Carroll et al., 2019), it was rapidly detected in several Enterobacteriaceae species worldwide (Osei Sekyere et al., 2020; Macesic et al., 2021a). Interestingly, few clinical isolates carrying the mcr-9 gene exhibit colistin resistance phenotype, which may be due to the low-level gene expression (Macesic et al., 2021a). Of note, increasing reports of E. cloacae isolates co-harboring MCR-9, carbapenemases, and extended-spectrum β-lactamase genes (ESBLs) are being reported worldwide due to the dissemination of self-transmissible IncHI2 plasmids (Haenni et al., 2020; Umeda et al., 2021). A recent study identified an mcr-9/blaNDM-1-co-harboring plasmid of E. cloacae from a lethal adult bloodstream infection (BSI) case (Lin et al., 2020). Severe invasive infections caused by such isolates definitely limit treatment options for clinicians and threaten public health. However, MCR-9-producing ECC infections in neonatal patients have never been reported.
In this study, we did a retrospective study to screen and examine the virulence profile and genetic context of the mcr-9-carrying ECC isolates from human, animal, and environmental samples in China from 2010 to 2020. The epidemiology, genetic features, antimicrobial profiles, virulence potential, and global phylogenomic analysis of the isolates were further investigated. In general, we describe for the first time the emergence of the mcr-9 colistin resistance gene in neonatal sepsis in China.
Materials and Methods
Study Design, Setting, and Bacterial Isolates
In this study, we screened MCR-9-producing ECC isolates recovered from human stool samples collected between 2010 and 2020, animal stool samples collected between 2016 and 2020, and bloodstream infection isolates from the Consortium of Blood Bacterial Resistant Investigation Collaborative Systems (BRICS) (Zheng et al., 2017b; Zheng et al., 2019a) collected between 2014 and 2021. ECC intestinal colonization isolates were collected from inpatients submitting specimens to the First Affiliated Hospital of Zhejiang University (Hangzhou, Zhejiang Province) and the First Affiliated Hospital of Zhengzhou University (Zhengzhou, Henan Province). Animal stool samples were retrieved from our previous studies (Zheng et al., 2017a; Feng et al., 2019; Zheng et al., 2019b; Zheng et al., 2019c). As a routine screening for ECC strains, the collected stool samples were plated on MacConkey agar (OXOID, Hampshire, UK) for 18-24h at 37°C. Preliminary bacterial identification was conducted by using MALDI-TOF MS (Bruker Daltonik GmbH, Bremen, Germany), and the identification of ECC species was confirmed by whole-genome sequencing-based genomic analysis. The mcr-9 gene was detected by using PCR and sequencing (Macesic et al., 2021a). Clinical data were extracted from medical record systems. Ethical approval was granted by the Ethics Committee of the First Affiliated Hospital of Zhejiang University. Individual consent was obtained for all patients.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of different antimicrobial agents were determined by the microdilution method in Mueller-Hinton broth for all isolates. The tested antibiotics were aminoglycosides (gentamicin and amikacin), β-lactams (amoxicillin/clavulanic acid, aztreonam, cefotaxime, ceftazidime, cefpirome, imipenem, meropenem, and piperacillin/tazobactam), fluoroquinolones (levofloxacin and ciprofloxacin), tetracyclines (tetracycline and tigecycline), colistin, chloramphenicol, florfenicol, fosfomycin, and trimethoprim/sulfamethoxazole. AST results were interpreted according to the standards of the Clinical Laboratory Standards Institute (Clinical and Laboratory Standards Institute (CLSI), 2021). Breakpoints of colistin and tigecycline were interpreted following guidelines of the European Committee on Antimicrobial Susceptibility Testing (The European Committee on Antimicrobial Susceptibility Testing, 2020) (http://www.eucast.org/).
Whole-Genome Sequencing
Total genomic DNA of ECC isolates was extracted using Gentra Puregene Yeast/Bact. Kit (Qiagen, Germany) according to the manufacturer’s protocol. Purified DNA was subjected to WGS on Illumina Novaseq (Illumina, Inc., CA, USA) system with the 150-bp paired-end approach and 150 × coverage. To further characterize the genetic environment of the mcr-9 genes, we performed long-read sequencing using Nanopore sequencing technology for five isolates according to the different sizes of mcr-9-harboring plasmids. MinION (Oxford Nanopore Technologies, Oxford, UK) sequencing was prepared for isolates 51118, 52744, 53287, 60403, and 61363. DNA Libraries were constructed by using Ligation Sequencing Kits 1D (SQK-LSK109) to generate long contigs.
In Silico MLST Analysis and Identification of Antimicrobial Resistance Genes and Plasmids
Raw reads were assembled into a number of scaffolds using Velvet 1.1 (Zerbino and Birney, 2008). Annotation was carried out by using Prokka (Seemann, 2014). Bacterial identification was conducted by average nucleotide identity (ANI) analysis with JSpeciesWS (Richter et al., 2016). Multi-locus sequence typing (MLST) analysis was performed as described previously (Cerqueira et al., 2017). The detection of antimicrobial resistance genes (ARGs) were identified using the ResFinder 2.1 database (Zankari et al., 2012) based on the identities (%). Plasmid replicons were identified using PlasmidFinder 2.0 (Carattoli and Hasman, 2020). Average nucleotide identity blast (ANIb) analysis was completed using pyani with default settings (https://github.com/widdowquinn/pyani). ANIb data for each strain were visualized by heatmaps.
Phylogenetic Analysis
To further characterize the phylogenetic structure of MCR-9-producing ECC isolates, our collection of MCR-9-carrying isolates was supplemented with strains from the global collection. Genomes of 70 MCR-9-carrying ECC isolates were downloaded from the National Center for Biotechnology Information (NCBI) for comparative genomic analysis (Table S1). The phylogenies of the MCR-9-producing strains were performed by ROARY software based on the core genome SNPs (Sitto and Battistuzzi, 2020). Core genes were defined as described previously (Holt et al., 2015). Phylogenetic tree visualizations were produced using the Interactive Tree of Life (https://itol.embl.de/).
Conjugation Assay
Conjugation experiments of all MCR-9-positive isolates were carried out using broth-based methods with E. coli J53 (azide-resistant) as the recipient strain. Transconjugants were selected on MH agar plates containing 150 μg/mL sodium azide plus 2 μg/mL colistin. The presence of mcr-9 in transconjugants was confirmed by PCR and Sanger sequencing.
Plasmid Analysis
Plasmid characterization was carried out by nuclease digestion pulsed-field gel electrophoresis (S1-PFGE), and Southern blot to estimate the size of the mcr-9-carrying plasmids (Zheng et al., 2016). The sequence of the target plasmids was assembled using plasmidSPAdes (Antipov et al., 2016). The sequences of representative plasmids were compared against other NCBI accessioned plasmid sequences using BLAST and plotted by BLAST Ring Image Generator (BRIG) (http://brig.sourceforge.net/). To identify targeted plasmids, a BLAST similarity search against the GenBank NT database was performed for each scaffold. Plasmid sequences were defined when > 70% of the scaffold length matched the plasmid sequences and < 30% compared either the mobile element or chromosomal sequences based on MUMMER alignment (Delcher et al., 2003). The best BLAST hit to each scaffold was manually inspected with the PLSDB database, a resource of complete bacterial plasmids (Galata et al., 2018).
Serum Killing Assay and In Vivo Galleria mellonella Infection Model
The virulence of MCR-9-producing ECC strains was tested in the serum killing assay and G. mellonella model of infection. Serum killing assay was performed as described previously (Shen et al., 2020). Briefly, fresh nonheated human serum (NHS) from different healthy individuals and frozen the samples at -80°C. The serum bactericidal effect was presented as survival curves and rates, using the following formula: Bacterial survival rate = (number of colonies with normal serum/number of colonies with inactivated serum) × 100%. Each experiment was performed in triplicate.
For G. mellonella in vivo infection assay, every 10 G. mellonella weighing about 250 mg were randomly divided into groups, and each G. mellonella was injected with 20ul 1 × 107 CFU/mL concentration of bacteria. In addition, the negative control group was injected with only 20ul PBS. All G. mellonella were cultured at 37°C, and the status of G. mellonella was observed every 12h for seven consecutive days (Niu et al., 2020). E. coli ATCC 25922 was used as the quality control strain.
Statistical Analysis
The R studio software was used for statistical analysis. A P-value of 0.05 was used as the significance level; P<0.05 indicated statistically significant differences.
Results
Bacterial Isolation and Identification
In general, we screened 1,782 stool samples from the First Affiliated Hospital of Zhengzhou University, 750 from the First Affiliated Hospital of Zhejiang University, 200 stool samples from cow, 303 from pig, 168 from fennec fox, and 170 from environment samples (Figure 1). Of the 3,373 samples from humans, animals, and the environment, a total of 136 ECC isolates were obtained in this study. Of 136 isolates, 56 recovered from stool sample, 71 from animals, and 9 from environment. Combined with 638 strains collected from the BRICS project, 774 ECC isolates were screened for MCR-9. As a result, eleven MCR-9 producing ECC strains (1.42%, 11/774) from 2010 to 2017 were identified in this work. Ten MCR-9-postive isolates were recovered from blood sample and one from stool sample.
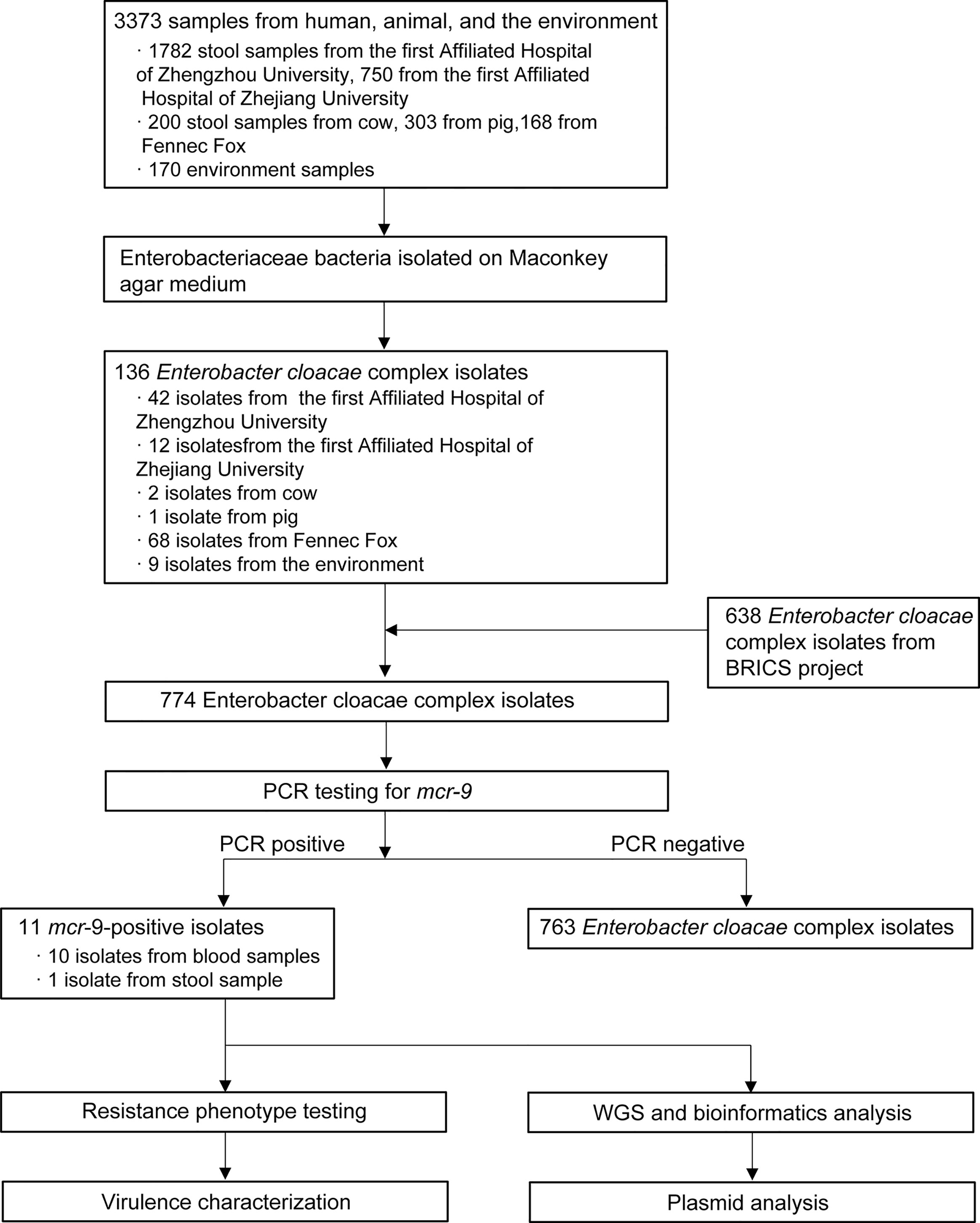
Figure 1 Schematic procedure of the study. BRICS, Blood Bacterial Resistant Investigation Collaborative Systems; WGS, whole-genome sequencing.
Emergence of MCR-9-Producing ECC in Neonatal Sepsis
Characteristics of the 11 ECC-associated cases are summarized in Figure 2. Of these cases, patients came from nine cities located in seven provinces (Figure 2A). It is worthy to note that three isolates were recovered from neonatal patients, one from child and seven from adults (Figure 2B). The isolates originated from patients with primary bloodstream infections (n = 10) and colonization (n = 1). The eleven isolates were all identified by MALDI-TOF MS as E. cloacae. The ANIb analysis revealed that nine isolates were Enterobacter hormaechei, and the other two were Enterobacter kobei (52744 and 58918) (Figure S1). Of note, two E. hormaechei isolates and one E. kobei were identified from neonatal sepsis cases.
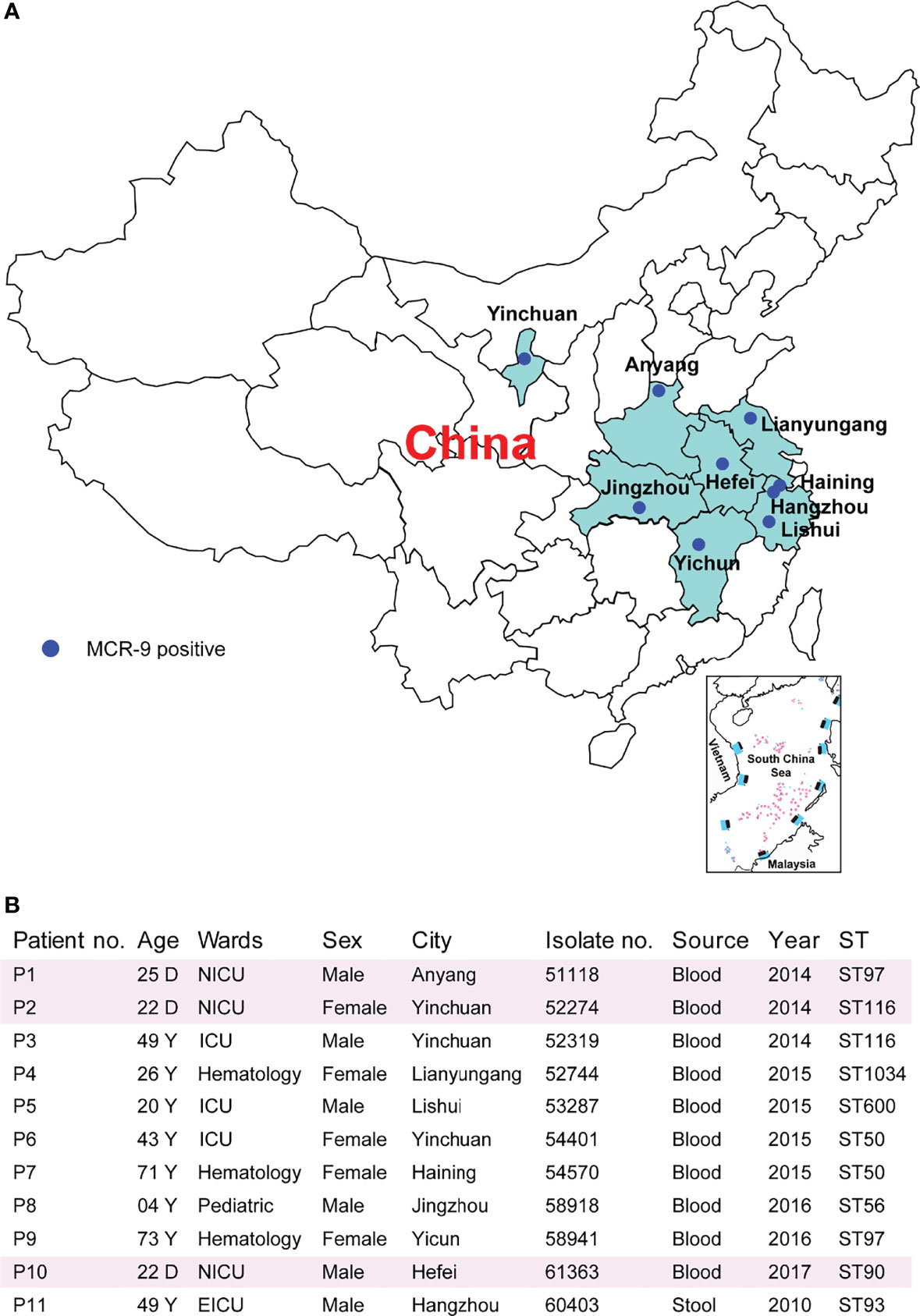
Figure 2 Characteristics of MCR-9-producing ECC-associated cases. (A) Study sites and geographical distribution of MCR-9-producing ECC isolates. Seven provinces shown in color were included in the surveillance for MCR-9. (B) Clinical characteristics and epidemiological characteristics of MCR-9-producing ECC-associated isolates.
Phenotypic and Molecular Characteristics of Eleven Isolates
Resistance determinants, plasmid replicons and STs are outlined in Table S1. The isolates were found to belong to ST50 (n = 2), ST97 (n = 2), ST116 (n = 2), ST56 (n = 1), ST90 (n = 1), ST93 (n = 1), ST600 (n = 1), and ST1034 (n = 1) (Figure 2B). Isolates were resistant to multiple antimicrobial classes, including amoxicillin/clavulanic acid (100%), cefotaxime (100%), ceftazidime (100%), and cefpirome (91%), but remained susceptible to amikacin (91%), tigecycline (91%) and fosfomycin (82%) (Table S2). Despite the presence of mcr-9, nine E. hormaechei isolates were colistin susceptible (MICs ≤ 2 mg/). In contrast, two E. kobei exhibit high level colistin resistance (MICs ≥ 64 mg/)
E. hormaechei Is Predominant in the MCR-9-Producing ECC Population
Core-genome phylogenetic analysis classified the 81 MCR-9-positive isolates into four clades (Figure 3). The 55 (57.9%) MCR-9-producing E. hormaechei strains constitute the largest clade, followed by E. cloaece (n = 12). Of note, two ST116 E. hormaechei isolates (52274 and 52319) recovered from the same hospital were phylogenetically closely related, indicating clonal dissemination in the region. Surprisingly, seven ST93 E. hormaechei isolates were identified from China, the USA, France, and Spain, suggesting global transmission.
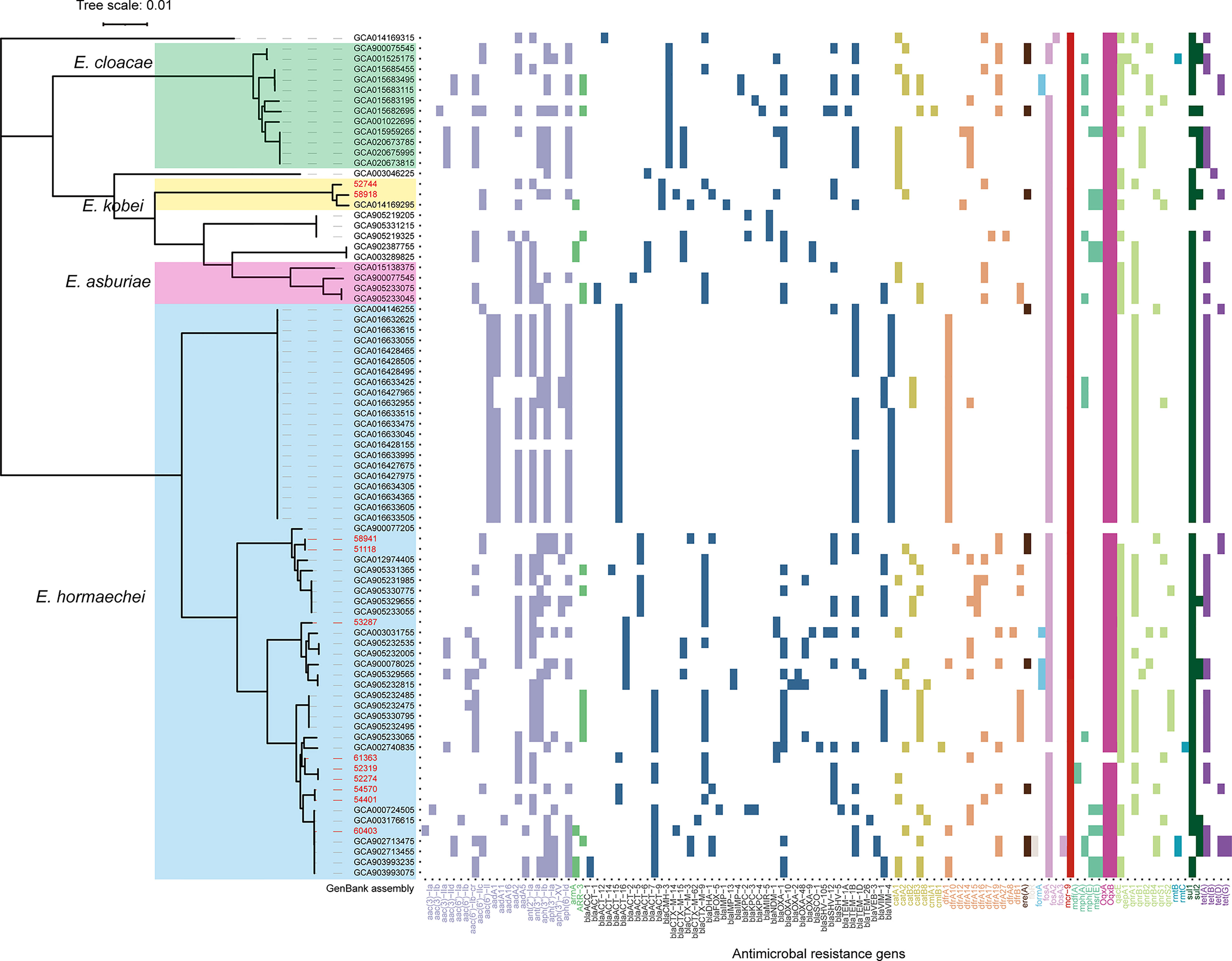
Figure 3 Core-genome phylogeny and resistome of 70 MCR-9-positive ECC isolates from NCBI genome database and the eleven isolates from this work. E. hormaechei, E. cloacae, E. kobei, and E. arburiae isolates are indicated in blue, green, yellow, and pink. Scale bars represent the number of substitutions per site. The ARGs distribution of MCR-9-positive ECC isolates is presented on the right. The ARGs groups are indicated with different colors. Isolates identified in this study are shown in red.
Co-Existence of MCR-9, CTX-M, and SHV-12 Encoding Genes Globally
ESBL genes were detected in all isolates identified in this work (Figure 3). Nine of the eleven isolates were positive for blaCTX-M, with blaCTX-M-9 the most prevalent (n = 6), followed by blaCTX-M-14 (n = 2) and blaCTX-M-3 (n = 1). Three isolates (58918, 53287, and 61363) were found to carry blaNDM-1 gene. WGS determined that blaNDM-1 was localized on a ~45 kb IncX3 plasmid and a ~60 kb IncN plasmid in isolates 53287 and 61363, respectively (Figure S2). The blaSHV-12 gene was detected in eight of the eleven ESBL-positive isolates. AmpC β-lactamase gene, blaATC, was detected in all ESBL-positive isolates. In silico analysis found that CTX-M-9/SHV-12/MCR-9 positive ECC isolates are prevalent in a global collection, which indicated that these three genes probably co-localized on a plasmid.
MCR-9 Was Mainly Carried by IncHI2 Plasmids
S1-PFGE and Southern blot revealed that mcr-9 was carried by plasmids in ten isolates, except isolate 58918, which is chromosome encoded (Figure 4A). We further identified ten contigs carrying mcr-9 gene from genome sequences. All were typed as IncHI2 plasmid ST1, except for p53287-MCR-9, which was classified as IncFII (Table S1). IncHI2 plasmid was ~244-330 kb in size and carried many mercury, tellurium, copper, and antimicrobial resistance genes (Figures 4, 5). These ARGs included resistance determinants for aminoglycosides, β-lactams, fluoroquinolones, macrolides, sulphonamides, tetracyclines, and trimethoprim. Conjugation experiments confirmed eight MCR-9-carrying plasmids were self-transmissible at 37 or 26°C (Table S1). In contrast, transconjugants containing p53287-MCR-9 and p60403-MCR-9 were not obtained, despite the filter-mating experiments were repeated at 25 to 37°C, suggesting that these plasmids could not be transferred. Examining the complete sequence of p53287-MCR-9 revealed that there were incomplete conjugative transfer loci on this plasmid (Figure 5A).
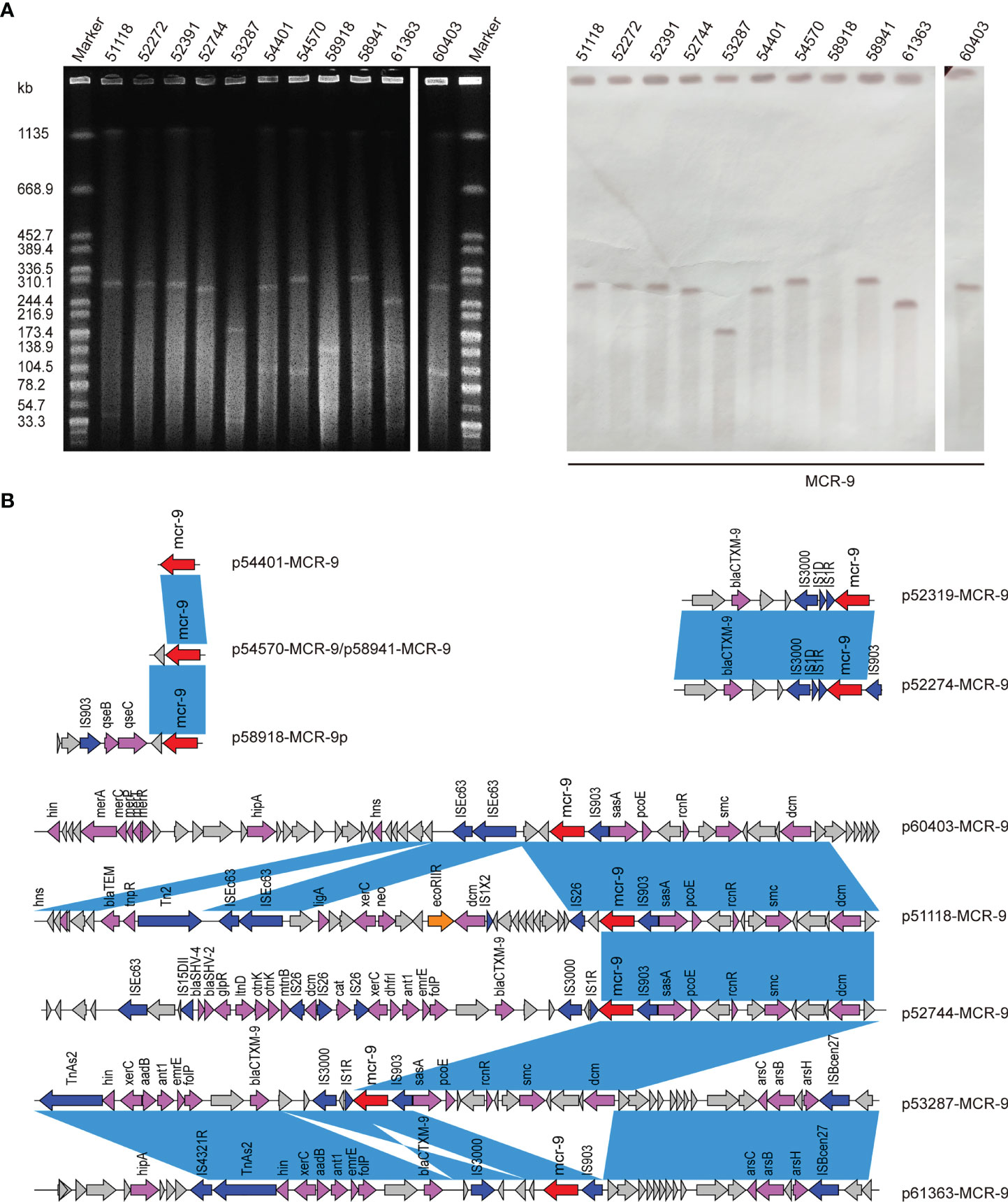
Figure 4 (A) S1-PFGE and Southern blot of eleven MCR-positive isolates. S1-PFGE patterns of Salmonella isolates and their relevant conjugants. Southern blot-hybridization of S1-nuclease digested DNA using a specific probe (mcr-9). M: XbaI digested total DNA of Salmonella enterica serotype Braenderup H9812 as a size marker. (B) Genetic contexts of mcr-9 genes in eleven ECC isolates. Blue rectangles highlight identical regions.
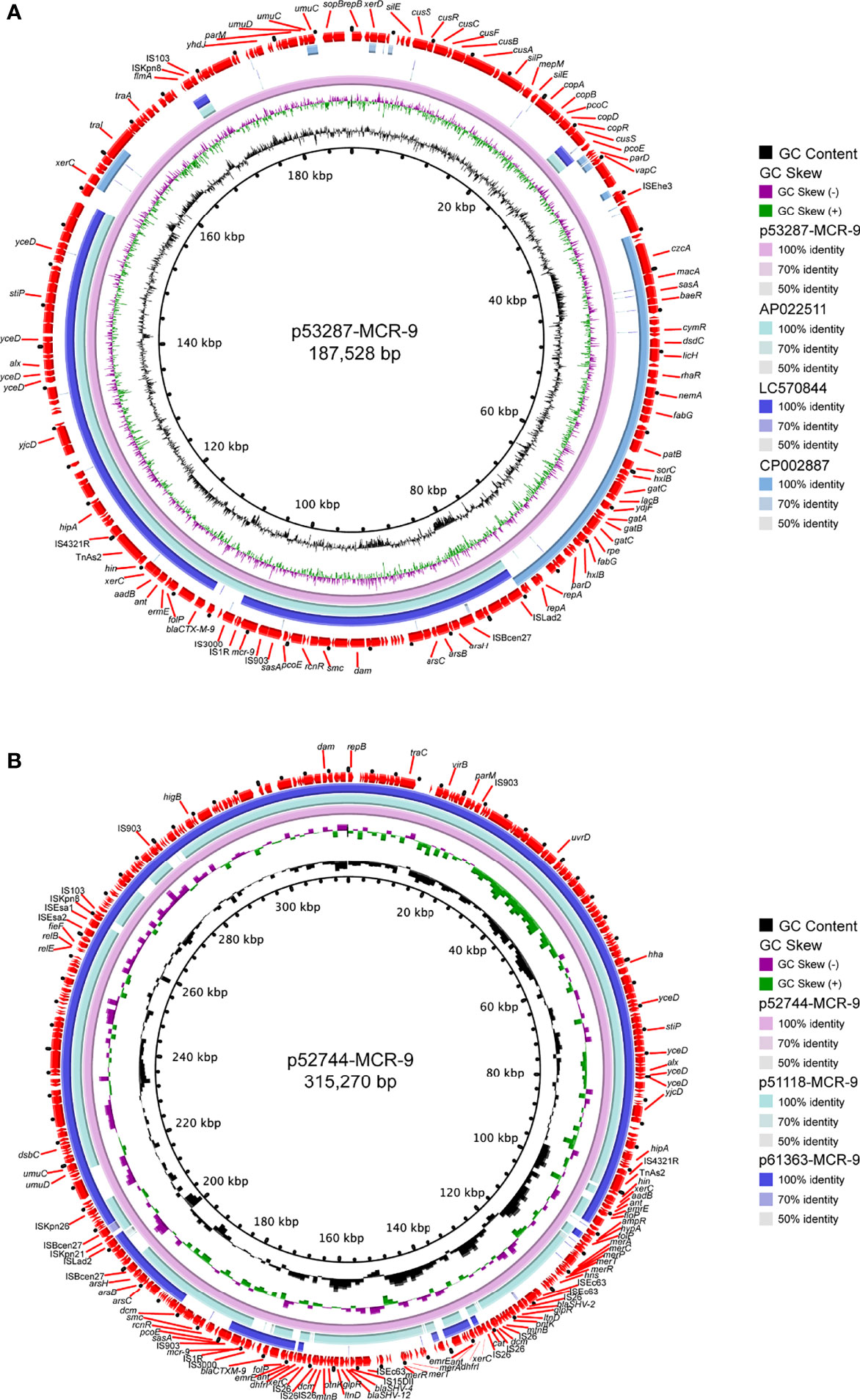
Figure 5 Circular representation of the studied mcr-9-carrying plasmids. (A) Analysis of the IncFII plasmid p53287-MCR-9 carried by the bacteremia associated E hormaechei strain 53287. (B) Sequence comparison of the mcr-9-encoding IncHI2 plasmids p57244-MCR-9, p51118-MCR-9, and p61363-MCR-9. GC content and GC Skew were represented on the inner map’s distance scale (kb). Each plasmid was compared to its most closely-related plasmid. The red arc around the map indicated ORFs. Certain important genes were also indicated on the ring.
Genetic Environment of MCR-9 Genes
The genetic environment of mcr-9 regions and plasmid backbone differed in our isolate collection, suggesting a diversity of MCR-9-carrying plasmids (Figure 4B). In p58918-MCR-9, two genes proposed to regulate the expression of colistin resistance, qseC and qseB, are localized downstream of mcr-9.1 and upstream of IS903. p52319-MCR-9 and p52274-MCR-9 shared a similar genetic context of mcr-9. Of note, the conserved region in mcr-9-harboring plasmids encoded the nickel-cobalt efflux transporters (rcnR/rcnS) and sensory protein kinases (pcoS/pcoE) (Figure 4B). However, the downstream region of mcr-9 was highly varied between plasmids (Figure 5B).
Virulence of MCR9-Producing ECC Isolates
Considering the emergence of MCR-9-producing ECC in neonatal sepsis cases, we further assessed the virulence profiles of these isolates. The serum survivability was >95% for three isolates and ranged from 65 to 83% for seven isolates (Figure 6A). In contrast, isolate 60403 showed low serum survivability. The G. mellonella infection assay revealed that seven isolates observed high survivability (> 84%) infection, while the other five isolates caused intermediate survivability (53–73%) (Figure 6B).
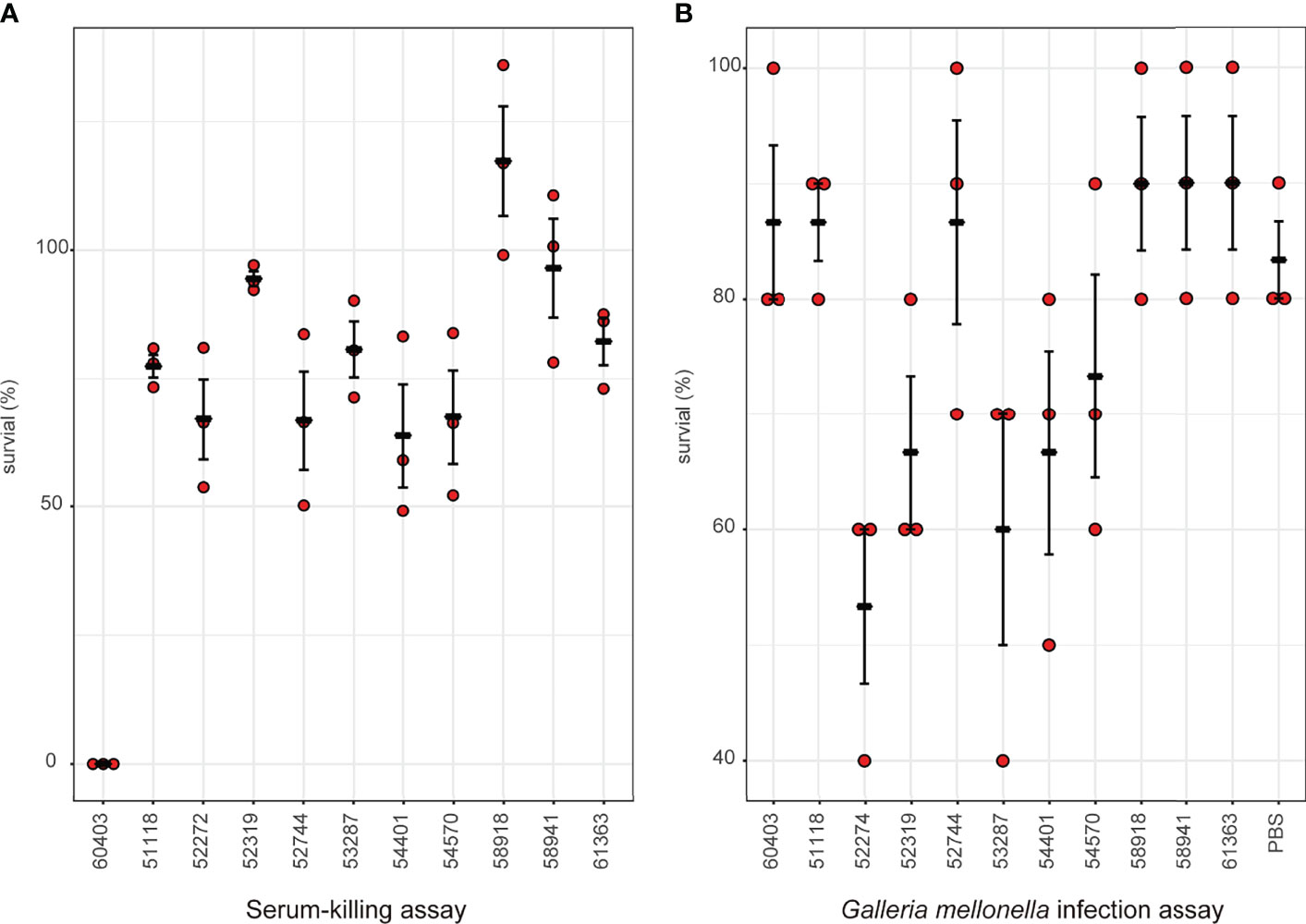
Figure 6 Virulence profiles of MCR-9-positive ECC isolates. (A) Serum-killing assay. Survival in a serum-killing assay of ECC strains. The survival is denoted in percentage. The bars denote means and standard errors of the mean. (B) Galleria mellonella infection assay. Survival at 144 h in a G mellonella assay of ECC strains. The survival is denoted in percentage. The bars denote means and standard errors of the mean.
Discussion
Thus far, the ECC species remain largely susceptible to several classes of antimicrobial agents used in China (Liu et al., 2015; Yang et al., 2018; Hu et al., 2020). However, the rapid increase of nosocomial infections caused by carbapenem-resistant ECC has raised growing worrisome worldwide, which result in the reuse of colistin to treat bacterial infections (Wang et al., 2018). Plasmid-mediated mcr colistin resistance genes can be transferred across Enterobacteriaceae species and result in global dissemination, representing a significant threat to public health (Zheng et al., 2018a; Zheng et al., 2018b). The putative colistin resistance gene mcr-9 was first identified by WGS to confer phenotypic resistance to colistin in Enterobacteriaceae (Carroll et al., 2019). Screening of the mcr-9 gene in isolates received from animals, the environment, and humans is necessary to understand its global or national spread. Before the current study, the prevalence, antimicrobial profiles, and clonality of MCR-9-producing ECC on a national scale in China were unclear. We screened our biobank over 3,000 various samples and identified mcr-9 in eleven isolates. Our study reveals widespread this newly identified colistin resistance gene, mcr-9, among bloodstream infection isolates.
We report here a relatively low prevalence of MCR-9 in ECC isolates, with a positivity rate of 1.42% (11/774), although it was higher than the 0.18% (60/33,205) rate in Salmonella enterica reported by Yanan Wang et al. (Wang et al., 2021). Since it was first detected in the USA (Carroll et al., 2019), MCR-9 has been found in five continents from various human, animal, environmental, and food samples (Bitar et al., 2020; Kamathewatta et al., 2020; Osei Sekyere et al., 2020; Elbediwi et al., 2021; Ha et al., 2021; Khodor et al., 2021). Our findings highlight the unexpected spread of MCR-9 and emphasize the need for close surveillance of antimicrobial resistance in bloodstream infection isolates.
Remarkably, among our mcr-9-positive isolates, all of them encoded aminoglycoside resistance, fosfomycin resistance, sulfonamide resistance, and β-lactamase genes, raising the concern that the spread of mcr-9 might also be related to the use of these antimicrobial agents in China. In this study, blaCTX-M-9 was co-existed with mcr-9 in the same plasmid. Interestingly, blaCTX-M-9 was previously found as the most prevalent ESBL gene in nosocomially acquired ECC in China (Zhou et al., 2018). The discovery of blaCTX-M-9- and mcr-9- co-harboring plasmid is of grave concern, as an epidemic could have potentially serious consequences.
Carbapenem-resistant Enterobacteriaceae (CRE) are important pathogens causing serious community-acquired and nosocomial infections globally (Zheng et al., 2019a; Zheng et al., 2020). Previously, MCR-9 has been identified in several CRE backgrounds since it was described in 2019 (Macesic et al., 2021b). Additionally, co-production of NDM-1 and MCR-9 in ECC isolates was mainly identified from China (Yuan et al., 2019; Faccone et al., 2020; Khalifa et al., 2020; Lin et al., 2020; Ai et al., 2021; Ding et al., 2021; Liu et al., 2021; Sun et al., 2021), these studies indicated that the extensive use of carbapenems in China might drive the occurrence of these multidrug-resistant ECC isolates. It is worthy to note that we first detected the NDM-1- and MCR-9-co-producing ECC isolates cause neonatal sepsis. Our data reveal the likelihood of wide dissemination of NDM-1 and MCR-9 in pediatric or neonatal intensive care units (PICU/NICU). This finding highlights the unexpected spread of NDM-1and MCR-9 and emphasizes the urgent need for effective surveillance of antimicrobial resistance in PICU/NICU. This linkage is a matter of concern since it could herald the possibility of a co-spread of the two genes, both involved in resistance to last-resort drugs (Lin et al., 2020).
IncHI2 plasmids were found to be the predominant replicon type carrying mcr-9, and the conserved rcnR-rcnA-pcoE-pcoS-IS903-mcr-9-wbuC structure exists in most mcr-9 cassettes (Macesic et al., 2021b). It was widely detected in various Enterobacteriaceae species (Bitar et al., 2020; Borjesson et al., 2020; Campos-Madueno et al., 2021; Liu et al., 2021; Simoni et al., 2021; Umeda et al., 2021). The observation in this work is consistent with previous investigations, and nine mcr-9 genes were carried by IncHI2 plasmid. Moreover, we identified a novel mcr-9-harboring IncFII plasmid in this work, which shared a relatively low similarity with any known plasmids. The spread of this plasmid in clinical settings should be closely monitored.
Conclusion
This is the first investigation aimed to screen the MCR-9-producing ECC isolates from clinical, healthy human, animal, and environment samples with one health perspective. This work also first detected the MCR-9-producing ECC from neonatal sepsis in China. We further reported the complete sequence of plasmids harboring the mcr-9 and blaNDM-1 genes. The mcr-9 gene was mainly located on IncHI2 plasmids. Our data highlights the risk for the hidden spread of the mcr-9 colistin resistance gene.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
Ethical approval was granted by the Ethics Committee of the First Affiliated Hospital of Zhejiang University. Individual consent was obtained for all patients.
Author Contributions
BZ and YX designed the study. BZ wrote the original manuscript. CC and HX collected the data and performed the data analyses. CC provided medication guidance. CC and RL involved in patient management and provided clinical information. HX, RL, XH, LW and HF performed laboratory tests. JH revised and supervised the paper. All authors contributed to the article and approved the final manuscript.
Funding
We gratefully acknowledge the financial support of the National Natural Science Foundation of China (82072314 and 81971984), the Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022006B and JNL-2022011B), and CAMS Innovation Fund for Medical Sciences (2019-I2M-5-045).
Conflict of Interest
Author JH was employed by Sansure Biotech Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.879409/full#supplementary-material
Supplementary Figure 1 | ANIb analysis of MCR-9-producing ECC isolates.
Supplementary Figure 2 | Circular representation of the studied blaNDM-1-carrying plasmids. (A) Sequence comparison of the IncX3 plasmid p53287-NDM-1carried by the bacteremia associated with E. hormaechei strain 53287. (B) Sequence comparison of the blaNDM-1-carrying IncN plasmid p61363-NDM-1. GC content and GC Skew were represented on the inner map’s distance scale (kb). Each plasmid was compared to its most closely-related plasmid. The red arc around the map indicated ORFs. Certain important genes were also indicated on the ring.
References
Ai, W., Zhou, Y., Wang, B., Zhan, Q., Hu, L., Xu, Y., et al. (2021). First Report of Coexistence of Bla SFO-1 and Bla NDM-1 Beta-Lactamase Genes as Well as Colistin Resistance Gene Mcr-9 in a Transferrable Plasmid of a Clinical Isolate of Enterobacter Hormaechei. Front. Microbiol. 12, 676113. doi: 10.3389/fmicb.2021.676113
Antipov, D., Hartwick, N., Shen, M., Raiko, M., Lapidus, A., Pevzner, P. A. (2016). Plasmidspades: Assembling Plasmids From Whole Genome Sequencing Data. Bioinformatics 32, 3380–3387. doi: 10.1093/bioinformatics/btw493
Bitar, I., Papagiannitsis, C. C., Kraftova, L., Chudejova, K., Mattioni Marchetti, V., Hrabak, J. (2020). Detection of Five Mcr-9-Carrying Enterobacterales Isolates in Four Czech Hospitals. mSphere 5, e01008–20. doi: 10.1128/mSphere.01008-20
Borjesson, S., Greko, C., Myrenas, M., Landen, A., Nilsson, O., Pedersen, K. (2020). A Link Between the Newly Described Colistin Resistance Gene Mcr-9 and Clinical Enterobacteriaceae Isolates Carrying blaSHV-12 From Horses in Sweden. J. Glob. Antimicrob. Resist. 20, 285–289. doi: 10.1016/j.jgar.2019.08.007
Campos-Madueno, E. I., Moser, A. I., Risch, M., Bodmer, T., Endimiani, A. (2021). Exploring the Global Spread of Klebsiella Grimontii Isolates Possessing blaVIM-1 and Mcr-9. Antimicrob. Agents Chemother. 65, e0072421. doi: 10.1128/AAC.00724-21
Carattoli, A., Hasman, H. (2020). PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol. Biol. 2075, 285–294. doi: 10.1007/978-1-4939-9877-7_20
Carroll, L. M., Gaballa, A., Guldimann, C., Sullivan, G., Henderson, L. O., Wiedmann, M. (2019). Identification of Novel Mobilized Colistin Resistance Gene Mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella Enterica Serotype Typhimurium Isolate. mBio 10, e00853–19. doi: 10.1128/mBio.00853-19
Cerqueira, G. C., Earl, A. M., Ernst, C. M., Grad, Y. H., Dekker, J. P., Feldgarden, M., et al. (2017). Multi-Institute Analysis of Carbapenem Resistance Reveals Remarkable Diversity, Unexplained Mechanisms, and Limited Clonal Outbreaks. Proc. Natl. Acad. Sci. U.S.A. 114, 1135–1140. doi: 10.1073/pnas.1616248114
Clinical and Laboratory Standards Institute (CLSI). (2021). Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. CLSI supplement M100. USA: Clinical and Laboratory Standards Institute.
Davin-Regli, A., Pages, J. M. (2015). Enterobacter Aerogenes and Enterobacter Cloacae; Versatile Bacterial Pathogens Confronting Antibiotic Treatment. Front. Microbiol. 6, 392. doi: 10.3389/fmicb.2015.00392
Delcher, A. L., Salzberg, S. L., Phillippy, A. M. (2003). Using MUMmer to Identify Similar Regions in Large Sequence Sets. Curr. Protoc. Bioinf. 10, 10 13. doi: 10.1002/0471250953.bi1003s00
Ding, M., Shi, J., Ud Din, A., Liu, Y., Zhang, F., Yan, X., et al. (2021). Co-Infections of Two Carbapenemase-Producing Enterobacter Hormaechei Clinical Strains Isolated From the Same Diabetes Individual in China. J. Med. Microbiol. 70. doi: 10.1099/jmm.0.001316
Elbediwi, M., Pan, H., Zhou, X., Rankin, S. C., Schifferli, D. M., Yue, M. (2021). Detection of Mcr-9-Harbouring ESBL-Producing Salmonella Newport Isolated From an Outbreak in a Large-Animal Teaching Hospital in the USA. J. Antimicrob. Chemother. 76, 1107–1109. doi: 10.1093/jac/dkaa544
Ellington, M. J., Heinz, E., Wailan, A. M., Dorman, M. J., De Goffau, M., Cain, A. K., et al. (2019). Contrasting Patterns of Longitudinal Population Dynamics and Antimicrobial Resistance Mechanisms in Two Priority Bacterial Pathogens Over 7 Years in a Single Center. Genome Biol. 20, 184. doi: 10.1186/s13059-019-1785-1
Faccone, D., Martino, F., Albornoz, E., Gomez, S., Corso, A., Petroni, A. (2020). Plasmid Carrying Mcr-9 From an Extensively Drug-Resistant NDM-1-Producing Klebsiella Quasipneumoniae Subsp. Quasipneumoniae Clinical Isolate. Infect. Genet. Evol. 81, 104273. doi: 10.1016/j.meegid.2020.104273
Feng, C., Wen, P., Xu, H., Chi, X., Li, S., Yu, X., et al. (2019). Emergence and Comparative Genomics Analysis of Extended-Spectrum-Beta-Lactamase-Producing Escherichia Coli Carrying Mcr-1 in Fennec Fox Imported From Sudan to China. mSphere 4, e00732–19. doi: 10.1128/mSphere.00732-19
Fernandez, J., Montero, I., Martinez, O., Fleites, A., Poirel, L., Nordmann, P., et al. (2015). Dissemination of Multiresistant Enterobacter Cloacae Isolates Producing OXA-48 and CTX-M-15 in a Spanish Hospital. Int. J. Antimicrob. Agents 46, 469–474. doi: 10.1016/j.ijantimicag.2015.07.003
Ferry, A., Plaisant, F., Ginevra, C., Dumont, Y., Grando, J., Claris, O., et al. (2020). Enterobacter Cloacae Colonisation and Infection in a Neonatal Intensive Care Unit: Retrospective Investigation of Preventive Measures Implemented After a Multiclonal Outbreak. BMC Infect. Dis. 20, 682. doi: 10.1186/s12879-020-05406-8
Galata, V., Fehlmann, T., Backes, C., Keller, A. (2018). PLSDB: A Resource of Complete Bacterial Plasmids. Nucleic Acids Res. 47, D195–D202. doi: 10.1093/nar/gky1050
Haenni, M., Metayer, V., Jarry, R., Drapeau, A., Puech, M. P., Madec, J. Y., et al. (2020). Wide Spread of Bla CTX-M-9/Mcr-9 IncHI2/ST1 Plasmids and CTX-M-9-Producing Escherichia Coli and Enterobacter Cloacae in Rescued Wild Animals. Front. Microbiol. 11, 601317. doi: 10.3389/fmicb.2020.601317
Ha, V. T. T., Tran, L. D., Mai, N. T. T., Hirabayashi, A., Nguyen, S. T., Tran, H. H., et al. (2021). Potential Spread of Mcr-9-Carrying IncHI2 Plasmids in Enterobacter Hormaechei in Vietnam. J. Glob. Antimicrob. Resist. 27, 332–334. doi: 10.1016/j.jgar.2021.09.012
Holt, K. E., Wertheim, H., Zadoks, R. N., Baker, S., Whitehouse, C. A., Dance, D., et al. (2015). Genomic Analysis of Diversity, Population Structure, Virulence, and Antimicrobial Resistance in Klebsiella Pneumoniae, an Urgent Threat to Public Health. Proc. Natl. Acad. Sci. U. S. A. 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Hu, K., Zeng, L., Zhang, J., Li, H., Su, S., Zhao, Y., et al. (2020). Antibiotic Susceptibility and Molecular Analyses of Clinical Enterobacter Cloacae Isolates in Eastern Heilongjiang Province, China. Ann. Palliat Med. 9, 1211–1219. doi: 10.21037/apm-20-1089
Kamathewatta, K., Bushell, R., Rafa, F., Browning, G., Billman-Jacobe, H., Marenda, M. (2020). Colonization of a Hand Washing Sink in a Veterinary Hospital by an Enterobacter Hormaechei Strain Carrying Multiple Resistances to High Importance Antimicrobials. Antimicrob. Resist. Infect. Control 9, 163. doi: 10.1186/s13756-020-00828-0
Khalifa, H. O., Soliman, A. M., Saito, T., Kayama, S., Yu, L., Hisatsune, J., et al. (2020). First Report of Foodborne Klebsiella Pneumoniae Coharboring Bla VIM-1, Bla NDM-1, and Mcr-9. Antimicrob. Agents Chemother. 64, e00882–20. doi: 10.1128/AAC.00882-20
Khodor, R., Salloum, T., El Jisr, T., El Chaar, M., Tokajian, S. (2021). Detection and Genomic Characterization of Mcr-9 in Enterobacter Hormaechei Recovered From a Pediatric Patient in Lebanon. Infect. Genet. Evol. 94, 105014. doi: 10.1016/j.meegid.2021.105014
Lin, M., Yang, Y., Yang, Y., Chen, G., He, R., Wu, Y., et al. (2020). Co-Occurrence of Mcr-9 and Bla NDM-1 in Enterobacter Cloacae Isolated From a Patient With Bloodstream Infection. Infect. Drug Resist. 13, 1397–1402. doi: 10.2147/IDR.S248342
Liu, Z., Hang, X., Xiao, X., Chu, W., Li, X., Liu, Y., et al. (2021). Co-Occurrence of Bla NDM-1 and Mcr-9 in a Conjugative IncHI2/HI2A Plasmid From a Bloodstream Infection-Causing Carbapenem-Resistant Klebsiella Pneumoniae. Front. Microbiol. 12, 756201. doi: 10.3389/fmicb.2021.756201
Liu, C., Qin, S., Xu, H., Xu, L., Zhao, D., Liu, X., et al. (2015). New Delhi Metallo-Beta-Lactamase 1(NDM-1), the Dominant Carbapenemase Detected in Carbapenem-Resistant Enterobacter Cloacae From Henan Province, China. PloS One 10, e0135044. doi: 10.1145/2818302
Macesic, N., Blakeway, L. V., Stewart, J. D., Hawkey, J., Wyres, K. L., Judd, L. M., et al. (2021a). Silent Spread of Mobile Colistin Resistance Gene Mcr-9.1 on IncHI2 'Superplasmids' in Clinical Carbapenem-Resistant Enterobacterales. Clin. Microbiol. Infect. 27, 1856.e7–1856.e13. doi: 10.1016/j.cmi.2021.04.020
Macesic, N., Blakeway, L. V., Stewart, J. D., Hawkey, J., Wyres, K. L., Judd, L. M., et al. (2021b). Silent Spread of Mobile Colistin Resistance Gene Mcr-9.1 on IncHI2 'Superplasmids' in Clinical Carbapenem-Resistant Enterobacterales. Clin. Microbiol. Infect. 27, e1857–1856 e1813, 1856. doi: 10.1016/j.cmi.2021.04.020
Mezzatesta, M. L., Gona, F., Stefani, S. (2012). Enterobacter Cloacae Complex: Clinical Impact and Emerging Antibiotic Resistance. Future Microbiol. 7, 887–902. doi: 10.2217/fmb.12.61
Niu, T., Guo, L., Luo, Q., Zhou, K., Yu, W., Chen, Y., et al. (2020). Wza Gene Knockout Decreases Acinetobacter Baumannii Virulence and Affects Wzy-Dependent Capsular Polysaccharide Synthesis. Virulence 11, 1–13. doi: 10.1080/21505594.2019.1700659
Osei Sekyere, J., Maningi, N. E., Modipane, L., Mbelle, N. M. (2020). Emergence of Mcr-9.1 in Extended-Spectrum-Beta-Lactamase-Producing Clinical Enterobacteriaceae in Pretoria, South Africa: Global Evolutionary Phylogenomics, Resistome, and Mobilome. mSystems 5, e00148–20. doi: 10.1128/mSystems.00148-20
Potron, A., Poirel, L., Rondinaud, E., Nordmann, P. (2013). Intercontinental Spread of OXA-48 Beta-Lactamase-Producing Enterobacteriaceae Over a 11-Year Period 2001 to 2011. Euro Surveill. 18, 20549. doi: 10.2807/1560-7917.ES2013.18.31.20549
Rahal, A., Andreo, A., Le Gallou, F., Bourigault, C., Bouchand, C., Ferriot, C., et al. (2021). Enterobacter Cloacae Complex Outbreak in a Neonatal Intensive Care Unit: Multifaceted Investigations and Preventive Measures Are Needed. J. Hosp. Infect. 116, 87–90. doi: 10.1016/j.jhin.2021.07.012
Richter, M., Rossello-Mora, R., Oliver Glockner, F., Peplies, J. (2016). JSpeciesWS: A Web Server for Prokaryotic Species Circumscription Based on Pairwise Genome Comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Seemann, T. (2014). Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, P., Berglund, B., Chen, Y., Zhou, Y., Xiao, T., Xiao, Y., et al. (2020). Hypervirulence Markers Among Non-ST11 Strains of Carbapenem- and Multidrug-Resistant Klebsiella Pneumoniae Isolated From Patients With Bloodstream Infections. Front. Microbiol. 11, 1199. doi: 10.3389/fmicb.2020.01199
Simoni, S., Mingoia, M., Brenciani, A., Carelli, M., Lleo, M. M., Malerba, G., et al. (2021). First IncHI2 Plasmid Carrying Mcr-9.1, blaVIM-1, and Double Copies of blaKPC-3 in a Multidrug-Resistant Escherichia Coli Human Isolate. mSphere 6, e0030221. doi: 10.1128/mSphere.00302-21
Sitto, F., Battistuzzi, F. U. (2020). Estimating Pangenomes With Roary. Mol. Biol. Evol. 37, 933–939. doi: 10.1093/molbev/msz284
Stoesser, N., Sheppard, A. E., Shakya, M., Sthapit, B., Thorson, S., Giess, A., et al. (2015). Dynamics of MDR Enterobacter Cloacae Outbreaks in a Neonatal Unit in Nepal: Insights Using Wider Sampling Frames and Next-Generation Sequencing. J. Antimicrob. Chemother. 70, 1008–1015. doi: 10.1093/jac/dku521
Sun, L., Zhao, X., Wang, L., Guo, X., Shi, X., Hu, L. (2021). Coexistence of Mcr-9 and blaNDM-1 in a Multidrug-Resistant Enterobacter Hormaechei Strain Recovered From a Bloodstream Infection in China. J. Glob. Antimicrob. Resist. 24, 440–442. doi: 10.1016/j.jgar.2021.02.011
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 10.0, 2020
Umeda, K., Nakamura, H., Fukuda, A., Matsumoto, Y., Motooka, D., Nakamura, S., et al. (2021). Genomic Characterization of Clinical Enterobacter Roggenkampii Co-Harbouring blaIMP-1- and blaGES-5-Encoding IncP6 and Mcr-9-Encoding IncHI2 Plasmids Isolated in Japan. J. Glob. Antimicrob. Resist. 24, 220–227. doi: 10.1016/j.jgar.2020.11.028
Wang, C., Feng, Y., Liu, L., Wei, L., Kang, M., Zong, Z. (2020). Identification of Novel Mobile Colistin Resistance Gene Mcr-10. Emerg. Microbes Infect. 9, 508–516. doi: 10.1080/22221751.2020.1732231
Wang, Y., Li, Z., Lyu, N., Ma, S., Liu, F., Hu, Y., et al. (2021). Comparative Genomic Analysis of Mobile Colistin Resistance Gene Mcr-9 in Salmonella Enterica. J. Infect. 82, e15–e17. doi: 10.1016/j.jinf.2020.12.029
Wang, Q., Wang, X., Wang, J., Ouyang, P., Jin, C., Wang, R., et al. (2018). Phenotypic and Genotypic Characterization of Carbapenem-Resistant Enterobacteriaceae: Data From a Longitudinal Large-Scale CRE Study in China, (2012-2016). Clin. Infect. Dis. 67, S196–S205. doi: 10.1093/cid/ciy660
Xu, T., Zhang, C., Ji, Y., Song, J., Liu, Y., Guo, Y., et al. (2021). Identi Fi Cation of Mcr-10 Carried by Self-Transmissible Plasmids and Chromosome in Enterobacter Roggenkampii Strains Isolated From Hospital Sewage Water. Environ. Pollut. 268, 115706. doi: 10.1016/j.envpol.2020.115706
Yaffee, A. Q., Roser, L., Daniels, K., Humbaugh, K., Brawley, R., Thoroughman, D., et al. (2016). Notes From the Field: Verona Integron-Encoded Metallo-Beta-Lactamase-Producing Carbapenem-Resistant Enterobacteriaceae in a Neonatal and Adult Intensive Care Unit–Kentucky 2015. MMWR Morb. Mortal. Wkly. Rep. 65, 190. doi: 10.15585/mmwr.mm6507a5
Yang, H., Chen, G., Hu, L., Liu, Y., Cheng, J., Ye, Y., et al. (2018). Enhanced Efficacy of Imipenem-Colistin Combination Therapy Against Multiple-Drug-Resistant Enterobacter Cloacae: In Vitro Activity and a Galleria Mellonella Model. J. Microbiol. Immunol. Infect. 51, 70–75. doi: 10.1016/j.jmii.2016.01.003
Yuan, Y., Li, Y., Wang, G., Li, C., Xiang, L., She, J., et al. (2019). Coproduction Of MCR-9 And NDM-1 By Colistin-Resistant Enterobacter Hormaechei Isolated From Bloodstream Infection. Infect. Drug Resist. 12, 2979–2985. doi: 10.2147/IDR.S217168
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zerbino, D. R., Birney, E. (2008). Velvet: Algorithms for De Novo Short Read Assembly Using De Bruijn Graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
Zheng, B., Chen, Y., Violetta, L., Xiao, Y., Li, L. (2019a). Bloodstream Infections Caused by Entero-Bacteriaceae in China. Lancet Infect. Dis. 19, 810–811. doi: 10.1016/S1473-3099(19)30352-4
Zheng, B., Dong, H., Xu, H., Lv, J., Zhang, J., Jiang, X., et al. (2016). Coexistence of MCR-1 and NDM-1 in Clinical Escherichia Coli Isolates. Clin. Infect. Dis. 63, 1393–1395. doi: 10.1093/cid/ciw553
Zheng, B., Feng, C., Xu, H., Yu, X., Guo, L., Jiang, X., et al. (2019b). Detection and Characterization of ESBL-Producing Escherichia Coli Expressing Mcr-1 From Dairy Cows in China. J. Antimicrob. Chemother. 74, 321–325. doi: 10.1093/jac/dky446
Zheng, B., Huang, C., Xu, H., Guo, L., Zhang, J., Wang, X., et al. (2017a). Occurrence and Genomic Characterization of ESBL-Producing, MCR-1-Harboring Escherichia Coli in Farming Soil. Front. Microbiol. 8, 2510. doi: 10.3389/fmicb.2017.02510
Zheng, B., Lv, T., Xu, H., Yu, X., Chen, Y., Li, J., et al. (2018a). Discovery and Characterisation of an Escherichia Coli ST206 Strain Producing NDM-5 and MCR-1 From a Patient With Acute Diarrhoea in China. Int. J. Antimicrob. Agents 51, 273–275. doi: 10.1016/j.ijantimicag.2017.09.005
Zheng, B., Xu, H., Huang, C., Yu, X., Guo, L., Han, H., et al. (2019c). Occurrence and Genomic Characterization of Two MCR-1-Producing Escherichia Coli Isolates From the Same Mink Farmer. mSphere 4, e00602–19. doi: 10.1128/mSphere.00602-19
Zheng, B., Xu, H., Lv, T., Guo, L., Xiao, Y., Huang, C., et al. (2020). Stool Samples of Acute Diarrhea Inpatients as a Reservoir of ST11 Hypervirulent KPC-2-Producing Klebsiella Pneumoniae. mSystems 5. doi: 10.1128/mSystems.00498-20
Zheng, B., Xu, H., Yu, X., Jiang, X., Zhang, J., Chen, Y., et al. (2018b). Low Prevalence of MCR-1-Producing Klebsiella Pneumoniae in Bloodstream Infections in China. Clin. Microbiol. Infect. 24, 205–206. doi: 10.1016/j.cmi.2017.08.004
Zheng, B., Yu, X., Xu, H., Guo, L., Zhang, J., Huang, C., et al. (2017b). Complete Genome Sequencing and Genomic Characterization of Two Escherichia Coli Strains Co-Producing MCR-1 and NDM-1 From Bloodstream Infection. Sci. Rep. 7, 17885. doi: 10.1038/s41598-017-18273-2
Keywords: MCR-9, Enterobacter cloacae complex, neonatal, sepsis, IncHI2
Citation: Chen C, Xu H, Liu R, Hu X, Han J, Wu L, Fu H, Zheng B and Xiao Y (2022) Emergence of Neonatal Sepsis Caused by MCR-9- and NDM-1-Co-Producing Enterobacter hormaechei in China. Front. Cell. Infect. Microbiol. 12:879409. doi: 10.3389/fcimb.2022.879409
Received: 19 February 2022; Accepted: 05 April 2022;
Published: 06 May 2022.
Edited by:
Muhammad Shafiq, Shantou University, ChinaReviewed by:
Jinhu Huang, Nanjing Agricultural University, ChinaSana Dhaouadi, University of Manouba, Tunisia
Copyright © 2022 Chen, Xu, Liu, Hu, Han, Wu, Fu, Zheng and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beiwen Zheng, zhengbw@zju.edu.cn; Yonghong Xiao, xiaoyonghong@zju.edu.cn
†These authors have contributed equally to this work
 Chunlei Chen
Chunlei Chen Hao Xu
Hao Xu Ruishan Liu
Ruishan Liu Xinjun Hu
Xinjun Hu Jianfeng Han5
Jianfeng Han5  Beiwen Zheng
Beiwen Zheng Yonghong Xiao
Yonghong Xiao