DNA mtCOI Barcodes for Maritime Biosecurity: A Proof of Concept in French Polynesia Ports
- 1Department of Functional Biology, University of Oviedo, Oviedo, Spain
- 2USR 3278 CNRS – EPHE, Centre de Recherche Insulaire et Observatoire de l’Environnement, Moorea, French Polynesia
- 3Laboratoire d’Excellence CORAIL, Centre de Recherche Insulaire et Observatoire de l’Environnement, Moorea, French Polynesia
DNA barcodes have been proposed for diverse applications as markers for species identification. One application that is not fully explored yet is their use for assessing the species biodiversity and presence of invasive alien species (IAS) in maritime biosecurity. The phylogeographical signals of the mitochondrial COI (mtCOI) gene have been sometimes used to infer the number of introductions and the origin of biological invasions. Here, we employed mtCOI barcodes of mollusks and acorn barnacles (N = 751) from ports of French Polynesia to infer the effect of port size, maritime traffic, and degree of openness in the risk of biological invasions. With 17.2% of non-indigenous species (NIS) recorded here, significant differences in diversity were found among docks and between long-time docked ships and their closest piers. A higher proportion of NIS was found from sheltered compared to open ports regardless of their size and traffic. Less frequent wave washing, a lower effect of currents, and partial isolation in sheltered ports could explain the difference. The results suggest that port biota surveys should focus first on ports sheltered from the open sea and emphasize the value of mtCOI barcodes for the early detection of potential invasive species and for prioritizing surveillance efforts.
Introduction
More than 90% of global trade goods are transported by ship1. This means the maritime ports convey most of the world trade traffic together with the organisms attached to the ships or transported in ballast water (Molnar et al., 2008). Merchandise imports are indeed significant in the introduction of biological invasions (Hulme, 2009). Ports are the hubs of marine invasions (e.g., Seebens et al., 2013; Bellard et al., 2016), and the factors that enhance their risk of biopollution should be identified as soon as possible. Among these, human population size explains biological invasions better than any other factor (Pyšek et al., 2010), so the size of port cities could increase biopollution risks. Ports located in estuaries—typically of low salinity—may have a higher risk of some biological pollutants, for example, Ponto-Caspian species (Paiva et al., 2018). Empty niches, suitable environmental conditions, and availability of vectors might be the most effective predictor for the invasibility of brackish water areas and estuaries (e.g., Paavola et al., 2005; Pejovic et al., 2016).
Early detection is the best tool to avoid establishment of new invasions (Gozlan et al., 2010; Blanchet, 2012). However, this is not always possible, not least because the need for accurate identification of species, as an essential component of this for biosecurity and conservation management strategies (Bax et al., 2001), is not able to be achieved. For example, traditional methods that rely on visual identification of specimens have been criticized for their poor ability to identify juvenile life stages that may be critically important in the establishment and spread of invasive populations. Also, there can be limited taxonomic resolution in many taxa, where morphologically cryptic species are difficult to distinguish, possibly confusing the distinction of exotic and native species (Caesar et al., 2006). Besides this, the samples need to be collected using specific manual sampling devices for different taxa (e.g., nets, electrofishing, filtering large water volumes, sediment cores, and SCUBA diving) and then sorted and individually taxonomically identified under the microscope in most cases. This limits how many samples and replicates can be collected and analyzed (Zaiko et al., 2018).
Zaiko et al. (2018) highlight the need to employ robust DNA-based tools, such as genetic barcoding, in aquatic biosecurity studies. Biosecurity not only prevents the arrival and establishing of new Invasive alien species (IAS), but it’s also for the management and analysis of existing pests, where, studying their entry retrospectively, we may have information that could help to prevent similar situations occurring again. DNA barcoding has been cited as a reliable, cheap, rapid, and accurate tool for non-indigenous species (NIS) identification and monitoring (Cross et al., 2010; Briski et al., 2011; Ardura et al., 2015a; Ardura and Planes, 2017). DNA-based tools, together with rapid assessment sampling, allow species identification at any life stage based on DNA extraction from a single individual, facilitating the early detection of new arriving species before an introduced population becomes fully established in a new habitat (Armstrong and Ball, 2005; Chown et al., 2008; Briski et al., 2011; Zhan and MacIsaac, 2015).
The use of mitochondrial COI (mtCOI) DNA barcodes (Hebert et al., 2003) for ascertaining the identity of species present in marine surveys is especially important for guaranteeing biosecurity in maritime ports (Madden et al., 2019). mtCOI also has a relatively low intraspecific variability, making it useful for species identification through DNA barcoding (Meyer and Paulay, 2005). In addition, its strong phylogeographical signal in some invertebrates make this region useful for various purposes related to biosecurity beyond exotic species gene detection. For example it has been used to trace the invasion paths of green crab Carcinus maenas in Australian shores (e.g., Burden et al., 2014), to infer the occurrence of multiple invasion hits of the pygmy mussel Xenostrobus securis in the Bay of Biscay (Devloo-Delva et al., 2016), and to identify geographic donor regions (Miralles et al., 2018). Conveniently it also has a substantial database with more than 3,000,000 sequences of species and populations from around the world2.
In this study, we employ mtCOI barcodes to ascertain NIS of Mollusca (mollusks) and Arthropoda: Crustacea (acorn barnacles) present in French Polynesia ports of different size that are connected by frequented or unfrequented maritime routes. We have chosen these taxonomic groups because they contain numerous highly invasive species that travel attached to hulls and also in ballast water (e.g., Molnar et al., 2008). We have considered port size, fresh water, sheltering level, human population nearby, and number of maritime routes (as international vs. local traffic) as key features that contribute to the arrival and establishment of marine NIS. The initial expectation was that big ports in a region have more NIS than small local ports, assuming homogeneity of the other factors considered in our study.
Materials and Methods
Study Locations
On the island of Moorea (French Polynesia), coastal NIS can be attributed to maritime traffic (e.g., Ardura et al., 2015a) with marine protection areas as a moderator (Ardura et al., 2016a). For this reason, we have targeted only samples taken within or close to ports located beyond protected areas (Figure 1). Three pairs of ports were considered, taking into account the distance between them (located at <30 km of coastline between each other) and connectivity (directly connected by regular lines). Therefore, the small ports of Afareitou and Vai’eane in the south part of the island, Papetoai and Pao-Pao in the north part, and the international Papeete harbor (in Tahiti) and Port Vai’are (Moorea island) that are connected by ferry with two companies operating several times a day all year round were analyzed.
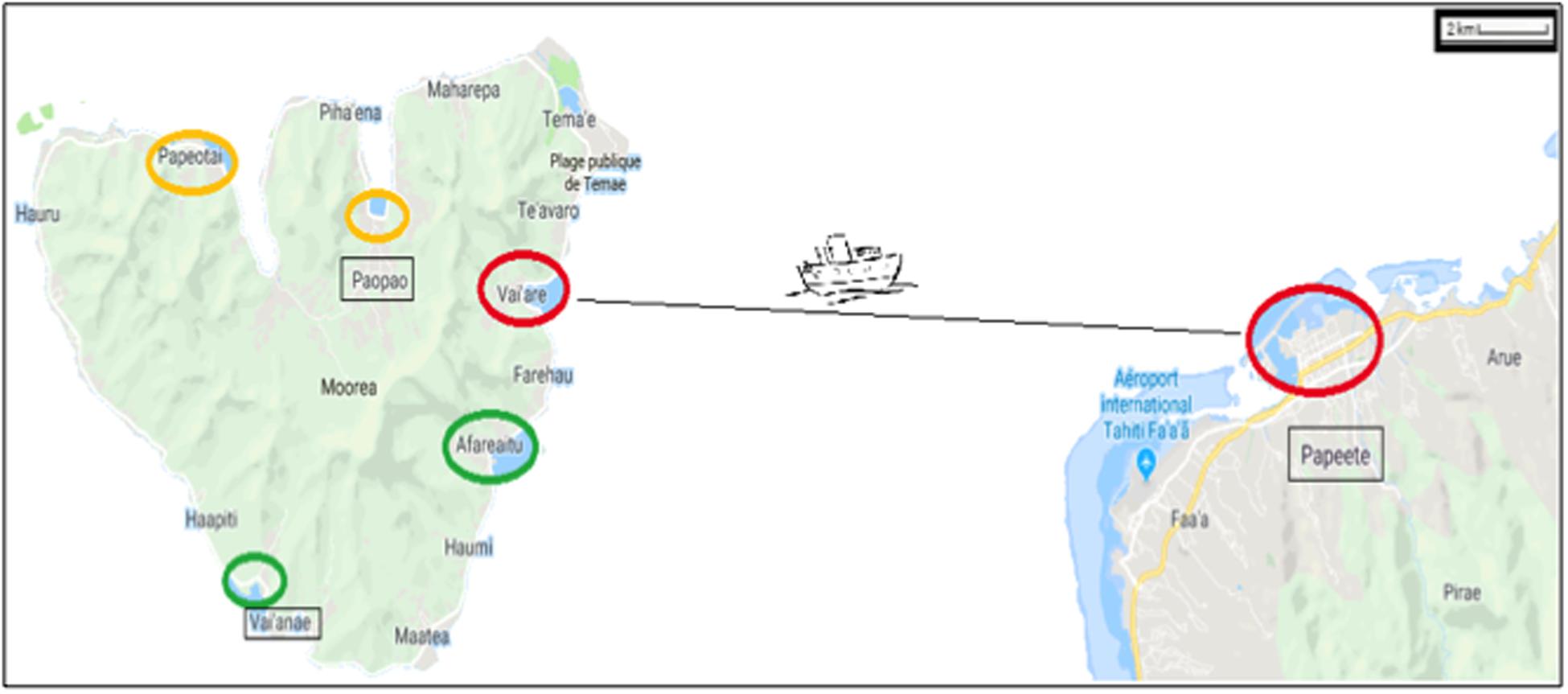
Figure 1. Map showing the regions studied within the Windward Islands (French Polynesia) at 17° 31–40′S/149° 25–50′W. The ports are marked with circles proportional to their size and colored according to their proportion of non-indigenous species as red > orange > green. Ships from Vai’are and Papeete ports (small and big red circle, respectively) were also sampled.
Port Features
The total length of the docks and piers was taken as a proxy of port size. It was estimated using the “distance measurement tool” in Google Maps (©2018 Google) with the maximum zooming possible. In addition to port size, the following features were considered: exposure to open sea (scored as sheltered, 0; semi-exposed, 1; open, 2), brackish water (vicinity of fresh water discharges as a proxy), and size of surrounding human population taken from national institute of statistics and economic studies3. These factors have been reported to be associated with marine biological invasions in other studies (e.g., Paavola et al., 2005; Molnar et al., 2008; Hulme, 2009; Pyšek et al., 2010).
Sampling
Mollusks were targeted for the regional study, and sessile crustaceans (acorn barnacles) were also considered for comparing docks and ships. For the non-native status, NIS are those species not listed or reported as a native to French Polynesia according to current inventories of Moorea fauna and the native distribution of each species (World Register of Marine Species, www.marinespecies.org; Encyclopedia of Life4). Invasive alien species status (IAS) are species listed in the globally invasive species database of the International Union for Nature Conservation5. Invasive alien species are highly invasive in several regions of the world and, thus, pose a real biosecurity risk to Moorea. For statistical analysis, they were considered with other NIS regardless of their invasive status.
The method of sampling employed in docks of small ports was described in Ardura et al. (2015a). Briefly, sampling was carried out picking (at random within species) mollusk and acorn barnacle individuals from the intertidal range (upper to lower), which is quite short in Moorea (maximum tidal range of 0.40 m), between August 26 and September 10, 2011 (Figure 1). An effort was made to obtain representative samples, proportional to the abundance of each species. The methodology described in Miralles et al. (2016) sampling from rectangles of approximately 200 m2 was followed in the larger ports: three rectangles in Papeete, two in Vai’are.
Mollusks and acorn barnacles were sampled from three ships docked in Papeete and one in Vai’are ports as well as from the closest dock. Fouling biota from three quadrats of approximately 30 cm × 30 cm was scratched with a spatula and then the mollusks and acorn barnacles were sorted, identified de visu with the help of taxonomic guides and voucher specimens from the collection of the CRIOBE in Moorea (French Polynesia) to species level when it was possible. A part of tissue (digestive tract was avoided to prevent possible contamination with gut content) was excised and stored in absolute ethanol (100%) for further DNA extraction and genetic identification from barcodes.
DNA Barcoding
DNA barcoding was carried out to ascertain de visu taxonomic identification as described in Ardura et al. (2015a). Total DNA was extracted from a small piece of tissue following the standard protocol described by Estoup et al. (1996), employing Chelex® resin (Bio-Rad Laboratories). The E.Z.N.A Mollusk DNA kit (IOMEGA, bio-tek) was used for the species with high content of mucopolysaccharides in muscle tissues, following the manufacturer’s instructions. In both cases, the tubes were stored at 4°C for immediate DNA analysis, and aliquots were frozen at −20°C for long-term preservation. A fragment within the mitochondrial Cytochrome oxidase I gene (COI) was PCR amplified and sequenced using Geller et al. (2013) primers. Some individuals were double-checked with a second marker, the 16S rRNA gene with the primers described by Palumbi (1996), to confirm the species when identification using COI was not sufficiently accurate (99% match, at least 450 nucleotides coverage). For species identification, the sequences were compared with international databases BOLD system for COI6 and the program BLAST within NCBI for 16S rRNA gene sequences7.
Statistics
Distribution normality of the different variables analyzed in the port data set was checked first, employing Shapiro–Whilk tests. Parametric or non-parametric tests were employed accordingly for further analysis.
In the exploratory analysis of the port data, pairwise correlations between habitat and community variables were done. Individual rarefaction curves for each port, and sample rarefaction curves for big and small ports of each region were constructed with the aim of estimating if the mollusk diversity was sufficiently represented in samples. Diversity of samples was estimated using the Shannon–Weaver index. Differences in diversity between samples were then estimated using permutations tests (n = 9 999). Two-sample paired tests (pairs of ports located in the same area but different in size) were performed to compare means (t tests) of big versus small ports for the percentage of NIS. For estimating the contribution of different independent variables to the variation of a dependent variable, a multiple linear regression model was applied.
All statistics were conducted using PAST version 3.8 (Hammer et al., 2001).
Results
Total Diversity Identified by DNA
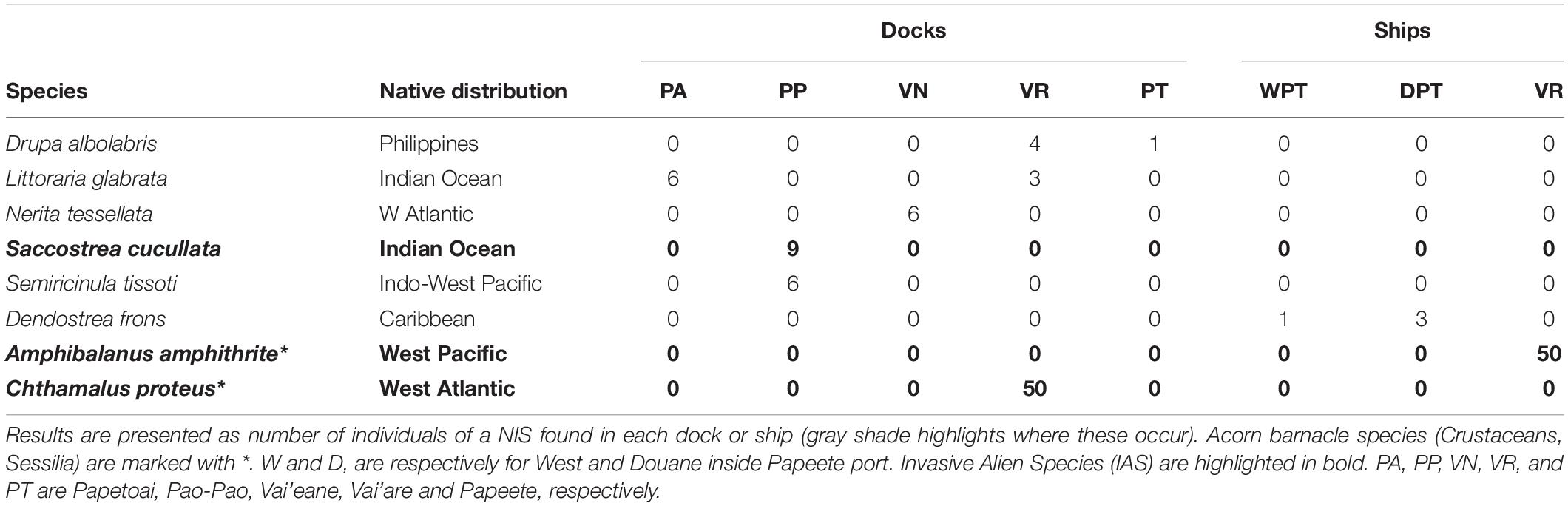
The port characteristics are in Supplementary Table 1, including the proportion of NIS. The ports considered were very different in size, exposure to open sea, fresh water proximity, and surrounding human population (Supplementary Table 1). The number of individuals for each species found in each port and ship are presented in Supplementary Table 2. A total of 751 mollusks were identified to species from the French Polynesia ports analyzed (excluding ships) and another 30 specimens from ships of Papeete and Vai’are ports. In addition to the mollusks, more than 100 acorn barnacles were found from Vai’are port (>50 attached on a ship, from which 50 were analyzed). The majority of species were able to be identified using the COI gene. Only two species required the additional 16S marker for their genetical identification: Nerita plicata and Pinctada maculata. COI and 16S sequences have been deposited in GenBank8 with the accession numbers KT149303 and KT149305 for 16S and KT149306, KT149308, KT149314-6, KT149319-23, KT290130, MH197042-4, and KJ663817-KJ663819 for COI.
Rarefaction curves for the mollusks (Supplementary Figure 1) generally reached a plateau, suggesting that the mollusk communities were representative, i.e., that no significant change to the species represented would occur with further sampling. In total, 155 NIS individuals (i.e., individuals of a species whose native distribution does not include the studied region) were found (17.34% of the samples; see Table 1). We found Drupa albolabris from the Philippines in Papeete (Tahiti Island) and Vai’are (Moorea Island) as well as the gastropods Nerita tessellata (Atlantic Ocean), Littoraria glabrata, and Semiricinula tissoti (Indian Ocean) and the invasive oyster of the Indian Ocean Saccostrea cucullata in Moorea Island. The Caribbean Dendostrea frons oyster was found in Papeete. To our knowledge, the last Polynesian mollusk inventory was published in 2009 (Tröndlé and Boutet, 2009). None of these species was described in that inventory. Therefore, from our knowledge, and taking into account the distribution described in the World Register of Marine Species (WORMS)9, none of the species listed here were previously recorded in this area. In addition to mollusks, the highly invasive West Pacific Amphibalanus amphitrites and West Atlantic Chthamalus proteus acorn barnacles were found from Vai’are port in Moorea Island. These two species were reported by Ardura et al. (2016a). For their relative abundance, most of the mollusk NIS were scarce except a few with >5% frequency: Saccrostrea cuculata (9.4%) and S. tissoti (6.3%) in Pao-Pao, L. glabrata in Papetoai (6.3%), N. tessellata in Vai’eane (8.1%). All the acorn barnacles analyzed, in contrast, were NIS.
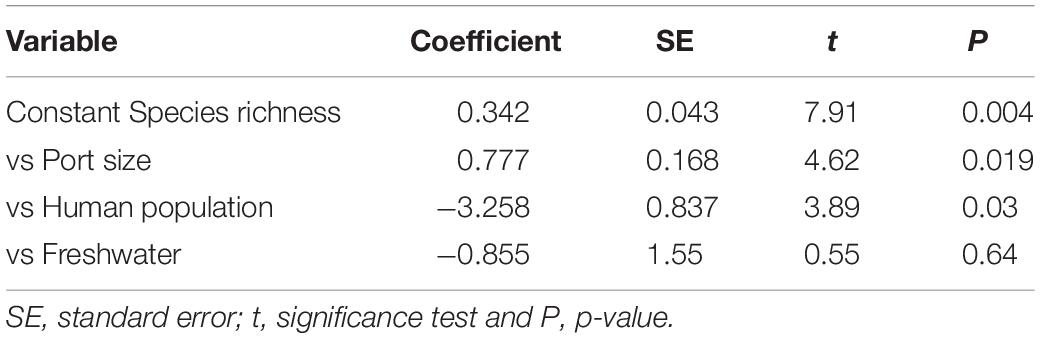
Analyzing all the ports together for the proportion of NIS, significant negative correlation was found only with exposure (i.e., how open is the entrance of the harbor): r = -0.82, 4 d.f., P = 0.041, the more exposed ports having a lower proportion of NIS than the more sheltered. Port size was positively correlated with the species richness (r = 0.83, 4 d.f., P = 0.040), and both port size and human population were significant in a multiple regression model (F = 27.9, df1 = 3, df2 = 2, P = 0.011) for explaining the species richness although the human population had a negative coefficient (Table 2). Fresh water was not significantly associated with any biotic measure.
At a subregional scale, the three pairs of ports considered (located at < 30 km of coastline between each other, or directly connected by regular lines as in the case of Papeete and Vai’are) coincided in a significantly higher proportion of NIS individuals occurring in the smaller port of the pair regardless of its degree of exposure (two-sample paired t test for differences between means with t = 5.500, P = 0.031 for a mean difference of 0.074 and 95% conf. 0.016–0.132) (Figure 2). Noteworthy, significant negative correlations were found between native biodiversity estimated from Shannon index and both%NIS individuals (tau = -0.6, 4 d.f., P = 0.038) and%NIS (tau = -0.867, 4 d.f., P = 0.014) in these Polynesian ports; this essentially means the higher the biodiversity, the lower the proportion of NIS and NIS individuals.
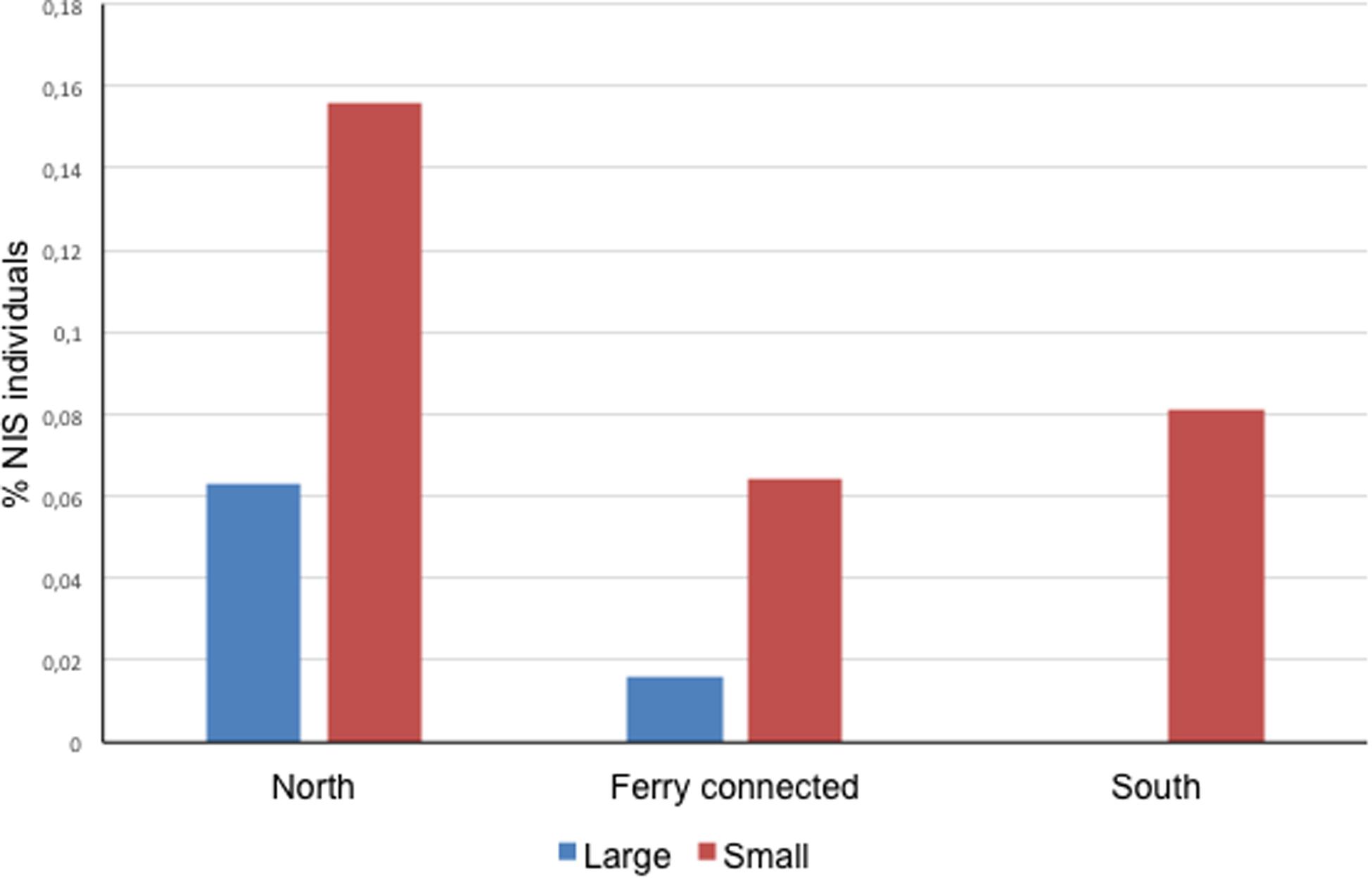
Figure 2. Proportion of NIS individuals in the ports sampled in this study organized by zones. Each zone contains a pair of ports, one bigger (large) and one smaller (small) (Zero NIS recorded in large port in the South area).
Small-Scale: Docks Versus Ships
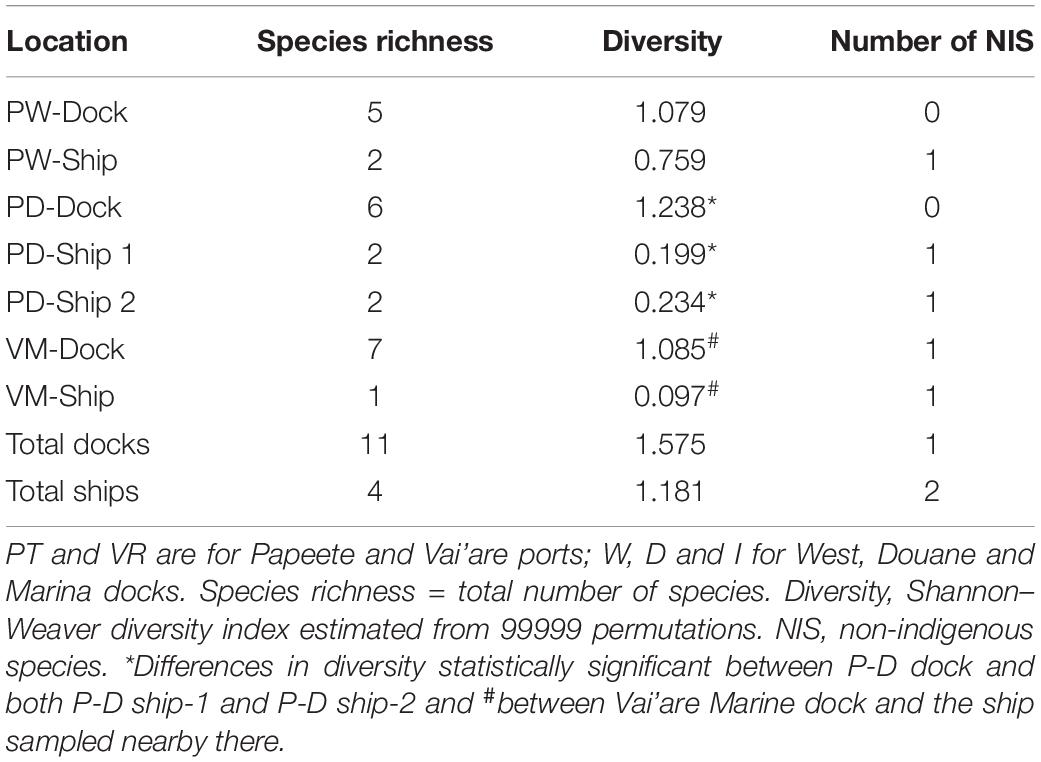
In the small-scale study with four ships, the most evident result was the difference between the mollusks and acorn barnacles attached on ships and those fouling on the very close docks (Supplementary Table 3). The total number of individuals was greater on the docks than on the ships. This was with the exception of the Vai’are Marina sites, where the numbers were similar and influenced heavily by the large numbers of barnacles at both. Second, the species occupying the two types of substrate diverged remarkably in all cases. For example, the native Littoraria species and Siphonaria normalis were prevalent on the docks, and the native P. maculata was prevalent on the ships. Of the IAS, both barnacle species were found at Vai’are, but C. proteus was only on the dock while A. amphithrite only on the ship. Moreover, the species richness and diversity estimates were obviously different, being much greater in the docks than on the ships, where only a few species occurred (Table 3). The differences in diversity were statistically significant between Papeete Douane (custom) dock and both ship 1 and ship 2 (permutation tests for Shannon–Weaver diversity indices with P value = 0.001 in the two cases) and between Vai’are Marine dock and the ship sampled nearby there (P = 0.0013); there was no significant difference between the Papeete West dock and the ship. For the difference in diversity between the Papeete West dock and the ship therein, it was not statistically significant (P = 0.385 in the permutation test).
Discussion
The results of this study confirmed that employing mtCOI as a genetic barcode is very useful for a rapid assessment of invertebrate species biodiversity when the individuals can be taken easily from the environment in general and from docks and ships in particular as here. It is a fast, cheap, and easy technique to inventory biodiversity and detect NIS as well as allowing morphological identifications to be confirmed in the case of cryptic or difficult to classify species (Lara et al., 2010; Williams et al., 2012). Previous substantial study to define Moorea’s biodiversity through barcodes was essential for the development of this new study about NIS presence in ports and nearby because all the species (native and NIS) found during the sampling were already in GenBank (Ardura et al., 2015a), making the use of the mtCOI marker very effective.
The use of the mtCOI gene as a main DNA barcode demonstrated the occurrence of NIS in ports of French Polynesia at a rate exceeding 17% of the total biodiversity. This is a level of biopollution higher than that found in more populated areas; for example, in some ports of Bay of Biscay with 9% of NIS and less than 15,000 inhabitants (Miralles et al., 2016). This should be considered a call for attention because Polynesian-Micronesian islands have been identified as a biodiversity hotspot and highly vulnerable to biological invasions if the current rates of global change persist over time (Bellard et al., 2014).
Another significant result for biosecurity was a higher frequency of NIS found in the smaller and sheltered ports. This was opposite to the expectation of more NIS in large ports anticipated due to major traffic associated with them. This could be related to the higher species richness found in larger ports in this study, where, following Stachowicz et al. (1999), biotic resistance could be involved; essentially, niches would be filled in large Polynesian ports with high native diversity, and new arrivals would have lesser opportunities to settle down. Despite the limited number of ports examined in this proof of concept, a negative correlation between native biodiversity and NIS (low native biodiversity, high proportion of NIS) suggests that biotic resistance is occurring as shown in ports of other regions (Miralles et al., 2016). In a previous study, this effect was not significant (Ardura et al., 2015a), probably because the ecosystems analyzed in that study were too heterogeneous (ports, protected areas, others), and here, we have considered only ports. As other authors point out (Shea and Chesson, 2002), biotic resistance may act at short or medium spatial scales, and its effect is likely diluted when ecologically distant sites are analyzed jointly. Further investigations could focus on protected Polynesian areas, including remote islands, where limited anthropogenic uses would favor native biodiversity (Ardura et al., 2016b) and less maritime traffic, which likely reduces the opportunities of new arrivals. Biotic resistance would accordingly be expected to be much higher there.
Port exposure was salient in our study for explaining the proportion of NIS over other factors of recognized effect on marine invasions, such as human population size (Pyšek et al., 2010) and freshwater discharge (Paavola et al., 2005). Port exposure was negatively correlated with NIS. This could be explained from the presence of waves that may disturb NIS settlement in open ports washing propagules away (unsettled larvae, young adults detaching from hulls or recently deposited on the rocks, etc.). Another spatial factor that influenced the level of NIS in our study was the location of the ports in the north or south coasts of the island with NIS being higher in the north. The explanation in this case is likely to be higher maritime traffic in the north than in the south because big cruise ships anchor in the profound Cook’s and Opunohu bays (next to Pao-Pao and Papetoai ports, respectively; see Figure 1).
Significant differences were found between ship hulls and their closely associated docks. The most evident result was the difference between the mollusks and acorn barnacles attached on ships and those fouling on the very close docks (Table 3). Ship hulls only had bivalves attached although gastropods were much more abundant than bivalve species on the rocks nearby; similarly, acorn barnacles A. amphitrites were found on a ship, and C. proteus was sampled from docks. These differences could be a matter of time because biofouling species diversity depends on the time a vessel remains in a recipient region (Hopkins and Forrest, 2008). However, here, the ships had been docked in the same place for a minimum of 3 months, a time that would be enough for their organisms to move to the closest rocks. Another explanation for the difference in species between ships and docks could be differential preferences of sessile animals for substrates and materials. As an example in mollusks, Rech et al. (2018) find many more bivalves than gastropods on artificial substrates; our results were clearly in concordance with this. Regarding acorn barnacles, there is genetic evidence of multiple introductions from different regions of the two species mentioned above (Ardura et al., 2016a), suggesting they are colonizing the island via maritime traffic. Why only one was attached to a ship? The explanation could be the same: acorn barnacles exhibit different substrate preferences depending on the species; for example, on Swedish shores Amphibalanus species were found preferentially on wood although other genera (e.g., Balanus) preferred harder substrates (Garcia-Vazquez et al., 2018). Our results of Amphibalanus attached to a ship hull and Chthamalus to dock rocks would be consistent with those findings in Sweden and also with other studies that have found big differences between ship hulls and harbor fouling organisms (e.g., Sylvester et al., 2011). In some ways, ships could be considered partially isolated habitats carrying their own fouling biota that, in some circumstances, may provide the pattern for them to move to surrounding habitats. The mixed origins of the NIS found in our study—none of them imported for aquaculture and the majority from the Indian Ocean and other regions of the Pacific, but also one Caribbean and another two from the West Atlantic—would confirm that the main vector of NIS in Polynesia is maritime traffic (Ardura et al., 2015a).
The study illustrates that local conditions can influence the nature of resident NIS and, presumably, help prioritize surveillance efforts. The risk of biological invasions is especially important in islands because they depend on maritime trade (Hulme, 2009), but not all areas seem to be equally vulnerable to them. From our results, the areas around the ports in the Windward Islands should be periodically monitored and samples from the different species mtCOI barcoded. A higher surveillance of the beaches nearby Vai’are and especially Pao-Pao, corresponding to sheltered ports and the second one being located in the north of the island, would be recommended because they have already a quite high frequency of NIS. As seen in this proof of concept, barcoding has considerable potential in port biosecurity, which emphasizes its use for early detection of potential invasive species.
Even with these positive results, this methodology implies a sampling, requiring a large effort of human resources and many specialists systematically sampling all ecosystems. The effort is greater in habitats with difficult physical accession that have to be reached from the sea and/or diving. In addition, some species cannot be detected when they are at a low density, in their first development stages, or have high mobility (not sessile species). In these cases, the use of environmental DNA and metabarcoding is useful to detect non-target species by traces of their DNA in the water (Ardura et al., 2015b; Zaiko et al., 2015, 2018). However, some drawbacks must be taken into account because environmental DNA techniques involve higher costs and substantial analytical effort (bioinformatics) to ensure efficient exploration of the sequence data obtained from multispecies communities (Blanchet, 2012). Therefore, the best methodology should be assessed for each particular study, depending on economic and material resources and the data available and necessary in each case (Ardura and Planes, 2017).
Finally, it is important to highlight that, although these molecular methods can answer some questions about biosecurity questions, a complete marine biosecurity program should integrate complementary scientific approaches, including traditional surveys, mathematical modeling, risk assessment frameworks, citizen collaboration, and molecular techniques.
Data Availability Statement
The datasets generated for this study can be found in the GenBank, KT149305, KT149315-6, KT149319-20, KT290130, MH197042-4, and KJ663817–KJ663819.
Author Contributions
AA performed laboratory analysis and collaborated in writing. SP collaborated in the study design and writing. EG-V designed the study, sampled and wrote the article draft.
Funding
This study was funded from the Spanish Ministry of Economy and Competitiveness under the Grant MINECO CGL2016-79209-R and the Government of Asturias Principality under the Grant IDI-2018-000201. This is a contribution from the Marine Observatory of Asturias (OMA), Asturias University Institute of Biotechnology, and the Spanish Research Group of Excellence ARENA. AA holds a postdoctoral Juan de la Cierva fellowship and received an IRCP-CRIOBE Grant (2018).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to two reviewers of Frontiers for their constructive comments on the manuscript that helped us so much to improving it.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2020.00179/full#supplementary-material
Footnotes
- ^ www.imo.org
- ^ https://www.ncbi.nlm.nih.gov/nuccore/?term=COI
- ^ https://www.insee.fr/fr/statistiques/3294362?sommaire=2122700
- ^ http://www.eol.org, accessed October 2019.
- ^ http://www.iucngisd.org/, accessed October 2019.
- ^ http://www.boldsystems.org/
- ^ http://www.ncbi.nlm.nih.gov/
- ^ https://www.ncbi.nlm.nih.gov/genbank/
- ^ http://www.marinespecies.org/index.php
References
Ardura, A., and Planes, S. (2017). Rapid assessment of non-indigenous species in the era of the eDNA barcoding: a mediterranean case study. Est. Coast. Shelf Sci. 188, 81–87. doi: 10.1016/j.ecss.2017.02.004
Ardura, A., Planes, S., and Garcia-Vazquez, E. (2015a). Aliens in paradise. boat density and exotic coastal mollusks in moorea Island (French Polynesia). Mar. Environ. Res. 112, 56–63. doi: 10.1016/j.marenvres.2015.08.007
Ardura, A., Zaiko, A., Martinez, J. L., Aurelija Samuiloviene, A., Borrell, Y., and Garcia-Vazquez, E. (2015b). Environmental DNA evidence of transfer of North Sea molluscs across tropical waters through ballast water. J. Mollusc. Stud. 81, 495–501. doi: 10.1093/mollus/eyv022
Ardura, A., Juanes, F., Planes, S., and Garcia-Vazquez, E. (2016a). Rate of biological invasions is lower in coastal marine protected areas. Sci. Rep. 6:33013. doi: 10.1038/srep33013
Ardura, A., Planes, S., and Garcia-Vazquez, E. (2016b). Phylogenetic analysis for detection of multiple fouling events: a pilot study of barnacles at Moorea Island (French Polynesia). Crustaceana 89, 863–875. doi: 10.1163/15685403-00003554
Armstrong, K., and Ball, S. (2005). DNA barcodes for biosecurity: invasive species identification. Phil. Trans. R. Soc. B Biol. Sci. 360, 1813–1823. doi: 10.1098/rstb.2005.1713
Bax, N., Carlton, J. T., Mathews-Amos, A., Haedrich, R. L., Howarth, F. G., Purcell, J. E., et al. (2001). The control of biological invasions in the world’s oceans. Conserv. Biol. 15, 1234–1246. doi: 10.1111/j.1523-1739.2001.99487.x
Bellard, C., Leclerc, C., Leroy, B., Bakkenes, M., Veloz, S., Thuiller, W., et al. (2014). Vulnerability of biodiversity hotspots to global change. Global Ecol. Biogeogr. 23, 1376–1386. doi: 10.1111/geb.12228
Bellard, C., Leroy, B., Thuiller, W., Rysman, J. F., and Courchamp, F. (2016). Major drivers of invasion risks throughout the world. Ecosphere 7, 1–14.
Blanchet, S. (2012). The use of molecular tools in invasion biology: an emphasis on freshwater ecosystems. Fisheries Manag. Ecol. 19, 120–132. doi: 10.1111/j.1365-2400.2011.00832.x
Briski, E., Cristescu, M. E., Bailey, S. A., and MacIsaac, H. J. (2011). Use of DNA barcoding to detect invertebrate invasive species from diapausing eggs. Biol. Inv. 13, 1325–1340. doi: 10.1007/s10530-010-9892-7
Burden, C. T., Stow, A. J., Hoggard, S. J., Coleman, M. A., and Bishop, M. J. (2014). Genetic structure of Carcinus maenas in southeast Australia. Mar. Ecol. Progr. Ser. 500, 139–147. doi: 10.3354/meps10704
Caesar, R. M., Sorensson, M., and Cognato, A. I. (2006). Integrating DNA data and traditional taxonomy to streamline biodiversity assessment: an example from edaphic beetles in the Klamath ecoregion. California USA. Divers. Distrib. 12, 483–489. doi: 10.1111/j.1366-9516.2006.00237.x
Chown, S. L., Sinclair, B. J., and van Vuuren, B. J. (2008). DNA barcoding and the documentation of alien species establishment on sub-Antarctic Marion Island. Polar Biol. 31, 651–655. doi: 10.1007/s00300-007-0402-z
Cross, H. B., Lowe, A. J., and Gurgel, F. D. (2010). “DNA barcoding of invasive species,” in Fifty Years of Invasion Ecology: The Legacy of Charles Elton, ed. D. Richardson (Oxford: Blackwells).
Devloo-Delva, F., Miralles, L., Ardura, A., Borrell, Y. J., Pejovic, I., Tsartsianidou, V., et al. (2016). Detection and characterisation of the biopollutant Xenostrobus securis (Lamarck 1819) Asturian population from DNA Barcoding and eBarcoding. Mar. Pol. Bull. 105, 23–29. doi: 10.1016/j.marpolbul.2016.03.008
Estoup, A., Largiader, C. R., Perrot, E., and Chourrout, D. (1996). Rapid one-tube DNA extraction for reliable PCR detection of fish polymorphic markers and transgenes. Mol. Mar. Biol. Biotech. 5, 295–298.
Garcia-Vazquez, E., Cani, A., Diem, A., Ferreira, C., Geldhof, R., Marquez, L., et al. (2018). Leave no traces – Beached marine litter shelters both invasive and native species. Mar. Pol. Bull. 131, 314–322. doi: 10.1016/j.marpolbul.2018.04.037
Geller, J., Meyer, C., Parker, M., and Hawk, H. (2013). Redesign of PCR primers for mitocondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Mol. Ecol. Res. 13, 851–861. doi: 10.1111/1755-0998.12138
Gozlan, R. E., Britton, J. R., Cowx, I., and Copp, G. H. (2010). Current knowledge on non-native freshwater fish introductions. J. Fish Biol. 76, 751–786. doi: 10.1111/j.1095-8649.2010.02566.x
Hammer, Ø, Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electronica 4, 1–9.
Hebert, P. D. N., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. R. Soc. Lon. B 270, 313–321. doi: 10.1098/rspb.2002.2218
Hopkins, G. A., and Forrest, B. M. (2008). Management options for vessel hull fouling: an overview of risks posed by in-water cleaning. ICES J. Mar. Sci. 65, 811–815. doi: 10.1093/icesjms/fsn026
Hulme, P. E. (2009). Trade, transport and trouble: managing invasive species pathways in an era of globalization. J. App. Ecol. 46, 10–18. doi: 10.1111/j.1365-2664.2008.01600.x
Lara, A., Ponce, J. L., Rodriguez, R., Casane, D., Côté, G., Bernatchez, L., et al. (2010). DNA barcoding of Cuban freshwater fishes: evidence for cryptic species and taxonomic conflicts. Mol. Ecol. Res. 10, 421–430. doi: 10.1111/j.1755-0998.2009.02785.x
Madden, M., Young, R. G., Brown, J. W., Miller, S. E., Frewin, A. J., and Hanner, R. H. (2019). Using DNA barcoding to improve invasive pest identification at U.S. ports-of-entry. PloS One 14:e0222291. doi: 10.1371/journal.pone.0222291
Meyer, C. P., and Paulay, G. (2005). DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 3:e422. doi: 10.1371/journal.pbio.0030422
Miralles, L., Ardura, A., Arias, A., Borrell, Y. J., Clusa, L., Dopico, E., et al. (2016). Barcodes of marine invertebrates from north Iberian ports: native diversity and resistance to biological invasions. Mar. Pol. Bull. 112, 183–188. doi: 10.1016/j.marpolbul.2016.08.022
Miralles, L., Ardura, A., Clusa, L., and Garcia-Vazquez, E. (2018). DNA barcodes of antipode marine invertebrates in bay of biscay and gulf of lion ports suggest new biofouling challenges. Sci. Rep. 8:16214.
Molnar, J. L., Gamboa, R. L., Revenga, C., and Spalding, M. D. (2008). Assessing the global threat of invasive species to marine biodiversity. Frontiers Ecol. Environ. 6:485–492. doi: 10.1890/070064
Paavola, M., Olenin, S., and Leppakoski, E. (2005). Are invasive species most successful in habitats of low native species richness across European brackish water seas? Est. Coast. Shelf Sci. 64, 738–750. doi: 10.1016/j.ecss.2005.03.021
Paiva, F., Barco, A., Chen, Y., Mirzajani, A., Chan, F. T., Lauringson, V., et al. (2018). Is salinity an obstacle for biological invasions? Global Change Biol. 24, 2708–2720. doi: 10.1111/gcb.14049
Palumbi, S. R. (1996). “Nucleic acids II: the polymerase chain reaction,” in Molecular Systematics, eds D. M. Hillis, C. Moritz, and B. K. Mable (Sunderland, MA: Sinauer Associates, Inc), 205–247.
Pejovic, I., Ardura, A., Miralles, L., Arias, A., Borrell, Y. J., and Garcia-Vazquez, E. (2016). DNA barcoding for assessment of exotic molluscs associated with maritime ports in northern Iberia. Mar. Biol. Res. 12, 168–176. doi: 10.1080/17451000.2015.1112016
Pyšek, P., Jarošík, V., Hulme, P. E., Kühn, I., Wild, J., Arianoutsou, M., et al. (2010). Disentangling the role of environmental and human pressures on biological invasions across Europe. Proc. Natl. Acad. Sci. U.S.A. 107, 12157–12162.
Rech, S., Borrell, Y. J., and Garcia-Vazquez, E. (2018). Anthropogenic marine litter composition in coastal areas may be a predictor of potentially invasive rafting fauna. PLoS One 13:e0191859. doi: 10.1371/journal.pone.0191859
Seebens, H., Gastner, M. T., and Blasius, B. (2013). The risk of marine bioinvasion caused by global shipping. Ecol. Lett. 16, 782–790. doi: 10.1111/ele.12111
Shea, K., and Chesson, P. (2002). Community ecology theory as a framework for biological invasions. Trends Ecol. Evol. 17, 170–176. doi: 10.1016/s0169-5347(02)02495-3
Stachowicz, J. J., Whitlatch, R. B., and Osman, R. W. (1999). Species diversity and invasion resistance in a marine ecosystem. Science 286, 1577–1579.
Sylvester, F., Kalaci, O., Leung, B., Lacoursière-Roussel, A., Clarke Murray, C., Choi, F., et al. (2011). Hull fouling as an invasion vector: can simple models explain a complex problem? J. Appl. Ecol. 48, 415–423. doi: 10.1111/j.1365-2664.2011.01957.x
Tröndlé, J., and Boutet, M. (2009). Inventory of Marine Molluscs of French Polynesia. Atoll Res. Bull. 570, 1–90. doi: 10.5479/si.00775630.570.1
Williams, P. H., Mark, B. J. F., Jamrs, C. C., Jiandong, A., Murat, A. A., Lincoln, B. R., et al. (2012). Unveiling cryptic species of the bumblebee subgenus Bombus s. str. worldwide with COI barcodes (Hymenoptera: Apidae). Syst. Biodivers. 10, 21–56. doi: 10.1080/14772000.2012.664574
Zaiko, A., Pochon, X., Garcia-Vazquez, E., Olenin, S., and Wood, S. (2018). Advantages and limitations of environmental DNA/RNA tools for marine biosecurity: management and surveillance of non-indigenous species. Front. Mar. Sci. 5:322. doi: 10.3389/fmars.2018.00322
Zaiko, A., Samuiloviene, A., Ardura, A., and Garcia-Vazquez, E. (2015). Metabarcoding approach for nonindigenous species surveillance in marine coastal waters. Mar. Poll. Bull. 100, 53–59. doi: 10.1016/j.marpolbul.2015.09.030
Keywords: biosecurity, French Polynesia, mtCOI barcode, barcode applications, maritime ports
Citation: Garcia-Vazquez E, Ardura A and Planes S (2020) DNA mtCOI Barcodes for Maritime Biosecurity: A Proof of Concept in French Polynesia Ports. Front. Ecol. Evol. 8:179. doi: 10.3389/fevo.2020.00179
Received: 03 October 2019; Accepted: 19 May 2020;
Published: 19 June 2020.
Edited by:
David S. Thaler, Biozentrum, Universität Basel, SwitzerlandReviewed by:
Yonas Isaak Tekle, Spelman College, United StatesKaren Frances Armstrong, Lincoln University, New Zealand
Copyright © 2020 Garcia-Vazquez, Ardura and Planes. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eva Garcia-Vazquez, egv@uniovi.es
 Eva Garcia-Vazquez
Eva Garcia-Vazquez Alba Ardura
Alba Ardura Serge Planes2,3
Serge Planes2,3