- 1College of Agriculture, South China Agricultural University, Laboratory of Bio-Pesticide Creation and Application of Guangdong Province, Guangzhou, China
- 2Beijing Genomic Institute, Shenzhen, China
- 3School of Life Sciences, Institute of Insect Science and Technology, South China Normal University, Guangzhou, China
Plutella xylostella, a global key pest, is one of the major lepidopteran pests of cruciferous vegetables owing to its strong ability of resistance development to a wide range of insecticides. Destruxin A, a mycotoxin of the entomopathogenic fungus, Metarhizium anisopliae, has broad-spectrum insecticidal effects and has been used as an alternative control strategy to reduce harmful effects of insecticides. However, microRNA (miRNA)-regulated reactions against destruxin A have not been elucidated yet. Therefore, here, to identify immunity-related miRNAs, we constructed four small RNA libraries from destruxin A-injected larvae of P. xylostella at three different time courses (2, 4, and 6 h) with a control, and sequenced by Illumina. Our results showed that totally 187 known and 44 novel miRNAs were identified in four libraries by bioinformatic analysis. Interestingly, among differentially expressed known miRNAs, some conserved miRNAs, such as miR-263, miR-279, miR-306, miR-2a, and miR-308, predicted to be involved in regulating immunity-related genes, were also identified. Worthy to mention, miR-306 and miR-279 were also listed as common abundantly expressed miRNA in all treatments. The Kyoto Encyclopedia of Genes and Genomes pathway analysis also indicated that differentially expressed miRNAs were involved in several immunity-related signaling pathways, including toll signaling pathway, IMD signaling pathway, JAK–STAT signaling pathway, and cell adhesion molecules signaling pathway. To the best of our knowledge, this is the first comprehensive report of destruxin A-responsive immunity-related miRNAs in P. xylostella. Our findings will improve in understanding the role of destruxin A-responsive miRNAs in the host immune system and would be useful to develop biological control strategies for controlling P. xylostella.
Introduction
In nature, with the passage of time, a number of new mechanisms of host defenses have been developed in the arms race of host–pathogen. Animals face pathogen challenge with innate and adaptive immune system whereas, invertebrates, unlike mammals, do not have an adaptive immune system, but instead they rely on a sophisticated innate immune system for defense against invading microbes (1). The innate immune system of insects is comprised of two main components, cellular and humoral immune responses (2). The former relies majorly on the action of hemocytes in the phagocytosis of pathogens (3), while the latter refers to the process of melanization with phenoloxidases (4) and synthesis of immune effector molecules (5). Upon microorganism invasion, the immune system reactions are initiated by the recognition proteins, including peptidoglycan recognition proteins, β-1,3-glucan recognition proteins (βGRPs), galectins, C-type lectins, and scavenger receptors (6). The recognition step then leads to amplification of signals by serine proteases and triggers the activation of immune signaling pathways followed by induction of antimicrobial peptides to clear the infection (7, 8).
MicroRNAs (miRNAs), a class of small RNAs (sRNAs), are reported to play a pivotal role in the gene expression regulation at the level of posttranscription in many organisms. Since the discovery of the first miRNA in Caenorhabditis elegans (9), many miRNAs have been identified in plants, animals, and insects (10), and our knowledge of miRNA interaction with targets is persistently increasing and developing with the passage of time. In insects, miRNAs have been demonstrated to be key mediators in various physiological processes, including embryonic development (11), apoptosis (12), morphogenesis (13), and cell differentiation (14), and a recent report demonstrated that miRNAs may also play crucial roles in the regulation of innate immunity (15).
The entomopathogenic fungi, such as Metarhizium anisopliae and Beauveria bassiana, are considered as an environmentally friendly approach for the control of insect pests. During pathogenesis, these entomopathogenic fungi secrete virulence factors to accelerate the death of infected host (16). Destruxins, the virulence factors of fungi, have been reported to exhibit high toxicity to various insect species when ingested or injected (17–19) and inhibit V-type ATPase hydrolytic activity, prompt oxidative stress, and affects the Ca2+ channel in muscle cells (20–22). Additionally, destruxins play a pivotal role in the immune system of insects, such as Drosophila melanogaster; innate immune response was suppressed by destruxin A following the inhibition of antimicrobial peptides (16), whereas the immune system of Bombyx mori was induced in response to destruxin A (23). Although recently, the number of studies reporting immunity-related genes in Plutella xylostella is growing (24–27), thanks to the availability of P. xylostella genome, however, information regarding miRNA-regulated reactions in insect–pathogen interactions is still in its infancy and, to the best of our knowledge, there is no information available on miRNA-regulated reactions against the secondary metabolites of fungi, especially destruxin A.
Keeping in view the importance of miRNAs in insect–pathogen interaction, as exhibited by previous reports, we hypothesized that the network of miRNA-guided gene expression regulation might play a pivotal role in the control of innate immunity in destruxin A-infected P. xylostella. Furthermore, we also intended to find out that how P. xylostella miRNAs respond to destruxin A at different time courses of infection and whether the immune system of P. xylostella has developed novel miRNAs to combat the infection of destruxin A. To address these questions, a high-throughput sequencing Illumina and real-time quantitative PCR (RT-qPCR) were carried out to identify unique, novel, and immune-responsive miRNAs in P. xylostella infected with destruxin A at three different time courses (2, 4, and 6 h).
Materials and Methods
Rearing of Insects and Destruxin A Preparation
A susceptible P. xylostella strain was obtained from the Engineering Research Centre of Biological Control, Ministry of Education, South China Agricultural University, China and was kept insecticide-free for 10 generations. P. xylostella were maintained at 25 ± 1°C, 65% relative humidity, and a 14:10 h (light: dark) photoperiod. Destruxin A was extracted from M. anisopliae strain MaQ-10. The purity of destruxin A was evaluated by high-performance liquid chromatography. Finally, destruxin A was diluted in phosphate buffered saline (PBS, PH 7.4).
Destruxin A Injection, sRNA Library Construction, and Sequencing
To inject destruxin A into the susceptible fourth instar larvae of P. xylostella, a stock solution of destruxin A (200 μg/mL) was prepared, and then 2 μL of that solution was injected to each larva. The control group was treated with PBS. A number of 30 larvae were collected from each treatment (2, 4, and 6 h post-injection) and control following instant freezing in liquid nitrogen. Trizol Total RNA Isolation Kit (Takara, Japan) was used to extract total RNA following manufacturer’s instructions. The concentrations of RNA were assessed using Nanodrop (Bio-Rad, USA) and its integrity was determined on Agilent 2100 Bioanalyzer (Agilent, USA).
Furthermore, RNAs were firstly ligated with 3′ adapter and after size fraction, ligated to 5′ adapter. The sRNA fractions were then used for reverse transcription following PCR. The final ligation PCR products, after purification, were sequenced using Illumina Genome Analyzer (San Diego, CA, USA) at the Beijing Genomics Institute (BGI, Shenzhen, China).
Bioinformatics of Destruxin A-Responsive sRNA Sequences
To obtain clean reads from raw data reads having low-quality, 5′ primer contaminants, without 3′ primers and insert tag, and sequences fewer than 18 nucleotides (nt), were filtered out. To analyze the distribution, the final clean reads of the four libraries were mapped to P. xylostella genome (GCA_000330985.1) using Bowtie program (28). All the remaining clean sequences were annotated into different classes to remove rRNA, scRNA, snoRNA, snRNA, and tRNA using Rfam database. Finally, the unannotated clean sequences were used to predict novel miRNAs using the miRDeep2 software.
Differential Expression Analysis of Destruxin A-Responsive miRNAs
The expression of miRNAs was compared between treatment and control to identify differentially expressed miRNAs. First, the expression of miRNA in the four libraries was normalized to transcripts per million. If the normalized expression of the miRNA was 0, it was modified to 0.01 to enable calculation. If the normalized expression of the miRNA was less than 1 in all libraries, it was ignored to compare for low expression. The normalization formula was
The normalized data were then used to calculate fold-change values and P-values, and a scatter plot of the fold-change values was generated. Fold-change was calculated as
The P-value was calculated by the following equation:
where x represents sRNA total clean reads in the control, y represents total clean reads in the treatment, N1 represents the normalized expression of a miRNA in library control, and N2 represents the normalized expression of the same miRNA in library treatment. The corrected P-value corresponds to differential gene expression test using Bonferroni method (29).
Genome-Wide Target Prediction for Destruxin A-Responsive miRNAs
To predict and analyze potential targets of differentially expressed miRNAs, three different software, including RNAhybrid (30), miRanda (31), and TargetScan (32) were used following the principles of target prediction as described previously elsewhere (33, 34). To increase the level of confidence and get more reliable results, we selected only those binding sites that were predicted by all three software.
Gene Ontology (GO) Enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of Predicted Targets of Destruxin A-Response miRNAs
The genome database of P. xylostella was used as the background to determine GO terms enriched within the predicted targets dataset using hypergeometric test and a corrected P-value (≤0.05) as a threshold in order to find out significantly enriched terms. Finally, KEGG pathway enrichment analysis was performed to identify significantly enriched pathways within the predicted targets datasets compared with the genome database using hypergeometric test and a corrected P-value (≤0.05) as a threshold.
Verification of Destruxin A-Responsive miRNAs by RT-qPCR
Real-time quantitative PCR is the method of choice for analyzing expression of genes and to confirm the results of RNA-Sequencing (35). The RT-qPCR analysis was conducted to ensure the expression levels of miRNAs displayed by Illumina sequencing results and 10 differentially expressed miRNAs were selected from the comparison of control vs. treatments. Total RNA was extracted from each sample as described earlier. The RNA sample (1 μg) was treated with DNaseI (Fermentas, Glen Burnie, MD, USA) following manufacturer’s protocol and then complementary DNA was synthesized using M-MLV reverse transcriptase (Promega, USA). The RT-qPCR was carried out on a Bio-Rad iQ2 optical system (Bio-Rad) using SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, USA) following the manufacturer’s guidelines. The amplification cycling parameters were: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 55°C for 10 s with a dissociation curve generated from 65–95°C to ensure the purity of PCR products (36). The U6 snRNA was used as an internal control for normalization and the relative expression of genes was calculated using the 2−ΔΔCT method (37). Each experiment was replicated in triplicate.
Statistical Analysis
Statistical analyses of the present study were performed using SPSS software (version 22.0; IBM Corp., Armonk, NY, USA). The differences between treatments were compared using Student’s t-test or one-way analysis of variance followed by Tukey’s test for multiple comparisons at the following significance levels: lowercase letters at P < 0.05 and capital case letters at P < 0.01. All results are expressed as means ± SEM.
Results
Construction of sRNA Libraries
Four sRNA libraries were constructed for control, 2, 4, and 6 h, and 11,586,075, 15,391,118, 12,893,039, and 13,876,345 high-quality reads were obtained, respectively. The low-quality sequences, reads with 5′ contaminants and without a 3′ primer or insert tag, and reads shorter than 18 nt, were eliminated; subsequently, 11,492,082 (99.19%), 15,048,896 (97.78%), 12,519,758 (97.1%), and 13,777,285 (99.29%) clean reads in the control, 2, 4, and 6 h were obtained for further analysis, respectively (Table S1 in Supplementary Material). The sRNA length distribution of the four libraries exhibited that most of the sRNAs ranged from 18 to 30 nt with 26 to 28 nt sRNAs being most abundant following 22 and 29 nt (Figure 1). Among the total and unique clean reads, 21,944,702 and 39,83,00 sRNAs between control and 2 h; 20,268,898 and 3,22,472 sRNAs between control and 4 h; 20,334,789 and 3,80,546 sRNAs between 6 h and control; 23,648,245 and 4,36,969 sRNAs between 2 and 4 h; 24,227,618 and 5,33,391 sRNAs between 6 and 2 h; and 22,256,011 and 4,35,747 sRNAs were common between 6 and 4 h, respectively (Figure S1 in Supplementary Material).
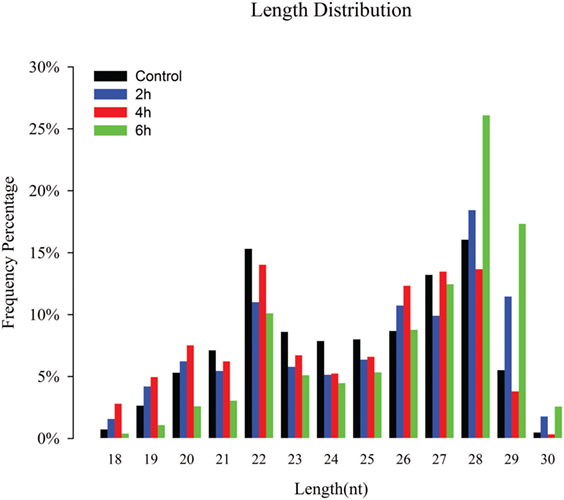
Figure 1. Size distribution of destruxin A-responsive small RNAs (sRNAs) in the libraries of Plutella xylostella. Different colors represent different libraries. x-Axis represents sRNA length distribution and y-axis represents frequency percentage.
Genome Mapping and sRNA Annotation
Of the clean reads, 7,359,318, 8,346,568, 6,685,514, and 7,855,270 reads from control, 2, 4, and 6 h accounted for 64.04, 55.46, 53.4, and 57.02%, respectively, and were mapped to the genome of P. xylostella (GCA_000330985.1) (Table S2 in Supplementary Material). The annotation of sRNAs was carried out by following priority rule of rRNA, etc.; (GenBank > Rfam) > known miRNA > repeat > exon > intron (38). The clean reads were categorized into miRNA, rRNA, snRNA, snoRNA, tRNA, and unannotated. The composition of the sRNA classes in each library is displayed in Figure S2 in Supplementary Material.
Identification of Destruxin A-Responsive Known miRNAs
The bioinformatic analysis was carried out to identify destruxin A-responsive known miRNAs in P. xylostella. Although miRNAs are among the most intensively studied molecules since last two decades, deciding what is and what is not a miRNA has been difficult and it has been argued that miRBase is riddled with false positives (sequences that are not derived from bona fide miRNA genes) (39–41). Recently, Etebari and Asgari (42) re-annotated miRNAs of P. xylostella and after removing previously reported low confidence precursor miRNAs (pre-miRNAs), finally, they confirmed 114 highly confident pre-miRNAs of P. xylostella following strict criteria. Therefore, in the present study, after successful mapping of clean reads against P. xylostella genome, the mapped miRNA sequences were matched to miRNAs reported by Etebari and Asgari (42). Our analysis initially identified, based on sequence similarity, in total, 187 mature miRNAs. Then, precursor sequences of these mature miRNAs were aligned to those reported by Etebari and Asgari (42), and 99 highly confident pre-miRNAs were confirmed, which produced 167 of 187 mature miRNAs. Our analysis indicated that pre-miRNA sequences of the remaining 20 conserved miRNAs were not detectable in the current assembly of P. xylostella genome. To obtain more reliable results, we removed those known miRNAs with read count < 10 in all libraries, and, finally, remaining 124 known miRNAs with precursor sequences (Table S3 in Supplementary Material), and 17 miRNAs without precursor sequences (Table S4 in Supplementary Material) were retained for further analysis. The remaining sequences that were not matched to conserved miRNAs were used to predict novel miRNAs by using the miRDeep2 program. The most abundant miRNA was pxy-miR-1-3p following pxy-miR-184-3p, pxy-let7-5p, and pxy-miR-31-5p. The top 10 most highly expressed miRNAs in the four libraries of P. xylostella are presented in Figure 2.
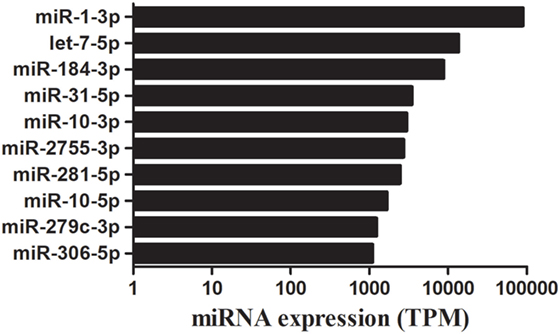
Figure 2. A time course of destruxin A-responsive top 10 abundantly expressed microRNA (miRNAs) in Plutella xylostella. Average expression value of all time courses is presented here. TPM, transcripts per million.
Identification of Destruxin A-Responsive Novel miRNAs
The novel miRNAs with predicted precursor and secondary structures were identified by using miRDeep2 software (43), and, in total, 44 potential novel miRNAs from all the libraries were identified (Table S5 in Supplementary Material) following the standard criteria of novel miRNA prediction with miRDeep score > 1, randfold P-value < 0.05, and MFE < -19 kcal/mol. The most abundant miRNA was pxy-novel-mir-1 following pxy-novel-mir-38, and pxy-novel-mir-3 (Table S5 in Supplementary Material).
Identification of Destruxin A-Responsive Differentially Expressed miRNAs
The differential expression analysis of known miRNAs among the treatments exhibited that 26, 53, and 24 miRNAs were identified as differentially expressed between control and 2, 4, and 6 h, respectively (Figure 3). Of these, 12, 48, and 18 miRNAs were upregulated while 14, 5, and 6 miRNAs were downregulated, respectively (Table S6 in Supplementary Material).
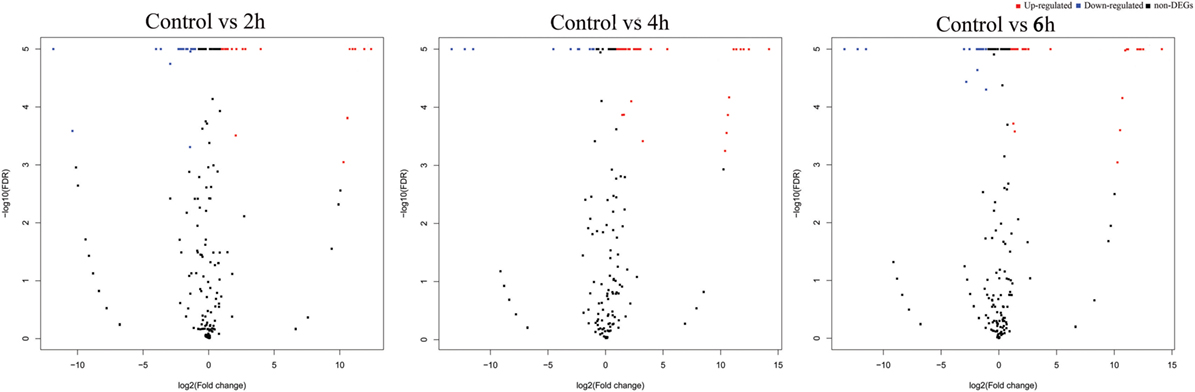
Figure 3. Volcano plots of destruxin A-responsive differentially expressed microRNA (miRNAs) in Plutella xylostella. The volcano plots represent differentially expressed miRNAs at different time courses (2, 4, and 6 h) compared to control.
The novel miRNA differential expression analysis indicated that 16, 17, and 18 miRNAs were identified as differentially expressed between control and 2, 4, and 6 h, respectively (Figure 3). Of these, 10, 12, and 11 miRNAs were upregulated whereas 6, 5, and 7 were downregulated, respectively (Table S7 in Supplementary Material). Overall, most of the differentially expressed miRNAs were upregulated in all the treatments.
Target Prediction of Destruxin A-Responsive Known and Novel miRNAs
The annotation of known and novel miRNA targets is necessary for defining their roles in response to destruxin A treatment. All the annotated genes of P. xylostella were screened using three different algorithms (RNAhybrid, miRanda, and TargetScan) to predict the potential binding sites for destruxin A-responsive miRNAs of P. xylostella. Our target prediction results identified 27,073 common spots between RNAhybrid and TargetScan, 26,989 between RNAhybrid and miRanda, and 28,183 between TargetScan and miRanda. When the target prediction results of all three software were combined, 26,874 common spots were detected and selected for further analysis (Figure 4).
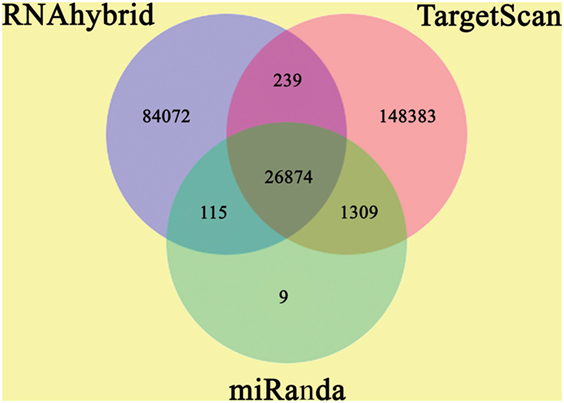
Figure 4. Target prediction of potential target genes of destruxin A-responsive microRNAs (miRNAs) in all libraries of Plutella xylostella. Venn diagram shows the number of miRNA targets and their overlapping spots predicted by the three programs (RNAhybrid, miRanda, and TargetScan).
GO Enrichment and KEGG Pathway Analysis of Destruxin A-Responsive miRNA Target Genes
The GO enrichment analysis was carried out to gain knowledge of the potential function of each putative target gene. The GO enrichment analysis exhibited that cellular process and metabolic process, cell and cell part, and catalytic activity and binding were the most enriched categories in the biological process, cellular component, and molecular function, respectively (Figure 5). The KEGG classification system categorized destruxin A-responsive miRNA target genes into different groups. In the gene repertoire of 2, 4, and 6 h, the top five enriched groups among KEGG categories included signal transduction, cancers, digestive system, immune system, and transport and catabolism (Figure 6).
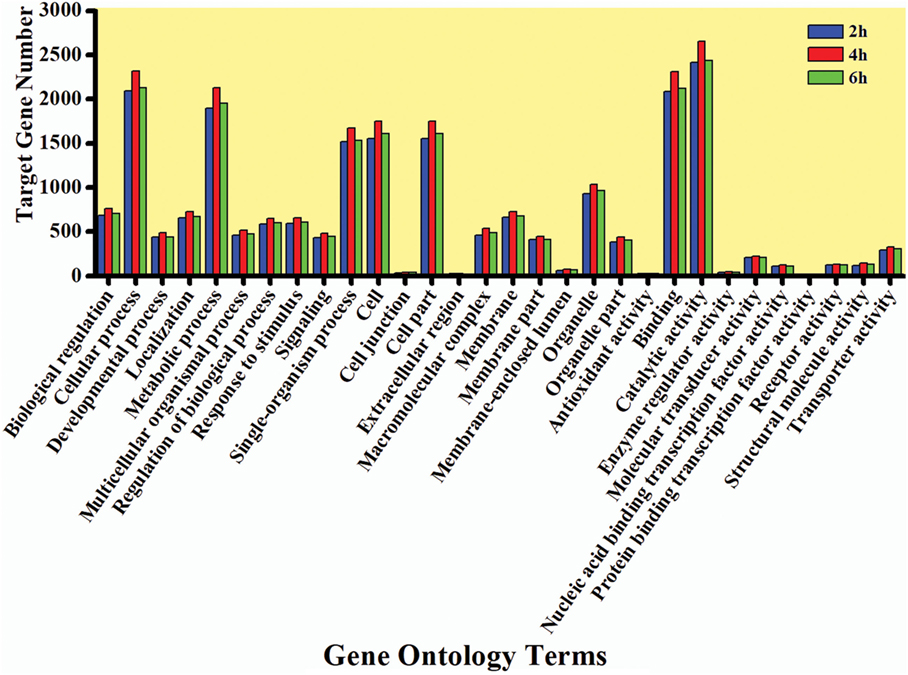
Figure 5. Gene ontology (GO) annotation of target genes of destruxin A-responsive microRNA in all libraries of Plutella xylostella. The abscissa is the GO annotation and the ordinate left is the gene number.
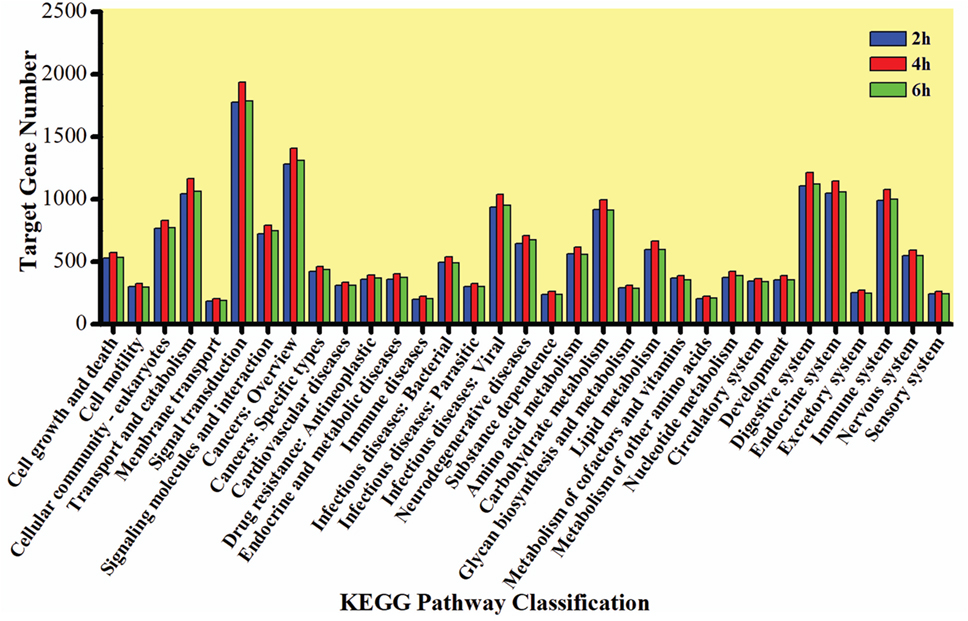
Figure 6. Kyoto Encyclopedia of Genes and Genomes (KEGG) classification of target genes of destruxin A-responsive microRNAs in all libraries of Plutella xylostella. The abscissa is the KEGG classification and the ordinate left is the gene number.
Moreover, KEGG pathway analysis of predicted target genes was carried out to identify potential pathways regulated by destruxin A-responsive differentially expressed miRNAs. The results exhibited that target genes were enriched in cell adhesion and focal adhesion (Figures S3 and S4 in Supplementary Material), as well as several immunity-related signaling pathways, including toll signaling pathway, IMD signaling pathway, and JAK–STAT signaling pathway (Figure 7 and Figure S5 in Supplementary Material).
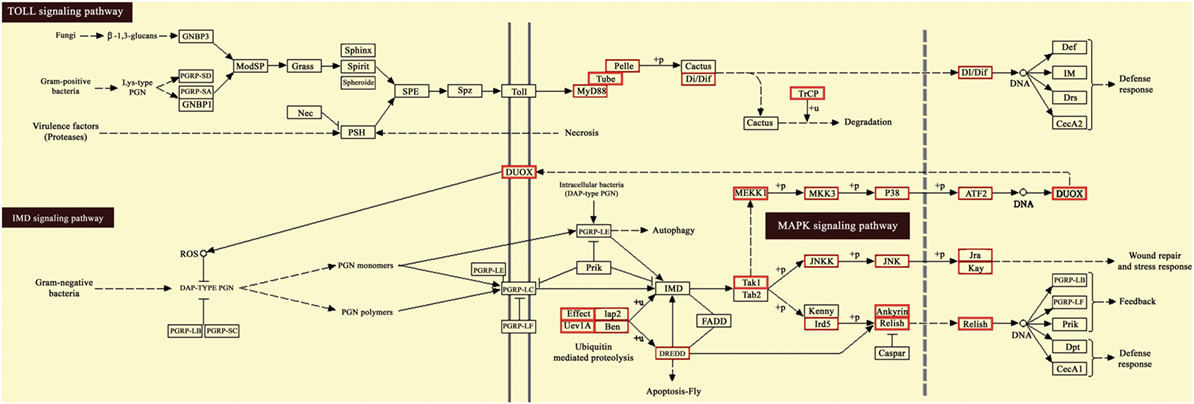
Figure 7. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway map for Toll and IMD signaling pathway. The red boxes indicate target genes of destruxin A-responsive microRNAs (miRNAs) in the particular pathways.
Validation of Differentially Expressed miRNAs by RT-qPCR
To validate sRNA sequencing results, 10 randomly selected differentially expressed miRNAs among control, 2, 4, and 6 h were analyzed by RT-qPCR (Figure 8). The results exhibited that the trend of expression level for the selected miRNAs showed a little discrepancy to that of RNA-Seq analysis, which might be due to the differences in the sensitivity, specificity, and algorithm between the two techniques.
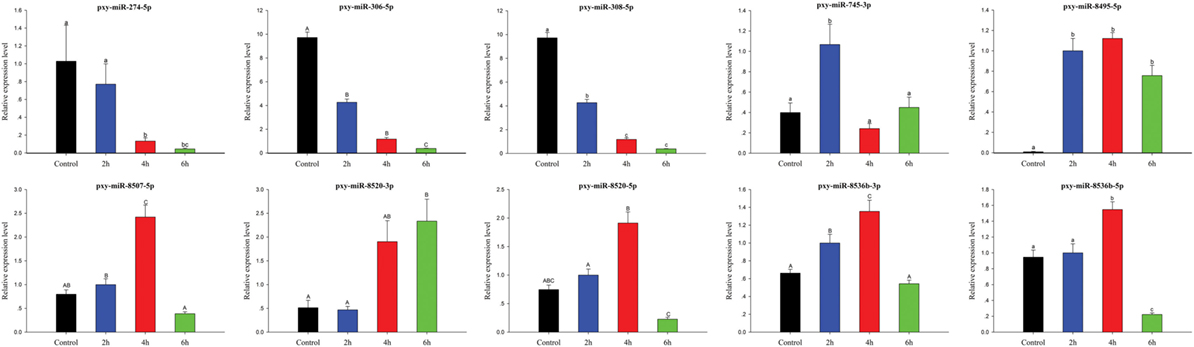
Figure 8. Expression of significantly differentially expressed microRNA (miRNAs) at different time courses after destruxin A injection. Each vertical bar represents the mean ± SEM (n = 3) for various time courses. Statistically significant differences in different groups are indicated by different letters (lowercase letters at P < 0.05 and capital case letters at P < 0.01).
Discussion
The diamondback moth, P. xylostella, has become the major lepidopteran pest of Brassica owing to its strong ability of resistance development to a wide range of insecticides. In the present scenario, there is a need to develop novel biological control methods, to reduce harmful effects of insecticides, as alternative control (44). The entomopathogenic fungi such as M. anisopliae, B. bassiana, and Isaria fumosorosea have gained an increased attention for controlling insect pests as they are considered to offer an environmentally friendly alternative to insecticides (45). The secondary metabolites of fungi, such as destruxins, are reported to have substantial insecticidal activity (46–48) and can destroy the immune system of insects (16, 23). miRNAs, the key regulators of gene expression at the posttranscriptional level, play a pivotal role in host–pathogen interaction. In recent years, the number of miRNAs isolated from insect species is increasing as new ones are deposited in the databases. Although the role of miRNAs in insect development is well understood, there is limited information available about the role of miRNAs in insect–pathogen interaction. Therefore, the present study aimed to acquire the immunity-related miRNAs in P. xylostella treated with destruxin A at three-time courses (2, 4, and 6 h). For this purpose, four sRNA libraries were constructed and sequenced using the high-throughput Illumina sequencing resulting in a total of 187 known and 44 novel miRNAs identified in the four libraries.
The sRNAs are classified into miRNAs, small interfering RNAs, and piwi-interacting RNAs (piRNAs) according to their size (49). Our length distribution results showed two peaks; one at 22 nt and the second at 28 nt, indicating to typical miRNAs and piRNAs. The piRNAs are commonly found in sRNA libraries of insects (50–52) and function as sequence-specific silencers in many organisms (53).
Some conserved miRNAs, such as miR-1, let-7, miR-10, and miR-306, showed abundant expression in the four libraries indicating that these miRNAs play vital regulatory roles in P. xylostella. These miRNAs were also discovered to be abundantly expressed in sRNA libraries of other insects (52, 54); however, a low copy number of miR-1, a conserved miRNA, was detected after parasitization in a previous report (55). Moreover, it is worth mentioning that the expression levels of miR-2755, miR-184, and miR-281 were also abundant in all the treatments indicating that these miRNAs might play a crucial role in P. xylostella immunity against microorganisms.
The differential expression analysis indicated that in total, 26, 53, and 24 known miRNAs were differentially expressed after treatment of P. xylostella with destruxin A at 2, 4, and 6 h, respectively. It is worth mentioning, of these, some conserved miRNAs such as miR-263, miR-279, miR-306, miR-2a, and miR-308 also showed differential expression in the present study. Previous studies reported that these miRNAs (miR-263, miR-279, miR-306, miR-2a, and miR-308) were also differentially expressed after treatment with pathogens indicating that these miRNAs play important role in the regulation of immunity-related genes in insects (52, 55, 56). Notably, miR-263 has been shown to regulate immunity-related signal transduction in Galleria mellonella by affecting the gene expression of tumor necrosis factor receptor superfamily (56). In addition, miR-263 was hypothesized to be involved in the regulation of signal modulation in Manduca sexta by affecting the gene expression of serine protease inhibitors (52). However, in the present study, a lower expression level, less than onefold, was observed. Similar to our findings, miR-279 was also upregulated when P. xylostella was parasitized with Diadegma semiclausum indicating that it plays an important role in the immune response of P. xylostella against parasitoids and microorganisms (55). Interestingly, pxy-miR-306 not only showed differential expression after treatment with destruxin A but was also listed in the top 10 highly expressed miRNAs in our study. It suggests that miR-306 family might be involved in the regulation of immunity-related genes. Previously, it has been found that miR-306 family is associated with Cry1Ab resistance in Ostrinia furnacallis (57). Of note, pxy-miR-308 was commonly differentially expressed in all the treatments indicating that it might play a vital role in immunity of P. xylostella against destruxin A by regulating immunity-related genes. According to a previous report, Mse-miR-308 was supposed to be involved in extracellular signal transduction and melanization (52).
To better understand the function of each putative target gene, GO annotation and KEGG pathway analysis were performed. According to GO annotation, the differentially expressed target genes were mainly classified in a cellular process, cell, catalytic activity, and metabolic process (Figure 5). Previously, similar GO annotation of differentially expressed target genes was obtained in O. furnacallis in response to Bacillus thuringiensis and Wolbachia-responsive miRNAs in Tetranychus urticae (57, 58).
It is of interest that KEGG pathway annotation of target genes resulted in the identification of several immunity-related signaling pathways, including toll signaling pathway, IMD signaling pathway, JAK–STAT signaling pathway, and cell adhesion molecules pathway. The toll signaling pathway is a crucial pathway in the innate immunity (59), and in insects, it is responsible for fungi and Gram-positive bacteria recognition (60). The presence of miRNA target genes in toll signaling pathway in the current study suggests that some miRNAs regulate innate immunity against the entomopathogenic fungi via toll pathway. Previous studies on the model insect D. melanogaster have also indicated the role of toll pathway in immunity against fungi and pointed out that toll-mutant flies were highly susceptible to fungi (61).
Interestingly, the cell adhesion molecules pathway was highly enriched in the current study. Cell adhesion molecules, glycoproteins, are expressed on the surface of a cell and are reported to play a vital role in biological processes, including immune response (62). These cell adhesion molecules are categorized as integrin family, selectins, immunoglobulin superfamily, and cadherins. The immunoglobulin superfamily proteins are reported to play an important role in facilitating specific interactions with particular pathogens (63). Taken together, the current study clearly presents patterns of differentially expressed miRNAs in P. xylostella treated with destruxin A at different time courses.
Conclusion
Concluding our findings, the present study adopted high-throughput sRNA sequencing to systematically screen out destruxin A-responsive immunity-related miRNAs in P. xylostella. According to our information, this is the first study about immunity-related miRNA profiles of P. xylostella in response to pathogens especially on secondary metabolites of entomopathogenic fungi like destruxin A. In the current study, several miRNAs that may regulate immunity through their targets and related pathways were identified. Among them, miR-263, miR-279, miR-306, miR-2a, and miR-308 are worthy to mention as these showed differential expression at different time courses and have also been predicted to regulate immunity in the other insects. Our findings will provide a strong foundation for further functional studies of miRNAs regulating immunity-related target genes of P. xylostella in response to microorganisms.
Ethics Statement
Our work confirms to the legal requirements of the country in which it was carried out.
Author Contributions
Conceived and designed the experiments: FJ, MS, and XiaoxX. Performed the experiments: MS, JX, and XiaoxX. Analyzed the data: MS, XiaojX, JY, XZ, and JX. Contributed reagents/materials/analysis tools: SL and QH. Wrote the manuscript: MS and XiaoxX. Revised the manuscript: FJ and YX.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. Urfa Bin Tahir (Department of Aquatic Animal Medicine, College of Fisheries, Huazhong Agricultural University, Wuhan, China) for preparation of figures.
Funding
This work was supported by a grant from The National Natural Science Foundation of China (31572069, 31371989), and Department of Science and Technology of Guangdong China (2015A020209128, 2016A020210080).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/article/10.3389/fimmu.2018.00185/full#supplementary-material.
References
1. Hussain M, Asgari S. MicroRNAs as mediators of insect host-pathogen interactions and immunity. J Insect Physiol (2014) 70:151–8. doi:10.1016/j.jinsphys.2014.08.003
2. Nappi AJ, Ottaviani E. Cytotoxicity and cytotoxic molecules in invertebrates. Bioessays (2000) 22:469–80. doi:10.1002/(SICI)1521-1878(200005)22:5<469:AID-BIES9>3.0.CO;2-4
3. Kanost MR, Jiang H, Yu XQ. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol Rev (2004) 198:97–105. doi:10.1111/j.0105-2896.2004.0121.x
4. Cerenius L, Söderhäll K. The prophenoloxidase-activating system in invertebrates. Immunol Rev (2004) 198:116–26. doi:10.1111/j.0105-2896.2004.00116.x
5. Steiner H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol Rev (2004) 198:83–96. doi:10.1111/j.0105-2896.2004.0120.x
6. Hultmark D. Drosophila immunity: paths and patterns. Curr Opin Immunol (2003) 15:12–9. doi:10.1016/S0952-7915(02)00005-5
7. Michel K, Kafatos F. Mosquito immunity against Plasmodium. Insect Biochem Mol Biol (2005) 35:677–89. doi:10.1016/j.ibmb.2005.02.009
8. Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol (2000) 30:95–105. doi:10.1016/S0965-1748(99)00113-7
9. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell (1993) 75:843–54. doi:10.1016/0092-8674(93)90529-Y
10. Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA (2006) 12:1161–7. doi:10.1261/rna.2322506
11. Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, et al. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell (2005) 121:1097–108. doi:10.1016/j.cell.2005.04.016
12. Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila microRNA targets. PLoS Biol (2003) 1:e60. doi:10.1371/journal.pbio.0000060
13. Legeai F, Rizk G, Walsh T, Edwards O, Gordon K, Lavenier D, et al. Bioinformatic prediction, deep sequencing of microRNAs and expression analysis during phenotypic plasticity in the pea aphid, Acyrthosiphon pisum. BMC Genomics (2010) 11:281. doi:10.1186/1471-2164-11-281
14. Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell (2003) 113:25–36. doi:10.1016/S0092-8674(03)00231-9
15. Harris JF, Micheva-Viteva S, Li N, Hong-Geller E. Small RNA-mediated regulation of host-pathogen interactions. Virulence (2013) 4:785–95. doi:10.4161/viru.26119
16. Pal S, Leger RJS, Wu LP. Fungal peptide destruxin A plays a specific role in suppressing the innate immune response in Drosophila melanogaster. J Biol Chem (2007) 282:8969–77. doi:10.1074/jbc.M605927200
17. Hu Q-B, An X-C, Jin F-L, Freed S, Ren S-X. Toxicities of destruxins against Bemisia tabaci and its natural enemy, Serangium japonicum. Toxicon (2009) 53:115–21. doi:10.1016/j.toxicon.2008.10.019
18. Meng X, Xu X, Hu J, Jin F, Hu Q, Sun Q, et al. Toxicity and differential protein analysis following destruxin A treatment of Spodoptera litura (Lepidoptera: Noctuidae) SL-1 cells. Toxicon (2011) 58:327–35. doi:10.1016/j.toxicon.2011.06.002
19. Ruiz-Sanchez E, Orchard I, Lange AB. Effects of the cyclopeptide mycotoxin destruxin A on the Malpighian tubules of Rhodnius prolixus (Stål). Toxicon (2010) 55:1162–70. doi:10.1016/j.toxicon.2010.01.006
20. Bandani AR, Amiri B, Butt TM, Gordon-Weeks R. Effects of efrapeptin and destruxin, metabolites of entomogenous fungi, on the hydrolytic activity of a vacuolar-type ATPase identified on the brush border membrane vesicles of Galleria mellonella midgut and on plant membrane-bound hydrolytic enzymes. Biochim Biophys Acta (2001) 1510:367–77. doi:10.1016/S0005-2736(00)00370-9
21. Samuels R, Charnley A, Reynolds S. The role of destruxins in the pathogenicity of 3 strains of Metarhizium anisopliae for the tobacco hornworm Manduca sexta. Mycopathologia (1988) 104:51–8. doi:10.1007/BF00437924
22. Samuels R, Reynolds S, Charnley A. Calcium channel activation of insect muscle by destruxins, insecticidal compounds produced by the entomopathogenic fungus Metarhizium anisopliae. Comp Biochem Physiol C Pharmacol (1988) 90:403–12. doi:10.1016/0742-8413(88)90018-7
23. Gong L, Chen X, Liu C, Jin F, Hu Q. Gene expression profile of Bombyx mori hemocyte under the stress of destruxin A. PLoS One (2014) 9:e96170. doi:10.1371/journal.pone.0096170
24. Lin H, Lin X, Zhu J, Yu X-Q, Xia X, Yao F, et al. Characterization and expression profiling of serine protease inhibitors in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). BMC Genomics (2017) 18:162. doi:10.1186/s12864-017-3583-z
25. Xia X, Yu L, Xue M, Yu X, Vasseur L, Gurr GM, et al. Genome-wide characterization and expression profiling of immune genes in the diamondback moth, Plutella xylostella (L.). Sci Rep (2015) 5:9877. doi:10.1038/srep09877
26. Han P, Jin F, Dong X, Fan J, Qiu B, Ren S. Transcript and protein profiling analysis of the destruxin A-induced response in larvae of Plutella xylostella. PLoS One (2013) 8:e60771. doi:10.1371/journal.pone.0060771
27. Shakeel M, Xu X, Xu J, Zhu X, Li S, Zhou X, et al. Identification of immunity-related genes in Plutella xylostella in response to fungal peptide destruxin A: RNA-Seq and DGE analysis. Sci Rep (2017) 7:10966. doi:10.1038/s41598-017-11298-7
28. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods (2012) 9:357–9. doi:10.1038/nmeth.1923
29. Abdi H. Bonferroni and Šidák corrections for multiple comparisons. In: Salkind N editor. Encyclopedia of Measurement and Statistics. (Vol. 3), Thousand Oaks, CA; London: SAGE (2007). p. 103–7.
30. Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res (2006) 34:W451–4. doi:10.1093/nar/gkl243
31. Enright AJ, John B, Sander C, Marks DS, Tuschl T, Gaul U. MicroRNA targets in Drosophila. Genome Biol (2003) 5:R1. doi:10.1186/gb-2003-5-1-r1
32. Agarwal V, Bell GW, Nam J-W, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. elife (2015) 4:e05005. doi:10.7554/eLife.05005
33. Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell (2005) 121:207–21. doi:10.1016/j.cell.2005.04.004
34. Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell (2005) 8:517–27. doi:10.1016/j.devcel.2005.01.018
35. Shakeel M, Rodriguez A, Tahir UB, Jin F. Gene expression studies of reference genes for quantitative real-time PCR: an overview in insects. Biotechnol Lett (2017). doi:10.1007/s10529-017-2465-4
36. Shakeel M, Zhu X, Kang T, Wan H, Li J. Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). J Asia Pac Entomol (2015) 18:123–30. doi:10.1016/j.aspen.2015.01.001
37. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods (2001) 25:402–8. doi:10.1006/meth.2001.1262
38. Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci U S A (2007) 104:18097–102. doi:10.1073/pnas.0709193104
39. Langenberger D, Bartschat S, Hertel J, Hoffmann S, Tafer H, Stadler P. MicroRNA or not microRNA? In: Norberto de Souza O, Telles GP, Palakal M, editors. Advances in Bioinformatics and Computational Biology. (Vol. 6832), Lecture Notes in Computer Science, Berlin, Germany: Springer, Heidelberg (2011). p. 1–9.
40. Meng Y, Shao C, Wang H, Chen M. Are all the miRBase-registered microRNAs true? A structure-and expression-based re-examination in plants. RNA Biol (2012) 9:249–53. doi:10.4161/rna.19230
41. Wang X, Liu XS. Systematic curation of miRBase annotation using integrated small RNA high-throughput sequencing data for C. elegans and Drosophila. Front Genet (2011) 2:25. doi:10.3389/fgene.2011.00025
42. Etebari K, Asgari S. Revised annotation of Plutella xylostella microRNAs and their genome-wide target identification. Insect Mol Biol (2016) 25:788–99. doi:10.1111/imb.12263
43. Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res (2012) 40:37–52. doi:10.1093/nar/gkr688
44. Shakeel M, Farooq M, Nasim W, Akram W, Khan FZA, Jaleel W, et al. Environment polluting conventional chemical control compared to an environmentally friendly IPM approach for control of diamondback moth, Plutella xylostella (L.), in China: a review. Environ Sci Pollut Res Int (2017) 24:14537–50. doi:10.1007/s11356-017-8996-3
45. Xu J, Xu X, Shakeel M, Li S, Wang S, Zhou X, et al. The entomopathogenic fungi Isaria fumosorosea plays a vital role in suppressing the immune system of Plutella xylostella: RNA-Seq and DGE analysis of immunity-related genes. Front Microbiol (2017) 8:1421. doi:10.3389/fmicb.2017.01421
46. Kershaw M, Moorhouse E, Bateman R, Reynolds S, Charnley A. The role of destruxins in the pathogenicity of Metarhizium anisopliae for three species of insect. J Invertebr Pathol (1999) 74:213–23. doi:10.1006/jipa.1999.4884
47. Pedras MSC, Zaharia LI, Ward DE. The destruxins: synthesis, biosynthesis, biotransformation, and biological activity. Phytochemistry (2002) 59:579–96. doi:10.1016/S0031-9422(02)00016-X
48. Liu B-L, Tzeng Y-M. Development and applications of destruxins: a review. Biotechnol Adv (2012) 30:1242–54. doi:10.1016/j.biotechadv.2011.10.006
49. Lucas K, Raikhel AS. Insect microRNAs: biogenesis, expression profiling and biological functions. Insect Biochem Mol Biol (2013) 43:24–38. doi:10.1016/j.ibmb.2012.10.009
50. Cristino AS, Tanaka ED, Rubio M, Piulachs M-D, Belles X. Deep sequencing of organ-and stage-specific microRNAs in the evolutionarily basal insect Blattella germanica (L.) (Dictyoptera, Blattellidae). PLoS One (2011) 6:e19350. doi:10.1371/journal.pone.0019350
51. Yu X, Zhou Q, Li S-C, Luo Q, Cai Y, Lin W-C, et al. The silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PLoS One (2008) 3:e2997. doi:10.1371/journal.pone.0002997
52. Zhang X, Zheng Y, Jagadeeswaran G, Ren R, Sunkar R, Jiang H. Identification and developmental profiling of conserved and novel microRNAs in Manduca sexta. Insect Biochem Mol Biol (2012) 42:381–95. doi:10.1016/j.ibmb
53. Kawaoka S, Arai Y, Kadota K, Suzuki Y, Hara K, Sugano S, et al. Zygotic amplification of secondary piRNAs during silkworm embryogenesis. RNA (2011) 17:1401–7. doi:10.1261/rna.2709411
54. Mehrabadi M, Hussain M, Asgari S. MicroRNAome of Spodoptera frugiperda cells (Sf9) and its alteration following baculovirus infection. J Gen Virol (2013) 94:1385–97. doi:10.1099/vir.0.051060-0
55. Etebari K, Hussain M, Asgari S. Identification of microRNAs from Plutella xylostella larvae associated with parasitization by Diadegma semiclausum. Insect Biochem Mol Biol (2013) 43:309–18. doi:10.1016/j.ibmb.2013.01.004
56. Mukherjee K, Vilcinskas A. Development and immunity-related microRNAs of the lepidopteran model host Galleria mellonella. BMC Genomics (2014) 15:705. doi:10.1186/1471-2164-15-705
57. Xu L-N, Ling Y-H, Wang Y-Q, Wang Z-Y, Hu B-J, Zhou Z-Y, et al. Identification of differentially expressed microRNAs between Bacillus thuringiensis Cry1Ab-resistant and-susceptible strains of Ostrinia furnacalis. Sci Rep (2015) 5:15461. doi:10.1038/srep15461
58. Rong X, Zhang Y-K, Zhang K-J, Hong X-Y. Identification of Wolbachia-responsive microRNAs in the two-spotted spider mite, Tetranychus urticae. BMC Genomics (2014) 15:1122. doi:10.1186/1471-2164-15-1122
59. Yamamoto M, Takeda K. Current views of toll-like receptor signaling pathways. Gastroenterol Res Pract (2010) 2010:240365. doi:10.1155/2010/240365
60. Stokes BA, Yadav S, Shokal U, Smith L, Eleftherianos I. Bacterial and fungal pattern recognition receptors in homologous innate signaling pathways of insects and mammals. Front Microbiol (2015) 6:19. doi:10.3389/fmicb.2015.00019
61. Lemaitre B, Reichhart J-M, Hoffmann JA. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci U S A (1997) 94:14614–9. doi:10.1073/pnas.94.26.14614
62. Elangbam C, Qualls C Jr, Dahlgren R. Cell adhesion molecules—update. Vet Pathol (1997) 34:61–73. doi:10.1177/030098589703400113
Keywords: microRNAs, destruxin A, immunity, innate, Plutella xylostella, differential expression analysis
Citation: Shakeel M, Xu X, Xu J, Li S, Yu J, Zhou X, Xu X, Hu Q, Yu X and Jin F (2018) Genome-Wide Identification of Destruxin A-Responsive Immunity-Related MicroRNAs in Diamondback Moth, Plutella xylostella. Front. Immunol. 9:185. doi: 10.3389/fimmu.2018.00185
Received: 31 August 2017; Accepted: 22 January 2018;
Published: 08 February 2018
Edited by:
Ram Savan, University of Washington, United StatesReviewed by:
Hai-peng Liu, Xiamen University, ChinaJesus Hernandez, Center for Research in Food and Development (CIAD), Mexico
Copyright: © 2018 Shakeel, Xu, Xu, Li, Yu, Zhou, Xu, Hu, Yu and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengliang Jin, jflbang@scau.edu.cn
†These authors have contributed equally to this work.
 Muhammad Shakeel
Muhammad Shakeel Xiaoxia Xu1†
Xiaoxia Xu1† Jin Xu
Jin Xu Jialin Yu
Jialin Yu Xianqiang Zhou
Xianqiang Zhou Qiongbo Hu
Qiongbo Hu Xiaoqiang Yu
Xiaoqiang Yu Fengliang Jin
Fengliang Jin