- Department of Microbiology and Immunology, The Peter Doherty Institute for Infection and Immunity, The University of Melbourne, Melbourne, VIC, Australia
Mucosal-associated invariant T (MAIT) cells are a subset of unconventional T cells that recognize the evolutionarily conserved major histocompatibility complex (MHC) class I-like antigen-presenting molecule known as MHC class I related protein 1 (MR1). Since their rise from obscurity in the early 1990s, the study of MAIT cells has grown substantially, accelerating our fundamental understanding of these cells and their possible roles in immunity. In the context of recent advances, we review here the relationship between MR1, antigen, and TCR usage among MAIT and other MR1-reactive T cells and provide a speculative discussion.
Introduction
Conventional T cells recognize peptide antigens presented by the major histocompatibility complex (MHC) class I or class II (MHC-I or MHC-II) molecules and elicit a cellular immune response to provide anti-microbial and anti-tumoral immunity for the host. Conventional T cells utilize a cell surface bound T cell receptor (TCR) to recognize peptide-MHC complexes presented on the surface of antigen-presenting cells (APCs). The TCR is a heterodimer comprised of a TCR α- and β-chain, each consisting of a variable (V) and constant (C) domain. Three sets of finger-like extensions on the TCRα- and β-chain variable domains, known as complementarity determining loops (CDR1, CDR2, and CDR3), are responsible for recognizing the peptide-MHC-complex and are collectively unique to each TCR. CDR1 and CDR2 loops are germline-encoded by the V gene segments, while the CDR3 loops are formed from somatic recombination of the V gene segments with a joining (J) (V-J) and/or diversity (D) gene segment (V-D-J) during T cell development (1). Together with random nucleotide additions and deletions in the CDR3 loop gene regions, this creates an enormous diversity of TCRs, clonally distributed amongst T cells (2).
Mucosal-associated invariant T (MAIT) cells are a subset of unconventional T cells that are restricted by a monomorphic MHC-I-like molecule, known as MHC class I related protein 1 (MR1) (3–6). In contrast to conventional T cells, MAIT cells express a “semi-invariant” αβ TCR, meaning they typically express the same TCRα chain paired to a preferred array of TCRβ chains (3). In humans, the “classical MAIT TCR” comprises a TCRα chain encoded by the V gene segment TRAV1-2 juxtaposed to the J gene segment TRAJ33, TRAJ20, or TRAJ12 (5, 7–9) that pairs preferentially with a TCRβ chain encoded by TRBV6 or TRBV20 V gene segment family members (7). In mice, the MAIT TCR is composed of homologous gene segments, a TRAV1-TRAJ33 TCRα chain, that pairs with a TCRβ chain composed of TRBV19 or TRBV13 V gene segments, both of which are murine orthologous segments of human TRBV6 (3, 5, 10). MAIT cells are abundant in healthy adults, representing on average 3% of blood T cells (11) and can be found throughout peripheral tissues (6, 7, 10, 12–23). The frequency of MAIT cells in laboratory mice is distinctly lower than in humans, although murine MAIT cells are also found in many peripheral organs (24, 25). The prototypical antigen presented by MR1 to MAIT cells is the small molecule 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU), an adduct of the riboflavin biosynthetic precursor 5-amino-6-D-ribitylaminouracil (5-A-RU) and methylglyoxal (26) (Figure 1). See recent reviews for details on the riboflavin biosynthesis and formation of 5-OP-RU from 5-A-RU (31, 32). Riboflavin biosynthesis is absent in mammals. Thus, by recognizing 5-OP-RU (25, 33, 34), and potentially other riboflavin-based ligands presented by MR1 (35), MAIT cells are able to sense a broad range of riboflavin biosynthesis proficient microbes in a highly conserved, innate-like manner, reviewed in (32). Human MAIT cells stimulated with 5-OP-RU rapidly secrete T helper (Th)1 and Th17 type cytokines (11, 36, 37) as well as cytotoxic granules (38). In mice, lung infection with riboflavin-synthesizing bacteria or co-administration of synthetic 5-OP-RU with adjuvant leads to a significant expansion of MAIT cells with Th1/17 cytokine secreting capacity (25, 34, 39), enabling MAIT cells to contribute to protection against several pathogens, including Klebsiella pneumoniae (40), Mycobacterium bovis BCG (41), Francisella tularensis (39), E. coli (42), Legionella longbeachae (34), and Clostridium difficile (43). Thus, observations to date suggest MAIT cells are poised, but perhaps not limited to, protecting peripheral tissues from microbial pathogen or commensal breach. In particular, MAIT cells have recently been shown to contribute to tissue repair at barrier sites (44–47). MAIT cells may also be involved in the tumoral immune response (48–52), however, elevated MAIT cell numbers at the tumor site in some cancers correlate with a poorer prognosis (49, 52). Notably, MAIT cells appear to be subject to a similar fate as conventional T cells during the anti-tumoral immune response, namely: T cell exhaustion, altered functional response, altered frequency, and drug sensitivity (50, 52–57). A cytokine-modulated (IL-7, IL-12, IL-18) tumor response that occurs independent of, or concurrent with, TCR stimulation should also be considered in the context of tumoral immunity, as MAIT cells are known to respond to inflammatory stimuli in this manner (15, 58, 59). Furthermore, MAIT cells from healthy donors can efficiently lyse MR1 proficient tumor cells presenting microbial agonists such as 5-OP-RU, suggested as a potential strategy to harness the MAIT cell response therapeutically (56). Perhaps similar in mechanism, disruption of barrier tissues (i.e., colorectal cancers) by tumors may allow invasive growth of commensal bacteria, providing a source of microbial ligand in the context of an inflammatory environment which may trigger anti-tumor MAIT cell responses (48–50, 60). Much is still unknown regarding the response by MAIT cells in the tumoral environment, particularly whether tumor associated, MAIT cell specific MR1 ligands exist and the factors that might drive MAIT cell to become pro- or anti-tumoral. MAIT cells have, however, attracted some interest as a potential immunotherapeutic target as they possess a number of favorable attributes such as a high precursor frequency, wide tissue distribution, potent cytokine response and cytotoxicity and a donor unrestricted nature (61).
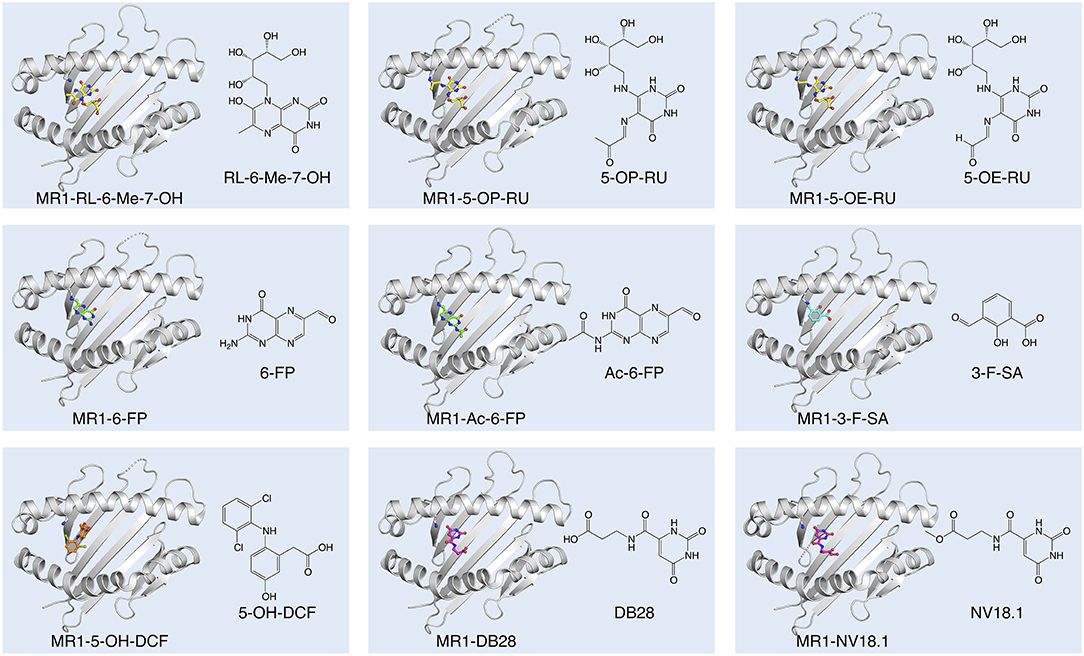
Figure 1. Diversity of small molecule ligands presented by MR1. Cartoon display (light gray) of the MR1 antigen-binding cleft (top-view) and ball-and-stick display of the antigen (colored) based on the protein data bank (PDB) deposited crystal structures, featuring the human A-F7 MAIT TCR in complex with human MR1-RL-6-Me-7-OH [PDB ID: 4L4V (27)], MR1-5-OP-RU and MR1-5-OE-RU [PDB IDs: 4NQC, 4NQE (26)], MR1-6-FP [PDB ID: 4L4T (27)], MR1-Ac-6-FP [PDB ID: 4PJF (28)], MR1-3-F-SA and MR1-5-OH-DCF [PDB IDs: 5U6Q, 5U72 (29)], and MR1-DB28 and MR1-NV18.1 [PDB IDs:6PVC and 6PVD (30)].
The Riboflavin-Based MR1 Ligands
Independent observations from Gold et al. and Bourhis et al. demonstrated that a wide range of bacteria and yeasts, and their supernatants, are capable of stimulating MAIT cells in an MR1-dependent manner (36, 62). On the assumption that MR1 would likely adopt a “MHC-I-fold” (63) in the presence of ligand, Kjer-Nielsen et al. folded soluble recombinant MR1 proteins in the presence of bacterial supernatant to capture ligands in the form of stable MR1-ligand-complexes (35). This approach of ligand-capture, combined with mass-spectrometry, and subsequent genetic manipulation of the riboflavin biosynthetic pathway in bacteria, led to the discovery of the pyrimidines; 5-OP-RU and 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil (5-OE-RU), and the substantially less potent, cyclised ribityllumazines; 7-hydroxy-6-methyl-8-D-ribityllumazine (RL-6-Me-7-OH); and 7-dimethyl-8-D-ribityllumazine (RL-6,7-diMe) as riboflavin-based, MR1-presented, MAIT cell stimulating antigens (26, 27, 32, 35) (Figure 1). While both 5-OP-RU and 5-OE-RU were detected in the supernatant of Escherichia coli and Salmonella enterica serovar Typhimurium, 5-OP-RU is the dominant MAIT cell antigen found in these bacterial supernatants (26). Nevertheless, MR1 tetramers loaded with 5-OE-RU stain a similar proportion of T cells compared to those loaded with 5-OP-RU (26) and synthetic versions of both antigens are similarly potent (64, 65). 5-A-RU co-incubated with methylglyoxal is, however, more potent in activating MAIT cells than 5-A-RU co-incubated with glyoxal (26, 66), presumably due to differences in the catabolic and metabolic kinetics of 5-OP-RU and 5-OE-RU (64). 5-OP-RU has emerged as the preferred antigen for research, both as synthetic antigen and loaded in MR1 tetramers. Accordingly, MR1-5-OP-RU tetramers have become the gold standard for identifying MAIT cells (67), enabling more accurate MAIT cell detection than using antibodies specific for surrogate markers (TRAV1-2, CD161, and CD8) (11). MR1 presentation of 5-OP-RU and 5-OE-RU involves the formation of a Schiff base between the Lys43 of MR1 and a residual carbonyl group on the ligand. This Schiff base, unique to MR1 ligand presentation amongst MHC molecules, stabilizes, and guides 5-OP-RU and 5-OE-RU into their position on MR1 in the A'-pocket amongst a cradle of aromatic residues (26). Notably, even in the absence of the Schiff base, by mutating Lys43 in MR1 to Ala (68), 5-OP-RU is presented by MR1 in the identical position (26), although the stability of this complex is markedly reduced (28). Unlike the pyrimidine ligands, RL-6-Me-7-OH does not form a Schiff base with MR1, however residues within the A'-pocket of MR1 help orient the ligand into a similar position (27). In a physiological context, neutralization of MR1 Lys43 by covalently bound ligand is essential for complete MR1 folding and trafficking from the endoplasmic reticulum (ER) to the cell surface (69).
All classical MAIT TCRs studied to date recognize MR1 presenting riboflavin-based antigens using a surprisingly consistent mode of docking, orthogonally over the cleft of MR1 and forming multiple contacts with aromatic residues that surround the ligand-binding site, notably residues Tyr62 and Tyr152 (26–28, 68, 70). The TCR also directly contacts the antigen, which occupies only 0.6% of the exposed surface in the MR1 cleft (70). Crucial to antigen recognition by classical MAIT TCRs is Tyr95α, a highly conserved residue in the CDR3α loop which directly contacts the 2′-OH group on the antigen ribityl moiety (Figure 2A) (26–28, 68). Structure-function studies with synthetic derivatives of 5-OP-RU and other riboflavin-based antigens have provided valuable insights on which chemical group modifications affect ligand antigenicity within the MAIT-MR1 axis (64, 65). While improving ligand stability, modification of the pyrimidine backbone of 5-OP-RU (C-5 and C-6) (Figure 2A), had only small effects on altering antigen potency (64), perhaps unsurprising as these atoms make no direct contact with the MAIT TCR (26, 28, 64, 65). In contrast, modifications to the ribityl moiety of 5-OP-RU and ribityllumazine ligands had profoundly negative effects on MAIT TCR recognition and antigen potency (65, 71). Particularly, removal of the ribityl 2′- and 3′-OH groups or the entire ribityl tail largely abolished MAIT TCR recognition of relevant 5-OP-RU analogs (65, 71). In contrast, shortening of the ribityl tail, while preserving the 2′- and 3′-OH groups (ethyl- or propyl-5-OP-RU analogs) did not appreciably affect MAIT TCR recognition (65, 71). Notably however, 2′-OH was not required for MAIT TCR recognition of a modified analog of the ribityllumazine RL-6-Me-7-OH (72). Structural modeling suggested the modified ribityl tail of the 2′-deoxy-RL-6-Me-7-OH is flexible enough to be oriented for recognition by the MAIT TCR Tyr95α via a hydrogen bond with the 5′-OH in place of the 2′-OH group (Figure 2A) (72). Although MAIT cells could recognize MR1 presenting the 2-deoxyribityllumazine, this interaction was not sufficient to lead to MAIT cell activation (72), stressing that distinct requirements exist for MAIT cell activation that are not satisfied by TCR engagement alone. Thus, the series of altered metabolite ligands helped to define the boundaries of riboflavin-based antigen diversity tolerated by the MAIT TCR and emphasized the importance of the ribityl tail and Tyr95α in MAIT TCR recognition. In light of this, new microbial ligands have been identified that share the ribityl moiety and some structural features with the riboflavin-based antigens (73). The lumazines 6-(2-carboxyethyl)-7-hydroxy-8-ribityllumazine (photolumazine I; PLI) and 6-(1H-indol-3-yl)-7-hydroxy-8-ribityllumazine (photolumazine III; PLIII) and the riboflavin analog 7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO) were isolated from soluble recombinant MR1 expressed in the context of live bacterial infection (Figure 2B) (73). Although all three ligands had ribityl moieties, only PLI and PLIII, possible adducts of 5-A-RU, were antigenic to MAIT cells, while FO was antagonistic (73). Similarly, three pyrimidines, mercaptopurine, floxuridine, and doxofylline (Figure 2C) were identified as putative MR1 ligands in an in silico screen of a range of small organic molecules, including drugs, drug metabolites, and drug-like molecules (29). In cellular assays, floxuridine and mercaptopurine were weakly agonistic but did not upregulate MR1 to detectable levels, while doxofylline weakly upregulated MR1 but was not agonistic (29). Further in silico screening of commercial compound libraries recently identified seven weakly agonistic MR1 ligands (Figure 2D) (30). None of the novel agonist ligands were as potent as 5-OP-RU in cellular assays or were able to upregulate MR1 surface expression, likely due to the inability to form a Schiff base with MR1 Lys43 (30). In contrast, in the same study, two pyrimidines were identified; 3-[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)formamido] propanoic acid (DB28) and its derivative methyl 3-[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)formamido] propanoate (NV18.1) that significantly modulated MR1 surface expression (30). These ligands bound to MR1 non-covalently (Figure 1), causing MR1 to be retained in the ER and prevented surface expression (30). Importantly, unlike the other ligands, DB28 and NV18.1 were non-stimulatory to PBMC-derived TRAV1-2+ MAIT cells (30).
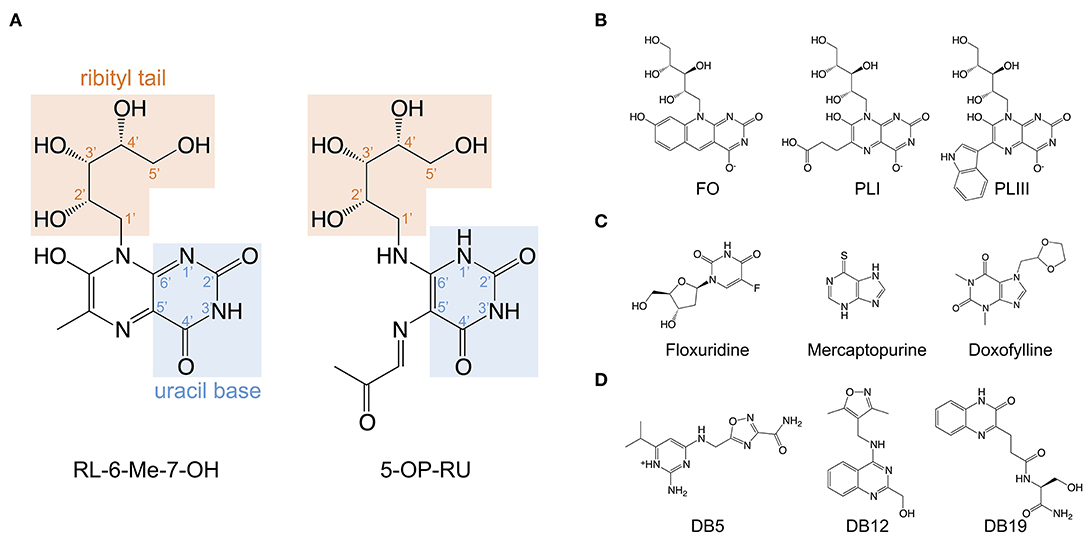
Figure 2. Structures of the riboflavin-based and related MR1 ligands. (A) Chemical structure of 7-hydroxy-6-methyl-8-D-ribityllumazine (RL-6-Me-7-OH) and 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) annotated with the position numbers for relevant functional groups. (B) MR1 ligands isolated from soluble recombinant MR1 expressed with E. coli or M. smegmatis; lumazines 6-(2-carboxyethyl)-7-hydroxy-8-ribityllumazine (photolumazine I; PLI) and 6-(1H-indol-3-yl)-7-hydroxy-8-ribityllumazine (photolumazine III; PLIII) and the riboflavin analog 7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO). (C,D). A selection of MAIT cell agonist and non-agonist MR1 ligands identified from in silico screens of multiple chemical libraries; mercaptopurine, floxuridine, doxofylline, and 2-amino-4-(((3-carbamoyl-1,2,4-oxadiazol-5-l)methyl)amino)-6-isopropylpyrimidin-1-ium (DB5), (4-(((3,5-dimethylisoxazol-4-yl)methyl)amino)quinazolin-2-yl)methanol (DB12) and (S)-N-(1-amino-3-hydroxy-1-oxopropan-2-yl)-3-(3-oxo-3,4-dihydroquinoxalin-2-yl)propenamide (DB19).
Characteristics of the MAIT TCR Repertoire
The classical MAIT TCRα rearrangements (TRAV1-2-TRAJ33/20/12) are highly conserved between genetically unrelated individuals, also referred to as public TCRα chain usage (3, 5). Collectively, these TCRα rearrangements account for the majority (~95%) of MAIT TCR clonotypes in human blood (7–9, 11, 62, 74). Nevertheless, variations of the classical MAIT TCR CDR3α region exist, typically at one or two non-germline-encoded residue positions (CAXXDSSYKLIF, CAVXXXDYKLSF, and CAXXDSNYQLIW for TRAJ12, TRAJ20, and TRAJ33 MAIT CDR3α rearrangements, respectively) (5, 11, 74). However, MR1-reactive clones responding to different microbes (bacteria and yeast) are characterized by more extensive non-germline-encoded differences within the TCRα chain, including what appeared to be microbe-specific MR1-reactive clonotypes in some instances (8). Similarly, systemic infection of healthy volunteers with S. enterica serovar Paratyphi A transiently altered the MAIT TCR repertoire within individuals at the clonotypic level (75). In this case, circulating, overrepresented MAIT clonotypes were reduced during infection but restored to homeostatic proportions at the resolution of infection (75). TCRs from MAIT clonotypes that expanded during infection showed greater stimulatory potential than TCRs from contracted MAIT clonotypes in response to synthetic riboflavin-based antigens or bacterial supernatant (S. enterica Paratyphi A or E. coli) (75), suggesting TCR-dependent but not microbe-specific differences in MAIT clonotypic responses. In mice, stimulation of circulating MAIT cells with the different riboflavin-based antigens induced activation of separate cell clusters based on their activated surface phenotype (66), perhaps involving preferential stimulation of clonal MAIT TCRs. Similarly, in mice, preferential accumulation of MAIT cells with divergent functional phenotypes occurs in response to acute infection by different bacterial species (S. enterica serovar Typhimurium or Legionella longbeachae) (34, 46, 76), however, divergence in the functional response of MAIT cells has not been correlated with TCR usage in mice. Interestingly, the TCRα chain TRAV1-2-TRAJ33 rearrangement is also found in some CD161−TRAV1-2+ T cells, and nearly one third of those TRAV1-2-TRAJ33 rearranged TCRs express Tyr95α (74). Whether these cells are also MR1-reactive is not known, although it should be noted that other MHC-reactive T cells and a subset of CD1b-restricted T cells also utilize the TRAV1-2 gene segment (77–79). A minority of MAIT TCRα TRAV1-2 rearrangements occur with non-classical junctional regions (non-TRAJ12/20/33), some of which do not contain the Tyr95α residue yet still confer MR1-5-OP-RU reactivity (11). These observations indicate that other factors must account for recognition of 5-OP-RU among “MAIT TCR outliers,” as in one case determined by crystallographic analysis (80).
Among MAIT cells, TCRβ chain composition provides the greatest source of variability within the TCR repertoire (7, 8, 10, 11, 28, 74). Despite this, TRBV6 is a dominant TRBV gene segment used amongst MAIT TCRs, although some variations exist and are dependent on the TCRα junctional segment (7, 74). For instance, TRAV1-2-TRAJ12 TCRα chains more commonly pair with TRBV6 (>80%), compared to TRAV1-2-TRAJ33 and TRAV1-2-TRAJ20 rearranged TCRα chains which pair roughly evenly with TCRβ chains encoded by TRBV6 and TRBV20 family members (7, 74). Up to one quarter of paired MAIT TCRs express TCRβ chains that are not from TRBV6 or TRBV20 V segment family members (7, 74). In line with the less stringent nature of TCRβ chain usage, the CDR3β region among classical MAIT TCRs are non-germline-encoded and quite hypervariable, ranging from 9 to 19 amino acids in length and containing no discernible sequence motifs (9, 28, 74). A comparison of seven canonical MAIT TCRs (TRAV1-2-TRAJ33 encoded TCRα chain paired with a TRBV6-1 encoded TCRβ chain) with similar CDR3α loops and highly variable CDR3β loop length and composition revealed how CDR3β hypervariability impacts on MAIT TCR recognition of MR1 (28). All of the TCRs bound to MR1-5-OP-RU within a similar range of affinities (Kdeq ~2 μM), with the exception of one TCR that bound MR1-5-OP-RU weaker (Kdeq ~9.1 μM) (28). Structural analysis of four of the TCRs bound to MR1-5-OP-RU, including the weaker affinity TCR, demonstrated that the overall TCR footprint onto MR1 was conserved (28). Not surprisingly, the greatest variability in MR1 engagement was from contacts by the TCRβ CDR loops and framework regions that differed between TCRs; yet an overall similar buried surface area (BSA) at the interface was achieved (28). CDR3β loop sequence impacted on conformational flexibility of this loop as well as MR1 residues contacted. Importantly, in some structures the CDR3β loop made direct contact with 5-OP-RU, thus fine-tuning MR1 recognition in an antigen-dependent manner (28).
The MAIT TCR repertoire varies throughout the course of life (fetal, neonate, young, and old) and has now been examined in some detail, revealing consistent changes in MAIT cell frequency as we age (7, 9, 74, 80, 81). The overall frequency of MAIT cells in blood increases rapidly from low numbers early in life (11, 82), typically over the first two-to-three decades, declining slowly thereafter (83, 84). In addition, the diversity in clonotypes decreases as certain MAIT cell clones expand over time (84). This is most pronounced in older adults (>65 years), where a small number of private (non-shared) MAIT cell clonotypes dominate the MAIT cell pool (84). It is likely that expansion of naïve MAIT cells in young individuals is a consequence of microbial exposure following thymic egress (36, 45, 82, 85, 86). Interestingly, a large proportion of CD161+ T cells found in cord blood express the canonical MAIT TCRα chain (TRAV1-2-TRAJ33) but do not recognize MR1-5-OP-RU tetramers (81). These non-reactive cells are not readily detectable in adults (11, 84), suggesting they do not expand akin to MR1-5-OP-RU-reactive MAIT cells. In this regard, some of these cells might feature a TCRβ chain which prohibits reactivity to MR1 presenting riboflavin-based antigens in favor of other antigens furnished by MR1, some of which may be less abundant or less stimulatory than the riboflavin-based antigens for MAIT cells expressing classical MAIT TCRs (28, 35, 80) or may trigger peripheral tolerance to avoid autoreactive or allergic responses (87). Accordingly, a pairwise comparison of MAIT TCR sequences in cord blood revealed much broader TCRβ diversity compared to those from MAIT cells from adult blood, further highlighting the effect of peripheral exposure on the narrowing of the MAIT TCR repertoire (84).
Non-Riboflavin Based MR1 Ligands
In addition to the well-defined riboflavin (vitamin B2)-based MR1-ligands which are classified as pyrimidines (5-OP-RU and 5-OE-RU) and ribityllumazines (RL-6-Me-7-OH and RL-6,7-diMe), MR1 also presents other classes of small molecules. They include a range of pterins, namely the photodegradation product of folic acid (vitamin B9), known as 6-formylpterin (6-FP) (35), the synthetically acetylated form of 6-FP, acetyl-6-formylpterin (Ac-6-FP) (28), the photodegradation product 2,4-diamino-6-formylpteridine (2,4-DA-6-FP) of the drugs aminopterin and methotrexate which are synthetic derivatives of folic acid (29), and a synthetic derivative of Ac-6-FP, 2-acetylamino-4-hydroxy-6-formylpteridine dimethyl acetal (33). See recent review for details on the photodegradation of folic acid (32). 6-FP, Ac-6-FP, and 2,4-DA-6-FP form a Schiff base with MR1 Lys43 based on crystal structures (28, 29, 35). Whilst the relevant formyl group of the synthetic derivative of Ac-6-FP, 2-acetylamino-4-hydroxy-6-formylpteridine dimethyl acetal, is blocked (33), partial hydrolysis of the acetal might generate an aldehyde capable of reacting with Lys43 of MR1. Indeed the latter, just like all other listed pterins, caused cell surface upregulation of MR1 (26, 28, 29, 33, 35). In contrast, an equivalently modified version of 6-FP, 2-amino-4-hydroxy-6-formylpteridine dimethyl acetale, did not cause MR1 surface upregulation (33). In peripheral blood of some healthy donors, rare subsets of MR1-Ac-6-FP and MR1-6-FP reactive T cells have been described based on tetramer staining (80), as detailed in the next section. Also, some canonical MAIT TCRs form notable interactions with MR1-6-FP (27) and Ac-6-FP (28), involving direct molecular contact between TCR and Ac-6-FP (28) but not 6-FP (27). However, 6-FP and Ac-6-FP typically do not stimulate MAIT cells (28, 35). Instead 6-FP and Ac-6-FP are mostly known as competitive inhibitors of activation by riboflavin-based antigens such as 5-OP-RU (27, 28). Also 2-acetylamino-4-hydroxy-6-formylpteridine dimethyl acetal acts as a potent competitive inhibitor (33). So far Ac-6-FP remains the most potent competitive inhibitor of MAIT cell activation by 5-OP-RU and other pyrimidine antigens, both in vitro in human and mouse cell line assays and in vivo in mice (29). The acetyl group in Ac-6-FP which is absent in 6-FP forms additional van der Waals interactions with MR1 as well as a hydrogen bond, correlating with an 8°C increase in the stability of MR1-Ac-6-FP in comparison to MR1-6-FP (28). This, as well as other factors, such as solubility, molecule stability, and capacity for cellular uptake likely contribute to the potency of Ac-6-FP as a competitive inhibitor.
The same small molecule drug screen that identified three pyrimidines as potential MR1 ligands (described above) also identified 19 non-pyrimidine MR1 ligands, which belonged to one or more of the following diverse classes of molecules: aromatic aldehyde, aromatic carboxylate, phenol alinine, flavone, isoflavone, enone, quinone (29). Most of these non-pyrimidine ligands could not stimulate a MAIT TCR reporter cell line, and in crystal structures analyzed, no TCR contacts with the MR1 ligands were observed. All of the non-stimulatory ligands tested, including the salicylic acid analog 3-formylsalicylic acid (3-F-SA) (Figure 1), competitively inhibited MAIT cell activation to a similar or lesser extent as 6-FP. Some non-pyrimidine ligands were able to stimulate a MAIT TCR reporter cell line to varying degrees, of which diclofenac was most potent and whose activity was assigned to one of its metabolites, 5-OH-diclofenac (29) (Figure 1). Unlike the pyrimidine and ribityllumazine antigens which are contacted in a hydrogen bond by a conserved residue in the MAIT TCRα chain (Tyr95α), the 5-OH group of 5-OH-diclofenac was contacted in a hydrogen bond by Glu99 from the CDR3β loop. This matched the observation that 5-OH-diclofenac only activated one of a panel of MAIT TCR reporter cell lines, indicating that the MAIT TCRβ chain was able to “fine-tune” responsiveness to certain ligands (29), in line with an emerging concept that some MAIT cells are capable of discriminating between different classes of antigens based on differences in the MAIT TCRβ chain (28, 73, 80, 88, 89). A weak stimulator, 5-formyl-salicylic acid, also only activated one of a panel of MAIT TCR reporter cell lines, suggestive of a similar mechanism (29). Except for diclofenac/5-OH-DCF, all MR1 ligands, of which crystal structure bound to MR1 and complexed with a MAIT TCR were determined as part of this study, featured a Schiff base with Lys43 of MR1 (26, 27, 29) (Figure 1).
In summary, accumulated data thus far (26–29, 35) indicate that MR1 has the capacity to bind structurally diverse small molecules (150–400 Da). Most of the MR1 ligands (except for RL-6-Me-7-OH and Diclofenac/5-OH-diclofenac) form a Schiff base with Lys43 of MR1. However, regardless of the Schiff base, all ligands are located broadly within a similar location in the A'-pocket of MR1, although some are displaced; or oriented differently, involving mostly rotations with broadly similar depositions of the planes of the aromatic rings (26, 27, 29, 35). One notable exception is diclofenac/5-OH diclofenac, whose ring was not deposed in the same plane, rather, the central ring was essentially perpendicular to that of any of the other MR1 ligands (Figure 1) (29). Both in the context of bacterial infections (73, 88), cancer and steady-state (44, 90) there is evidence for the existence of additional MR1 ligands, including some that do not appear to be pyrimidines or ribityllumazines (73, 88), although the chemical identities of these remain to be defined. Some of these ligands appear to be recognized by T cells that do not necessarily share the phenotype of MR1-5-OP-RU specific MAIT cells, described in detail in the next section.
Diverse MR1-Reactive T Cells
A range of MR1-antigen reactivity patterns have emerged for individual T cell clones and the TCRs they express. These reactivity patterns were identified based on (i) binding of T cell clones or TCR reporter cell lines to MR1-antigen tetramer (involving riboflavin-based antigens or other antigens) or mutated MR1 Lys43 to Ala (MR1-K43A) tetramers; and (ii) activation of T cell clones or TCR reporter cell lines in cellular assays with antigen presenting cells expressing physiological levels of MR1 or overexpressing MR1 or MR1-K43A. Notably, this is an active area of research and it is likely that additional reactivity patterns will emerge. Whilst the patterns observed so far are not always absolute, in that one reactivity might be less dominant, for simplicity they can be grouped as outlined below and illustrated in Figure 3 and Table 1. Similarly, a particular T cell phenotype (TCR usage, surface markers, transcription factors) can be exclusively or more frequently associated with one particular antigen reactivity pattern, although in some cases significant overlaps in TCR usage (and other phenotypic characteristics) have been observed for TCRs with different MR1-antigen reactivity patterns.
(1) Reactivity to MR1-5-OP-RU/5-OE-RU (and less potent ribityllumazines) but not to other MR1-ligands. This reactivity is mediated by the population of cells referred to as MAIT cells, expressing the canonical MAIT TCR and phenotypic markers associated with the MAIT cell lineage in humans and mice (7, 24, 26, 28, 80). The same reactivity can also be mediated by a small fraction of human, TRAV1-2− T cells with diverse TCR usage (e.g., clone MAV36 that expresses a TRAV36+ TCR), including some cells that feature a MAIT-like phenotype (CD161+, IL-18Rα+, CD218a+, CD26+) (80, 89). Notably, a follow-up study revealed that TRAV36+ MR1 reactive T cells are possibly a second public TCR family, alongside MAIT cells, capable of specifically recognizing riboflavin-based antigens in the context of MR1 (80, 89). The TRAV36 gene segment of these TCRs was not rearranged with the classical MAIT TRAJ gene segments (TRAJ33, TRAJ12, TRAJ20), and hence lacked a Tyr95α (80, 89). Reminiscent of the classical MAIT TCRα chain, they displayed a largely germline-encoded CDR3α loop. In stark contrast to canonical MAIT TCRs, these TCRs featured nearly invariant CDR3β loops of constrained length (14 amino acids) (89). Structural studies on the MAV36 TCR revealed the CDR1a loop was predominant at the MR1 interface, including contacting the antigen directly, analogous to the role of the CDR3α loop of classical MAIT TCRs (80). Not surprisingly, among TRAV1-2+ T cells, MR1-5-OP-RU tetramer+ cells predominate, the majority of which express a classical MAIT phenotype, while amongst TRAV1-2− cells, the frequency of MR1-5-OP-RU tetramer+ cells is similar to that of MR1-pterin tetramer+ cells [<0.2% of circulating T cells (80)], although it is perhaps higher among CD8+ T cells (88). Regardless of antigen recognition pattern, generally TRAV1-2− MR1-reactive T cells are heterogeneous in surface markers and transcription factor profile, with the majority lacking CD45RO, PLZF, RORγt, and being heterogenous in T-bet expression, in line with developmental and functional differences compared to MAIT cells (80). Recently, novel MR1-reactive TCR rearrangements were identified from T cells in C57BL/6 mice deficient in the canonical MAIT junctional gene segment TRAJ33 (89). In spite of genetic deletion of the TRAJ33 gene segment, MAIT cells were detected using MR1-5-OP-RU tetramer, albeit at a dramatically reduced frequency (50-fold less) compared to wildtype mice (89). MR1-reactive T cells from TRAJ33 knockout (KO) mice formed two distinct subsets based on TCR usage: those that expressed classical MAIT TCRs (TRAV1, Tyr95α) with more diverse junctional gene segments; most notably TRAV1-TRA12/TRAJ9/TRAJ40 rearrangements, or MR1-reactive T cells with more diverse TCR usage altogether (non-TRAV1) (89). T cell clones that expressed the classical MAIT TCRs, derived from the TRAJ33 KO mice, recognized MR1-5-OP-RU tetramers but not MR1-Ac-6-FP tetramers, verifying their specificity for the riboflavin-based antigens. In contrast, amongst the more diverse TRAV1− MR1-reactive T cell clones, none were solely MR1-5-OP-RU specific (see recognition pattern 2) (89).
(2) Reactivity to MR1-5-OP-RU/5-OE-RU (and less potent ribityllumazines) and other MR1-ligands. This reactivity involves ligand-dependent crossreactivity, a well-known concept in T cell biology. Examples include the cross-recognition of pterins (6-FP and Ac-6-FP) and pyrimidines (5-OP-RU) by <5% of human TRAV1-2+ T cells (e.g., clones AM1, AM2, AM3) that expressed a classical MAIT TCR, typically featuring the TRAV1-2-TRAJ33 rearrangement and no discernible TCRβ motifs (80). Only a very small fraction of MR1-reactive TRAV1-2− T cells (e.g., clone MAV14, TRAV14) in humans were identified to be crossreactive (80, 89). All of these TRAV1-2− T cells expressed TCRs with diverse usage and only one TCR sequence featured the Tyr95α (80). Differential antigen recognition was a common feature among both TRAV1-2+ and TRAV1-2− crossreactive T cells, whereby some TCRs preferred antigens of a specific class (riboflavin-based or folate-derived), while others were capable of distinguishing between antigens of the same class (6-FP or Ac-6-FP) (80). A TRAV1-2− T cell clone (D462-E4, TRAV12-2) reacted to MR1-5-OP-RU tetramer and responded to RL-6-Me-7-OH but not RL-6,7-diMe and in addition to infection with Streptococcus pyogenes which lacks the riboflavin biosynthetic pathway, suggestive of an undefined riboflavin-independent MR1 antigen (88). The same clone also responded moderately to PLI, to a level similar as compared to RL-6-Me-7-OH (73). This clone did not express the MAIT cell marker CD161, although it might have been downregulated as a result of T cell expansion with anti-CD3 (88). The same research team also identified another TRAV1-2+ T cell clone (D481-C7) which also reacted with MR1-5-OP-RU tetramer, preferentially recognized RL-6,7-diMe over RL-6-Me-7-OH, and potently responded to PLI and PLIII (73). Some MAIT TCR expressing reporter cell lines and subsets of primary TRAV1-2+ MAIT cells also displayed crossreactivity to chemically diverse drugs, drug metabolites and drug-like molecules (29, 30).
(3) Auto-reactivity to MR1, a more MR1-centric reactivity compared to antigen-centric reactivity. This type of reactivity was found amongst human TRAV1-2+ T cells, where clone M33-64, which expressed a classical MAIT TCR, stained with MR1 tetramer loaded with 5-OP-RU or pterin antigens as well as MR1-K43A tetramer. A matching TCR reporter cell line displayed MR1-dose dependent activation and limited antigen dose dependency (80). Surprisingly, a mutation of Tyr95α to Phe in the M33-64 TCR did not abolish MR1-5-OP-RU reactivity, in stark contrast to all previously studied MAIT TCRs, which was attributed to a dominant role of the CDR3β loop in both stabilizing the TCR and in making considerable contacts with MR1 (80). Autoreactivity in the context of MR1 overexpressing cell lines was also observed in <15% of mouse hybridomas derived from TAP−/− C57BL/6 mice (clones 6C2, 6H2, 18G7, 8D12) (5, 6, 92–95) and <10% of hybridomas from Vα19Tg Cα−/− mice, all featuring canonical MAIT TCRs (94). Interestingly, based on extensive mutagenesis, the 6C2 hybridoma used overlapping but distinct TCR residues for the recognition of MR1 on E. coli infected cells vs. MR1 overexpressing cells in the absence of infection (95). Recently MR1-autoreactivity was also found to be mediated by γδ TCR+ T cells, present in human PBMCs (<0.001 to 0.1% of CD3+ T cells and from <0.1 to 5% of γδ T cells) and in tissues (liver, stomach, lung, and duodenum), the frequencies of which may be enriched in association with some diseases (96). Some MR1-reactive γδ TCRs bound underneath the antigen binding cleft (contacting primarily the α3 domain of MR1), while others interacted with the antigen binding cleft of MR1, akin to the classical MAIT αβ TCR. The former type was observed in blood of 6 out of 20 individuals. Activation of γδ TCR expressing reporter cell lines varied and was dependent on MR1 with some potential for modulation by MR1-bound antigen. MR1-reactive γδ TCRs were diverse in gene usage. Most (72%) used TRDV1; the remainders, aside from one TRDV5+ clone, expressed TRDV3. Whilst all functional TRGV genes including TRGV2, 3, 4, 5, 8, and 9 were found amongst MR1-reactive γδ T cells, TRDV1+ and TRDV3+ TCRs predominantly paired with TRGV8. MR1-5-OP-RU tetramer+ γδ T cells were mostly CD4−CD8α− or CD8α+ with variable CD161 expression. Thus, they resembled other cells of the γδ T cell lineage and appeared phenotypically diverse (96). Whether MR1-reactive γδ T cells exist in mice or other species remains to be investigated.
(4) Reactivity to other MR1-ligands but not MR1-5-OP-RU/5-OE-RU (and less potent ribityllumazines). Such reactivity pattern was first observed for the TRAV1-2− MAV21 TCR (TRAV21) which specifically recognized pterins but not 5-OP-RU (80). Recently, Crowther et al. identified another TRAV1-2− T cell clone in blood of a healthy individual (MC.7.G5, TRAV38.2/DV8-TRAJ31, TRBV25.1 TRBJ2.3) which specifically recognized an antigen/s that was expressed or upregulated in cancerous cells but not non-cancerous cells (resting, activated, stressed or infected), in an MR1-dependent manner (90). Interestingly, the observed T cell reactivity was donor-unrestricted, with multiple HLA-mismatched tumor cells recognized in an MR1-dependent manner, whilst not exerting allo-reactivity, making this T cell clone particularly interesting for immunotherapies (90). Potent activation, including cytotoxicity, was achieved with physiological levels of MR1 and high effector (T cell) to target (APC) ratios. Both Ac-6-FP and bacterial riboflavin-based antigens were not recognized and acted as competitive inhibitors of activation. Whilst a clone with a similar reactivity pattern was also isolated from a second donor, the frequency of similar clones within an individual and their prevalence in the general population and in the context of cancers are unknown. Also, the phenotype of the clones (other than the TCR of one of them) was not described which may provide insights into its developmental origin. Previously, Lepore et al. (44) had identified a population of T cells in healthy individuals that comprised 1 in 2,500–5,000 of circulating T cells. Designated “MR1T” cells, clones of these cells recognized, in the context of physiological levels of MR1 and dependent on MR1, endogenous cellular antigens from various tissues, including cancer tissues, and thus were not cancer specific. One MR1T cell clone was TRAV1-2+ (clone DGA4), but most were TRAV1-2−, expressing diverse TCRα and β chains (e.g., clone DGB129; TRAV29/DV5-TRAJ23; and TRBV12-4-TRBJ1-1) (44). Like the MC.7.G5 clone, MR1T cell clones (e.g., DGB129) did not recognize 6-FP and their activation was inhibited by 6-FP. However, unlike the MC.7.G5 clone, which did not bind MR1-K43A tetramer or respond to MR1-K43A overexpressing cells (90), MR1T cell clones equally recognized wild type and MR1-K43A overexpressing cells. This suggested to the authors that relevant antigens that were recognized did not depend on Schiff base formation with MR1 for TCR recognition but may also suggest an MR1-centric recognition or MR1 autoreactivity by some MR1T cells for which 5-OP-RU/ribityllumazine are permissive antigens but pterins are not. At least two clones (DGB129 and DGB70) did however, differentially recognize fractions of cell/tumor lysates, demonstrating differential antigen specificity for at least some clones. With the exception for one MR1T clone, all TRAV1-2− clones were CD161− and all, except for one, were CD8α+ (DGB129, TRAV1-2−, CD161−, CD8α+; DGB70, TRAV1-2− [TRAV5], CD161−, CD8−). DGA4, the TRAV1-2+ clone, was CD161+, CD8−. Phenotypic and functional characterization of MR1T cell clones showed multiple chemokine receptor expression profiles and secretion of diverse effector molecules, suggesting functional heterogeneity within this population that was also distinct from that of MAIT cells. In a T-helper like function, clone DGB129 induced maturation of monocyte derived dendritic cells and another clone suggested the possible contribution of MR1T cells to intestinal epithelial barrier homeostasis.
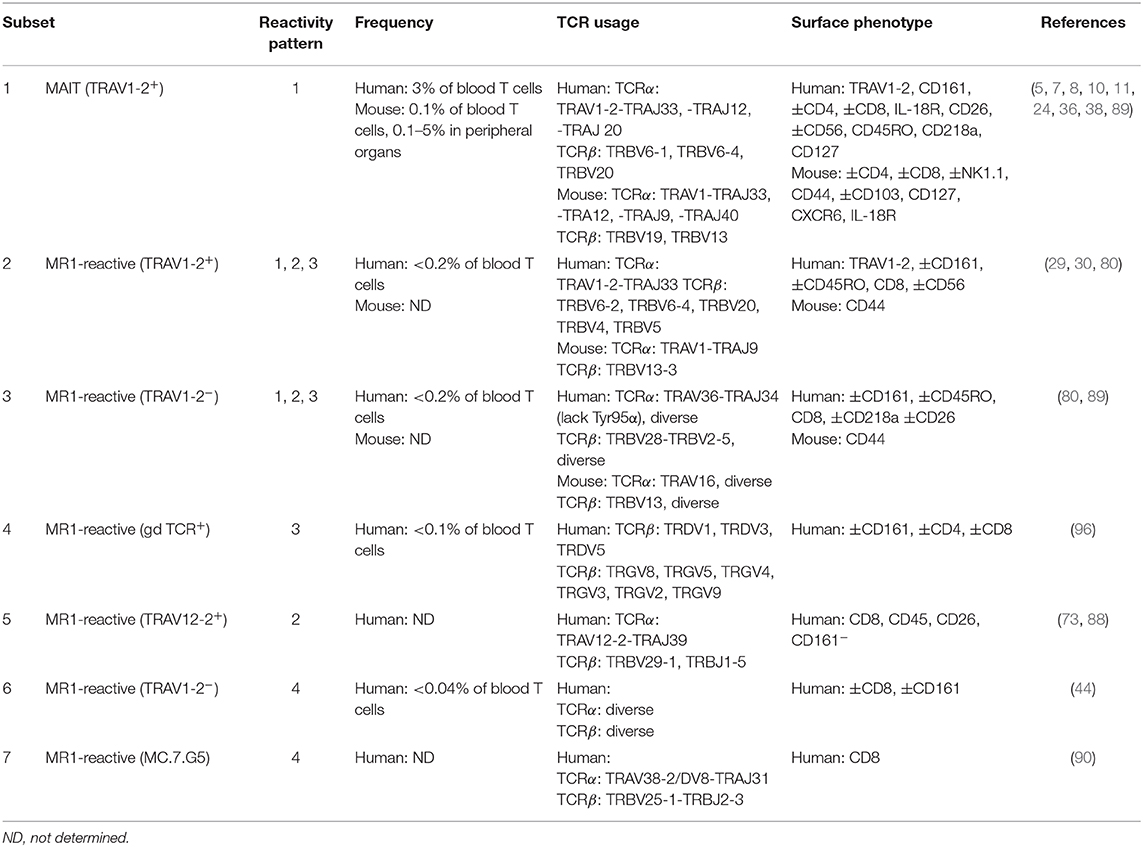
Table 1. Frequency, TCR usage (where known) and surface phenotype of each subset of MR1-reactive T cells as depicted in Figure 3.
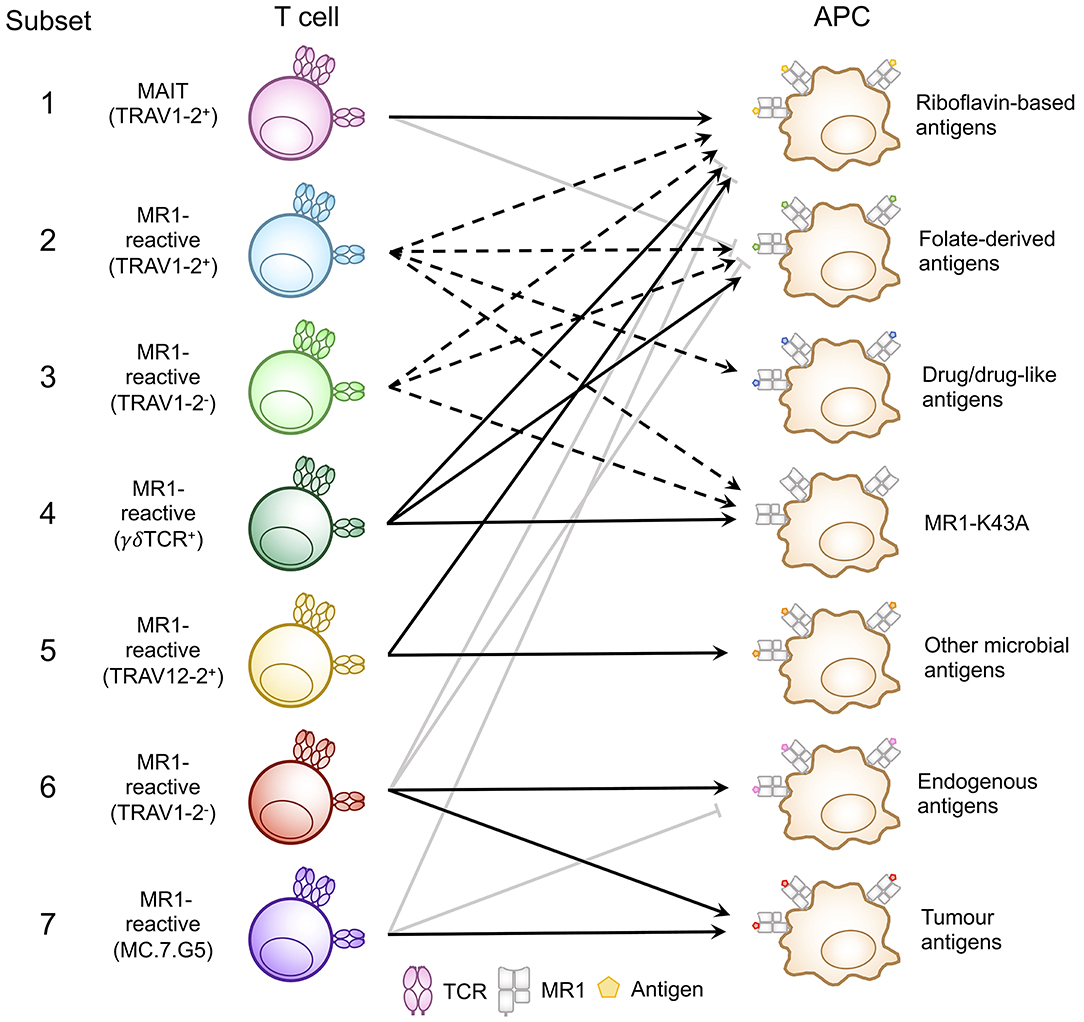
Figure 3. Classification of known MR1-reactive T cells based on MR1-antigen reactivity pattern. MR1-reactive T cells bear an αβ or γδ T cell receptor (TCR) that recognizes one or more classes of antigen presented by MR1 on antigen presenting cells (APCs). Solid point arrows indicate that all T cells within the subset recognize the specified antigen, broken point arrows indicate that some T cells within the subset recognize the specified antigen and gray block arrows indicate no recognition of the class of antigen. Table 1 outlines the frequency, TCR usage (where known) and surface characteristics of each subset using this classification. This schematic was inspired by Figure 1 from a recent commentary piece (91).
Discussion
We are beginning to appreciate the diversity of microbial and endogenous (including cancer) antigens presented by MR1 and the heterogenous populations of T cells which recognize them. Independent and repeated identification of these cells has led to the classification “MR1-reactive T cells,” illustrated in Figure 3 and Table 1, inclusive of MR1-restricted MAIT cells, and more phenotypically and functionally heterogenous T cells, some of which are MAIT-like in their phenotype, grouped together as “other MR1-reactive T cells” (44, 80, 89, 91). Whilst TRAV1-2+ T cells are dominant amongst MAIT cells, other MR1-reactive T cells mostly include TRAV1-2− αβ TCR+ T cells (44, 73, 80, 89, 90) as well as γδ TCR+ T cells (96). Thus, the current stage of research suggests that most MR1-reactive T cells, namely MAIT cells, feature a biased TCR α-chain, and react with a limited repertoire of pyrimidine and ribityllumazine antigens derived from riboflavin biosynthesis. There are also several examples of TCR β-chain usage influencing antigen preference and allowing for antigen crossreactivity by classical MAIT TCRs, and future work might identify additional examples of physiological relevance. The emerging descriptions of other MR1-reactive T cells highlight that the classical MAIT TCR α-chain is not the only solution that warrants MR1-reactivity, however, more work is needed to validate and corroborate the TCR usage and antigen specificities of other, more diverse T cells. In particular, determining the chemical identities and biosynthetic origins of the tumor-associated and endogenous antigens recognized by some of the other MR1-reactive T cells will allow the generation of tetramers which will greatly assist in determining the prevalence of these cells as well as their potentially important role in anti-tumor immunity and other immune-functions. It will also help to better characterize the phenotype (including the TCR usage) and of this diverse set of T cells including public as compared to private existence of individual clones, recently described for TRAV36+ MR1-reactive T cells (89). Another interesting question concerns the lineage development of other MR1-reactive T cells that feature a non-MAIT phenotype. Is it possible that these cells might have originated from another T cell lineage and cross-react across MHC families, including classical MHC molecules (MHC-I and MHC-II) and MHC-I-like molecules? In TCR α-chain transgenic mice that lack MR1 (Vα19iCα−/−MR1−/−) MR1-5-OP-RU tetramer+ T cells make up ~32% of T cells in the spleen as compared to ~50% in mice proficient in MR1 (Vα19iCα−/−) (97). The population of these cells mediated both MR1-dependent and MHC-I (H2-Kb/H2-Db) dependent reactivity in vitro (97), although it was not investigated whether these specificities were mediated by single T cells and single TCRs. More recently, MR1-5-OP-RU tetramer+ T cells featuring the canonical MAIT TCRα chain (TRAV1-TRAJ33) but not the typical TRBV13 bias were also found in very low percentages in the thymus of non-transgenic mice, Mus musculus castaneus (CAST mice), modified to lack MR1 (86). Thus, MR1-5-OP-RU tetramer+ T cells can develop in the absence of MR1, inferring selection may occur on other MHC molecules. Indeed, MR1 is highly conserved with HLA-A2 (39% sequence conservation), suggesting mimicry as a potential mode of crossreactivity by these cells. So far evidence of peripheral T cells that crossreact between classical MHC molecules and MHC-I-like molecules remains elusive.
Author Contributions
MS and SE wrote and revised the manuscript, read, and approved the submitted version. All authors contributed to the article and approved the submitted version.
Funding
SE was supported by a fellowship from the Australian Research Council (ARC), (ARC DECRA DE170100407) and a project grant from the Australian National Health and Medical Research Council (NHMRC project Grant No. GNT1157388).
Conflict of Interest
SE is an inventor on patents describing MR1 antigens and MR1 tetramers.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Jia Q, Zhou J, Chen G, Shi Y, Yu H, Guan P, et al. Diversity index of mucosal resident T lymphocyte repertoire predicts clinical prognosis in gastric cancer. Oncoimmunology. (2015) 4:e1001230. doi: 10.1080/2162402X.2014.1001230
2. Shcherbinin DS, Belousov VA, Shugay M. Comprehensive analysis of structural and sequencing data reveals almost unconstrained chain pairing in TCRαβ complex. PLoS Comput Biol. (2020) 16:e1007714. doi: 10.1371/journal.pcbi.1007714
3. Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. (1993) 178:1–16. doi: 10.1084/jem.178.1.1
4. Hashimoto K, Hirai M, Kurosawa Y. A gene outside the human MHC related to classical HLA class I genes. Science. (1995) 269:693–5. doi: 10.1126/science.7624800
5. Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, De La Salle H, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. (1999) 189:1907–21. doi: 10.1084/jem.189.12.1907
6. Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. (2003) 422:164–9. doi: 10.1038/nature01433
7. Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. (2013) 210:2305–20. doi: 10.1084/jem.20130958
8. Gold MC, Mclaren JE, Reistetter JA, Smyk-Pearson S, Ladell K, Swarbrick GM, et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med. (2014) 211:1601–10. doi: 10.1084/jem.20140507
9. Lepore M, Kalinichenko A, Colone A, Paleja B, Singhal A, Tschumi A, et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRβeta repertoire. Nat Commun. (2014) 5:3866. doi: 10.1038/ncomms5493
10. Lepore M, De Lalla C, Gundimeda SR, Gsellinger H, Consonni M, Garavaglia C, et al. A novel self-lipid antigen targets human T cells against CD1c(+) leukemias. J Exp Med. (2014) 211:1363–77. doi: 10.1084/jem.20140410
11. Gherardin NA, Souter MN, Koay HF, Mangas KM, Seemann T, Stinear TP, et al. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol Cell Biol. (2018) 96:507–25. doi: 10.1111/imcb.12021
12. Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. (2011) 117:1250–9. doi: 10.1182/blood-2010-08-303339
13. Cosgrove C, Ussher JE, Rauch A, Gärtner K, Kurioka A, Hühn MH, et al. Early and nonreversible decrease of CD161++/MAIT cells in HIV infection. Blood. (2013) 121:951–61. doi: 10.1182/blood-2012-06-436436
14. Leeansyah E, Ganesh A, Quigley MF, Sönnerborg A, Andersson J, Hunt PW, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. (2013) 121:1124–35. doi: 10.1182/blood-2012-07-445429
15. Tang XZ, Jo J, Tan AT, Sandalova E, Chia A, Tan KC, et al. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J Immunol. (2013) 190:3142–52. doi: 10.4049/jimmunol.1203218
16. Fergusson JR, Smith KE, Fleming VM, Rajoriya N, Newell EW, Simmons R, et al. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep. (2014) 9:1075–88. doi: 10.1016/j.celrep.2014.09.045
17. Serriari NE, Eoche M, Lamotte L, Lion J, Fumery M, Marcelo P, et al. Innate mucosal-associated invariant T (MAIT) cells are activated in inflammatory bowel diseases. Clin Exp Immunol. (2014) 176:266–74. doi: 10.1111/cei.12277
18. Jeffery HC, Van Wilgenburg B, Kurioka A, Parekh K, Stirling K, Roberts S, et al. Bacteria exposed biliary epithelium and liver B cells activate intrahepatic mait cells in an MR1-dependent manner. J Hepatol. (2015) 64:1118–27. doi: 10.1016/j.jhep.2015.12.017
19. Hinks TS, Wallington JC, Williams AP, Djukanović R, Staples KJ, Wilkinson TM. Steroid-induced deficiency of mucosal-associated invariant T cells in the chronic obstructive pulmonary disease lung. Implications for nontypeable Haemophilus influenzae infection. Am J Respir Crit Care Med. (2016) 194:1208–18. doi: 10.1164/rccm.201601-0002OC
20. Gibbs A, Leeansyah E, Introini A, Paquin-Proulx D, Hasselrot K, Andersson E, et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. (2017) 10:35–45. doi: 10.1038/mi.2016.30
21. Hama I, Tominaga K, Yamagiwa S, Setsu T, Kimura N, Kamimura H, et al. Different distribution of mucosal-associated invariant T cells within the human cecum and colon. Centr Euro J Immunol. (2019) 44:75. doi: 10.5114/ceji.2019.84020
22. Sobkowiak MJ, Davanian H, Heymann R, Gibbs A, Emgard J, Dias J, et al. Tissue-resident MAIT cell populations in human oral mucosa exhibit an activated profile and produce IL-17. Eur J Immunol. (2019) 49:133–43. doi: 10.1002/eji.201847759
23. Rha MS, Han JW, Kim JH, Koh JY, Park HJ, Kim SI, et al. Human liver CD8+ MAIT cells exert TCR/MR1-independent innate-like cytotoxicity in response to IL-15. J Hepatol. (2020). doi: 10.1016/j.jhep.2020.03.033. [Epub ahead of print].
24. Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med. (2015) 212:1095–108. doi: 10.1084/jem.20142110
25. Chen Z, Wang H, D'souza C, Sun S, Kostenko L, Eckle SB, et al. Mucosal-associated invariant T-cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol. (2017) 10:58–68. doi: 10.1038/mi.2016.39
26. Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. (2014) 509:361–5. doi: 10.1038/nature13160
27. Patel O, Kjer-Nielsen L, Le Nours J, Eckle SB, Birkinshaw R, Beddoe T, et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat Commun. (2013) 4:2142. doi: 10.1038/ncomms3142
28. Eckle SB, Birkinshaw RW, Kostenko L, Corbett AJ, Mcwilliam HE, Reantragoon R, et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J Exp Med. (2014) 211:1585–600. doi: 10.1084/jem.20140484
29. Keller AN, Eckle SB, Xu W, Liu L, Hughes VA, Mak JY, et al. Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat Immunol. (2017) 18:402–11. doi: 10.1038/ni.3679
30. Salio M, Awad W, Veerapen N, Gonzalez-Lopez C, Kulicke C, Waithe D, et al. Ligand-dependent downregulation of MR1 cell surface expression. Proc Natl Acad Sci USA. (2020) 117:10465–75. doi: 10.1073/pnas.2003136117
31. Eckle SB, Corbett AJ, Keller AN, Chen Z, Godfrey DI, Liu L, et al. Recognition of vitamin B precursors and byproducts by mucosal associated invariant T cells. J Biol Chem. (2015) 290:30204–11. doi: 10.1074/jbc.R115.685990
32. Kjer-Nielsen L, Corbett AJ, Chen Z, Liu L, Mak JY, Godfrey DI, et al. An overview on the identification of MAIT cell antigens. Immunol Cell Biol. (2018) 96:573–87. doi: 10.1111/imcb.12057
33. Soudais C, Samassa F, Sarkis M, Le Bourhis L, Bessoles S, Blanot D, et al. In vitro and in vivo analysis of the gram-negative bacteria-derived riboflavin precursor derivatives activating mouse MAIT cells. J Immunol. (2015) 194:4641–9. doi: 10.4049/jimmunol.1403224
34. Wang H, D'souza C, Lim XY, Kostenko L, Pediongco TJ, Eckle SBG, et al. MAIT cells protect against pulmonary legionella longbeachae infection. Nat Commun. (2018) 9:3350. doi: 10.1038/s41467-018-05202-8
35. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. (2012) 491:717–23. doi: 10.1038/nature11605
36. Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. (2010) 11:701–8. doi: 10.1038/ni.1890
37. Kurioka A, Jahun AS, Hannaway RF, Walker LJ, Fergusson JR, Sverremark-Ekstrom E, et al. Shared and distinct phenotypes and functions of human CD161++ Valpha7.2+ T cell subsets. Front Immunol. (2017) 8:1031. doi: 10.3389/fimmu.2017.01031
38. Kurioka A, Ussher JE, Cosgrove C, Clough C, Fergusson JR, Smith K, et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. (2015) 8:429–40. doi: 10.1038/mi.2014.81
39. Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc Natl Acad Sci USA. (2013) 110:E3119–28. doi: 10.1073/pnas.1302799110
40. Georgel P, Radosavljevic M, Macquin C, Bahram S. The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Mol Immunol. (2011) 48:769–75. doi: 10.1016/j.molimm.2010.12.002
41. Chua WJ, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun. (2012) 80:3256–67. doi: 10.1128/IAI.00279-12
42. Cui Y, Franciszkiewicz K, Mburu YK, Mondot S, Le Bourhis L, Premel V, et al. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J Clin Invest. (2015) 125:4171–85. doi: 10.1172/JCI82424
43. Smith AD, Foss ED, Zhang I, Hastie JL, Giordano NP, Gasparyan L, et al. Microbiota of MR1 deficient mice confer resistance against Clostridium difficile infection. PLoS ONE. (2019) 14:e0223025. doi: 10.1371/journal.pone.0223025
44. Lepore M, Kalinichenko A, Calogero S, Kumar P, Paleja B, Schmaler M, et al. Functionally diverse human T cells recognize non-microbial antigens presented by MR1. Elife. (2017) 6:e24476. doi: 10.7554/eLife.24476.020
45. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. (2019) 366:eaax6624. doi: 10.1126/science.aax6624
46. Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. (2019) 28:3249–62.e5. doi: 10.1016/j.celrep.2019.07.039
47. Leng T, Akther HD, Hackstein CP, Powell K, King T, Friedrich M, et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. (2019) 28:3077–91 e3075. doi: 10.1016/j.celrep.2019.08.050
48. Sundström P, Ahlmanner F, Akéus P, Sundquist M, Alsén S, Yrlid U, et al. Human mucosa-associated invariant T cells accumulate in colon adenocarcinomas but produce reduced amounts of IFN-γ. J Immunol. (2015) 195:3472–81. doi: 10.4049/jimmunol.1500258
49. Zabijak L, Attencourt C, Guignant C, Chatelain D, Marcelo P, Marolleau JP, et al. Increased tumor infiltration by mucosal-associated invariant T cells correlates with poor survival in colorectal cancer patients. Cancer Immunol Immunother. (2015) 64:1601–8. doi: 10.1007/s00262-015-1764-7
50. Ling L, Lin Y, Zheng W, Hong S, Tang X, Zhao P, et al. Circulating and tumor-infiltrating mucosal associated invariant T (MAIT) cells in colorectal cancer patients. Sci Rep. (2016) 6:20358. doi: 10.1038/srep20358
51. Won EJ, Ju JK, Cho YN, Jin HM, Park KJ, Kim TJ, et al. Clinical relevance of circulating mucosal-associated invariant T cell levels and their anti-cancer activity in patients with mucosal-associated cancer. Oncotarget. (2016) 7:76274. doi: 10.18632/oncotarget.11187
52. Duan M, Goswami S, Shi JY, Wu LJ, Wang XY, Ma JQ, et al. Activated and exhausted MAIT cells foster disease progression and indicate poor outcome in hepatocellular carcinoma. Clin Cancer Res. (2019) 25:3304–16. doi: 10.1158/1078-0432.CCR-18-3040
53. Favreau M, Venken K, Faict S, Maes K, De Veirman K, De Bruyne E, et al. Both mucosal-associated invariant and natural killer T-cell deficiency in multiple myeloma can be countered by PD-1 inhibition. Haematologica. (2017) 102:e266. doi: 10.3324/haematol.2017.163758
54. Shaler CR, Tun-Abraham ME, Skaro AI, Khazaie K, Corbett AJ, Mele T, et al. Mucosa-associated invariant T cells infiltrate hepatic metastases in patients with colorectal carcinoma but are rendered dysfunctional within and adjacent to tumor microenvironment. Cancer Immunol Immunother. (2017) 66:1563–75. doi: 10.1007/s00262-017-2050-7
55. Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. (2017) 169:1342–56.e1316. doi: 10.1016/j.cell.2017.05.035
56. Gherardin NA, Loh L, Admojo L, Davenport AJ, Richardson K, Rogers A, et al. Enumeration, functional responses and cytotoxic capacity of MAIT cells in newly diagnosed and relapsed multiple myeloma. Sci Rep. (2018) 8:4159. doi: 10.1038/s41598-018-22130-1
57. Mitchell J, Kvedaraite E, Von Bahr Greenwood T, Henter J-I, Pellicci DG, Berzins SP, et al. Altered populations of unconventional T cell lineages in patients with langerhans cell histiocytosis. Sci Rep. (2018) 8:16506. doi: 10.1038/s41598-018-34873-y
58. Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, De Lara C, et al. CD 161++ CD 8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+ IL-18 in a TCR-independent manner. Eur J Immunol. (2014) 44:195–203. doi: 10.1002/eji.201343509
59. Loh L, Wang Z, Sant S, Koutsakos M, Jegaskanda S, Corbett AJ, et al. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18–dependent activation. Proc Natl Acad Sci USA. (2016) 113:10133–8. doi: 10.1073/pnas.1610750113
60. Kelly J, Minoda Y, Meredith T, Cameron G, Philipp MS, Pellicci DG, et al. Chronically stimulated human MAIT cells are unexpectedly potent IL-13 producers. Immunol Cell Biol. (2019) 97:689–99. doi: 10.1111/imcb.12281
61. Godfrey DI, Koay HF, Mccluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol. (2019) 20:1110–28. doi: 10.1038/s41590-019-0444-8
62. Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. (2010) 8:e1000407. doi: 10.1371/journal.pbio.1000407
63. Garboczi DN, Hung DT, Wiley DC. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci USA. (1992) 89:3429–33. doi: 10.1073/pnas.89.8.3429
64. Mak JY, Xu W, Reid RC, Corbett AJ, Meehan BS, Wang H, et al. Stabilizing short-lived schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nat Commun. (2017) 8:14599. doi: 10.1038/ncomms14599
65. Awad W, Ler GJM, Xu W, Keller AN, Mak JYW, Lim XY, et al. The molecular basis underpinning the potency and specificity of MAIT cell antigens. Nat Immunol. (2020) 21:400–11. doi: 10.1038/s41590-020-0616-6
66. Schmaler M, Colone A, Spagnuolo J, Zimmermann M, Lepore M, Kalinichenko A, et al. Modulation of bacterial metabolism by the microenvironment controls MAIT cell stimulation. Mucosal Immunol. (2018) 11:1060–70. doi: 10.1038/s41385-018-0020-9
67. Souter MNT, Loh L, Li S, Meehan BS, Gherardin NA, Godfrey DI, et al. Characterization of Human Mucosal-associated Invariant T (MAIT) Cells. Curr Protoc Immunol. (2019) 127:e90. doi: 10.1002/cpim.90
68. Reantragoon R, Kjer-Nielsen L, Patel O, Chen Z, Illing PT, Bhati M, et al. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med. (2012) 209:761–74. doi: 10.1084/jem.20112095
69. Mcwilliam HE, Eckle SB, Theodossis A, Liu L, Chen Z, Wubben JM, et al. The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat Immunol. (2016) 17:531–7. doi: 10.1038/ni.3416
70. Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, Mccluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. (2015) 33:169–200. doi: 10.1146/annurev-immunol-032414-112334
71. Braganza CD, Motozono C, Sonoda K-H, Yamasaki S, Shibata K, Timmer MSM, et al. Agonistic or antagonistic mucosal-associated invariant T (MAIT) cell activity is determined by the 6-alkylamino substituent on uracil MR1 ligands. Chem Commun. (2020) 56:5291–4. doi: 10.1039/D0CC00247J
72. Braganza CD, Shibata K, Fujiwara A, Motozono C, Sonoda KH, Yamasaki S, et al. The effect of MR1 ligand glyco-analogues on mucosal-associated invariant T (MAIT) cell activation. Org Biomol Chem. (2019) 17:8992–9000. doi: 10.1039/C9OB01436E
73. Harriff MJ, Mcmurtrey C, Froyd CA, Jin H, Cansler M, Null M, et al. MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci Immunol. (2018) 3:eaao2556. doi: 10.1126/sciimmunol.aao2556
74. Held K, Beltran E, Moser M, Hohlfeld R, Dornmair K. T-cell receptor repertoire of human peripheral CD161hiTRAV1-2+ MAIT cells revealed by next generation sequencing and single cell analysis. Hum Immunol. (2015) 76:607–14. doi: 10.1016/j.humimm.2015.09.002
75. Howson LJ, Napolitani G, Shepherd D, Ghadbane H, Kurupati P, Preciado-Llanes L, et al. MAIT cell clonal expansion and TCR repertoire shaping in human volunteers challenged with Salmonella Paratyphi A. Nat Commun. (2018) 9:253. doi: 10.1038/s41467-017-02540-x
76. Chen X, Wang X, Besra GS, Gumperz JE. Modulation of CD1d-restricted NKT cell responses by CD4. J Leukoc Biol. (2007) 82:1455–65. doi: 10.1189/jlb.0307163
77. Tynan FE, Borg NA, Miles JJ, Beddoe T, El-Hassen D, Silins SL, et al. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J Biol Chem. (2005) 280:23900–9. doi: 10.1074/jbc.M503060200
78. Tynan FE, Reid HH, Kjer-Nielsen L, Miles JJ, Wilce MC, Kostenko L, et al. AT cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. (2007) 8:268–76. doi: 10.1038/ni1432
79. Van Rhijn I, Kasmar A, De Jong A, Gras S, Bhati M, Doorenspleet ME, et al. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol. (2013) 14:706–13. doi: 10.1038/ni.2630
80. Gherardin NA, Keller AN, Woolley RE, Le Nours J, Ritchie DS, Neeson PJ, et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity. (2016) 44:32–45. doi: 10.1016/j.immuni.2015.12.005
81. Ben Youssef G, Tourret M, Salou M, Ghazarian L, Houdouin V, Mondot S, et al. Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. J Exp Med. (2018) 215:459–79. doi: 10.1084/jem.20171739
82. Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. (2016) 17:1300–11. doi: 10.1038/ni.3565
83. Godfrey DI, Le Nours J, Andrews DM, Uldrich AP, Rossjohn J. Unconventional T cell targets for cancer immunotherapy. Immunity. (2018) 48:453–73. doi: 10.1016/j.immuni.2018.03.009
84. Loh L, Gherardin NA, Sant S, Grzelak L, Crawford JC, Bird NL, et al. Human mucosal-associated invariant T cells in older individuals display expanded TCRαlphabeta clonotypes with potent antimicrobial responses. J Immunol. (2020) 204:1119–33. doi: 10.4049/jimmunol.1900774
85. Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. (2009) 7:e54. doi: 10.1371/journal.pbio.1000054
86. Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. (2019) 366:494–9. doi: 10.1126/science.aaw2719
87. De Lima Moreira M, Souter MNT, Chen Z, Loh L, Mccluskey J, Pellicci DG, et al. Hypersensitivities following allergen antigen recognition by unconventional T cells. Allergy. (2020). doi: 10.1111/all.14279. [Epub ahead of print].
88. Meermeier EW, Laugel BF, Sewell AK, Corbett AJ, Rossjohn J, Mccluskey J, et al. Human TRAV1-2-negative MR1-restricted T cells detect S. pyogenes and alternatives to MAIT riboflavin-based antigens. Nat Commun. (2016) 7:12506. doi: 10.1038/ncomms12506
89. Koay HF, Gherardin NA, Xu C, Seneviratna R, Zhao Z, Chen Z, et al. Diverse MR1-restricted T cells in mice and humans. Nat Commun. (2019) 10:2243. doi: 10.1038/s41467-019-10198-w
90. Crowther MD, Dolton G, Legut M, Caillaud ME, Lloyd A, Attaf M, et al. Genome-wide CRISPR–Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat Immunol. (2020) 21:178–85. doi: 10.1038/s41590-019-0578-8
91. Mori L, De Libero G. ‘Bohemian rhapsody'of MR1T cells. Nat Immunol. (2020) 21:108–10. doi: 10.1038/s41590-019-0588-6
92. Huang S, Gilfillan S, Cella M, Miley MJ, Lantz O, Lybarger L, et al. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J Biol Chem. (2005) 280:21183–93. doi: 10.1074/jbc.M501087200
93. Huang S, Gilfillan S, Kim S, Thompson B, Wang X, Sant AJ, et al. MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J Exp Med. (2008) 205:1201–11. doi: 10.1084/jem.20072579
94. Huang S, Martin E, Kim S, Yu L, Soudais C, Fremont DH, et al. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci USA. (2009) 106:8290–5. doi: 10.1073/pnas.0903196106
95. Young MH, U'ren L, Huang S, Mallevaey T, Scott-Browne J, Crawford F, et al. MAIT cell recognition of MR1 on bacterially infected and uninfected cells. PLoS ONE. (2013) 8:e53789. doi: 10.1371/journal.pone.0053789
96. Le Nours J, Gherardin NA, Ramarathinam SH, Awad W, Wiede F, Gully BS, et al. A class of gammadelta T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science. (2019) 366:1522–7. doi: 10.1126/science.aav3900
Keywords: MR1, MAIT cells, antigen diversity, TCR repertoire diversity, T cell subsets
Citation: Souter MNT and Eckle SBG (2020) Biased MAIT TCR Usage Poised for Limited Antigen Diversity? Front. Immunol. 11:1845. doi: 10.3389/fimmu.2020.01845
Received: 10 April 2020; Accepted: 09 July 2020;
Published: 18 August 2020.
Edited by:
James E. Ussher, University of Otago, New ZealandReviewed by:
Dhruv Sethi, Obsidian Therapeutics, United StatesEfstratios Stratikos, National Centre of Scientific Research Demokritos, Greece
Copyright © 2020 Souter and Eckle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sidonia B. G. Eckle, seckle@unimelb.edu.au
 Michael N. T. Souter
Michael N. T. Souter Sidonia B. G. Eckle
Sidonia B. G. Eckle