- 1Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, The University of Melbourne, Melbourne, VIC, Australia
- 2Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, VIC, Australia
- 3ARC Centre of Excellence in Advanced Molecular Imaging, Monash University, Clayton, VIC, Australia
- 4State Key Laboratory of Respiratory Disease, Guangzhou Institute of Respiratory Disease, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
Mucosal-associated Invariant T (MAIT) cells recognize vitamin B-based antigens presented by the non-polymorphic MHC class I related-1 molecule (MR1). Both MAIT T cell receptors (TCR) and MR1 are highly conserved among mammals, suggesting an important, and conserved, immune function. For many years, the antigens they recognize were unknown. The discovery that MR1 presents vitamin B-based small molecule ligands resulted in a rapid expansion of research in this area, which has yielded information on the role of MAIT cells in immune protection, autoimmune disease and recently in homeostasis and cancer. More recently, we have begun to appreciate the diverse nature of the small molecule ligands that can bind MR1, with several less potent antigens and small molecule drugs that can bind MR1 being identified. Complementary structural information has revealed the complex nature of interactions defining antigen recognition. Additionally, we now view MAIT cells (defined here as MR1-riboflavin-Ag reactive, TRAV1-2+ cells) as one subset of a broader family of MR1-reactive T cells (MR1T cells). Despite these advances, we still lack a complete understanding of how MR1 ligands are generated, presented and recognized in vivo. The biological relevance of these MR1 ligands and the function of MR1T cells in infection and disease warrants further investigation with new tools and approaches.
Introduction
The enormous diversity of possible T cell receptors, combined with the presentation of antigens on polymorphic MHC molecules, enables the detection of a vast number of foreign or altered-self molecules by T cells. Most well-characterized is the specific recognition of peptide antigens by conventional CD4+ and CD8+ T cells, when presented on MHC Class II and Class I molecules, respectively. Recently, there have been significant advances in understanding antigen recognition by unconventional T cells, in particular natural-killer T (NKT) cells, which recognize lipid-based antigens in the context of CD1 molecules, as well as γδ-T cells, most of which respond to phosphoantigens from infected cells and cancer cells in the context of butyrophilin molecules (1–3). For NKT cells, which share many characteristics with MAIT cells, the first antigen was described in 1997 to be α-galactosylceramide (α-Galcer), derived from a marine sponge (4). Since then, it has become clear that greater diversity exists both in the array of ligands presented by CD1 molecules, and the subsets of NKT cells capable of recognizing these (5, 6).
MAIT cells are a highly conserved unconventional T cell subset, which are abundant in humans and recognize antigens in the context of MR1. MAIT cells express a semi-invariant TCR, comprising TRAV1-2-TRAJ33/12/20 α-chains, paired with a limited array of TCR-β chains (typically TRBV6-1, TRBV6-4, or TRBV20) in humans, and homologous receptors (TRAV1-TRAJ33 paired with TRBV19 or TRBV13) in mice (7–9). The constrained nature of the MAIT TCR repertoire and monomorphic antigen presentation molecule suggested a more limited array of antigens than for conventional T cells. Similar to other unconventional T cell subsets, the MR1T-MR1 axis is being revealed as more complex than initially believed. The first MR1-ligand, the non-agonist 6-formyl pterin (6-FP) was identified in 2012 (10), then some transitory pyrimidine-based MAIT cell antigens were identified in 2014 (11), among which 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) represents the most potent MAIT cell agonist to date. Since then several more MR1 ligands have been described, as well as an increased definition of subsets of MR1-reactive cells, beyond the recognition of riboflavin-based antigens by MAIT cells. Thus, MR1-antigen recognition and MR1-reactive T cell responses in immunity are emerging fields. Like conventional and other unconventional T cell subsets, there is a great promise for developing MR1T-cell-based therapies in several contexts. The definition of the scope of antigens that they recognize, the fine detail of their antigen specificity, and the factors that govern their activation and function will be crucial for achieving this goal.
MAIT Cell Recognition of a new Class of T Cell Antigen
MAIT cells were initially described over 25 years ago as an abundant CD4−CD8− (DN) T cell subset in human blood (8), and later dubbed Mucosal-associated Invariant T (MAIT) cells, due to their conserved TCR usage and enrichment in mucosal tissues, such as the small intestine, in mice and humans (9, 12, 13). It was subsequently shown that MAIT cells reside broadly in tissues like conventional T cells (14) and are restricted to MR1 (12, 15), which has been highly conserved through evolution (16). For several years the antigens recognized through the MR1-MAIT axis were unknown, although early studies suggested they were non-peptide-based molecules (9, 17). The key breakthrough; MR1-ligand identification, was triggered by two key publications from 2010, which demonstrated that MAIT cells could be activated, in an MR1 dependent manner, by a wide range of bacteria and yeasts, but that viruses were non-stimulatory (18, 19). This suggested that the potent activating ligands may come from a conserved biosynthetic pathway and, after careful detective work, this source of antigen was discovered to be microbial biosynthesis of riboflavin (vitamin B2) (10). For a detailed history of the discovery of vitamin B based MAIT cell antigens, readers are referred to previous review articles (20–22) and original research publications (10, 11).
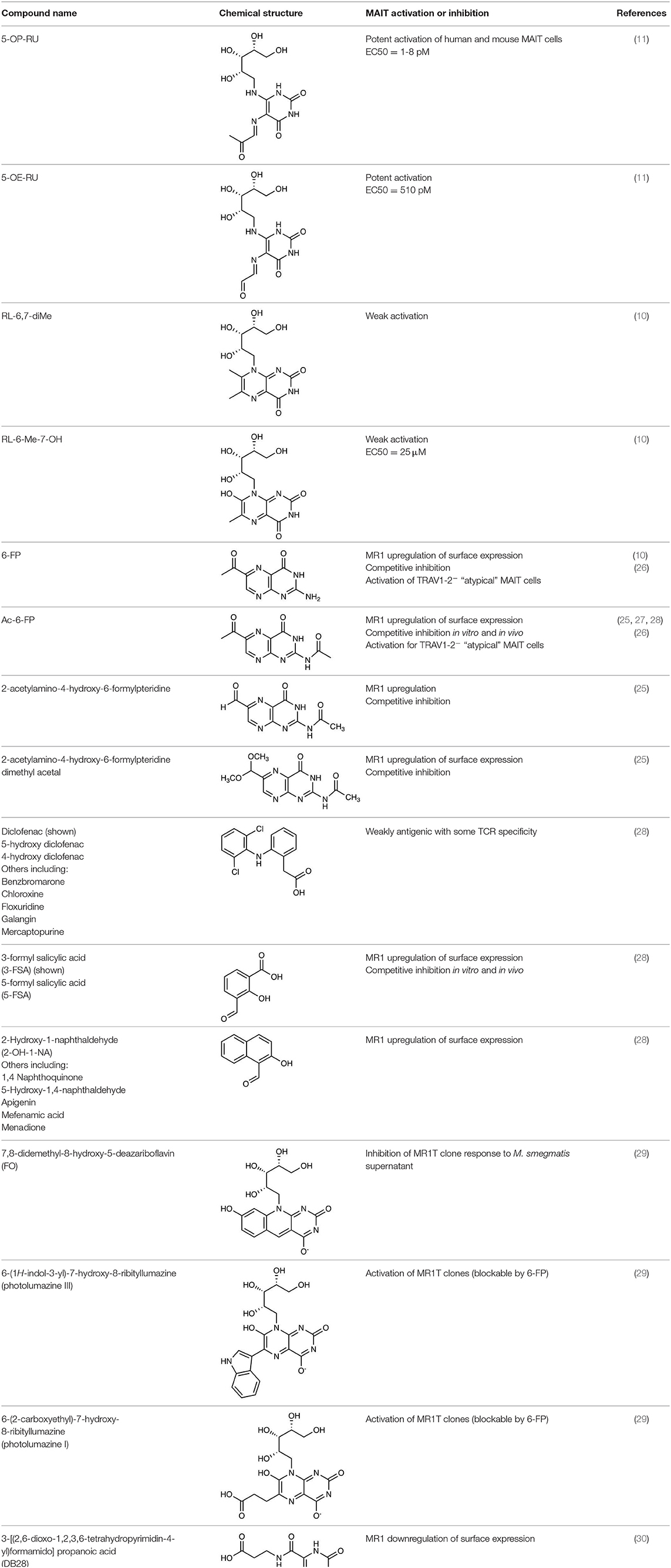
Riboflavin synthesis is a highly conserved biosynthetic pathway, which is essential for many bacteria and yeasts (23, 24). Some organisms cannot synthesize riboflavin but have transporters to take it up from their environment (24), and importantly, several of these microbes (including Enterococcus faecalis and Listeria monocytogenes) are non-stimulatory for MAIT cells (18, 19). A series of enzymes drive each step in the riboflavin biosynthetic pathway, with ribA and ribG (alternatively named ribD in some microorganisms) being essential for the production of a key intermediate 5-amino-6-D-ribitylaminouracil (5-A-RU). Condensation of 5-A-RU with small carbon metabolites, including glyoxal and methylglyoxal, results in the formation of highly potent pyrimidine MAIT cell antigens 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil (5-OE-RU) and 5-OP-RU, respectively (11) (Figure 1). These pyrimidine antigens are highly unstable, thus are further converted to lumazine derivatives unless trapped by MR1. The derived lumazines, RL-6,7-diMe and RL-6-Me-7-OH, are also capable of activation of human and murine MAIT cells, albeit with reduced potency (10, 25) (Table 1). Studies that identified these novel small molecule antigens utilized an MR1-capture approach, in which recombinant human MR1 was refolded, with human β2 microglobulin, in the presence of culture supernatant from bacteria, such as Salmonella Typhimurium, or media controls. Importantly, these metabolite antigens could be detected by liquid chromatography-mass spectrometry (LC-MS) of MR1 refolded in the presence of supernatant from riboflavin-producing bacteria capable of activating MAIT cells, but not from the MAIT cell non-stimulatory bacteria Enterococcus faecalis, or Lactococcus lactis mutants lacking individual rib enzymes (11).
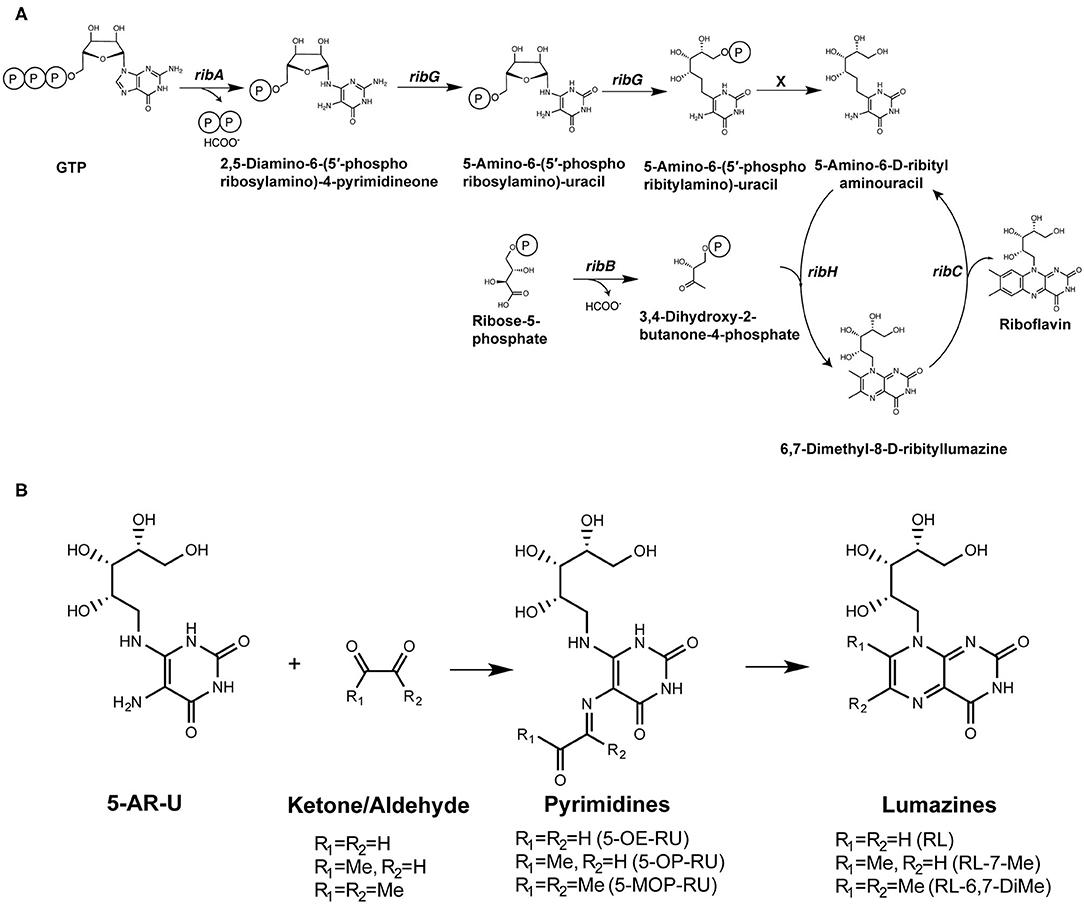
Figure 1. Formation of riboflavin based MAIT antigens. (A) Riboflavin biosynthesis pathway. (B) The riboflavin biosynthesis intermediate 5-A-RU non-enzymatically reacts with small metabolites to form pyrimidine antigens 5-OP-RU and 5-OE-RU. These can be captured by MR1, or alternatively cyclize to form lumazines, some of which are also weakly antigenic [modified from (11)].
The derivation of the potent antigens, 5-OP-RU and 5-OE-RU, from a metabolic intermediate in a conserved biosynthetic riboflavin pathway, 5-A-RU, appeared to answer the question of why the MR1-MAIT axis has been so highly conserved through mammalian evolution (12, 16, 31) since it suggested that MAIT cells would play an important immune role in protecting against diverse microbial pathogens. The idea that MAIT cells may have evolved to directly detect pathogens (by sensing 5-A-RU-derived molecules) and rapidly respond to metabolically active microorganisms that breach the mucosal barriers was then pursued by many researchers in the field, including our own group. Indeed, the presence of MAIT cells has now been shown to contribute to protective immunity against several pathogens capable of riboflavin synthesis (32–34). More recently, roles in barrier function and tissue repair have also been described (35–40), and this would be consistent with the possible sensing of antigens from microflora, which may indicate a breach of barrier function.
Although the most potent MAIT cell agonist known to date, 5-OP-RU, is often studied or cited in isolation, several MR1 ligands, including both MAIT cell agonists and non-agonists, have now been described (Table 1). Even in the initial discovery studies it was evident that, like for conventional T cells and other unconventional T cells, there was not just one antigen, but a family of related molecules that could bind MR1 and potentially interact with MAIT cells. The question of just how large this MR1-ligand family is, remains. MR1 bound antigens are recognized by MAIT cells through their TCR, which is conserved, but not completely invariant. Thus, it has been hypothesized that different antigens are differentially recognized by MAIT cells expressing different TCRs. Indeed structural studies have demonstrated a role for TCRβ chain in antigen recognition, suggesting that certain subsets of MAIT cells may be enriched in response to different antigens (27). We will address both the diversity of MR1 ligands and the recognition of MR1-antigens by MAIT cells and other MR1-reactive T cells in the following sections.
Expansion of the MR1 Ligand Family
The first identified MR1-ligand, 6-FP, is a photosynthetic breakdown product of folic acid. This pterin-based ligand is non-stimulatory for most MAIT cells (10) and exhibits a competitive inhibitory effect on MAIT cell activation by 5-OP-RU (25, 27, 28, 41). Acetyl-6-FP (Ac-6-FP) and acetylamino-4-hydroxy-6-formylpteridine dimethyl acetal, synthetic derivatives of 6-FP, are similarly capable of binding to MR1 (as judged by an increase in cell surface expression), but do not activate MAIT cells and, like 6-FP, competitively inhibit activation by 5-OP-RU (25, 27, 28, 41) or E. coli (25, 42).
Recently, the family of MR1 ligands has grown significantly, with several groups identifying compounds capable of binding to MR1 and/or stimulating MAIT cells (Table 1). Using in silico screens of chemical libraries followed by in vitro functional testing, Keller et al. showed that over 20 compounds, with different chemical scaffolds, could bind to MR1 and modulate MAIT cell activity (28). Interestingly, these included some common drugs and drug metabolites such as diclofenac and the salicylates 3-formylsalicylic acid and 5-formylsalicylic acid. This study also demonstrated competitive inhibition of MAIT cell activation in vivo, suggesting potentially important physiological effects. Additionally, there was some selectivity in the ability of diclofenac metabolites to activate cell lines expressing MAIT TCRs with the same TCR α-chain but different β-chains, consistent with the role of the β-chain in antigen recognition shown in an earlier study (27).
It is considered likely that additional riboflavin-related antigens may exist. Soudais et al. showed that, as well as methylglyoxal and glyoxal, the small molecule di-hydroxy acetone (DHA) in combination with 5-A-RU, caused activation of mouse MAIT cells, in vitro. However, it is unclear whether this was due to the generation of a novel antigen, or conversion of DHA to methylglyoxal (25). In contrast, the same small molecules, glyoxal, methylglyoxal, and DHA, when mixed with 5-nitro-6-D-ribitylaminouracil (5-N-RU) did not significantly activate mouse MAIT cells (25). In 2018, Harriff et al. identified several additional MR1 ligands. These included photolumazines I and III, each capable of activating TRAV1-2+ T cell clones and, more weakly, a TRAV1− T cell clone in an MR1-dependent manner and 7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO), which could competitively inhibit activation. They also described the capture by MR1 of the plant flavonoid hesperidin, although this did not appear to either activate or inhibit MAIT cells (29). The reduced potency of these MR1-ligands for MAIT cells (or reporter cells expressing MAIT TCRs) (Table 1), and the selective activation of subsets of MR1T cells, mean it is currently unclear what the physiological role of T cell detection of such molecules plays in immunity.
Most recently, using an in silico screen based on the MR1 binding pocket, Salio et al. identified an intriguing effect of a novel MR1 ligand (3-[(2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl)formamido]propanoic acid “DB28,” which appeared to decrease, rather than increase cell surface expression of MR1, as well as competitively inhibiting activation of MAIT cells by agonist ligands (30).
Although many do not appear to be antigenic to MR1-5-OP-RU reactive MAIT cells, it is now widely accepted that the MR1 ligands encompass more than those originally described. Intriguingly, crystal structures (10, 11, 27, 41, 43, 44) show that the known ligands do not fully occupy the MR1 antigen-binding cleft, suggesting that, in addition to small variations, much larger and structurally different scaffolds of MR1-ligands may be possible. Thus, it is likely that the list of described MR1 ligands will continue to grow, and these may also represent antigens for MAIT cells and other MR1T cells.
Understanding MAIT Cell Recognition of MR1-Presented Riboflavin-Based Antigens
A remarkable characteristic of the potent pyrimidine antigens, 5-OP-RU and 5-OE-RU, and the pterin-based ligands, such as 6-FP, is that these are capable of forming a covalent bond to the MR1 antigen-presenting molecule via a Schiff base formed between the ligand and a Lys residue at position 43 of MR1, which sits in the base of the antigen-binding pocket (10, 11, 45). Whilst several MR1-ligands have now been described, the ability to form this linkage appears critical for stable binding to MR1, as detected by egress of loaded MR1 molecules from the ER and upregulation on the cell surface (44–46). The lower potency of the related lumazine antigens also appears to be due to their inability to form a Schiff base with the Lys 43 residue. Interestingly, the MR1 downregulating ligand, DB28, was unable to form a Schiff based with the Lys 43 residue (30). Within MAIT TCRs, there was the conservation of key residues including a highly conserved Tyr at position 95 of the CDR3 loop of the TCR α-chain. X-ray crystallographic analyses of MAIT TCR-MR1 complexes revealed that this Tyr95α “reaches” down toward the small molecule antigens, potentially explaining the selective TCR usage among MR1-5-OP-RU reactive MAIT cells (11, 27, 41, 47), with the MAIT TCR β chain also playing a role in antigen recognition (27).
Despite these advances, our understanding of the high potency of 5-OP-RU remains incomplete. A panel of 20 analogs of 5-OP-RU was recently developed (44, 48) (Figure 3) in order to better understand the intricate factors for MR1 and TCR binding. These “altered metabolite ligands” (AML) are equivalent to altered peptide ligands (APL) which have been instrumental in defining the rules governing classical MHC I and II peptide recognition by conventional T cells. Using the complementary approaches of chemical, functional and high-resolution structural analyses, Ler et al. established a set of molecular rules governing ligand binding to MR1 and interactions with the TCR, driving activation. The impact of various modifications to the antigen correlated with the extent to which they disrupted the formation of an “interaction triad,” a network of hydrogen bonds between the conserved Tyr95α in the CDRα loop of MAIT TCRs, the ribityl moiety of the antigen and the Tyr152 residue of MR1. MAIT cell activation potency was found to be orchestrated by dynamic compensatory interactions within this “interaction triad”. Among all tested ligands, 5-OP-RU, the most potent MAIT agonist identified to date, was found the most capable of establishing a strong and stable interaction triad. Even small modifications to the ribityl chain resulted in profoundly reduced potency in cellular assays.
Similar conclusions were reached by a second group, who produced a partially overlapping set of 5-OP-RU analogs (Figure 2A) by different methodologies (49). In both studies, removing the terminal hydroxyl group of the ribityl chain significantly reduced the ability of the ligand to engage the TCR and activate MAIT cells. With this interaction removed, the ligands could act as competitive inhibitors (49). In another study, sugar analogs of the weaker lumazine ligands RL-6-Me-7-OH were also shown to bind MR1 and tetramers loaded with these analogs could stain a cell line expressing a MAIT TCR (50) (Figure 2B). However, their potency in stimulating bone-fide MAIT cells has not yet been examined.
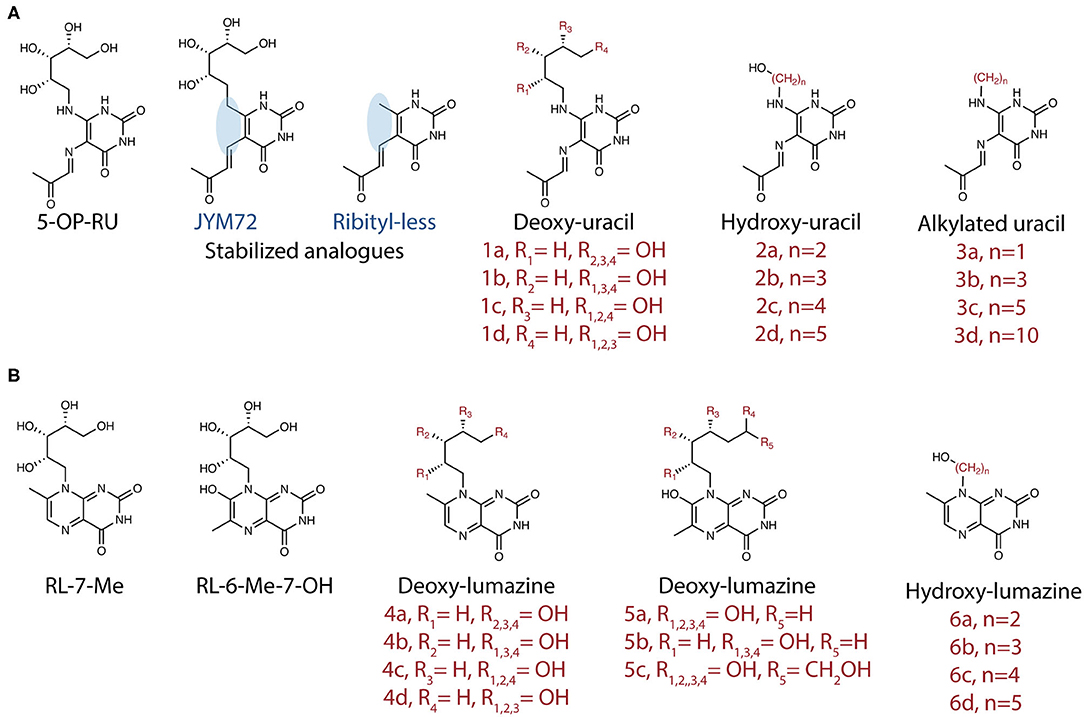
Figure 2. Synthetic analogs of (A) pyrimidine antigen 5-OP-RU or (B) lumazines RL-7-Me and RL-6-Me-7-OH have been created to understand how different modifications impact MR1 binding and MAIT cell activation or inhibition.
More than MAITS; The Increasing Diversity of MR1-Reactive T Cells
MAIT cells were initially described as a large population of cells with restricted TCR α- and β-chain usage (8, 9). This, together with the high conservation of MR1 (51) suggested a capacity for recognition of a single or limited set of antigens. However, as described above, the array of MR1-ligands that may serve as T cell antigens is now acknowledged to be larger than first believed (Table 1). Development of MR1-tetramers in 2013 (K43A mutant MR1) and 2014 (WT mouse and human MR1) enabled sorting and TCR sequencing of MR1-5-OP-RU reactive cells. This revealed greater heterogeneity of TCR usage than previously described (7); a finding later supported by other studies (52–54). We define MAIT cells here as TRAV1-2+ MR1-riboflavin-Ag-reactive T cells (Figure 3). These cells can be detected using MR1-5-OP-RU tetramers, are restricted to MR1, acquire the hallmark promyelocytic leukemia zinc finger (PLZF) molecule during thymic development (55), and display an effector-memory phenotype (CD44hi and CD62Llo).
Figure 3. Diversity and characteristics of MR1-reactive T cells. % refer to those reported in human PBMCs. For subsets with diverse TCRs examples are listed. The lower panel shows cartoon representations of the crystal structures of ternary TCR-MR1-Ag complexes: The typical MAIT A-F7 TRAV1-2-TRBV6-1 TCR-MR1-5-OP-RU (PDB; 6PUC); the atypical TRAV36-TRBV28 TCR-MR1-5-OP-RU (PDB; 5D7L); MR1-autorective M33.64 (TRAV1-2/TRBV6-4) TCR-MR1-5-OP-RU (PDB; 5D5M); and G7 γδ TCR with MR1-5-OP-RU complex (PDB; 6MWR). The MR1 and β2-microglobulin molecules are colored white and pale-cyan, respectively. TRAV1-2 TCRα, light-blue; TRAV6 TCRβ, light-pink; TRAV36 TCRα, violet-purple; TRBV28 TCRβ, sky-blue; TRGV9 TCRγ; lemon, TRDV1 TCRδ, orange. Structural illustration was created using PyMOL Molecular Graphics System, Version 1.8.6, Schrodinger and BioRender.com.
In addition to the riboflavin-reactive TRAV1-2+ MAIT cells, populations of TRAV1-2− MR1-reactive T (MR1T) cells have been described by several groups. In 2016, Gherardin et al. described “atypical MAIT cells,” which were variably riboflavin- or folate- reactive or MR1 autoreactive cells (26), with a role for the CDR3β loops of the TCRs in determining reactivity. These cells were relatively rare, comprising <0.1% of αβ-T cells in human blood. Additionally, more diverse TRAV1-2− cells were described, which were phenotypically distinct from TRAV1-2+ MAIT cells. X-ray crystallographic analyses of one TRAV1-2− (TRAV36-TRBV28) TCR in complex with-MR1-5-OP-RU revealed a different mode of recognition, whereby the TCR docked more centrally on MR1 compared to the TRAV1-2+ TCRs [reviewed in detail (56)] (Figure 3). Broadly consistent with these studies, another group reported the existence of MR1-5-OP-RU reactive TRAV1-2− cells, with at least one clone instead expressing TRAV12-2, which lacks the Tyr95 residue previously found to be conserved in TRAV1-2+ MAIT cells (57). Furthermore, this clone displayed distinct antigen specificity, detecting infection with Streptococcus pyogenes (group A strep), which does not have the capacity to produce riboflavin, suggested recognition of microbial non-ribityl-based antigens (57). Similarly, a proportion of cells staining with MR1-tetramers generated with E. coli-derived ligands were TRAV1-2−, again indicating that not all MR1T cells are TRAV1-2+ MAIT cells (29). Lepore et al. and Koay et al. have also identified MR1T cells in healthy individuals, which were capable of reacting to MR1-expressing cells in the absence of microbial ligands. These MR1T cells were found to have a diverse TCR repertoire and functional capacity (58, 59).
In addition to αβ-T cells, γδ-T cell populations displaying reactivity to MR1 have recently been described. Interestingly, a distinct population of these γδ-T cells recognized MR1 via a novel binding mode, whereby the TCR binds the underside of the MR1 molecule (60) (Figure 3). Thus, in addition to its role in presenting microbial antigens for specific recognition by MAIT cells, it appears that MR1 may act as a pattern recognition receptor, with its cell surface upregulation potentially triggering a more innate-like immune response. It is unclear whether MR1 molecules are present at the cell surface in an empty form, but it is considered likely that they contain endogenous (self) ligands. Indeed, recent studies suggest MR1 can present endogenous or tumor antigens to MR1T cells other than MAIT cells (58, 61). However, these antigens have not yet been identified.
Thus, our definition of MR1-reactive T cells has expanded to encompass not just MR1-5-OP-RU reactive TRAV1-2+ MAIT cells, but folate-reactive and autoreactive atypical MAIT cells, as well as γδ-T cells (Figure 3). Thus, the term MR1T cells is now used to denote all MR1-reactive cells, of which MAIT cells represent the majority. In contrast, other subsets of MR1T identified to date are PLZF negative. While the co-evolution of MR1 with the TRAV1 invariant TCR alpha chain (16, 62) suggests that presentation of microbial signature Ag to MAIT cells is the primary function of MR1, questions remain regarding the thymic development, functional capability and physiological relevance of the other MR1T cell populations. Although some subsets, such as the folate-reactive “atypical MAIT cells” represent only a very small percentage, as pointed out by Gherardin et al., these are present in similar numbers in humans to type I NKT cells, and may be expanded in disease settings (26). The selective recognition of antigens by various subsets of MR1T cells expressing different TCRs suggests that these unconventional T cells may generate display specific memory cell pools similar to conventional T cells. Indeed, a recent study by Loh et al. demonstrated that older adults have large clonal expansions of MAIT cells, similar to those seen in conventional virus-specific CD8+ T cells (63). Clonal expansion of MAIT cells has also been reported (64) and these may suggest differential recognition of diverse MR1-ligands, or expansion of higher affinity clones in some individuals, perhaps due to a history of infection. A recent study of a small cohort of patients with multiple sclerosis reported oligoclonality, with limited TCRβ repertoires, which remained stable over 3 years (65). Stable oligoclonality was confirmed in healthy individuals by Howson et al., who also found in six individuals the MAIT TCRβ repertoire maintained oligoclonality following S. Paratyphi infection, but with expansion and contraction of particular clonotypes, resulting in an altered composition. Cell lines expressing TCRs from expanded clones were more responsive to MAIT cell riboflavin-based antigens (66) suggesting preferential expansion of activated MAIT cells. The TCR repertoire of expanded cells has not yet been assessed in experimental settings such as in mouse models using defined antigens. Since the selective recognition of MR1 ligands by subsets of MR1T cells, potentially with functional differences, present opportunities for selective targeting, more research is needed into this area.
MR1-Antigen Recognition in Physiological Settings
Although several advances have been made in defining MR1-ligands and their recognition by MR1T cells, many questions remain about the physiological relevance of each antigen. As outlined recently by Ler et al. (44), the ability of ligands to generate a MAIT cell response is the result of the combination of several factors, including chemical properties of the ligands, the ability to bind strongly to MR1 resulting in egress to the cell surface, and the avidity of T cell recognition. The expression and regulation of MR1-ligand presentation remain incompletely understood, and this will impact the activation of MR1-reactive T cells in vivo. It is clear from studies comparing different natural ligands or altered metabolite ligands (AML), that the recognition and subsequent response depends on many factors, including chemical stability, binding to MR1 (significantly dependent on the ability to form a Schiff base covalent bond) and TCR recognition (27, 41, 44, 46). To date, 5-OP-RU is the most potent MAIT antigen in any assay, activating MAIT cells even at pmol concentrations. It has also been shown to be relevant to MAIT cell thymic development and activation of MAIT cells in vivo. On the other hand, a key question remaining in the field is whether any of the less potent antigens, antigens that activate smaller subsets of MR1T cells, or MR1 ligands that inhibit MAIT cell activation, are physiologically relevant. Here, we will focus not on the MRIT cell response in these settings, but clues to the in vivo production and MR1-presentation of antigens in different contexts.
Physiological Expression and Impact of MAIT Antigens
Riboflavin-based antigens that stimulate MAIT cells include the potent pyrimidine-based compounds 5-OP-RU and 5-OE-RU, have now been identified from several bacterial species, including Salmonella Typhimurium, Escherichia coli, Lactococcus lactis, and were not detected from riboflavin deficient bacteria (E. faecalis) (10, 11). Indeed, bacteria with deletions in the rib genes necessary for this biosynthetic pathway are incapable of a full in vivo response, as MAIT cells accumulated in the lungs after intranasal infection with Salmonella Typhimurium but not a ribDH-deficient mutant (67). Many other microbes possessing the riboflavin pathway have now been shown to activate MAIT cells [summarized in a previous review (20)] and the requirement for the presence of the riboflavin pathway in this activation was confirmed for E. coli (25) and S. pneumoniae (68), as well as in a study screening 47 microbiota-associated bacterial species. In the latter study, differences in activation capacity were observed between different phyla, and levels of riboflavin production correlated with MAIT cell activity (69). The presence of shared antigens, 5-OP-RU and 5-OE-RU by these microbes is likely, although so far these have only been detected for a handful of bacterial species. Additionally, the physiological relevance of the less potent, but related, lumazines particularly during infection, has not been fully addressed.
MAIT cells have been analyzed in several mouse models of bacterial infection, demonstrating their role in protecting against pathogenic organisms such as Francisella tularensis (33, 70), Klebsiella pneumoniae (32), Mycobacterium bovis BCG (54), and Legionella longbeachae (34). Riboflavin-based antigens are likely produced not only by pathogenic microorganisms during infection, but also by commensal microorganisms. Indeed, the microbiota is important for the development of MAIT cells, as they are almost completely absent in the periphery in germ-free mice (12, 55). The re-introduction of bacteria, even with single strains, was shown to restore MAIT cells (19). Recently it was demonstrated that the ability of microflora to support thymic development of MAIT cells was dependent on riboflavin biosynthesis, and that providing exogenous 5-OP-RU was sufficient to reconstitute MAIT cell development (71). Recent studies also demonstrate the importance of gut microbiota and the presence of rib genes, on MAIT cell reconstitution after allogenic haematopoietic cell transplantation in humans (72, 73). Dysbiosis in viral infections or changes to the gut microbiota have been shown to be associated with impaired MAIT cell responses (74) and it is hypothesized that dysbiosis in metabolic conditions also affects MAIT cells, altering their capacity to promote barrier integrity, and instead triggering a pathogenic, inflammatory MAIT cell response (75). Understanding microenvironmental factors controlling the production of antigen by both pathogens and commensal microorganisms will be important for a complete understanding of MAIT cell responses.
Despite an abundance of information on the regulation of riboflavin production in several bacteria and fungi (23, 24), and the identification of 5-A-RU as the key chemical building block of these molecules, the in vivo production of riboflavin-based antigens, including 5-OP-RU and 5-OE-RU (11), remains poorly understood. For example, recently Kurioka et al. showed that the S. pneumoniae riboflavin operon genes, which are highly conserved between Streptoccci, were upregulated with heat stress, followed by later downregulation (68). It is thus probable that MR1-antigen production, and the resultant MAIT cell activation, may be enhanced under different conditions that may be encountered during the course of infection. Schmaler et al. showed that the growth of bacteria in conditions designed to resemble the human colon (e.g., low oxygen) or on different sugar sources affected bacterial metabolism and the subsequent ability of bacterial samples to activate MAIT cells (76). Intriguingly, the ability of E. coli to activate CD8+ human peripheral blood MAIT cells, in vitro, was increased by treatment with the pesticides chlorpyrifos and glyphosate, which also were found to cause changes in the riboflavin and folate biosynthesis pathways (42). Moreover, folate producing bacteria that didn't stimulate MAIT cells (Lactobacillus reuteri and Bifidobacterium adolescentis) were shown to inhibit activation of human MAIT cells by E. coli, as measured by the production of IFNγ and TNF (42), suggesting the balance of activating and inhibitory MR1-ligands may be important for the generation of MAIT cell responses, in vivo.
In humans, MAIT cell responses during infection have been suggested by studies of MAIT cells in patient cohorts, including in the blood and an increasing selection of tissues, paired with subsequent in vitro analysis. These studies included patients with tuberculosis, Helicobacter infection, cholera, and undefined infections such as sepsis and community-acquired pneumonia (77–80). MAIT cells were also activated in human volunteers infected with live Salmonella enterica Paratyphi A vaccine (66). The role of human MAIT cells in infection and diseases has been reviewed elsewhere (14, 81, 82) so will not be covered in detail here.
In addition to the setting of infection with pathogens producing riboflavin-based antigens, MAIT cells have been studied in the context of viral infections, autoimmune diseases including diabetes, multiple sclerosis systemic lupus erythematosus and arthritis, and have most recently gained attention as potentially important in cancer. There have been several recent reviews covering the findings in these areas as well as animal models developed, which allow the study of MAIT cells in these settings (75, 83–87). The function of MAIT cells in these settings is complex and differs from the direct anti-bacterial role seen in infection models. For example, in the non-obese mouse model of diabetes, deletion of MR1, and thus MAIT cells, resulted in a loss of gut integrity, indicating a protective function, but MAIT cell alterations, including increase in cytotoxic effectors were also observed in mice and human type-1 diabetic patients at the onset of disease suggesting a pathogenic role (35). A role for MAIT cells in tissue repair or inflammatory conditions such as inflammatory bowel disease would be consistent with their enrichment at barrier sites where they may encounter metabolically active pathogenic or commensal microorganisms. However, the recognition of riboflavin-based or other MR1-ligands in these settings has not yet been confirmed.
MR1 Expression and Regulation
MR1 is highly conserved and shows low genetic polymorphism (16). The MR1 protein is expressed, along with β2m, at low to undetectable levels on a range of cell types (88). MR1 is ubiquitously expressed at the transcript level in mouse and human cells (31, 89), although recent studies point to different levels of expression in different tissues (90, 91). Like MHC Class I molecules, MR1 forms stable complexes on the cell surface only when stably folded in the presence of a ligand (88, 92) and upon ligand binding MR1 is released from the ER and traffics to the cell surface (45), a characteristic exploited for detecting the presence of ligands in cellular assays of MR1 upregulation (10, 11, 45). However, MR1 remains difficult to analyse due to its low expression, particularly in primary murine cells, despite evidence for an its function both in MAIT cell development (12) and impressive MAIT cell accumulation in vivo (67).
Several cell types have been used as MR1 antigen-presenting cells (APCs) in in vitro assays. In vivo, one recent study showed that both bone marrow-derived professional APC and non-bone marrow-derived cells (epithelial or stromal cells) can present antigen to activate a MAIT cell response, with the relative importance determined by the biology of the infecting pathogen (93). Thus, it remains to be fully understood which cells are responsible for MAIT cell activation in physiological settings such as during bacterial infection.
Downregulation of MR1 by HSV-1 and CMV suggests that TCR-dependent MAIT cell activation may also be impaired in the context of viral infection (94). Given that MAIT cell numbers and function are altered in human cohorts including bacterial infection or sepsis, viral infection and autoimmune disease, it remains to be shown whether there is a direct link to MR1 expression. Seshadri et al. identified a single nucleotide polymorphism in an intronic region of MR1, which was associated with susceptibility to tuberculosis in a Vietnamese adult cohort. This SNP was also associated with MR1 expression in cell lines, and thus may act to modulate the MR1-dependent response of MAIT cells (95). MR1 also exists in multiple isoforms (31) but the biological relevance of these is unclear.
The Context of Antigen Recognition Determines MAIT Cell Responses
MAIT cells have been described as “innate-like” rapid responders, with a memory-like phenotype. However, like conventional T cells, MR1-antigen alone is insufficient to drive an optimal MAIT cell response, with co-stimuli including cytokines and/or surface co-receptors required (67) and able to enhance MAIT responses to antigen stimulation (96–99), although in vitro findings have not always translated to in vivo studies (70). Additionally, polarizing effects on MAIT cell function have been demonstrated. In particular, priming with IL-7 or treatment with IL-23 significantly increases IL-17 secretion, which otherwise is minimally induced in MAIT cells (93, 100, 101). IL-7 has been linked to effects on MAIT cell function and polarization in disease settings such as multiple sclerosis (102) and primary biliary cholangitis (103). Recently, it was shown that ICOS and IL-23 are important for MAIT cell in vivo responses in mouse infection models with Salmonella and Legionella (93). In genetically deficient mice lacking these co-stimulators, expansion of MAIT cells was impaired and the profiles of responding pulmonary MAIT cells shifted from RORγt+ to T-bet+ phenotypes (93).
Different cytokine signals synergize to control MAIT responses (104), and thus, the MAIT cell functional capacity may differ in tissue settings compared to blood MAIT cells (105). In humans, most peripheral blood MAIT cells display type-1 responses upon stimulation, whereas in tissues a IL-17-secreting population is more readily detected (80, 105), suggesting there may be functional differences. Following infection with S. Paratyphi, the MAIT cell response was partially IL-12-dependent, suggesting the quality and quantity of MAIT cell immunity is driven by the combination of pathogen signals and host cytokines (66). The augmenting effects of cytokines may act via increasing expression of the MAIT TCR (101) or upregulating cellular components related to TCR signaling (100).
In the absence of TCR-dependent antigen recognition, MAIT cells can be activated by cytokines, including IL-12 and IL-18 (54, 106–108), and by super-antigens (109). In one study, the MAIT cell response in group A streptococcus (GAS) infection appeared to contain both TCRβ-dependent (super-antigen) and independent activation (110). Cytokines likely drive the MAIT cell activation observed in viral infections and contribute to their response during bacterial infection (111). Importantly, there are qualitative and quantitative differences in the MAIT cell responses triggered by TCR-dependent and TCR-independent mechanisms (38). However, the implications of these differences in various immune settings are yet to be determined.
In settings of chronic inflammation, infection or cancer, MAIT cells display altered phenotypes (112), can produce Th2 cytokines (113), and can contribute to pathology (114). MAIT cell function may be impaired in settings such as chronic infection (115–117) and cancer (118). Various studies have implicated immune checkpoints (116, 119, 120), cytokines such as IL-10 (121) and suppression of MAIT cell function by hydrogen peroxide released from neutrophils (122). Thus, in addition to recognition of MR1-bound antigen, the context of other signals in different setting is important for MAIT cell activation and function.
Tools for the Assessment of MR1 Ligand Production and Recognition
The discovery of a new class of antigens, while providing exciting opportunities, also presents challenges when seeking to understand their biological significance. The study of metabolite-based ligands presents new and different challenges to the better-known peptide and lipid-based antigens. The MR1-binding and MAIT cell activation by metabolites of drugs such as diclofenac (28), as well as the recently described breakdown of 5-A-RU in air (123), highlights the importance of confirming the precise identity of MR1 captured ligands by a combination of assays such as high accuracy mass spectroscopy, X-ray crystallography, NMR spectroscopy, functional assays and ultimately in vivo studies. Identification of 5-OP-RU and 5-OE-RU was originally achieved by mass spectrometric analysis of recombinantly expressed MR1 protein refolded in the presence of bacterial supernatant to “capture” the small molecule ligands, in combination with X-ray crystallography and functional validation (10, 11, 20). These techniques do not easily lend themselves to the detection of MR1-bound ligands from complex biological samples, and to date, the field has not developed a simple assay to confirm antigen identity. Thus, many studies did not confirm the identity of the antigens involved. Evidence of riboflavin pathway involvement provided by detecting or deleting rib genes has been used as an indirect confirmation. However, it is possible that 5-A-RU recombines with other small molecules to form alternate antigens. For example, Harriff et al. suggest that novel antigens photolumazines I and III may form from 5-A-RU and α-ketoglutarate, the availability of which may be altered in nitrosative stress during intracellular infection (29).
The MR1 autoreactivity shown by some MR1T cells also reveals the importance of validating reactivity with specific controls (26, 60). In this section we present a summary of the basic tools and approaches used to develop our current understanding of the immune response to metabolite antigens.
Detection of MAIT Cells Using Antibodies or Tetramers
The precise identification of MAIT cells was hampered for several years, particularly in mice and other non-human mammals, by the lack of specific reagents for their detection. Human MAIT cells can be identified by co-staining with a panel of antibodies. Typically, CD3, CD161, and TRAV1-2 (Vα7.2) are used (124, 125), although some groups have also used the high expression levels on MAIT cells of surrogate markers such as CD26 and IL-18R (13, 57, 126, 127). The CD4+, CD8+, and double-negative (DN) MAIT cell subsets have been variably included or excluded. However, it is now clear that MAIT cells contain all of these subsets (128). Complicating these approaches, it is apparent that the expression of some markers may change in different physiological settings. For example, CD161 expression decreased in rheumatoid arthritis (129) and HIV infection (121). In mice, there are currently no available antibodies recognizing the TCR Vα19 utilized by mouse MAIT cells. This, combined with the low MAIT cell numbers in naïve laboratory mice, prevented the systematic assessment of MAIT cells in wild-type laboratory mice prior to the generation of MR1-tetramer reagents (7, 11, 54, 67, 130).
The first generation of MR1-tetramers (7) utilized a modified human MR1 containing a Lys to Ala mutation at position 43 at the base of the MR1 A' binding pocket. This mutation enabled the refolding of “empty” MR1-β2m molecules in the absence of an added ligand, which would otherwise be needed to stabilize the MR1-β2m complex. These K43A MR1-β2m monomers could then be loaded with an antigen of choice and tetramerised with fluorochrome-coupled streptavidin, enabling the detection of MAIT cells in human blood and intestinal cell preparations (7). The elucidation of the formation of 5-OP-RU and 5-OE-RU from 5-A-RU and methylglyoxal or glyoxal, respectively, enabled the subsequent generation of mouse and human [and later macaque (131)] wild-type MR1 tetramers loaded with each of these molecules. These were formed by refolding the MR1 and β2m proteins in the presence of the precursor molecules, followed by biotinylation and tetramerization by standard methods (11). The resultant new generation tetramers were far more stable and easier to use in standard flow cytometry protocols. These reagents have now enabled the detection and study of MAIT cells in many contexts, similarly to the use over many years of MHC Class I (132) and CD1d tetramers (133). MR1 tetramers loaded with 6-FP and Ac-6-FP were similarly generated. These are typically used as control reagents but have been shown to detect a small number of reactive or MR1-autoreactive cells (26), the biological relevance of which is currently unclear.
Despite the clear overlap between MAIT cells detected using MR1-5-OP-RU tetramers and using antibodies to TRAV1-2 [~95% in human blood (11)], these reagents will not detect all MR1T cells, since some of these may have different antigen specificities. This may be particularly true in tissues, or in different mammalian species, where analysis has been less extensive. Thus, similar to the methods for detecting MR1-ligands discussed above, the generation of MR1-tetramers loaded with different sources of metabolite molecules (29) will likely be useful in understanding MR1T cells other than 5-OP-RU-reactive MAIT cells. For small modifications to 5-OP-RU, such as with the altered metabolite ligands discussed above (44), the identified populations almost completely overlap, suggesting that either MR1T cells recognizing ligands other than 5-OP-RU are rare, or were excluded from this analysis by prior gating. Thus, removing the assumption of shared markers, and the use of tetramers loaded with more complex sources of potential ligands is expected to broaden the scope of detection of MR1T cells.
In vitro Cellular Assays
In order to identify MR1-bound ligands that are recognized by MAIT or other MR1T cells, several groups have developed cellular assays for screening. These utilize either T cell clones or T cell lines (such as Jurkat, SKW3, or mouse 6C2) engineered to express a MAIT TCR, which accordingly acts as a reporter cell line. These are co-cultured with cell lines expressing or overexpressing MR1, which act as antigen-presenting cells. Again, various cell lines have been utilized by different groups, in some cases with MR1 deficient cell lines acting as controls (25, 34, 49, 57, 76, 134). Alternatively, gated MAIT cells within PBMCs are analyzed after stimulation, either alone, or with cell lines or dendritic cells as APCs (11, 28, 46, 124, 135–137). Although a number of studies have examined the in vitro activation responses of MAIT cells sourced from human tissues (77, 100, 138, 139) to our knowledge these have not been used for MR1 ligand identification purposes. The activation of cells in these assays has been assessed by upregulation of markers [e.g., CD69, CD25, CD137(41-BB)], down-regulation of CD3 and cytokine production. These readouts give an indication of the presence of activating MR1-ligands, but do not reveal their identity. The presence of MR1-ligands is also indicated by the upregulation of cell surface MR1, as detected by conformational monoclonal antibodies 26.5 or 8F2.F9 (15, 140). However, this measure has some caveats, including lower sensitivity than MAIT cell activation, and the increase in surface expression can be missed depending on the timing of the assay due to the instability of the compounds or MR1-ligand complexes.
Interestingly, 5-A-RU, which is not believed to be a MAIT cell antigen in its own right, can activate MAIT reporter cells in culture, indicating the formation of antigens, such as 5-OP-RU, by reacting with aldehydes or ketones from the medium (such as methylglyoxal) or within cells (11). This highlights the danger of inferring antigen identity when using whole-cell assays to test for MAIT cell activation. To limit the complexity relative to the cell systems, plate-bound MR1 has also been utilized in a similar activation assay (141). The direct elution of MR1-bound ligands from immunoprecipitated cell surface MR1 for identification by mass spectrometry is technically challenging, but has been shown using C1R cells overexpressing MR1 (C1R.MR1 cells) for Ac-6-FP, 3-F-SA and diclofenac metabolites (28).
Chemical and Biochemical Investigations of MR1 Antigens and Analogs
The small molecules 5-OP-RU and 5-OE-RU are formed as chemical intermediates in the reaction of 5-A-RU with methylglyoxal and glyoxal, respectively. These open-ring compounds rapidly cyclise to form relatively weak lumazine antigen RL-7-Me, and RL (Figure 1). Their original identification as MR1 ligands was achieved by refolding recombinantly expressed MR1 in the presence of synthetic compounds or complex sources of antigen, such as bacterial culture supernatant (10, 11, 28), which was demonstrated to be sufficient to stimulate a MAIT cell response (47). This method enabled the MR1 molecules to “fish out” ligands that were capable of binding in a relatively unbiased approach, for subsequent analysis by LC-MS, which was complemented by high-resolution crystal structures. Bacteria with rib gene deletions (S. Typhimurium, L. lactis) were generated to confirm the involvement of the riboflavin synthesis pathway (10, 11). The relatively short half-life of 5-OP-RU in aqueous solution [t1/2 half-life time (t1/2) ~1.5 h, 37°C] (11, 46) suggests that related compounds, if they exist in nature, may be similarly difficult to identify. More recently the Lewinsohn group used a modified procedure to assess the capture of MR1-ligands from mammalian or insect cells infected with E.coli and M. smegmatis (29). LC-MS combined with molecular networking analysis revealed many candidate ligands, with a subset of these functionally validated using MR1T cell clones (29) (Table 1).
The chemical instability of MAIT antigens poses difficulties for the potential development of therapies targeting MAIT cells. On the other hand, this also provides us an insight into an important aspect of the biology of these cells. MAIT cells appear to be precisely poised to detect metabolically active microbes by recognizing antigens that are swiftly captured by MR1. Nevertheless, a few groups have attempted to develop tools in the form of more stable analogs, with the hope that these may better stimulate MAIT cell responses. Mak et al. reported that 5-OP-RU synthesized in DMSO had improved stability. Stabilized compounds (Figure 3) could be generated by replacing the exocyclic nitrogen atoms with carbon atoms (44, 46). However, whilst replicating a similar MAIT recognition and response, this analog was ~1,000-fold less efficient than 5-OP-RU in boosting MAIT cells numbers when delivered intranasally with CpG (46). This again, tells us something of the exquisite specificity of MR1-MAIT recognition, selected through the co-evolution of mammalian hosts and pathogenic or commensal microorganisms.
Using a different approach to tackle the chemical instability of MAIT antigens, the Painter group recently created a stabilized 5-A-RU “pro-drug” which releases 5-A-RU upon enzymatic cleavage in the recycling endosomes and which was active in vivo (123). Another group reported that the stability of the precursor 5-A-RU, long known as a chemically unstable precursor of riboflavin synthesis is increased by synthesizing and storing it as its HCl-salt (142). The recent availability of 5-A-RU from commercial sources should further facilitate its study in this context.
Mouse Models
The ability of compounds to bind MR1, and to activate or inhibit MAIT cells, has been tested in various assays. However, understanding their physiological relevance requires in vivo models. Historically, the study of MAIT cells in mice has been difficult, due to their low numbers, and lack of specific reagents. Four main types of mice have been employed for their study; gnotobiotic (germ-free) mice, in which MAIT cells are barely detectable (12, 55), MR1−/− mice, which do not develop MAIT cells (12), and conversely, transgenic mice expressing the Vα19i invariant TCR α chain (12, 143, 144), and CAST/EiJ mice (145), which have higher numbers of MAIT cells. The use of these and other tools to study MAIT cells in mice has been previously reviewed (83). With our recent understanding that MR1T cells exhibit greater diversity than originally believed, it is important to note that many studies use MR1−/− mice as a “MAIT-cell deficient” model, but these will also lack MR1-dependent responses by non-MAIT MR1T cells. Additionally, the Vα19iTg mice, are not a clear-cut “MAIT cell transgenic.” In these mice, MR1-tetramer-reactive cells were of high frequency, however about one third of these cells did not express the promyelocytic leukemia zinc finger (PLZF) protein, a hallmark of classical MAIT cells. Additionally, a high frequency of MR1-tetramer-reactive cells could be detected even in the absence of MR1 (Vα19iTg.MR1−/− mice) (54), and presumably these were selected by conventional MHC molecules during their development in the thymus. More recently, it has been described that MAIT cells are deficient in TCR Jα18 knockout mice (146), and also in Jα33 knockout mice, which revealed a residual population of MAIT-like TRAV1-2+ or TRAV1-2− cells that were responsive to antigen or bacterial infection (59).
Models of infection with bacterial pathogens in mice (32–34, 54) and non-human primates (147) have revealed much about MAIT cell biology and their role in protective immunity. The development of MR1-5-OP-RU-tetramer reagents (7, 11) has enabled the specific assessment of MAIT cells in naïve SPF-housed mice (67, 130, 148), in disease-relevant models including infection, autoimmune disease (35, 149, 150), transplantation (151) and cancer (152), and MAIT cell development (55, 71, 153). MR1-tetramers also allow the assessment of the ability of defined compounds to act as antigens, with 5-OP-RU (67, 93), premixed 5-A-RU and methylglyoxal (25, 123) and a pro-drug designed to release 5-A-RU (123) all shown to boost MAIT cell numbers in mice, in the presence of TLR agonists or other co-stimuli. However, the direct assessment of MAIT cell responses to MR1 ligands in vivo is still a relatively under-explored area with more research needed.
Conclusions
The MR1-MR1T cell field has progressed rapidly in the short time since the discovery of the first MR1-ligands. We now understand that both MR1-ligands and MR1-reactive T cells are more diverse than first believed. MR1T, particularly MAIT cells, have been studied in physiological settings such as homeostasis, infection, autoimmune disease and cancer, sometimes with contradictory findings in both human cohorts and animal studies. There remain several questions and challenges in the field. For example, we do not fully understand the production of ligands from microbes, how these enter cells, how they are processed and presented, or the full range of signals determining the quality and quantity of the TCR-MR1-dependent response. Deciphering the complexity of antigen presentation and recognition, and the relative importance of other signals contributing to the MR1T response, particularly within the local tissue environment, is essential to understanding their biology. For instance, how do these cells differentiate between pathogens and commensal microorganisms allowing them to play diverse roles including aiding the clearance of pathogens, tissue repair and gut homeostasis? It is possible that there is a threshold amount of antigen that needs to penetrate the mucosal barriers before they will respond, and thus the abundant microbes in the intestines, which can synthesize riboflavin, are normally ignored. However, we consider it likely that the context of antigen and other signals drives different responses, including in a tissue-specific manner, whereby these cells contribute to homeostasis or inflammation and immune protection. Answering these questions will require the analysis of clearly defined MR1T subsets, and likely new tools to combat the issues of chemical instability and ligand identification. Additionally, the definition of an increasing number of MR1T subsets with different Ag-recognition and phenotypic features raises many questions about the physiological relevance and therapeutic potential of these cells. MR1T cells offer great opportunities as attractive targets in vaccination (154) and immunotherapy (61). We hope that further advances in understanding T cell recognition of MR1-ligands using definitive tools and approaches such as those described above, will enable the MR1-MR1T axis to be harnessed to combat infection and disease.
Author Contributions
WA and AC generated the figures and tables. All authors wrote and edited the manuscript.
Funding
AC was the recipient of a Future Fellowship (FT160100083) from the Australian Research Council.
Conflict of Interest
AC and ZC are inventors on patents describing MR1 ligands and MR1-tetramer reagents.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Tanaka Y, Morita CT, Tanaka Y, Nieves E, Brenner MB, Bloom BR. Natural and synthetic non-peptide antigens recognized by human gamma delta T cells. Nature. (1995) 375:155–8. doi: 10.1038/375155a0
2. Constant P, Davodeau F, Peyrat MA, Poquet Y, Puzo G, Bonneville M, et al. Stimulation of human gamma delta T cells by nonpeptidic mycobacterial ligands. Science. (1994) 264:267–70. doi: 10.1126/science.8146660
3. Rigau M, Ostrouska S, Fulford TS, Johnson DN, Woods K, Ruan Z, et al. Butyrophilin 2A1 is essential for phosphoantigen reactivity by gammadelta T cells. Science. (2020) 367:aay5516. doi: 10.1126/science.aay5516
4. Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. (1997) 278:1626–9. doi: 10.1126/science.278.5343.1626
5. Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. (2015) 16:1114–23. doi: 10.1038/ni.3298
6. Moody DB, Cotton RN. Four pathways of CD1 antigen presentation to T cells. Curr Opin Immunol. (2017) 46:127–33. doi: 10.1016/j.coi.2017.07.013
7. Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. (2013) 210:2305–20. doi: 10.1084/jem.20130958
8. Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J Exp Med. (1993) 178:1–16. doi: 10.1084/jem.178.1.1
9. Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. (1999) 189:1907–21. doi: 10.1084/jem.189.12.1907
10. Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. (2012) 491:717–23. doi: 10.1038/nature11605
11. Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. (2014) 509:361–5. doi: 10.1038/nature13160
12. Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. (2003) 422:164–9. doi: 10.1038/nature01433
13. Dusseaux M, Martin E, Serriari N, Peguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. (2011) 117:1250–9. doi: 10.1182/blood-2010-08-303339
14. Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol. (2019) 20:1110–28. doi: 10.1038/s41590-019-0444-8
15. Huang S, Gilfillan S, Cella M, Miley MJ, Lantz O, Lybarger L, et al. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J Biol Chem. (2005) 280:21183–93. doi: 10.1074/jbc.M501087200
16. Boudinot P, Mondot S, Jouneau L, Teyton L, Lefranc MP, Lantz O. Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc Natl Acad Sci USA. (2016) 113, E2983–92. doi: 10.1073/pnas.1600674113
17. Huang S, Gilfillan S, Kim S, Thompson B, Wang X, Sant, et al. MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J Exp Med. (2008) 205:1201–11. doi: 10.1084/jem.20072579
18. Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. (2010) 8:e1000407. doi: 10.1371/journal.pbio.1000407
19. Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. (2010) 11:701–8. doi: 10.1038/ni.1890
20. Kjer-Nielsen L, Corbett AJ, Chen Z, Liu L, Mak JY, Godfrey DI, et al. An overview on the identification of MAIT cell antigens. Immunol Cell Biol. (2018) 96:573–87. doi: 10.1111/imcb.12057
21. Eckle SB, Corbett AJ, Keller AN, Chen Z, Godfrey DI, Liu L, et al. Recognition of vitamin B precursors and byproducts by mucosal associated invariant T cells. J Biol Chem. (2015) 290:30204–11. doi: 10.1074/jbc.R115.685990
22. Keller AN, Corbett AJ, Wubben JM, McCluskey J, Rossjohn J. MAIT cells and MR1-antigen recognition. Curr Opin Immunol. (2017) 46:66–74. doi: 10.1016/j.coi.2017.04.002
23. Bacher A, Eberhardt S, Fischer M, Kis K, Richter G. Biosynthesis of vitamin b2 (riboflavin). Annu Rev Nutr. (2000) 20:153–67. doi: 10.1146/annurev.nutr.20.1.153
24. Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. (2002) 30:3141–51. doi: 10.1093/nar/gkf433
25. Soudais C, Samassa F, Sarkis M, Le Bourhis L, Bessoles S, Blanot D, et al. In vitro and in vivo analysis of the gram-negative bacteria-derived riboflavin precursor derivatives activating mouse MAIT cells. J Immunol. (2015) 194:4641–9. doi: 10.4049/jimmunol.1403224
26. Gherardin NA, Keller AN, Woolley RE, Le Nours J, Ritchie DS, Neeson PJ, et al. Diversity of T cells restricted by the MHC class I-related molecule MR1 facilitates differential antigen recognition. Immunity. (2016) 44:32–45. doi: 10.1016/j.immuni.2015.12.005
27. Eckle SB, Birkinshaw RW, Kostenko L, Corbett AJ, McWilliam HE, Reantragoon R, et al. A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells. J Exp Med. (2014) 211:1585–600. doi: 10.1084/jem.20140484
28. Keller AN, Eckle SB, Xu W, Liu L, Hughes VA, Mak JY, et al. Drugs and drug-like molecules can modulate the function of mucosal-associated invariant T cells. Nat Immunol. (2017) 18:402–411. doi: 10.1038/ni.3679
29. Harriff MJ, McMurtrey C, Froyd CA, Jin H, Cansler M, Null M, et al. MR1 displays the microbial metabolome driving selective MR1-restricted T cell receptor usage. Sci Immunol. (2018) 3:eaao2556. doi: 10.1126/sciimmunol.aao2556
30. Salio M, Awad W, Veerapen N, Gonzalez-Lopez C, Kulicke C, Waithe D, et al. Ligand-dependent downregulation of MR1 cell surface expression. Proc Natl Acad Sci USA. (2020) 117:10465–75. doi: 10.1073/pnas.2003136117
31. Riegert P, Wanner V, Bahram S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J Immunol. (1998) 161:4066–77.
32. Georgel P, Radosavljevic M, Macquin C, Bahram S. The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Mol Immunol. (2011) 48:769–75. doi: 10.1016/j.molimm.2010.12.002
33. Meierovics A, Yankelevich WJ, Cowley SC. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. PNAS. (2013) 110, E3119–28. doi: 10.1073/pnas.1302799110
34. Wang H, D'Souza C, Lim XY, Kostenko L, Pediongco TJ, Eckle SBG, et al. MAIT cells protect against pulmonary Legionella longbeachae infection. Nat Commun. (2018) 9:3350. doi: 10.1038/s41467-018-05202-8
35. Rouxel O, Da Silva J, Beaudoin L, Nel I, Tard C, Cagninacci L, et al. Cytotoxic and regulatory roles of mucosal-associated invariant T cells in type 1 diabetes. Nat Immunol. (2017) 18:1321–31. doi: 10.1038/ni.3854
36. Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. (2019) 28:3249–62 e5. doi: 10.1016/j.celrep.2019.07.039
37. Leng T, Akther HD, Hackstein CP, Powell K, King T, Friedrich M, et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. (2019) 28:3077–91 e5. doi: 10.1016/j.celrep.2019.08.050
38. Lamichhane R, Schneider M, de la Harpe SM, Harrop TWR, Hannaway RF, Dearden PK, et al. TCR- or cytokine-activated CD8(+) mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep. (2019) 28:3061–76 e5. doi: 10.1016/j.celrep.2019.08.054
39. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. (2019) 366:6464. doi: 10.1126/science.aax6624
40. Salou M, Lantz O. A TCR-dependent tissue repair potential of MAIT cells. Trends Immunol. (2019) 40:975–7. doi: 10.1016/j.it.2019.09.001
41. Patel O, Kjer-Nielsen L, Le Nours J, Eckle SB, Birkinshaw R, Beddoe T, et al. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat Commun. (2013) 4:2142. doi: 10.1038/ncomms3142
42. Mendler A, Geier F, Haange SB, Pierzchalski A, Krause JL, Nijenhuis I, et al. Mucosal-associated invariant T-Cell (MAIT) activation is altered by chlorpyrifos- and glyphosate-treated commensal gut bacteria. J Immunotoxicol. (2020) 17:10–20. doi: 10.1080/1547691X.2019.1706672
43. Lopez-Sagaseta J, Dulberger CL, Crooks JE, Parks CD, Luoma AM, McFedries A, et al. The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proc Natl Acad Sci USA. (2013) 110:E1771–8. doi: 10.1073/pnas.1222678110
44. Awad W, Ler GJM, Xu W, Keller AN, Mak JYW, Lim XY, et al. The molecular basis underpinning the potency and specificity of MAIT cell antigens. Nat Immunol. (2020) 21:400–11. doi: 10.1038/s41590-020-0616-6
45. McWilliam HE, Eckle SB, Theodossis A, Liu L, Chen Z, Wubben JM, et al. The intracellular pathway for the presentation of vitamin B-related antigens by the antigen-presenting molecule MR1. Nat Immunol. (2016) 17:531–7. doi: 10.1038/ni.3416
46. Mak JY, Xu W, Reid RC, Corbett AJ, Meehan BS, Wang H, et al. Stabilizing short-lived Schiff base derivatives of 5-aminouracils that activate mucosal-associated invariant T cells. Nat Commun. (2017) 8:14599. doi: 10.1038/ncomms14599
47. Reantragoon R, Kjer-Nielsen L, Patel O, Chen Z, Illing PT, Bhati M, et al. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J Exp Med. (2012) 209:761–74. doi: 10.1084/jem.20112095
48. Ler GJ M, Xu W, Mak JYW, Liu L, Bernhardt PV, Fairlie DP. Computer modelling and synthesis of deoxy and monohydroxy analogues of a ribitylaminouracil bacterial metabolite that potently activates human T cells. Chemistry. (2019) 25:15594–608. doi: 10.1002/chem.201903732
49. Braganza CD, Motozono C, Sonoda KH, Yamasaki S, Shibata K, Timmer MSM, et al. Agonistic or antagonistic mucosal-associated invariant T (MAIT) cell activity is determined by the 6-alkylamino substituent on uracil MR1 ligands. Chem Commun. (2020) 56:5291–94. doi: 10.1039/D0CC00247J
50. Braganza CD, Shibata K, Fujiwara A, Motozono C, Sonoda KH, Yamasaki S, et al. The effect of MR1 ligand glyco-analogues on mucosal-associated invariant T (MAIT) cell activation. Org Biomol Chem. (2019) 17:8992–9000. doi: 10.1039/C9OB01436E
51. Huang S, Martin E, Kim S, Yu L, Soudais C, Fremont, et al. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc Natl Acad Sci USA. (2009) 106:8290–5. doi: 10.1073/pnas.0903196106
52. Lepore M, Kalinichenko A, Colone A, Paleja B, Singhal A, Tschumi A, et al. Parallel T-cell cloning and deep sequencing of human MAIT cells reveal stable oligoclonal TCRbeta repertoire. Nat Commun. (2014) 5:3866. doi: 10.1038/ncomms4866
53. Gold MC, McLaren JE, Reistetter JA, Smyk-Pearson S, Ladell K, Swarbrick GM, et al. MR1-restricted MAIT cells display ligand discrimination and pathogen selectivity through distinct T cell receptor usage. J Exp Med. (2014) 211:1601–10. doi: 10.1084/jem.20140507
54. Sakala IG, Kjer-Nielsen L, Eickhoff CS, Wang X, Blazevic A, Liu L, et al. Functional heterogeneity and antimycobacterial effects of mouse mucosal-associated invariant T cells specific for riboflavin metabolites. J Immunol. (2015) 195:587–601. doi: 10.4049/jimmunol.1402545
55. Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. (2016) 17:1300–11. doi: 10.1038/ni.3565
56. Awad W, Le Nours J, Kjer-Nielsen L, McCluskey J, Rossjohn J. Mucosal-associated invariant T cell receptor recognition of small molecules presented by MR1. Immunol Cell Biol. (2018) 96:588–597. doi: 10.1111/imcb.12017
57. Meermeier EW, Laugel BF, Sewell AK, Corbett AJ, Rossjohn J, McCluskey J, et al. Human TRAV1-2-negative MR1-restricted T cells detect S. pyogenes and alternatives to MAIT riboflavin-based antigens. Nat Commun. (2016) 7:12506. doi: 10.1038/ncomms12506
58. Lepore M, Kalinichenko A, Calogero S, Kumar P, Paleja B, Schmaler M, et al. Functionally diverse human T cells recognize non-microbial antigens presented by MR1. Elife. (2017) 6:24476. doi: 10.7554/eLife.24476
59. Koay HF, Gherardin NA, Xu C, Seneviratna R, Zhao Z, Chen Z, et al. Diverse MR1-restricted T cells in mice and humans. Nat Commun. (2019) 10:2243. doi: 10.1038/s41467-019-10198-w
60. Le Nours J, Gherardin NA, Ramarathinam SH, Awad W, Wiede F, Gully BS, et al. A class of gammadelta T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science. (2019) 366:1522–7. doi: 10.1126/science.aav3900
61. Crowther MD, Dolton G, Legut M, Caillaud ME, Lloyd A, Attaf M, et al. Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat Immunol. (2020) 21:178–85. doi: 10.1038/s41590-019-0578-8
62. Mondot S, Boudinot P, Lantz O. MAIT, MR1, microbes and riboflavin, a paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules? Immunogenetics. (2016) 68:537–48. doi: 10.1007/s00251-016-0927-9
63. Loh L, Gherardin NA, Sant S, Grzelak L, Crawford JC, Bird NL, et al. Human mucosal-associated invariant T cells in older individuals display expanded tcralphabeta clonotypes with potent antimicrobial responses. J Immunol. (2020) 204:1119–33. doi: 10.4049/jimmunol.1900774
64. Wong EB, Gold MC, Meermeier EW, Xulu BZ, Khuzwayo S, Sullivan ZA, et al. TRAV1-2(+) CD8(+) T-cells including oligoconal expansions of MAIT cells are enriched in the airways in human tuberculosis. Commun Biol. (2019) 2:203. doi: 10.1038/s42003-019-0442-2
65. Carnero Contentti E, Farez MF, Correale J. Mucosal-associated invariant T cell features and TCR repertoire characteristics during the course of multiple sclerosis. Front Immunol. (2019) 10:2690. doi: 10.3389/fimmu.2019.02690
66. Howson LJ, Napolitani G, Shepherd D, Ghadbane H, Kurupati P, Preciado-Llanes L, et al. MAIT cell clonal expansion and TCR repertoire shaping in human volunteers challenged with Salmonella Paratyphi. Nat Commun A. (2018) 9:253. doi: 10.1038/s41467-017-02540-x
67. Chen Z, Wang H, D'Souza C, Sun SKL, Eckle SBG, Meehan BS, et al. Mucosal-associated Invariant T cell activation and accumulation after in vivo infection depends on microbial riboflavin synthesis and co-stimulatory signals. Mucosal Immunol. (2017) 10:58–68. doi: 10.1038/mi.2016.39
68. Kurioka A, van Wilgenburg B, Javan RR, Hoyle R, van Tonder AJ, Harrold CL, et al. Diverse streptococcus pneumoniae strains drive a mucosal-associated invariant T-cell response through major histocompatibility complex class i-related molecule-dependent and cytokine-driven pathways. J Infect Dis. (2018) 217:988–99. doi: 10.1093/infdis/jix647
69. Tastan C, Karhan E, Zhou W, Fleming E, Voigt AY, Yao X, et al. Tuning of human MAIT cell activation by commensal bacteria species and MR1-dependent T-cell presentation. Mucosal Immunol. (2018) 11:1591–605. doi: 10.1038/s41385-018-0072-x
70. Jesteadt E, Zhang I, Yu H, Meierovics A, Chua Yankelevich WJ, Cowley S. Interleukin-18 is critical for mucosa-associated invariant T cell gamma interferon responses to francisella species in vitro but not in vivo. Infect Immun. (2018) 86:e00117–18. doi: 10.1128/IAI.00117-18
71. Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. (2019) 366:494–9. doi: 10.1126/science.aaw2719
72. Bhattacharyya A, Hanafi LA, Sheih A, Golob JL, Srinivasan S, Boeckh MJ, et al. Graft-derived reconstitution of mucosal-associated invariant T cells after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. (2018) 24:242–51. doi: 10.1016/j.bbmt.2017.10.003
73. Konuma T, Kohara C, Watanabe E, Takahashi S, Ozawa G, Suzuki K, et al. Reconstitution of circulating mucosal-associated invariant T cells after allogeneic hematopoietic cell transplantation, its association with the riboflavin synthetic pathway of gut microbiota in cord blood transplant recipients. J Immunol. (2020) 204:1462–73. doi: 10.4049/jimmunol.1900681
74. Merlini E, Cerrone M, van Wilgenburg B, Swadling L, Cannizzo ES, d'Arminio Monforte A, et al. Association between impaired Valpha7.2+CD161++CD8+ (MAIT) and Valpha7.2+CD161-CD8+ T-cell populations and gut dysbiosis in chronically HIV- and/or HCV-infected patients. Front Microbiol. (2019) 10:1972. doi: 10.3389/fmicb.2019.01972
75. Bertrand L, Lehuen A. MAIT cells in metabolic diseases. Mol Metab. (2019) 27S:S114–21. doi: 10.1016/j.molmet.2019.06.025
76. Schmaler M, Colone A, Spagnuolo J, Zimmermann M, Lepore M, Kalinichenko A, et al. Modulation of bacterial metabolism by the microenvironment controls MAIT cell stimulation. Mucosal Immunol. (2018) 11:1060–70. doi: 10.1038/s41385-018-0020-9
77. Booth JS, Salerno-Goncalves R, Blanchard TG, Patil SA, Kader HA, Safta AM, et al. Mucosal-associated invariant T cells in the human gastric mucosa and blood, role in helicobacter pylori infection. Front Immunol. (2015) 6:466. doi: 10.3389/fimmu.2015.00466
78. Leung DT, Bhuiyan TR, Nishat NS, Hoq MR, Aktar A, Rahman MA, et al. Circulating mucosal associated invariant T cells are activated in Vibrio cholerae O1 infection and associated with lipopolysaccharide antibody responses. PLoS Negl Trop Dis. (2014) 8:e3076. doi: 10.1371/journal.pntd.0003076
79. Grimaldi D, Le Bourhis L, Sauneuf B, Dechartres A, Rousseau C, Ouaaz F, et al. Specific MAIT cell behaviour among innate-like T lymphocytes in critically ill patients with severe infections. Intensive Care Med. (2014) 40:192–201. doi: 10.1007/s00134-013-3163-x
80. Lu B, Liu M, Wang J, Fan H, Yang D, Zhang L, et al. IL-17 production by tissue-resident MAIT cells is locally induced in children with pneumonia. Mucosal Immunol. (2020) doi: 10.1038/s41385-020-0273-y
81. Reantragoon R, Boonpattanaporn N, Corbett AJ, McCluskey J. Mucosal-associated invariant T cells in clinical diseases. Asian Pac J Allergy Immunol. (2016) 34:3–10.
82. Rouxel O, Lehuen A. Mucosal-associated invariant T cells in autoimmune and immune-mediated diseases. Immunol Cell Biol. (2018) 96:618–29. doi: 10.1111/imcb.12011
83. D'Souza C, Chen Z, Corbett AJ. Revealing the protective and pathogenic potential of MAIT cells. Mol Immunol. (2018) 103:46–54. doi: 10.1016/j.molimm.2018.08.022
84. Provine NM, Klenerman P. MAIT cells in health and disease. Annu Rev Immunol. (2020) 38:203–28. doi: 10.1146/annurev-immunol-080719-015428
85. Lukasik Z, Elewaut D, Venken K. MAIT Cells Come to the Rescue in Cancer Immunotherapy? Cancers. (2020) 12:413. doi: 10.3390/cancers12020413
86. Zumwalde NA, Gumperz JE. Mucosal-associated invariant T cells in tumors of epithelial origin. Adv Exp Med Biol. (2020) 1224:63–77. doi: 10.1007/978-3-030-35723-8_5
87. Wong EB, Ndung'u Kasprowicz VOT. The role of mucosal-associated invariant T cells in infectious diseases. Immunology. (2017) 150:45–54. doi: 10.1111/imm.12673
88. Abos B, Gomez Del Moral M, Gozalbo-Lopez B, Lopez-Relano J, Viana V, et al. Human MR1 expression on the cell surface is acid sensitive, proteasome independent and increases after culturing at 26 degrees C. Biochem Biophys Res Commun. (2011) 411:632–6. doi: 10.1016/j.bbrc.2011.07.007
89. Hashimoto K, Hirai M, Kurosawa Y. A gene outside the human MHC related to classical HLA class I genes. Science. (1995) 269:693–5. doi: 10.1126/science.7624800
90. Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. (2010) 28:1248–50. doi: 10.1038/nbt1210-1248
91. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. (2015) 347:1260419. doi: 10.1126/science.1260419
92. Miley MJ, Truscott SM, Yu YY, Gilfillan S, Fremont DH, Hansen TH, et al. Biochemical features of the MHC-related protein 1 consistent with an immunological function. J Immunol. (2003) 170:6090–8. doi: 10.4049/jimmunol.170.12.6090
93. Wang H, Kjer-Nielsen L, Shi M, D'Souza C, Pediongco TJ, Cao H, et al. IL-23 costimulates antigen-specific MAIT cell activation and enables vaccination against bacterial infection. Sci Immunol. (2019) 4:eaaw0402. doi: 10.1126/sciimmunol.aaw0402
94. McSharry BP, Samer C, McWilliam H, Ashley CL, Yee MB, Steain M, et al. Virus-mediated suppression of the antigen presentation molecule MR1. Cell Rep. (2020) 30:2948–62 e4. doi: 10.1016/j.celrep.2020.02.017
95. Seshadri C, Thuong NT, Mai NT, Bang ND, Chau TT, Lewinsohn DM, et al. A polymorphism in human MR1 is associated with mRNA expression and susceptibility to tuberculosis. Genes Immun. (2017) 18:8–14. doi: 10.1038/gene.2016.41
96. Slichter CK, McDavid A, Miller HW, Finak G, Seymour BJ, McNevin JP, et al. Distinct activation thresholds of human conventional and innate-like memory T cells. JCI Insight. (2016) 1:e86292. doi: 10.1172/jci.insight.86292
97. Jiang J, Chen X, An H, Yang B, Zhang F, Cheng X. Enhanced immune response of MAIT cells in tuberculous pleural effusions depends on cytokine signaling. Sci Rep. (2016) 6:32320. doi: 10.1038/srep32320
98. Zhang M, Ming S, Gong S, Liang S, Luo Y, Liang Z, et al. Activation-induced cell death of mucosal-associated invariant T cells is amplified by OX40 in type 2 diabetic patients. J Immunol. (2019) 203:2614–20. doi: 10.4049/jimmunol.1900367
99. Lamichhane R, Galvin H, Hannaway RF, de la Harpe SM, Munro F, Tyndall JD, et al. Type I interferons are important co-stimulatory signals during T cell receptor mediated human MAIT cell activation. Eur J Immunol. (2020) 50:178–91. doi: 10.1002/eji.201948279
100. Tang XZ, Jo J, Tan AT, Sandalova E, Chia A, Tan KC, et al. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J Immunol. (2013) 190:3142–52. doi: 10.4049/jimmunol.1203218
101. Gracey E, Qaiyum Z, Almaghlouth I, Lawson D, Karki S, Avvaru N, et al. D, IL-7 primes IL-17 in mucosal-associated invariant T (MAIT) cells, which contribute to the Th17-axis in ankylosing spondylitis. Ann Rheum Dis. (2016) 75:2124–32. doi: 10.1136/annrheumdis-2015-208902
102. Willing A, Jager J, Reinhardt S, Kursawe N, Friese MA. Production of IL-17 by MAIT cells is increased in multiple sclerosis and is associated with IL-7 receptor expression. J Immunol. (2018) 200:974–82. doi: 10.4049/jimmunol.1701213
103. Jiang X, Lian M, Li Y, Zhang W, Wang Q, Wei Y, et al. The immunobiology of mucosal-associated invariant T cell (MAIT) function in primary biliary cholangitis, Regulation by cholic acid-induced Interleukin-7. J Autoimmun. (2018) 90:64–75. doi: 10.1016/j.jaut.2018.01.007
104. Wallington JC, Williams AP, Staples KJ. IL-12 and IL-7 synergize to control mucosal-associated invariant T-cell cytotoxic responses to bacterial infection. J Allergy Clin Immunol. (2018) 41:2182–95. doi: 10.1016/j.jaci.2017.08.009
105. Gibbs A, Leeansyah E, Introini A, Paquin-Proulx D, Hasselrot K, Andersson E, et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. (2017) 10:35–45. doi: 10.1038/mi.2016.30
106. Ussher JE, Bilton M, Attwod E, Shadwell J, Richardson R, de Lara C, et al. CD161(++) CD8(+) T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur J Immunol. (2014) 44:195–203. doi: 10.1002/eji.201343509
107. van Wilgenburg B, Scherwitzl I, Hutchinson EC, Leng T, Kurioka A, Kulicke C, et al. MAIT cells are activated during human viral infections. Nat Commun. (2016) 7:11653. doi: 10.1038/ncomms11653
108. Sattler A, Dang-Heine C, Reinke P, Babel N. IL-15 dependent induction of IL-18 secretion as a feedback mechanism controlling human MAIT-cell effector functions. Eur J Immunol. (2015) 45:2286–98. doi: 10.1002/eji.201445313
109. Shaler CR, Choi J, Rudak PT, Memarnejadian A, Szabo PA, Tun-Abraham ME, et al. MAIT cells launch a rapid, robust and distinct hyperinflammatory response to bacterial superantigens and quickly acquire an anergic phenotype that impedes their cognate antimicrobial function, Defining a novel mechanism of superantigen-induced immunopathology and immunosuppression. PLoS Biol. (2017) 15:e2001930. doi: 10.1371/journal.pbio.2001930
110. Emgard J, Bergsten H, McCormick JK, Barrantes I, Skrede S, Sandberg JK, et al. MAIT cells are major contributors to the cytokine response in group A streptococcal toxic shock syndrome. Proc Natl Acad Sci USA. (2019) 116:25923–31. doi: 10.1073/pnas.1910883116
111. Suliman S, Murphy M, Musvosvi M, Gela A, Meermeier EW, Geldenhuys H, et al. MR1-independent activation of human mucosal-associated invariant T cells by mycobacteria. J Immunol. (2019) 203:2917–27. doi: 10.4049/jimmunol.1900674
112. Kefalakes H, Rehermann B. Inflammation drives an altered phenotype of mucosal-associated invariant T cells in chronic hepatitis D virus infection. J Hepatol. (2019) 71:237–9. doi: 10.1016/j.jhep.2019.05.024
113. Kelly J, Minoda Y, Meredith T, Cameron G, Philipp MS, Pellicci DG, et al. Chronically stimulated human MAIT cells are unexpectedly potent IL-13 producers. Immunol Cell Biol. (2019) 97:689–99. doi: 10.1111/imcb.12281
114. D'Souza C, Pediongco T, Wang H, Scheerlinck JY, Kostenko L, Esterbauer R, et al. Mucosal-associated invariant T cells augment immunopathology and gastritis in chronic helicobacter pylori infection. J Immunol. (2018) 200:1901–16. doi: 10.4049/jimmunol.1701512
115. Bolte FJ, O'Keefe AC, Webb LM, Serti E, Rivera E, Liang TJ, et al. Intra-hepatic depletion of mucosal-associated invariant T cells in hepatitis C virus-induced liver inflammation. Gastroenterology. (2017) 153:1392–403 e2. doi: 10.1053/j.gastro.2017.07.043
116. Leeansyah E, Ganesh A, Quigley MF, Sonnerborg A, Andersson J, Hunt PW, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. (2013) 121:1124–35. doi: 10.1182/blood-2012-07-445429
117. Leeansyah E, Svard J, Dias J, Buggert M, Nystrom J, Quigley, et al. Arming of MAIT cell cytolytic antimicrobial activity is induced by IL-7 and defective in HIV-1 Infection. PLoS Pathog. (2015) 11:e1005072. doi: 10.1371/journal.ppat.1005072
118. Melo AM, O'Brien AM, Phelan JJ, Kennedy SA, Wood NAW, Veerapen N, et al. Mucosal-associated invariant T cells display diminished effector capacity in oesophageal adenocarcinoma. Front Immunol. (2019) 10:1580. doi: 10.3389/fimmu.2019.01580
119. Jiang J, Wang X, An H, Yang B, Cao Z, Liu Y, et al. Mucosal-associated invariant T-cell function is modulated by programmed death-1 signaling in patients with active tuberculosis. Am J Respir Crit Care Med. (2014) 190:329–39. doi: 10.1164/rccm.201401-0106OC
120. Yong YK, Saeidi A, Tan HY, Rosmawati M, Enstrom PF, Batran RA, et al. Hyper-expression of PD-1 is associated with the levels of exhausted and dysfunctional phenotypes of circulating CD161(++)TCR iValpha7.2(+) mucosal-associated invariant T cells in chronic hepatitis B virus infection. Front Immunol. (2018) 9:472. doi: 10.3389/fimmu.2018.00472
121. Tang X, Zhang S, Peng Q, Ling L, Shi H, Liu Y, et al. Sustained IFN-I stimulation impairs MAIT cell responses to bacteria by inducing IL-10 during chronic HIV-1 infection. Sci Adv. (2020) 6:eaaz0374. doi: 10.1126/sciadv.aaz0374
122. Schneider M, Hannaway RF, Lamichhane R, de la Harpe SM, Tyndall JDA, Vernall AJ, et al. Neutrophils suppress mucosal-associated invariant T cells in humans. Eur J Immunol. (2020) 50:643–55. doi: 10.1002/eji.201948394
123. Lange J, Anderson RJ, Marshall AJ, Chan STS, Bilbrough TS, Gasser O, et al. The chemical synthesis, stability, and activity of MAIT cell prodrug agonists that access MR1 in recycling endosomes. ACS Chem Biol. (2020) 15:437–45. doi: 10.1021/acschembio.9b00902
124. Souter MN, Loh L, Li S, Meehan B, Gherardin NA, Godfrey DI, et al. Characterization of human mucosal-associated invariant T (MAIT) cells. Curr Prot Immunol. (2019) 127:e90. doi: 10.1002/cpim.90
125. Cossarizza A, Chang HD, Radbruch A, Acs A, Adam D, Adam-Klages S, et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur J Immunol. (2019) 49:1457–973. doi: 10.1002/eji.201970107
126. Sharma PK, Wong EB, Napier RJ, Bishai WR, Ndung'u T, Kasprowicz VO, et al. High expression of CD26 accurately identifies human bacteria-reactive MR1-restricted MAIT cells. Immunology. (2015) 145:443–53. doi: 10.1111/imm.12461
127. Eberhard JM, Hartjen P, Kummer S, Schmidt RE, Bockhorn M, Lehmann C, et al. CD161+ MAIT cells are severely reduced in peripheral blood and lymph nodes of HIV-infected individuals independently of disease progression. PLoS ONE. (2014) 9:e111323. doi: 10.1371/journal.pone.0111323
128. Gherardin NA, Souter MN, Koay HF, Mangas KM, Seemann T, Stinear TP, et al. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol Cell Biol. (2018) 96:507–25. doi: 10.1111/imcb.12021
129. Koppejan H, Jansen D, Hameetman M, Thomas R, Toes REM, van Gaalen FA. Altered composition and phenotype of mucosal-associated invariant T cells in early untreated rheumatoid arthritis. Arthritis Res Ther. (2019) 21:3. doi: 10.1186/s13075-018-1799-1
130. Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SBG, Meehan B, et al. Identification of phenotypically and functionally heterogeneous mouse Mucosal Associated Invariant T cells using MR1 tetramers. J Exp Med. (2015) 20:1095–108. doi: 10.1084/jem.20142110
131. Juno JA, Wragg KM, Amarasena T, Meehan BS, Mak JYW, Liu L, et al. MAIT cells upregulate alpha4beta7 in response to acute simian immunodeficiency virus/simian HIV infection but are resistant to peripheral depletion in pigtail macaques. J Immunol. (2019) 202:2105–20. doi: 10.4049/jimmunol.1801405
132. Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. (1996) 274:94–6. doi: 10.1126/science.274.5284.94
133. Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. (2000) 191:1895–903. doi: 10.1084/jem.191.11.1895
134. Laugel B, Lloyd A, Meermeier EW, Crowther MD, Connor TR, Dolton G, et al. Engineering of isogenic cells deficient for MR1 with a CRISPR/Cas9 lentiviral system, tools to study microbial antigen processing and presentation to human MR1-restricted T cells. J Immunol. (2016) 197:971–82. doi: 10.4049/jimmunol.1501402
135. Ussher JE, van Wilgenburg B, Hannaway RF, Ruustal K, Phalora P, Kurioka A, et al. TLR signaling in human antigen-presenting cells regulates MR1-dependent activation of MAIT cells. Eur J Immunol. (2016) 46:1600–14. doi: 10.1002/eji.201545969
136. Kurioka A, Jahun AS, Hannaway RF, Walker LJ, Fergusson JR, Sverremark-Ekstrom E, et al. Shared and distinct phenotypes and functions of human CD161++ Valpha7.2+ T cell subsets. Front Immunol. (2017) 8:1031. doi: 10.3389/fimmu.2017.01031
137. Salio M, Gasser O, Gonzalez-Lopez C, Martens A, Veerapen N, Gileadi U, et al. Activation of human mucosal-associated invariant T cells induces CD40L-dependent maturation of monocyte-derived and primary dendritic cells. J Immunol. (2017) 199:2631–8. doi: 10.4049/jimmunol.1700615
138. Rha MS, Han JW, Kim JH, Koh JY, Park HJ, Kim SI, et al. Human liver CD8+ MAIT cells exert TCR/MR1-independent innate-like cytotoxicity in response to IL-15. J Hepatol. (2020) doi: 10.1016/j.jhep.2020.03.033. [Epub ahead of print].
139. Berkson JD, Slichter CK, DeBerg HA, Delaney MA, Woodward-Davis AS, Maurice NJ, et al. Inflammatory cytokines induce sustained CTLA-4 cell surface expression on human MAIT cells. Immunohorizons. (2020) 4:14–22. doi: 10.4049/immunohorizons.1900061
140. Chua WJ, Kim S, Myers N, Huang S, Yu L, Fremont DH, et al. Endogenous MHC-related protein 1 is transiently expressed on the plasma membrane in a conformation that activates mucosal-associated invariant T cells. J Immunol. (2011) 186:4744–50. doi: 10.4049/jimmunol.1003254
141. Hagel JP, Garner LC, Bilton M, Mehta H, Leng T, Hackstein CP, et al. Human MAIT cell activation in vitro. Methods Mol Biol. (2020) 2098:97–124. doi: 10.1007/978-1-0716-0207-2_7
142. Li K, Vorkas CK, Chaudhry A, Bell DL, Willis RA, Rudensky A, et al. Synthesis, stabilization, and characterization of the MR1 ligand precursor 5-amino-6-D-ribitylaminouracil (5-A-RU). PLoS ONE. (2018) 13:e0191837. doi: 10.1371/journal.pone.0191837
143. Kawachi I, Maldonado J, Strader C, Gilfillan S. MR1-restricted V alpha 19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J Immunol. (2006) 176:1618–27. doi: 10.4049/jimmunol.176.3.1618
144. Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. (2009) 7:e54. doi: 10.1371/journal.pbio.1000054
145. Cui Y, Franciszkiewicz K, Mburu YK, Mondot S, Le Bourhis L, Premel V, et al. Mucosal-associated invariant T cell-rich congenic mouse strain allows functional evaluation. J Clin Invest. (2015) 125:4171–85. doi: 10.1172/JCI82424
146. Xie J, Pan Y, Tao H, Wang P, Chen Y, Gao J, et al. Deficiency of mucosal-associated invariant T cells in TCRJalpha18 germline knockout mice. Immunohorizons. (2019) 3:203–7. doi: 10.4049/immunohorizons.1900035
147. Greene JM, Dash P, Roy S, McMurtrey C, Awad W, Reed JS, et al. MR1-restricted mucosal-associated invariant T (MAIT) cells respond to mycobacterial vaccination and infection in nonhuman primates. Mucosal Immunol. (2017) 10:802–813. doi: 10.1038/mi.2016.91
148. Chen Z, Wang H, D'Souza C, Koay HF, Meehan B, Zhao Z, et al. Characterization and Purification of Mouse Mucosal-Associated Invariant T (MAIT) Cells. Curr Protoc Immunol. (2019) 127:e89. doi: 10.1002/cpim.89
149. Murayama G, Chiba A, Suzuki H, Nomura A, Mizuno T, Kuga T, et al. A critical role for mucosal-associated invariant T cells as regulators and therapeutic targets in systemic lupus erythematosus. Front Immunol. (2019) 10:2681. doi: 10.3389/fimmu.2019.02681
150. Chiba A, Murayama G, Miyake S. Mucosal-associated invariant T cells in autoimmune diseases. Front Immunol. (2018) 9:1333. doi: 10.3389/fimmu.2018.01333
151. Varelias A, Bunting MD, Ormerod KL, Koyama M, Olver SD, Straube J, et al. Recipient mucosal-associated invariant T cells control GVHD within the colon. J Clin Invest. (2018) 128:1919–36. doi: 10.1172/JCI91646
152. Yan J, Allen S, McDonald E, Das I, Mak JYW, Liu L, et al. MAIT cells promote tumor initiation, growth, and metastases via tumor MR1. Cancer Discov. (2020) 10:124–41. doi: 10.1158/2159-8290.CD-19-0569
153. Legoux F, Gilet J, Procopio E, Echasserieau K, Bernardeau K, Lantz O. Molecular mechanisms of lineage decisions in metabolite-specific T cells. Nat Immunol. (2019) 20:1244–55. doi: 10.1038/s41590-019-0465-3
Keywords: mucosal-associated invariant T cell, MR1, antigen, ligand, MR1T, MAIT
Citation: Corbett AJ, Awad W, Wang H and Chen Z (2020) Antigen Recognition by MR1-Reactive T Cells; MAIT Cells, Metabolites, and Remaining Mysteries. Front. Immunol. 11:1961. doi: 10.3389/fimmu.2020.01961
Received: 29 May 2020; Accepted: 21 July 2020;
Published: 27 August 2020.
Edited by:
Emmanuel Treiner, INSERM U1043 Centre de Physiopathologie de Toulouse Purpan, FranceReviewed by:
Laurent Gapin, University of Colorado Denver, United StatesMegan K. L. MacLeod, University of Glasgow, United Kingdom
Francois Legoux, Institut Curie, France
Copyright © 2020 Corbett, Awad, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandra J. Corbett, corbetta@unimelb.edu.au
 Alexandra J. Corbett
Alexandra J. Corbett Wael Awad
Wael Awad Huimeng Wang
Huimeng Wang Zhenjun Chen
Zhenjun Chen