- 1School of Pharmacy, Hengyang Medical College, University of South China, Hengyang, China
- 2Hunan Provincial Key Laboratory of Tumor Microenvironment Responsive Drug Research, University of South China, Hengyang, China
At present, the incidence of cancer is becoming more and more common, but its treatment has always been a problem. Although a small number of cancers can be treated, the recurrence rates are generally high and cannot be completely cured. At present, conventional cancer therapies mainly include chemotherapy and radiotherapy, which are the first-line therapies for most cancer patients, but there are palliatives. Approaches to cancer treatment are not as fast as cancer development. The current cancer treatments have not been effective in stopping the development of cancer, and cancer treatment needs to be imported into new strategies. Non-coding RNAs (ncRNAs) is a hot research topic at present. NcRNAs, which include microRNAs (miRNAs), circular RNAs (circRNAs), and long non-coding RNAs (lncRNAs), participate in all aspects of cancer biology. They are involved in the progression of tumors into a new form, including B-cell lymphoma, glioma, or the parenchymal tumors such as gastric cancer and colon cancer, among others. NcRNAs target various immune checkpoints to affect tumor proliferation, differentiation, and development. This might represent a new strategy for cancer treatment.
Introduction
The current treatments for tumors are not suitable for the increasingly evolving forms of cancer. Although new therapies that target proteins such as antibodies against programmed cell death 1 (PD-1), programmed death-ligand 1 (PD-L1), and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) have ushered in a new era in cancer pharmacotherapy and drug development, some oncogene-encoded protein targets are intractable or insufficient for achieving remission. In addition, cancer cells can develop drug resistance (1). As a result, successful cancer treatment necessitates several types of targets implicated in carcinogenic pathways.
Non-coding RNAs (ncRNAs) are transcripts that do not encode proteins, except the annotated messenger RNAs (mRNAs) (2). NcRNAs play a role in various aspects of cancer biology, including cell death, proliferation, and treatment resistance (3). There are three different types of ncRNAs, namely, microRNAs (miRNAs), circular RNAs (circRNAs), and long non-coding RNAs (lncRNAs). MiRNAs are short non-coding regulatory RNAs that regulate protein expression by binding to the 3′-untranslated region (3′-UTR) of mRNA and limiting mRNA translation while also altering transcription (4). Some concrete miRNA systems are eminently related to the initiation and progression of cancer, as well as drug resistance (1, 5). CircRNAs are covalently closed-loop ncRNAs without the 5′ and 3′ ends, and they are created by the back-splicing of pre-mRNAs (6). Additionally, they measure a category of ncRNAs or protein-coding RNAs derived from one coding DNA or multiple exons and are located within the living substance (7, 8). CircRNAs act as miRNA sponges to modulate the organic phenomenon and have an effect on the biological processes of cancers (9). Previous studies showed that circRNAs can take part in many physiological and pathological courses, including tumorigenesis and the progression of cancer (10–12). LncRNAs are RNAs with more than 200 nucleic acids and have the specialty of evolution and expression of tissue. They are abnormal in a lot of carcinomas and participate in post-transcriptional shearing, editing, and in the transportation, translation, and degradation of post-transcription (13, 14).
Immune checkpoints can measure the level of immune activation. They are a class of molecules that are expressed on immune cells. The growth and the progression of cancer are related to immune suppression (15). Over the past decade, the inhibition of immune checkpoint inhibitors has increasingly propelled the event of cancer medical specialty (16–18).
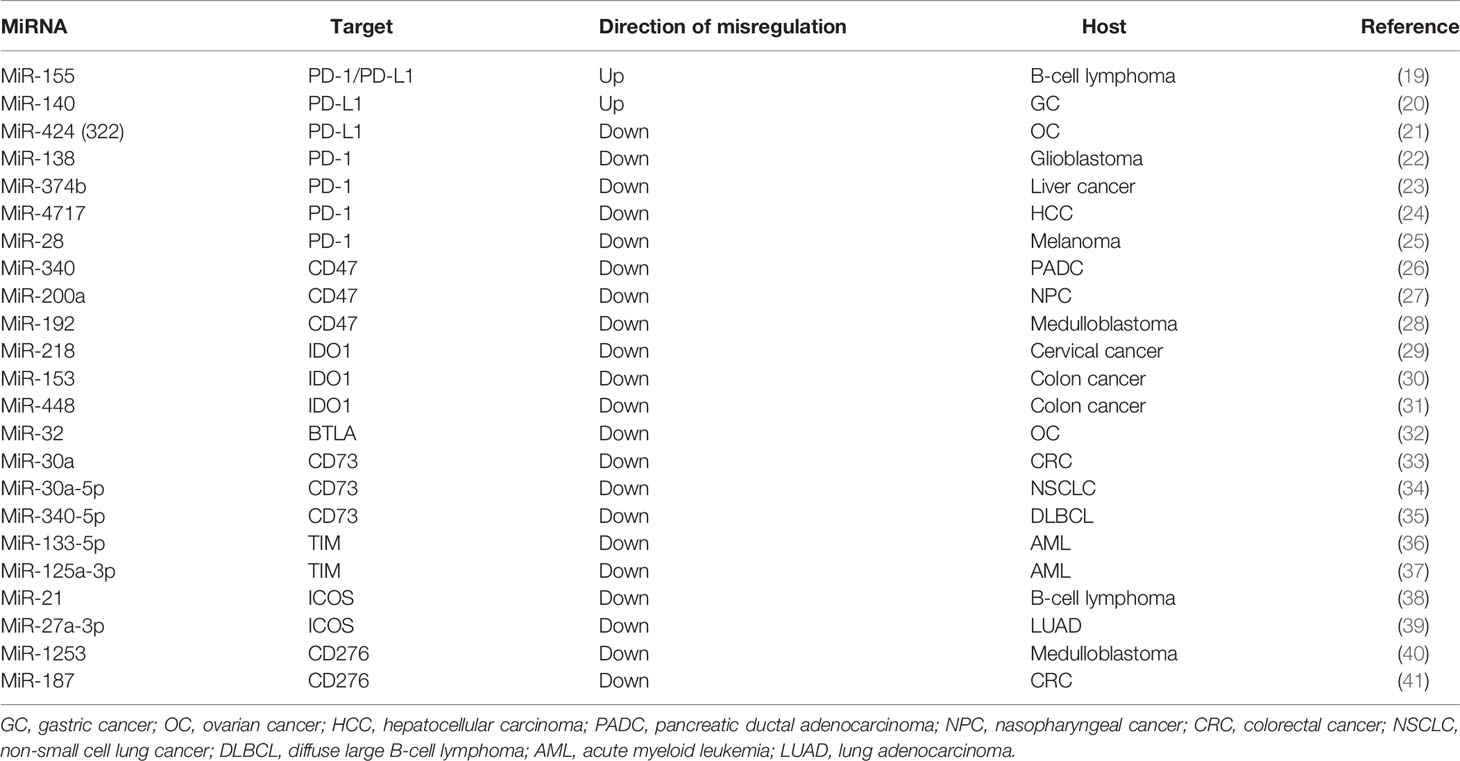
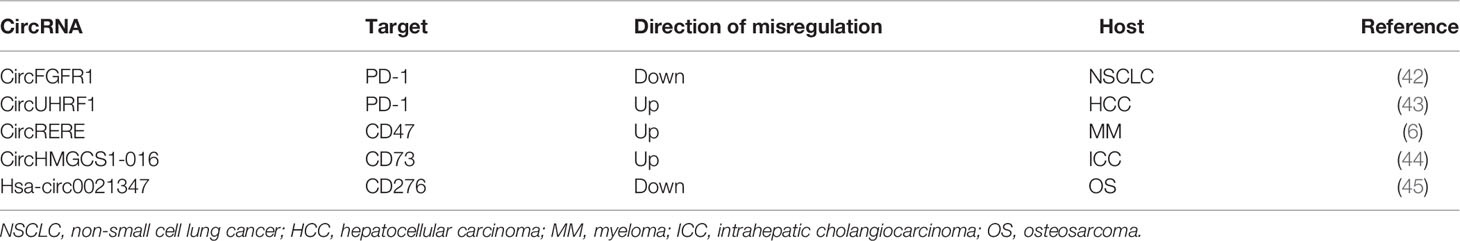
There are reports showing that blocking the expression of immune checkpoints can have an impact on the development of cancer (15), but the current research works have not been clear on its specific mechanism. This paper mainly expounds on the role of ncRNAs in immune checkpoints the specific mechanisms of microRNAs target immune checkpoints in cancer can be showed in Table 1, the mechanisms of circular RNAs target it in cancer can be showed in Table 2, and that of Long non-coding RNAs target in cancer can be showed in Table 3, thus making explicit the incidence and development of human cancers in order to provide new perspectives for their treatment. The specific mechanisms of the three types of ncRNAs in different immune checkpoints are shown in Figure 1.
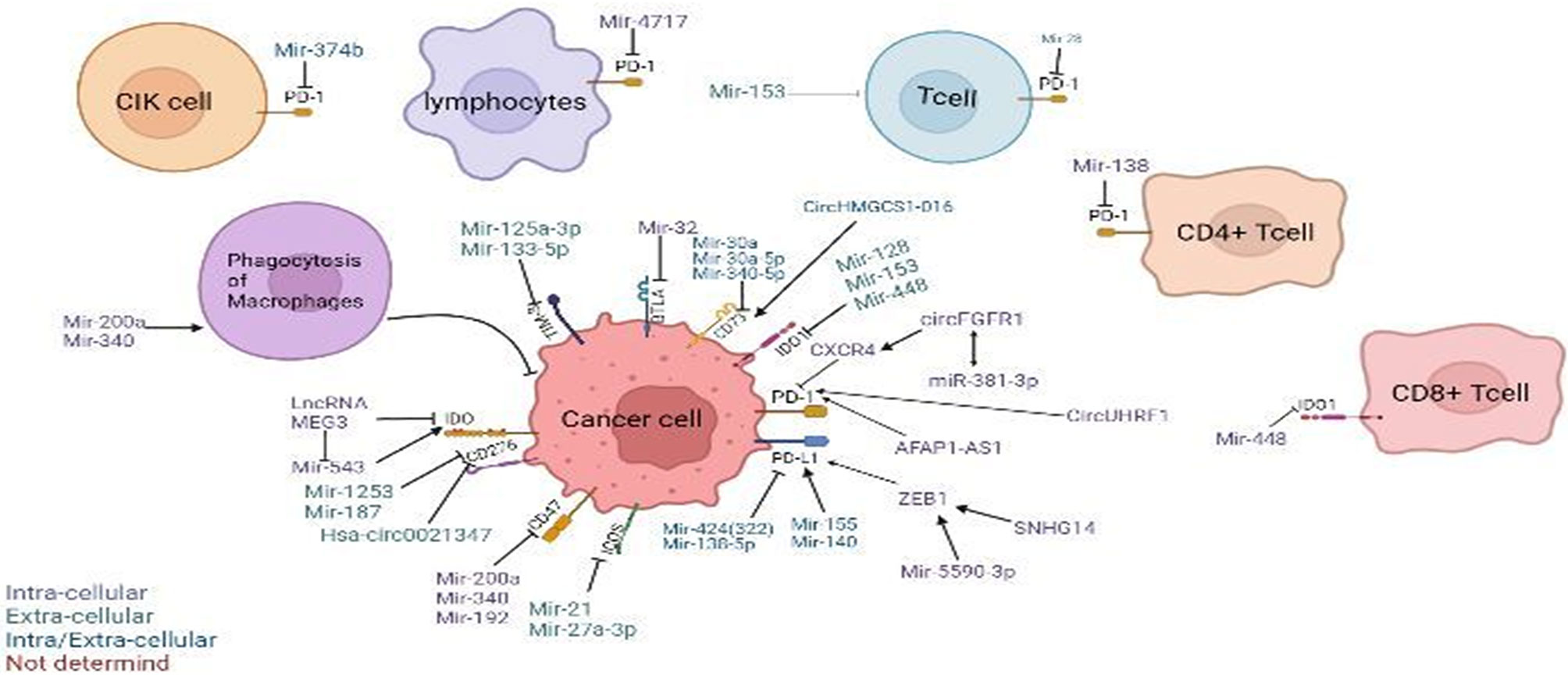
Figure 1 Regulation of non-coding RNAs (ncRNAs) in the tumor microenvironment. Modulatory effects of ncRNAs on cytokine-induced killer (CIK) cells and phagocytosis of macrophages. ncRNAs regulate different immune checkpoints, including PD-1, PD-L1, CD47, CD73, IDO, IDO1, TIM-3, BTLA, ICOS, and B7-H3(CD276), in cancer cells, T cells, T lymphocytes, CD4+ T cells, and CD8+ T cells. Arrow denotes stimulatory regulation, while T denotes inhibitory regulation. The colored legend on the bottom left panel represents the sites of action of the depicted immunomodulatory ncRNAs.
The Impact of NcRNAs Targeting PD-1/PD-L1 on Cancer
PD-1 and PD-L1 intercede the immunological disorder effects of tumors through encouraging T-cell apoptosis and the induction of regulatory T cells (Tregs) (49). The anticancer effects of antibody treatments targeting PD-L1 and PD-1 have been demonstrated, which are similar to those of CTLA-4 by reversing the immunosuppressive effects of PD-1 (50).
MiRNAs Target PD-1/PD-L1
MiR-155 is closely related to the progression of B-cell lymphoma. In the tumor microenvironment, it enhances the interaction between B-lymphoma cells and CD8+ T cells, which may be directly targeted by the inhibition of PD-1/PD-L1 (19). In the spleen and bone marrow, miR-155 transgenic mice exhibited pre-B-cell proliferation and also showed malignant B-cell transformation (51). In B-lymphoma cells, miR-155 increased the expression of PD-L1 through directly combining with the 3′-UTR, aggravating CD8+ T-cell necrobiosis, and preserving neoplasm immunity in a particularly PD-1/PD-L1-dependent manner (19). The AKT and ERK pathway is the key regulator of PD-1-mediated CD8+ T-cell function (52). In DB cells, ectopic expression of miR-155 caused an increased sensitivity to anti-PD-L1 and anti-PD-1 antibodies (19). In nude mice, the downregulation of miR-155 promoted the apoptosis of B-lymphoma cells and delayed xenograft tumor formation (53). In summary, miR-155 may be a potential target for anti-PD-L1 protein treatment of malignant B-cell neoplastic diseases.
MiR-140 played a suppressive role in the growth of gastric cancer in vitro and in vivo (54, 55). It also strengthened antitumor immunity in gastric cancer (20). PD-L1 was directly targeted by miR-140 in gastric cancer cells (20). Previous studies showed that cell-intrinsic PD-L1/PD-1 signaling promoted tumor development by activating downstream mammalian target of rapamycin (mTOR) signaling (56). Previous research also showed that miR-140 played a suppressive role in mTOR signaling in gastric cancer (57). Moreover, miR-140 can reduce the expression of PD-L1 (20). Signaling inhibition of PD-L1/PD-1 was able to stop immune suppression and promote antitumor response (58). By targeting PD-L1 in vitro and in vivo, the upregulation of miR-140 can inhibit gastric cancer growth; it is also related to an enhanced antitumor immunity (20). Consequently, miR-140 may represent a new therapeutic target for the treatment of gastric cancer through targeting PD-L1.
MiR-424(322) was conversely correlated with the expressions of PD-L1, PD-1, CD80, and CTLA-4 in a clinical gene expression array dataset (21). Through direct binding to the 3′-UTR of these genes, the overexpression of miR-424(322) inhibited the expressions of PD-L1 and CD80 (21). It has been proven that the PD-L1/PD-1 immune checkpoint pathway can inhibit the T-cell antitumor immune response (59), and PD-L1 preferentially modulates the secretion of regulator cytokines in T cells (60). Restoration of the expression of miR-424(322) in female internal reproductive organ cancer chemoresistant cells caused a decrease in the expression of PD-L1 (21). Blockade of PD-L1 restored immune suppression and caused the expansion of tumor-infiltrating lymphocytes (TILs) (61). Restoration of the expression of miR-424(322) increased the sensitivity of cancer cells to drug treatment and activated T cells through blocking PD-L1 in in vivo and in vitro models (21). All the above indicate that miR-424(322) can target PD-1 and be involved in the treatment of ovarian cancer.
With PD-1 and CTLA-4, various miRNA-targeting algorithms have indicated the possible interactions of miR-138 (22). There is an explicit variety of miRNAs within the brain, and miR-138 is said to regulate nerve fiber spine formation in rat hippocampal neurons (62). The accumulation of relevant information cumulatively revealed that, in vivo, miR-138 preferentially modulates the immune system, especially interacting with PD-1 and CTLA-4 to inhibit tumor-infiltrating Tregs and later alleviating the damage by immunological disorder cells in the tumor microenvironment (22). In glioma, miR-124 enhanced the T-cell-mediated immune clearance by inhibiting STAT3 signaling (63). Therefore, the hypothesis that miR-138 could regulate Tregs has been proven and that the administration of miR-138 in vivo could exert potent antitumor immune effects through interacting with CTLA-4 and PD-1 (22). This could be a promising idea for glioma treatment.
The transforming gene PD-1 is related to the suppression of proliferation and protein production of T cells in the progression of liver disease (23). The downregulation of PD-1 was revealed to be able to reverse immune dysfunction (64–66). In T cells, PD-1 acts as a regulator inhibiting the proliferation and cytokine production (67). Relevant incontestable knowledge revealed that metallic element one could be a real target of miR-374b (23). In B cells, miR-374b is associated with the acceleration of cell proliferation and, therefore, the production of aberrant glycosylated antibody (immunoglobulin, Ig) A1 (68). One study significantly showed that miR-374b mimics played a suppressive role in the mRNA and macromolecule expression of PD-1 in cytokine-induced killer (CIK) cells and that it also has influence on the tumor-targeting capability of CIK cells (23). The interaction between PD-1 and miR-374b may offer some new ideas on the design of new therapeutic modalities for liver cancer treatment.
PD-1 plays a prominent role in the regulation of T-lymphocyte function and could be a main issue of cell death sensitivity (69, 70). More and more research studies have demonstrated that the PD-1 pathway and its ligands to be basically concerned in virus infection, as well as HBV infection (24). PD-L1 is induced by viral infection, interferon (IFN)-α, and IFN-γ of hepatocytes and mediates T-cell apoptosis (71). The SNP rs10204525 has been connected with chronic HBV infection (25, 72, 73), and the PD-1 gene is located in the 3′-UTR (24). Additionally, miR-4717 allele-specifically regulates the expression of PD-1 through a totally different interaction with its polymorphic target within its 3′-UTR. This also means that miR-4717 plays a role in the susceptibility to chronic HBV infection (24). The relationship between PD-1 and miR-4717 may provide new ideas for HBV treatment.
Experimental data from melanoma-bearing mice suggested that miR-28 has the potential to regulate the PD-1, TIM3, and BTLA gene in T cells (74). In the cancer environment, high levels of overexpressed inhibitory receptors are among the main features of T-cell exhaustion (75). Dual-luciferase assay of the PD-1 3′-UTR showed that miR-28 directly silenced PD-1 via binding to its 3′-UTR (74). Moreover, PD-1 has been recognized because of the primary biomarker for T cells that are used up (76). It can be inferred that miR-28 depletes T cells by silencing PD-1 and that it has an antitumor effect.
CircRNAs Target PD-1
Fibroblast protein receptor 1 (FGFR1) is a type of oncogene that may promote the progression and tumorigenesis of non-small cell lung cancer (NSCLC) cells using totally different strategies (77). CircFGFR1 can also promote resistance of NSCLC cells to anti-PD-1 agents. In xenograft mice, overexpression of circFGFR1 could significantly reduce PD-1 in Lewis lung carcinoma (LLC) cells. The tumor growth in LLC-circFGFR1 cell recipient xenograft mice showed obvious phenotypic resistance to anti-PD-1 therapy compared with that of the mock cell group, and xenograft mice had a shorter survival time (42). Previous research studies showed that forced circFGFR3 expression competitively binds to miR-22-3p to promote NSCLC cell invasion and proliferation and facilitate galectin-1 activity (78). CXCR4 could be a possible intermediator that guides cytotoxic T-lymphocyte exclusion and participates in the resistance to anti-PD-1 cancer therapeutic methods (79, 80). The expression of circFGFR1/CXCR4 and the frequency of CD8+ T cells showed a negative correlation in NSCLC tissues (42). Furthermore, NSCLC cells are sensitive to anti-PD-1 immunology therapy after knockout of CXCR4 (42). Therefore, inhibiting the circFGFR1/CXCR4 pathways in NSCLC cells may provide a novel opportunity to inhibit resistance to anti-PD-1 immunotherapy in the treatment of NCSLC.
Researchers have found that forced UHRF1 expression can promote tumorigenesis and the progression of hepatocellular carcinoma (HCC) (81). It was indicated that circRNA from the expression of UHRF1 can be upregulated and could possibly promote the progression of HCC (43). In HCC patients, overexpression of PD-1 in natural killer (NK) cells can promote the functional dysregulation of activated NK cells (82, 83). Exosomal circUHRF1 secreted by HCC cells induced NK-cell exhaustion and mediated the resistance to anti-PD-1 therapy (43). In another study, it was shown that, compared to HCC patients susceptible to anti-PD-1 therapy, the number of NKG2D-positive cells in HCC patients resistant to anti-PD-1 therapy was significantly reduced. In addition, the results of a xenograft model by subcutaneous implantation showed that circUHRF1 knockdown cells led to sensitivity to anti-PD-1 treatment and to an increase in the overall survival rate (43). All these show that forced circUHRF1 expression might prevent the response of HCC to anti-PD-1 therapy.
LncRNAs Target PD-1/PD-L1
By utilizing the Gene Expression Omnibus (GEO) dataset, a previous study found that the key molecular marker of growth immune evasion PD-1 was completely related to the expression of AFAP1-AS1 in nasopharyngeal cancer (NPC) and that they are co-expressed in infiltrating lymphocytes in NPC tissues (47). In recent studies, PD-L1 and PD-1 have shown high expression levels in NPC (84–86). In order to regulate the expression of PD-1, AFAP1-AS1 functions as a competing endogenous RNA (ceRNA) (87–89). The expression level of the lncRNA AFAP1-AS1 was high in NPC, and it also promoted the violation and metastasis of cancer (47). All the above show that AFAP1-AS1 and PD-1 are great therapeutic targets for suppressing the metastasis of tumor and activating antitumor immunity.
Zinc finger E-box-binding homeobox gene 1 (ZEB1) is a transcriptional factor (TF) demonstrated to be associated with gene control in numerous varieties of cancer cells regarding invasion, migration, epithelial–mesenchymal translation (EMT), and proliferation (90–92). Small nucleolar RNA host gene 14 (SNHG14) elicits oncogenic functions through modulating proliferation, migration, and invasion to confer chemoresistance in different malignancies (48). Previous research works showed that ZEB1 can improve immune evasion by upregulating the expression of PD-L1 in cancer cells (93, 94). The ribonucleic acid expression of ZEB1, and therefore the macromolecule expressions of ZEB1 and PD-L1, in diffuse large B-cell lymphoma (DLBCL) cells was reduced under SNHG14 silencing (48). Through PD-1/PD-L1, SNHG14/ZEB1 evoked the interaction of DLBCL cells with CD8+ T cells and triggered apoptosis (48). This means that the interaction of SNHG14/ZEB1 and PD-1/PD-L1 might be a promising target in DLBCL treatment.
The Impact of NcRNAs Targeting CD47 on Cancer
CD47 is a transmembrane supermolecule that functions as a ligand for signal regulatory protein alpha (SIRPα) and is additionally widely expressed on all traditional cells and overexpressed on growth cells (95).
MiRNAs Target CD47
MiR-340 has been demonstrated to regulate the expressions of genes related to tumor progression and is involved in tumor suppression (96, 97). A study showed that, at both the mRNA and protein levels in Panc02 cells, the overexpression of miR-340 by mimic transfection significantly decreased CD47, especially in pancreatic ductal carcinoma (PDAC) (26). CD47 is a direct target of miR-340 and is downregulated by miR-340 through a binding site in its 3′-UTR (26). The functions of anti-CD47 include the inhibition and causation of efficient “self” signal uptake of cancer cells (98). Phagocytosis by macrophages was greatly enhanced in miR-340-overexpressed tumor-bearing mice, and the overexpression of miR-340 further increased with CD47 blockade (26). Therefore, miR-340 targeting CD47 may be a good option for PDAC treatment.
MiR-200a is one of the important factors in the miR-200 unit, which has five other members: miR-200a/b/c, miR-141, and miR-429 (99–101). MiR-200a is downregulated while CD47 is upregulated in NPC. The expression of miR-200a was negatively correlated with the levels of CD47 using Spearman’s correlation analysis (27). MiR-200a inhibited the expression of CD47, similar to small interfering RNA (siRNA) CD47, and restrained the macromolecular translation and reduced the expression of CD47 (27). CD47–SIRPα blockade can enhance the phagocytic activity of phagocytes to eliminate tumor cells (102). Long-standing research has provided proof that, with the activation of CD47 signaling, cancer cells will impair the immune system and evade the antitumor function of macrophages (103, 104). Studies also showed that miR-200a suppressed the proliferation, colony formation, migration, and invasion of NPC cells by downregulating CD47 (27). All these can provide potential novel therapy for NPC with the use of miR-200a/CD47.
MiRs that play suppressive roles in tumors are downregulated in cancer, contrary to the oncogenic ones (105). Compared to the normal cerebellum, the expression level of miR-192 in medulloblastoma cells is lower, and studies indicated that miR-192 suppresses the expressions of ITGAV, ITGB1, ITGB3, and CD47 in medulloblastoma (28). Previous studies showed that miR-192 is a metastasis suppressor gene (106, 107). ITGAVB3 has shown positive correlation with the adhesion ability of cells, which means that it has increased metastatic potential (108, 109). In addition, research works showed that the suppression of miR-192-mediated CD47 restrained leptomeningeal dissemination of medulloblastoma with molecular crosstalk with ITGAVB3 (28). All of these provide new perspectives in medulloblastoma treatment.
CircRNAs Target CD47
CircRNA arginine–glutamic acid dipeptide repeat (circRERE) is a natural sponge of miR-152-3p, and the miR-152-3p inhibitor attenuated the suppressive effect of circRERE on bortezomib (BTZ) resistance (6). CD47 is associated with the degree of immune deactivation, and the therapeutic strategy concerning anti-CD47 has been employed to kill myeloma (MM) cells (110, 111). In accordance with its sponge effects on miR-152-3p, circRERE modulates the resistance of BTZ in MM cells through suppressing miR-152-3p (6). Overexpression of miR-152-3p resulted in the repression of the IC50 of BTZ and its cell proliferative ability, but accelerated cell apoptosis, which was then abolished by promotion of the expression of CD47. Protein detection by Western blot showed that, in 8226R5 and MM1, the upregulation of CD47 counteracted the miR-152-3p mimic-induced pro-apoptotic and anti-proliferative effects (6). BTZ is a first-line chemotherapeutic agent for MM, but treatment frequently fails due to the resistance to BTZ (112). Moreover, CD47 is a downstream target of miR-152-3p, and miR-152-2p inhibits BTZ resistance by reducing the levels of CD47 in MM cells (6). Downregulation of circRERE may be a new therapeutic target by inducing BTZ resistance in MM treatment.
The Impact of NcRNAs Targeting IDO1 on Cancer
Indoleamine 2,3-dioxygenase 1 (IDO1), IDO2, and tryptophan 2,3-dioxygenase (TDO) are the initial and rate-limiting enzymes of the tryptophan metabolism pathway (113, 114). These enzymes convert essential amino acids into kynurenine and 3-hydroxyanthranilic acid. Essential amino acids are vital aminoalkanoic acids for the survival, proliferation, and activation of nerve fiber cells and lymphatic cells (30).
MiRNAs Target IDO1
A study has shown miR-218 directly targeting the 3′-UTR of IDO1, and in cervical cancer tissues and cells, miR-218 was downregulated and negatively related to IDO1 (29). T cells in the G1 phase have high sensitivity to tryptophan deficiency, and IDO1 can lead to a significant lack of tryptophan. IDO overexpression may suppress the proliferation of T cells (115, 116). The overexpression of miR-218 considerably suppressed the expression of IDO1, inhibited cell viability, and promoted the caspase-mediated cell death of cervical cancer cells through the transfection of miR-218 mimics into HeLa cervical cancer cells (29). All the above can provide novel measures for the treatment of cervical cancer and some other tumors.
Cancer cells express higher levels of immunosuppressive molecules after interferon gamma (IFN-γ) induction and then escape immune elimination (117). It is believed that the upregulation of IDO1 by IFN-γ in neoplasm cells can cause immunological disorder in the tumor microenvironment, thereby restricting cytotoxic T cells (118). MiR-153 plays a suppressive role in tumors (119), which can directly target and downregulate the expression of IDO1 in colon cancer cells (30). It also limits the upregulation of IFN-γ on IDO1 and displays an antitumor effect.
IFN-γ strongly induced IDO1 in tumor cells (117). MiR-448 targeted IDO1, as shown by the results of dual-luciferase reporter assay and WB assay (31). MiR-448 inhibited the protein expression of IDO1 in human colon cancer (31), while CD8+ T cells greatly induced IDO1 (117). In addition, CD8+ T cells circulate in the blood and play a cytotoxic role (120). Previous studies showed that miR-448 blocked the expression of IDO1 to suppress the apoptosis of CD8+ T cells (31). This might provide insights for the treatment of colon cancer.
LncRNAs Target IDO
LncRNAs are molecular markers of tumor diagnosis and prognosis (46, 121). A study showed that the lncRNA MEG3 had a low expression and functioned as a tumor suppresser in NSCLC (32). In NSCLC cells overexpressing MEG3, the levels of IDO1, IDO2, TDO, and the autophagy-related Beclin-1 and LC3-II proteins were apparently reduced, whereas that of the autophagy-related LC3-I supermolecule was obviously enhanced (122). MEG3 regulates the miR-543/IDO signaling pathway to influence the autophagy of lung carcinoma cells (32). Regulation of the miR-543/IDO signaling pathway by MEG3 can be a treatment option for NSCLC.
The impact of MiRNA Targeting BTLA on Cancer
The expression of MiR-32 is markedly lower in ovarian cancer tissues than that in adjacent normal tissues; miR-32 also directly targets the 3′-UTR of BTLA (123). BTLA is a new member of the CD28 family (34). Previous research showed that miR-32 can suppress the malignant behavior of HeLa cells (124). Moreover, the high expression level of BTLA could significantly play an anticancer role in miR-32, while the overexpression of miR-32 could greatly inhibit the mRNA and protein expressions of BTLA (123). A negative feedback loop was formed between miR-32 and BTLA, regulating the malignant behavior of ovarian cancer cells (123). The low expression levels of MiR-32 and its target on BTLA in ovarian cancer cells can help inspire treatment options for ovarian cancer.
The Impact of NcRNAs Targeting CD73 on Cancer
Ecto5′-nucleotidase (CD73), a component of the purinergic signaling pathway, is a 70-kDa glycosylphosphatidylinositol-anchored cell surface supermolecule encoded by the NT5E cistron that is crucial in adenosinergic signaling (125).
MiRNAs Target CD73
It was shown that miRNAs are closely related to the pathogenesis of cancers, such as cancer proliferation (33). CD73 can play an important part in cancer development (126). CD73 is a direct target gene of miR-30a in colorectal cancer (CRC) cells, while miR-30a showed a negative effect in the regulation of the expression levels of CD73 mRNA and protein (127). The effect of miR-30a overexpression can be reversed by the reexpression of CD73, and miR-30a inhibits the cell proliferation and tumor growth of body part cancer through targeting CD73 (127). The interaction between CD73 and miR-30a may be a promising approach for CRC treatment.
In NSCLC tissues, the expression of miR-30a-5p is considerably downregulated (128). Related studies showed that loss of CD73 significantly suppressed the flexibility of NSCLC cells in migrating, and CD73 repressed cell proliferation in NSCLC cells by moving speeding up cell cycle (125). CD73 has an important effect on tumor cells through the EGFR signaling pathway (35, 129). In NSCLC cells, miR-30a-5p can inhibit the expression of CD73 by directly binding to its 3′-UTR, and the ectopic expression of miR-30a-5p was significantly reduced while the expression of CD73 was increased (125). MiR-30a-5p targets the CD73 3′-UTR to inhibit its expression, which may be a good approach for NSCLC treatment.
Researchers have discovered that KMT5A, a downstream target of miR-340-5p, regulated CD8+ T cells and facilitated the immunosuppressive ability of DLBCL cells (130). The functional avidity of CD8+ T cells is associated with superior control of tumor growth (131). CD73-expressing tumor cells showed negative effects on the regulation of the antitumor T-cell response, which could also increase T-cell apoptosis (132). In LY-1 cells, silencing KMT5A enhanced the ubiquitination of CD73, which was reduced by COP1 knockdown, downregulating CD73 (130). The miR-340-5p/KMT5A axis plays an antitumor role on DLBCL cells independent of immune regulation (130). This provides a new perspective for the treatment of DLBCL.
CircRNAs Target CD73
CircRNAs often act as miRNA sponges (133). CD73 has been demonstrated to promote tumor progression by suppressing the antitumor immune response, which is a new immune checkpoint related to adenosine metabolism (44). GAL-8, a member of the glycan-binding protein unit, was proposed to be able to induce the apoptosis of activated T cells and is involved with Th17 cells. It also promotes the differentiation of immunosuppressive Tregs, therefore showing immunosuppressive effects (134). CD73 and GAL-8 are targets of circHMGCS1-016 and miR-1236-3p in intrahepatic cholangiocarcinoma (ICC). A study showed that the proteins of CD73 and GAL-8 were highly expressed in ICC tissues and that they showed a negative correlation with CD8+ T cells (135). Furthermore, circHMGCS1-016 promoted ICC progression through miR-1236-3p/CD73 and GAL-8, acting as a sponge for miR-1236-3p (135). The regulation of CD73 and GAL-8 expressions by circHMGCS1-016 may be a novel therapy for ICC.
The Impact of MicroRNA Targeting TIM-3 on Cancer
Human TIM-3 (T-cell immunoglobulin mucin receptor 3) is a specific supermolecule along with 301 amino acids and is encoded by HAVCR2 (136).
Leukemic stem cells (LSCs) are abnormally proliferated in acute myeloid leukemia (AML), which is a complicated blood malignancy (137, 138). Both LSCs and biological process stem cells (hematopoietic stem cells, HSCs) are found in the bone marrow of AML patients. Among them, a characteristic of TIM-3 is its particular expression on LSCs (36, 37). TIM-3 may be considered as an acceptable candidate to eradicate AML LSCs (36). Based on flow cytometry analyses, miR-125a-3P and miR-133-5p showed a silencing phenomenon on the protein expression of TIM-3 in the HL-60 cell line (39, 139). The significant distinction of the silencing impact between the levels of informational RNAs and proteins of TIM-3 indicated that miR-125a-3p and miR-133a-5p may play restrictive roles in TIM-3 expression at the informational RNA change of location level by steric hindrance rather than informational RNA degradation to provide antitumor effects (39, 139).
The Impact of MicroRNA Targeting ICOS on Cancer
Inducible co-stimulator (ICOS) and its ligand (ICOSL) operate as very important regulatory mechanism between immune and epithelium cells (140). ICOS is also an essential co-stimulatory molecule for T-cell function (141).
MiR-21 plays a key role in the regulation of the disease progression of B-cell lymphoma (38, 142). An experiment discovered a pre-B malignant lymphoid-like phenotype caused by the overexpression of miR-21 (143). MiR-21 induced the expression of ICOS on Tregs through the upregulation of p-STAT3, increased the interaction of Tregs with epithelial cells via the ICOS/ICOSL axis, stimulated neoplasm ontogenesis, and also led to the chemoresistance of B-cell lymphoma (144). Mir21 can also regulate the expressions of ICOS/ICOSL to affect the progression of CRC [36]. All these may provide novel treatment options for B-cell lymphoma.
ICOS is a marker of T-cell activation (145), and it was upregulated in the tumor tissues of obese patients with lung adenocarcinoma (LUAD) (141). Obesity mediates immune exhaustion through leptin (146). Studies have shown that miR-27a-3p could directly interact with ICOS and downregulate its expression in LUAD (141). IFN-γ is a significant activator of innate and adaptive immunity (147). However, the level of miR-27a-3p was obviously reduced in adipose-derived exosomes from obese individuals, which promoted ICOS+ T-cell proliferation, thereby influencing IFN-γ secretion and, hence, inhibiting the occurrence and development of LUAD (141). This provides new concept for the clinical treatment of LUAD.
The Impact of NcRNAs Targeting CD276 on Cancer
Human B7-H3 (CD276) is a member of the B7/CD28 antibody taxonomic group that provides crucial co-stimulatory signals that regulate the functions of T-lymphocytes in growth regulation, infections, and immune response (148). 2Ig and 4Ig of human B7-H3 both repressed lymphocyte proliferation and protein production (149).
MiRNAs Target CD276
CD276/B7-H3 has a high expression in solid tumors and leads to immune evasion and metastasis (40, 150). High expressions of CDK6 and CD276 have a negative influence on medulloblastoma survival, meaning that they play pro-oncogenic roles in medulloblastoma, supporting similar findings from prior studies (151). On the other hand, miR-1253 transfection plays a downregulated role in the expressions of CDK6 and CD276 (152). Silencing miR-1253 targets CDK6 and CD276 to suppress the growth of medulloblastoma (152). This provides good guidance to medulloblastoma therapy.
Previous studies indicated that CD276 appears to be involved in CRC progression and metastasis (41, 153). Moreover, CD276 is able to participate in miRNA-related regulation (154). CD276 is an immunoregulatory molecule that is directly targeted by miR-187 and is negatively related to miR-187 levels in CRC cells (45). The downregulation of miR-187 correlates with higher tumor grade and stage and functions as a tumor suppressor (155). Studies have shown that miR-187 was significantly downregulated in CRC tissues and its cell lines, and the overexpression of miR-187 can inhibit the proliferation, migration, and invasion of CRC cells and promote CRC cell apoptosis (45). The miR-187/CD276 axis provides a new potential therapeutic target for the treatment of CRC.
CircRNAs Target CD276
Hsa_circ0021347 is a possible target regulated by B7-H3 in osteosarcoma (OS) cells, and its expression is reciprocally associated with the expression of B7-H3 (156). Analysis results have provided proof that B7-H3 accelerates growth cell immune escape through suppressing T-cell-mediated cellular immunity (157, 158). A previous study showed that miR-646 was involved in promoting the formation of OS (159). Bioinformatics analysis revealed that hsa_circ0021347 and miR-646 are involved in OS and that miR-646 potentially interacted with NOB1 (156). It can be inferred that the hsa_circ0021347–miR-646–NOB1 axis may be a good research direction for osteosarcoma treatment.
Conclusion and Perspectives
The role of ncRNAs on immune checkpoints is complex. PD-1, PD-L1, CD47, BTLA, and the other immune checkpoints are regulated by miRNAs through directly combining with the 3′-UTR. CircRNAs can directly act on immune checkpoints, but most act as “miRNA sponge” to indirectly adjust them. LncRNAs and immune checkpoints interact to regulate the development of cancers. The three types of ncRNAs (miRNAs, circRNAs, and lncRNAs) have certain roles in promoting the study of tumor resistance and the development of new drug targets or immunotherapy options. Also, some of the specific roles of circRNAs and lncRNAs on immune checkpoints are not clearly elucidated and require further study. The targeted effect of ncRNAs on immune checkpoints is indicative of the development of immunotherapy for cancers.
Author Contributions
YJ wrote the original draft and contributed to drawing. XY revised the manuscripts. XL provided ideas. LZ, YW, SD, and PC read and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Hengyang City Science and Technology Planning Project (grant no. 202150063473), the Scientific Research Project of Hunan Provincial Health Commission (grant no. 202202044140), the Scientific Research Project of Hunan Provincial Education Department (grant no. 21B0438), and the Hunan Province Cooperative Innovation Center for Molecular Target New Drug Study (grant no. 2014-405).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ni SJ, Zhao LQ, Wang XF, Wu ZH, Hua RX, Wan CH, et al. CBX7 Regulates Stem Cell-Like Properties of Gastric Cancer Cells via P16 and AKT-NF-κb-miR-21 Pathways. J Hematol Oncol (2018) 11(1):17. doi: 10.1186/s13045-018-0562-z
2. Eddy SR. Non-Coding RNA Genes and the Modern RNA World. Nat Rev Genet (2001) 2(12):919–29. doi: 10.1038/35103511
3. Liu Y, Liu X, Lin C, Jia X, Zhu H, Song J, et al. Noncoding RNAs Regulate Alternative Splicing in Cancer. J Exp Clin Cancer Res (2021) 40(1):11. doi: 10.1186/s13046-020-01798-2
4. Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell (2004) 116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5
5. Wang WT, Han C, Sun YM, Chen TQ, Chen YQ. Noncoding RNAs in Cancer Therapy Resistance and Targeted Drug Development. J Hematol Oncol (2019) 12(1):55. doi: 10.1186/s13045-019-0748-z
6. Fang W, Mu J, Yang Y, Liu L. CircRERE Confers the Resistance of Multiple Myeloma to Bortezomib Depending on the Regulation of CD47 by Exerting the Sponge Effect on miR-152-3p. J Bone Oncol (2021) 30:100381. doi: 10.1016/j.jbo.2021.100381
7. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature (2012) 489(7414):57–74. doi: 10.1038/nature11247
8. Zhao J, Lee EE, Kim J, Yang R, Chamseddin B, Ni C, et al. Transforming Activity of an Oncoprotein-Encoding Circular RNA From Human Papillomavirus. Nat Commun (2019) 10(1):2300. doi: 10.1038/s41467-019-10246-5
9. Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN. Circular RNA Splicing. Adv Exp Med Biol (2018) 1087:41–52. doi: 10.1007/978-981-13-1426-1_4
10. Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC, et al. Circular RNA Circtrim33-12 Acts as the Sponge of MicroRNA-191 to Suppress Hepatocellular Carcinoma Progression. Mol Cancer (2019) 18(1):105. doi: 10.1186/s12943-019-1031-1
11. Zhang J, Hou L, Liang R, Chen X, Zhang R, Chen W, et al. CircDLST Promotes the Tumorigenesis and Metastasis of Gastric Cancer by Sponging miR-502-5p and Activating the NRAS/MEK1/ERK1/2 Signaling. Mol Cancer (2019) 18(1):80. doi: 10.1186/s12943-019-1015-1
12. Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA Circrhot1 Promotes Hepatocellular Carcinoma Progression by Initiation of NR2F6 Expression. Mol Cancer (2019) 18(1):119. doi: 10.1186/s12943-019-1046-7
13. Peng WX, Koirala P, Mo YY. LncRNA-Mediated Regulation of Cell Signaling in Cancer. Oncogene (2017) 36(41):5661–7. doi: 10.1038/onc.2017.184
14. Hu X, Zhang L, Tian J, Ma J. Long Non-Coding RNA PART1 Predicts a Poor Prognosis and Promotes the Malignant Progression of Pancreatic Cancer by Sponging miR-122. World J Surg Oncol (2021) 19(1):122. doi: 10.1186/s12957-021-02232-3
15. Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune Checkpoint Inhibitors: Recent Progress and Potential Biomarkers. Exp Mol Med (2018) 50(12):1–11. doi: 10.1038/s12276-018-0191-1
16. Eggermont AMM, Blank CU, Mandala M, Long GV, Atkinson V, Dalle S, et al. Adjuvant Pembrolizumab Versus Placebo in Resected Stage III Melanoma. N Engl J Med (2018) 378(19):1789–801. doi: 10.1056/NEJMoa1802357
17. Forde PM, Chaft JE, Smith KN, Anagnostou V, Cottrell TR, Hellmann MD, et al. Neoadjuvant PD-1 Blockade in Resectable Lung Cancer. N Engl J Med (2018) 378(21):1976–86. doi: 10.1056/NEJMoa1716078
18. Wang D, Lin J, Yang X, Long J, Bai Y, Yang X, et al. Combination Regimens With PD-1/PD-L1 Immune Checkpoint Inhibitors for Gastrointestinal Malignancies. J Hematol Oncol (2019) 12(1):42. doi: 10.1186/s13045-019-0730-9
19. Zheng Z, Sun R, Zhao HJ, Fu D, Zhong HJ, Weng XQ, et al. MiR155 Sensitized B-Lymphoma Cells to Anti-PD-L1 Antibody via PD-1/PD-L1-Mediated Lymphoma Cell Interaction With CD8+T Cells. Mol Cancer (2019) 18(1):54. doi: 10.1186/s12943-019-0977-3
20. Zhao M, Liu Q, Liu W, Zhou H, Zang X, Lu J. MicroRNA−140 Suppresses Helicobacter Pylori−Positive Gastric Cancer Growth by Enhancing the Antitumor Immune Response. Mol Med Rep (2019) 20(3):2484–92. doi: 10.3892/mmr.2019.10475
21. Xu S, Tao Z, Hai B, Liang H, Shi Y, Wang T, et al. miR-424(322) Reverses Chemoresistance via T-Cell Immune Response Activation by Blocking the PD-L1 Immune Checkpoint. Nat Commun (2016) 7:11406. doi: 10.1038/ncomms11406
22. Wei J, Nduom EK, Kong LY, Hashimoto Y, Xu S, Gabrusiewicz K, et al. MiR-138 Exerts Anti-Glioma Efficacy by Targeting Immune Checkpoints. Neuro Oncol (2016) 18(5):639–48. doi: 10.1093/neuonc/nov292
23. Huang F, Wang B, Zeng J, Sang S, Lei J, Lu Y. MicroRNA-374b Inhibits Liver Cancer Progression via Down Regulating Programmed Cell Death-1 Expression on Cytokine-Induced Killer Cells. Oncol Lett (2018) 15(4):4797–804. doi: 10.3892/ol.2018.7951
24. Zhang G, Li N, Li Z, Zhu Q, Li F, Yang C, et al. microRNA-4717 Differentially Interacts With Its Polymorphic Target in the PD1 3’ Untranslated Region: A Mechanism for Regulating PD-1 Expression and Function in HBV-Associated Liver Diseases. Oncotarget (2015) 6(22):18933–44. doi: 10.18632/oncotarget.3662
25. Zhang G, Liu Z, Duan S, Han Q, Li Z, Lv Y, et al. Association of Polymorphisms of Programmed Cell Death-1 Gene With Chronic Hepatitis B Virus Infection. Hum Immunol (2010) 71(12):1209–13. doi: 10.1016/j.humimm.2010.08.014
26. Xi Q, Zhang J, Yang G, Zhang L, Chen Y, Wang C, et al. Restoration of miR-340 Controls Pancreatic Cancer Cell CD47 Expression to Promote Macrophage Phagocytosis and Enhance Antitumor Immunity. J Immunother Cancer (2020) 8(1):e00253. doi: 10.1136/jitc-2019-000253
27. Zhao Y, Yu X, Tang H, Han R, Wang X, Wang J, et al. MicroRNA-200a Promotes Phagocytosis of Macrophages and Suppresses Cell Proliferation, Migration, and Invasion in Nasopharyngeal Carcinoma by Targeting Cd47. BioMed Res Int (2020) 2020:3723781. doi: 10.1155/2020/3723781
28. Yang SY, Choi SA, Lee JY, Park AK, Wang KC, Phi JH, et al. miR-192 Suppresses Leptomeningeal Dissemination of Medulloblastoma by Modulating Cell Proliferation and Anchoring Through the Regulation of DHFR, Integrins, and CD47. Oncotarget (2015) 6(41):43712–30. doi: 10.18632/oncotarget.6227
29. Zhu L, Tu H, Liang Y, Tang D. MiR-218 Produces Anti-Tumor Effects on Cervical Cancer Cells In Vitro. World J Surg Oncol (2018) 16(1):204. doi: 10.1186/s12957-018-1506-3
30. Huang Q, Xia J, Wang L, Wang X, Ma X, Deng Q, et al. miR-153 Suppresses IDO1 Expression and Enhances CAR T Cell Immunotherapy. J Hematol Oncol (2018) 11(1):58. doi: 10.1186/s13045-018-0600-x
31. Lou Q, Liu R, Yang X, Li W, Huang L, Wei L, et al. miR-448 Targets IDO1 and Regulates CD8(+) T Cell Response in Human Colon Cancer. J Immunother Cancer (2019) 7(1):210. doi: 10.1186/s40425-019-0691-0
32. Wang C, Tao X, Wei J. Effects of LncRNA MEG3 on Immunity and Autophagy of Non-Small Cell Lung Carcinoma Through IDO Signaling Pathway. World J Surg Oncol (2021) 19(1):244. doi: 10.1186/s12957-021-02346-8
33. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA Genes Are Frequently Located at Fragile Sites and Genomic Regions Involved in Cancers. Proc Natl Acad Sci USA (2004) 101(9):2999–3004. doi: 10.1073/pnas.0307323101
34. Imai Y, Hasegawa K, Matsushita H, Fujieda N, Sato S, Miyagi E, et al. Expression of Multiple Immune Checkpoint Molecules on T Cells in Malignant Ascites From Epithelial Ovarian Carcinoma. Oncol Lett (2018) 15(5):6457–68. doi: 10.3892/ol.2018.8101
35. Zhi X, Wang Y, Yu J, Yu J, Zhang L, Yin L, et al. Potential Prognostic Biomarker CD73 Regulates Epidermal Growth Factor Receptor Expression in Human Breast Cancer. IUBMB Life (2012) 64(11):911–20. doi: 10.1002/iub.1086
36. Kikushige Y, Shima T, Takayanagi S, Urata S, Miyamoto T, Iwasaki H, et al. TIM-3 Is a Promising Target to Selectively Kill Acute Myeloid Leukemia Stem Cells. Cell Stem Cell (2010) 7(6):708–17. doi: 10.1016/j.stem.2010.11.014
37. Jan M, Chao MP, Cha AC, Alizadeh AA, Gentles AJ, Weissman IL, et al. Prospective Separation of Normal and Leukemic Stem Cells Based on Differential Expression of TIM3, a Human Acute Myeloid Leukemia Stem Cell Marker. Proc Natl Acad Sci USA (2011) 108(12):5009–14. doi: 10.1073/pnas.1100551108
38. Baraniskin A, Kuhnhenn J, Schlegel U, Chan A, Deckert M, Gold R, et al. Identification of microRNAs in the Cerebrospinal Fluid as Marker for Primary Diffuse Large B-Cell Lymphoma of the Central Nervous System. Blood (2011) 117(11):3140–6. doi: 10.1182/blood-2010-09-308684
39. Hojati Z, Ganjalikhani-Hakemi M, Ameri M, Alimohammadi-Jelodar SF, Dehbashi M, Mohammad Ganji M, et al. Evaluation of Silencing Effect of miR-133a-5p Mimic on TIM-3 Expression in AML (HL-60) Cell Line. Indian J Clin Biochem (2020) 35(3):359–66. doi: 10.1007/s12291-019-00834-z
40. Picarda E, Ohaegbulam KC, Zang X. Molecular Pathways: Targeting B7-H3 (CD276) for Human Cancer Immunotherapy. Clin Cancer Res (2016) 22(14):3425–31. doi: 10.1158/1078-0432.Ccr-15-2428
41. Ingebrigtsen VA, Boye K, Tekle C, Nesland JM, Flatmark K, Fodstad O. B7-H3 Expression in Colorectal Cancer: Nuclear Localization Strongly Predicts Poor Outcome in Colon Cancer. Int J Cancer (2012) 131(11):2528–36. doi: 10.1002/ijc.27566
42. Zhang PF, Pei X, Li KS, Jin LN, Wang F, Wu J, et al. Circular RNA Circfgfr1 Promotes Progression and Anti-PD-1 Resistance by Sponging miR-381-3p in Non-Small Cell Lung Cancer Cells. Mol Cancer (2019) 18(1):179. doi: 10.1186/s12943-019-1111-2
43. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer Cell-Derived Exosomal Circuhrf1 Induces Natural Killer Cell Exhaustion and may Cause Resistance to Anti-PD1 Therapy in Hepatocellular Carcinoma. Mol Cancer (2020) 19(1):110. doi: 10.1186/s12943-020-01222-5
44. Ghalamfarsa G, Kazemi MH, Raoofi Mohseni S, Masjedi A, Hojjat-Farsangi M, Azizi G, et al. CD73 as a Potential Opportunity for Cancer Immunotherapy. Expert Opin Ther Targets (2019) 23(2):127–42. doi: 10.1080/14728222.2019.1559829
45. Wang ZS, Zhong M, Bian YH, Mu YF, Qin SL, Yu MH, et al. MicroRNA-187 Inhibits Tumor Growth and Invasion by Directly Targeting CD276 in Colorectal Cancer. Oncotarget (2016) 7(28):44266–76. doi: 10.18632/oncotarget.10023
46. Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, et al. The NORAD lncRNA Assembles a Topoisomerase Complex Critical for Genome Stability. Nature (2018) 561(7721):132–6. doi: 10.1038/s41586-018-0453-z
47. Tang Y, He Y, Shi L, Yang L, Wang J, Lian Y, et al. Co-Expression of AFAP1-AS1 and PD-1 Predicts Poor Prognosis in Nasopharyngeal Carcinoma. Oncotarget (2017) 8(24):39001–11. doi: 10.18632/oncotarget.16545
48. Zhao L, Liu Y, Zhang J, Liu Y, Qi Q. LncRNA SNHG14/miR-5590-3p/ZEB1 Positive Feedback Loop Promoted Diffuse Large B Cell Lymphoma Progression and Immune Evasion Through Regulating PD-1/PD-L1 Checkpoint. Cell Death Dis (2019) 10(10):731. doi: 10.1038/s41419-019-1886-5
49. Intlekofer AM, Thompson CB. At the Bench: Preclinical Rationale for CTLA-4 and PD-1 Blockade as Cancer Immunotherapy. J Leukoc Biol (2013) 94(1):25–39. doi: 10.1189/jlb.1212621
50. Hirano F, Kaneko K, Tamura H, Dong H, Wang S, Ichikawa M, et al. Blockade of B7-H1 and PD-1 by Monoclonal Antibodies Potentiates Cancer Therapeutic Immunity. Cancer Res (2005) 65(3):1089–96.
51. Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, et al. Pre-B Cell Proliferation and Lymphoblastic Leukemia/High-Grade Lymphoma in E(mu)-Mir155 Transgenic Mice. Proc Natl Acad Sci USA (2006) 103(18):7024–9. doi: 10.1073/pnas.0602266103
52. Boussiotis VA. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N Engl J Med (2016) 375(18):1767–78. doi: 10.1056/NEJMra1514296
53. Zhu FQ, Zeng L, Tang N, Tang YP, Zhou BP, Li FF, et al. MicroRNA-155 Downregulation Promotes Cell Cycle Arrest and Apoptosis in Diffuse Large B-Cell Lymphoma. Oncol Res (2016) 24(6):415–27. doi: 10.3727/096504016x14685034103473
54. Yan X, Zhu Z, Xu S, Yang LN, Liao XH, Zheng M, et al. MicroRNA-140-5p Inhibits Hepatocellular Carcinoma by Directly Targeting the Unique Isomerase Pin1 to Block Multiple Cancer-Driving Pathways. Sci Rep (2017) 7:45915. doi: 10.1038/srep45915
55. Zou J, Xu Y. MicroRNA-140 Inhibits Cell Proliferation in Gastric Cancer Cell Line HGC-27 by Suppressing Sox4. Med Sci Monit (2016) 22:2243–52. doi: 10.12659/msm.896633
56. Kleffel S, Posch C, Barthel SR, Mueller H, Schlapbach C, Guenova E, et al. Melanoma Cell-Intrinsic PD-1 Receptor Functions Promote Tumor Growth. Cell (2015) 162(6):1242–56. doi: 10.1016/j.cell.2015.08.052
57. Ji X, Wang E, Tian F. MicroRNA-140 Suppresses Osteosarcoma Tumor Growth by Enhancing Anti-Tumor Immune Response and Blocking mTOR Signaling. Biochem Biophys Res Commun (2018) 495(1):1342–8. doi: 10.1016/j.bbrc.2017.11.120
58. Goodman A, Patel SP, Kurzrock R. PD-1-PD-L1 Immune-Checkpoint Blockade in B-Cell Lymphomas. Nat Rev Clin Oncol (2017) 14(4):203–20. doi: 10.1038/nrclinonc.2016.168
59. Xu C, Fillmore CM, Koyama S, Wu H, Zhao Y, Chen Z, et al. Loss of Lkb1 and Pten Leads to Lung Squamous Cell Carcinoma With Elevated PD-L1 Expression. Cancer Cell (2014) 25(5):590–604. doi: 10.1016/j.ccr.2014.03.033
60. Blank C, Gajewski TF, Mackensen A. Interaction of PD-L1 on Tumor Cells With PD-1 on Tumor-Specific T Cells as a Mechanism of Immune Evasion: Implications for Tumor Immunotherapy. Cancer Immunol Immunother (2005) 54(4):307–14. doi: 10.1007/s00262-004-0593-x
61. Duraiswamy J, Freeman GJ, Coukos G. Therapeutic PD-1 Pathway Blockade Augments With Other Modalities of Immunotherapy T-Cell Function to Prevent Immune Decline in Ovarian Cancer. Cancer Res (2013) 73(23):6900–12. doi: 10.1158/0008-5472.Can-13-1550
62. Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, et al. A Functional Screen Implicates microRNA-138-Dependent Regulation of the Depalmitoylation Enzyme APT1 in Dendritic Spine Morphogenesis. Nat Cell Biol (2009) 11(6):705–16. doi: 10.1038/ncb1876
63. Wei J, Wang F, Kong LY, Xu S, Doucette T, Ferguson SD, et al. miR-124 Inhibits STAT3 Signaling to Enhance T Cell-Mediated Immune Clearance of Glioma. Cancer Res (2013) 73(13):3913–26. doi: 10.1158/0008-5472.Can-12-4318
64. Bengsch B, Martin B, Thimme R. Restoration of HBV-Specific CD8+ T Cell Function by PD-1 Blockade in Inactive Carrier Patients Is Linked to T Cell Differentiation. J Hepatol (2014) 61(6):1212–9. doi: 10.1016/j.jhep.2014.07.005
65. Boni C, Fisicaro P, Valdatta C, Amadei B, Di Vincenzo P, Giuberti T, et al. Characterization of Hepatitis B Virus (HBV)-Specific T-Cell Dysfunction in Chronic HBV Infection. J Virol (2007) 81(8):4215–25. doi: 10.1128/jvi.02844-06
66. Peng G, Li S, Wu W, Tan X, Chen Y, Chen Z. PD-1 Upregulation Is Associated With HBV-Specific T Cell Dysfunction in Chronic Hepatitis B Patients. Mol Immunol (2008) 45(4):963–70. doi: 10.1016/j.molimm.2007.07.038
67. Spranger S, Koblish HK, Horton B, Scherle PA, Newton R, Gajewski TF. Mechanism of Tumor Rejection With Doublets of CTLA-4, PD-1/PD-L1, or IDO Blockade Involves Restored IL-2 Production and Proliferation of CD8(+) T Cells Directly Within the Tumor Microenvironment. J Immunother Cancer (2014) 2:3. doi: 10.1186/2051-1426-2-3
68. Hu S, Bao H, Xu X, Zhou X, Qin W, Zeng C, et al. Increased miR-374b Promotes Cell Proliferation and the Production of Aberrant Glycosylated IgA1 in B Cells of IgA Nephropathy. FEBS Lett (2015) 589(24 Pt B):4019–25. doi: 10.1016/j.febslet.2015.10.033
69. Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 Immunoinhibitory Receptor by a Novel B7 Family Member Leads to Negative Regulation of Lymphocyte Activation. J Exp Med (2000) 192(7):1027–34. doi: 10.1084/jem.192.7.1027
70. Chen L. Co-Inhibitory Molecules of the B7-CD28 Family in the Control of T-Cell Immunity. Nat Rev Immunol (2004) 4(5):336–47. doi: 10.1038/nri1349
71. Mühlbauer M, Fleck M, Schütz C, Weiss T, Froh M, Blank C, et al. PD-L1 Is Induced in Hepatocytes by Viral Infection and by Interferon-Alpha and -Gamma and Mediates T Cell Apoptosis. J Hepatol (2006) 45(4):520–8. doi: 10.1016/j.jhep.2006.05.007
72. Zhang G, Li Z, Han Q, Li N, Zhu Q, Li F, et al. Altered TNF-α and IFN-γ Levels Associated With PD1 But Not TNFA Polymorphisms in Patients With Chronic HBV Infection. Infect Genet Evol (2011) 11(7):1624–30. doi: 10.1016/j.meegid.2011.06.004
73. Zhang G, Li N, Zhang P, Li F, Yang C, Zhu Q, et al. PD-1 mRNA Expression Is Associated With Clinical and Viral Profile and PD1 3’-Untranslated Region Polymorphism in Patients With Chronic HBV Infection. Immunol Lett (2014) 162(1 Pt A):212–6. doi: 10.1016/j.imlet.2014.09.001
74. Li Q, Johnston N, Zheng X, Wang H, Zhang X, Gao D, et al. miR-28 Modulates Exhaustive Differentiation of T Cells Through Silencing Programmed Cell Death-1 and Regulating Cytokine Secretion. Oncotarget (2016) 7(33):53735–50. doi: 10.18632/oncotarget.10731
75. Jiang Y, Li Y, Zhu B. T-Cell Exhaustion in the Tumor Microenvironment. Cell Death Dis (2015) 6(6):e1792. doi: 10.1038/cddis.2015.162
76. Merelli B, Massi D, Cattaneo L, Mandalà M. Targeting the PD1/PD-L1 Axis in Melanoma: Biological Rationale, Clinical Challenges and Opportunities. Crit Rev Oncol Hematol (2014) 89(1):140–65. doi: 10.1016/j.critrevonc.2013.08.002
77. Zhang R, Dong Y, Sun M, Wang Y, Cai C, Zeng Y, et al. Tumor-Associated Inflammatory Microenvironment in Non-Small Cell Lung Cancer: Correlation With FGFR1 and TLR4 Expression via PI3K/Akt Pathway. J Cancer (2019) 10(4):1004–12. doi: 10.7150/jca.26277
78. Qiu BQ, Zhang PF, Xiong D, Xu JJ, Long X, Zhu SQ, et al. CircRNA Fibroblast Growth Factor Receptor 3 Promotes Tumor Progression in Non-Small Cell Lung Cancer by Regulating Galectin-1-AKT/ERK1/2 Signaling. J Cell Physiol (2019) 234(7):11256–64. doi: 10.1002/jcp.27783
79. Chen IX, Chauhan VP, Posada J, Ng MR, Wu MW, Adstamongkonkul P, et al. Blocking CXCR4 Alleviates Desmoplasia, Increases T-Lymphocyte Infiltration, and Improves Immunotherapy in Metastatic Breast Cancer. Proc Natl Acad Sci USA (2019) 116(10):4558–66. doi: 10.1073/pnas.1815515116
80. Seo YD, Jiang X, Sullivan KM, Jalikis FG, Smythe KS, Abbasi A, et al. Mobilization of CD8(+) T Cells via CXCR4 Blockade Facilitates PD-1 Checkpoint Therapy in Human Pancreatic Cancer. Clin Cancer Res (2019) 25(13):3934–45. doi: 10.1158/1078-0432.Ccr-19-0081
81. Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, et al. UHRF1 Overexpression Drives DNA Hypomethylation and Hepatocellular Carcinoma. Cancer Cell (2014) 25(2):196–209. doi: 10.1016/j.ccr.2014.01.003
82. Waidmann O. Recent Developments With Immunotherapy for Hepatocellular Carcinoma. Expert Opin Biol Ther (2018) 18(8):905–10. doi: 10.1080/14712598.2018.1499722
83. Sun H, Liu L, Huang Q, Liu H, Huang M, Wang J, et al. Accumulation of Tumor-Infiltrating CD49a(+) NK Cells Correlates With Poor Prognosis for Human Hepatocellular Carcinoma. Cancer Immunol Res (2019) 7(9):1535–46. doi: 10.1158/2326-6066.Cir-18-0757
84. Zhang J, Fang W, Qin T, Yang Y, Hong S, Liang W, et al. Co-Expression of PD-1 and PD-L1 Predicts Poor Outcome in Nasopharyngeal Carcinoma. Med Oncol (2015) 32(3):86. doi: 10.1007/s12032-015-0501-6
85. Hsu MC, Hsiao JR, Chang KC, Wu YH, Su IJ, Jin YT, et al. Increase of Programmed Death-1-Expressing Intratumoral CD8 T Cells Predicts a Poor Prognosis for Nasopharyngeal Carcinoma. Mod Pathol (2010) 23(10):1393–403. doi: 10.1038/modpathol.2010.130
86. Fang W, Zhang J, Hong S, Zhan J, Chen N, Qin T, et al. EBV-Driven LMP1 and IFN-γ Up-Regulate PD-L1 in Nasopharyngeal Carcinoma: Implications for Oncotargeted Therapy. Oncotarget (2014) 5(23):12189–202. doi: 10.18632/oncotarget.2608
87. Wang P, Guo Q, Gao Y, Zhi H, Zhang Y, Liu Y, et al. Improved Method for Prioritization of Disease Associated lncRNAs Based on ceRNA Theory and Functional Genomics Data. Oncotarget (2017) 8(3):4642–55. doi: 10.18632/oncotarget.13964
88. Sun Y, Cheng H, Wang G, Yu G, Zhang D, Wang Y, et al. Deregulation of miR-183 Promotes Melanoma Development via lncRNA MALAT1 Regulation and ITGB1 Signal Activation. Oncotarget (2017) 8(2):3509–18. doi: 10.18632/oncotarget.13862
89. Yang S, Ning Q, Zhang G, Sun H, Wang Z, Li Y. Construction of Differential mRNA-lncRNA Crosstalk Networks Based on ceRNA Hypothesis Uncover Key Roles of lncRNAs Implicated in Esophageal Squamous Cell Carcinoma. Oncotarget (2016) 7(52):85728–40. doi: 10.18632/oncotarget.13828
90. Katsura A, Tamura Y, Hokari S, Harada M, Morikawa M, Sakurai T, et al. ZEB1-Regulated Inflammatory Phenotype in Breast Cancer Cells. Mol Oncol (2017) 11(9):1241–62. doi: 10.1002/1878-0261.12098
91. Su W, Xu M, Chen X, Chen N, Gong J, Nie L, et al. Long Noncoding RNA ZEB1-AS1 Epigenetically Regulates the Expressions of ZEB1 and Downstream Molecules in Prostate Cancer. Mol Cancer (2017) 16(1):142. doi: 10.1186/s12943-017-0711-y
92. De Craene B, Berx G. Regulatory Networks Defining EMT During Cancer Initiation and Progression. Nat Rev Cancer (2013) 13(2):97–110. doi: 10.1038/nrc3447
93. Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, et al. Metastasis Is Regulated via microRNA-200/ZEB1 Axis Control of Tumour Cell PD-L1 Expression and Intratumoral Immunosuppression. Nat Commun (2014) 5:5241. doi: 10.1038/ncomms6241
94. Noman MZ, Janji B, Abdou A, Hasmim M, Terry S, Tan TZ, et al. The Immune Checkpoint Ligand PD-L1 Is Upregulated in EMT-Activated Human Breast Cancer Cells by a Mechanism Involving ZEB-1 and miR-200. Oncoimmunology (2017) 6(1):e1263412. doi: 10.1080/2162402x.2016.1263412
95. Brown EJ, Frazier WA. Integrin-Associated Protein (CD47) and Its Ligands. Trends Cell Biol (2001) 11(3):130–5. doi: 10.1016/s0962-8924(00)01906-1
96. Qin Y, Zhou X, Huang C, Li L, Liu H, Liang N, et al. Lower miR-340 Expression Predicts Poor Prognosis of Non-Small Cell Lung Cancer and Promotes Cell Proliferation by Targeting CDK4. Gene (2018) 675:278–84. doi: 10.1016/j.gene.2018.06.062
97. Huang K, Tang Y, He L, Dai Y. MicroRNA-340 Inhibits Prostate Cancer Cell Proliferation and Metastasis by Targeting the MDM2-P53 Pathway. Oncol Rep (2016) 35(2):887–95. doi: 10.3892/or.2015.4458
98. Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, et al. The CD47-Signal Regulatory Protein Alpha (SIRPa) Interaction Is a Therapeutic Target for Human Solid Tumors. Proc Natl Acad Sci USA (2012) 109(17):6662–7. doi: 10.1073/pnas.1121623109
99. O’Brien SJ, Carter JV, Burton JF, Oxford BG, Schmidt MN, Hallion JC, et al. The Role of the miR-200 Family in Epithelial-Mesenchymal Transition in Colorectal Cancer: A Systematic Review. Int J Cancer (2018) 142(12):2501–11. doi: 10.1002/ijc.31282
100. Jurmeister S, Baumann M, Balwierz A, Keklikoglou I, Ward A, Uhlmann S, et al. MicroRNA-200c Represses Migration and Invasion of Breast Cancer Cells by Targeting Actin-Regulatory Proteins FHOD1 and PPM1F. Mol Cell Biol (2012) 32(3):633–51. doi: 10.1128/mcb.06212-11
101. Senol O, Schaaij-Visser TB, Erkan EP, Dorfer C, Lewandrowski G, Pham TV, et al. miR-200a-Mediated Suppression of Non-Muscle Heavy Chain IIb Inhibits Meningioma Cell Migration and Tumor Growth In Vivo. Oncogene (2015) 34(14):1790–8. doi: 10.1038/onc.2014.120
102. Veillette A, Chen J. Sirpα-CD47 Immune Checkpoint Blockade in Anticancer Therapy. Trends Immunol (2018) 39(3):173–84. doi: 10.1016/j.it.2017.12.005
103. Yoshida K, Tsujimoto H, Matsumura K, Kinoshita M, Takahata R, Matsumoto Y, et al. CD47 Is an Adverse Prognostic Factor and a Therapeutic Target in Gastric Cancer. Cancer Med (2015) 4(9):1322–33. doi: 10.1002/cam4.478
104. Jaiswal S, Chao MP, Majeti R, Weissman IL. Macrophages as Mediators of Tumor Immunosurveillance. Trends Immunol (2010) 31(6):212–9. doi: 10.1016/j.it.2010.04.001
105. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs With a Role in Cancer. Nat Rev Cancer (2006) 6(4):259–69. doi: 10.1038/nrc1840
106. Khella HW, Bakhet M, Allo G, Jewett MA, Girgis AH, Latif A, et al. miR-192, miR-194 and miR-215: A Convergent microRNA Network Suppressing Tumor Progression in Renal Cell Carcinoma. Carcinogenesis (2013) 34(10):2231–9. doi: 10.1093/carcin/bgt184
107. Feng S, Cong S, Zhang X, Bao X, Wang W, Li H, et al. MicroRNA-192 Targeting Retinoblastoma 1 Inhibits Cell Proliferation and Induces Cell Apoptosis in Lung Cancer Cells. Nucleic Acids Res (2011) 39(15):6669–78. doi: 10.1093/nar/gkr232
108. Mizejewski GJ. Role of Integrins in Cancer: Survey of Expression Patterns. Proc Soc Exp Biol Med (1999) 222(2):124–38. doi: 10.1046/j.1525-1373.1999.d01-122.x
109. Li X, Regezi J, Ross FP, Blystone S, Ilić D, Leong SP, et al. Integrin Alphavbeta3 Mediates K1735 Murine Melanoma Cell Motility In Vivo and In Vitro. J Cell Sci (2001) 114(Pt 14):2665–72. doi: 10.1242/jcs.114.14.2665
110. Sun J, Muz B, Alhallak K, Markovic M, Gurley S, Wang Z, et al. Targeting CD47 as a Novel Immunotherapy for Multiple Myeloma. Cancers (Basel) (2020) 12(2):305. doi: 10.3390/cancers12020305
111. Storti P, Vescovini R, Costa F, Marchica V, Toscani D, Dalla Palma B, et al. CD14(+) CD16(+) Monocytes Are Involved in Daratumumab-Mediated Myeloma Cells Killing and in Anti-CD47 Therapeutic Strategy. Br J Haematol (2020) 190(3):430–6. doi: 10.1111/bjh.16548
112. Liu J, Du F, Chen C, Li D, Chen Y, Xiao X, et al. CircRNA ITCH Increases Bortezomib Sensitivity Through Regulating the miR-615-3p/PRKCD Axis in Multiple Myeloma. Life Sci (2020) 262:118506. doi: 10.1016/j.lfs.2020.118506
113. Hennequart M, Pilotte L, Cane S, Hoffmann D, Stroobant V, Plaen E, et al. Constitutive IDO1 Expression in Human Tumors Is Driven by Cyclooxygenase-2 and Mediates Intrinsic Immune Resistance. Cancer Immunol Res (2017) 5(8):695–709. doi: 10.1158/2326-6066.Cir-16-0400
114. Li F, Zhang R, Li S, Liu J. IDO1: An Important Immunotherapy Target in Cancer Treatment. Int Immunopharmacol (2017) 47:70–7. doi: 10.1016/j.intimp.2017.03.024
115. Davar D, Bahary N. Modulating Tumor Immunology by Inhibiting Indoleamine 2,3-Dioxygenase (IDO): Recent Developments and First Clinical Experiences. Target Oncol (2018) 13(2):125–40. doi: 10.1007/s11523-017-0547-9
116. Folgiero V, Miele E, Carai A, Ferretti E, Alfano V, Po A, et al. IDO1 Involvement in mTOR Pathway: A Molecular Mechanism of Resistance to mTOR Targeting in Medulloblastoma. Oncotarget (2016) 7(33):52900–11. doi: 10.18632/oncotarget.9284
117. Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-Regulation of PD-L1, IDO, and T(regs) in the Melanoma Tumor Microenvironment Is Driven by CD8(+) T Cells. Sci Transl Med (2013) 5(200):200ra116. doi: 10.1126/scitranslmed.3006504
118. Zhai L, Spranger S, Binder DC, Gritsina G, Lauing KL, Giles FJ, et al. Molecular Pathways: Targeting IDO1 and Other Tryptophan Dioxygenases for Cancer Immunotherapy. Clin Cancer Res (2015) 21(24):5427–33. doi: 10.1158/1078-0432.Ccr-15-0420
119. Zhao XS, Wang YN, Lv M, Kong Y, Luo HX, Ye XY, et al. miR-153-3p, a New Bio-Target, Is Involved in the Pathogenesis of Acute Graft-Versus-Host Disease via Inhibition of Indoleamine- 2,3-Dioxygenase. Oncotarget (2016) 7(30):48321–34. doi: 10.18632/oncotarget.10220
120. Rosenberg SA. Progress in Human Tumour Immunology and Immunotherapy. Nature (2001) 411(6835):380–4. doi: 10.1038/35077246
121. Gao M, Guo Y, Xiao Y, Shang X. Comprehensive Analyses of Correlation and Survival Reveal Informative lncRNA Prognostic Signatures in Colon Cancer. World J Surg Oncol (2021) 19(1):104. doi: 10.1186/s12957-021-02196-4
122. Dong Z, Zhang A, Liu S, Lu F, Guo Y, Zhang G, et al. Aberrant Methylation-Mediated Silencing of lncRNA MEG3 Functions as a ceRNA in Esophageal Cancer. Mol Cancer Res (2017) 15(7):800–10. doi: 10.1158/1541-7786.Mcr-16-0385
123. Zhang RR, Wang LM, Shen JJ. Overexpression of miR-32 Inhibits the Proliferation and Metastasis of Ovarian Cancer Cells by Targeting BTLA. Eur Rev Med Pharmacol Sci (2020) 24(9):4671–8. doi: 10.26355/eurrev_202005_21154
124. Liu YJ, Zhou HG, Chen LH, Qu DC, Wang CJ, Xia ZY, et al. MiR-32-5p Regulates the Proliferation and Metastasis of Cervical Cancer Cells by Targeting HOXB8. Eur Rev Med Pharmacol Sci (2019) 23(1):87–95. doi: 10.26355/eurrev_201901_16752
125. Zhu J, Zeng Y, Li W, Qin H, Lei Z, Shen D, et al. CD73/NT5E Is a Target of miR-30a-5p and Plays an Important Role in the Pathogenesis of Non-Small Cell Lung Cancer. Mol Cancer (2017) 16(1):34. doi: 10.1186/s12943-017-0591-1
126. Zhang B. CD73 Promotes Tumor Growth and Metastasis. Oncoimmunology (2012) 1(1):67–70. doi: 10.4161/onci.1.1.18068
127. Xie M, Qin H, Luo Q, Huang Q, He X, Yang Z, et al. MicroRNA-30a Regulates Cell Proliferation and Tumor Growth of Colorectal Cancer by Targeting CD73. BMC Cancer (2017) 17(1):305. doi: 10.1186/s12885-017-3291-8
128. Zhu J, Zeng Y, Xu C, Qin H, Lei Z, Shen D, et al. Expression Profile Analysis of microRNAs and Downregulated miR-486-5p and miR-30a-5p in Non-Small Cell Lung Cancer. Oncol Rep (2015) 34(4):1779–86. doi: 10.3892/or.2015.4141
129. Wu R, Chen Y, Li F, Li W, Zhou H, Yang Y, et al. Effects of CD73 on Human Colorectal Cancer Cell Growth In Vivo and In Vitro. Oncol Rep (2016) 35(3):1750–6. doi: 10.3892/or.2015.4512
130. Xu Y, Liu Z, Lv L, Li P, Xiu B, Qian W, et al. MiRNA-340-5p Mediates the Functional and Infiltrative Promotion of Tumor-Infiltrating CD8(+) T Lymphocytes in Human Diffuse Large B Cell Lymphoma. J Exp Clin Cancer Res (2020) 39(1):238. doi: 10.1186/s13046-020-01752-2
131. Bobisse S, Genolet R, Roberti A, Tanyi JL, Racle J, Stevenson BJ, et al. Sensitive and Frequent Identification of High Avidity Neo-Epitope Specific CD8 (+) T Cells in Immunotherapy-Naive Ovarian Cancer. Nat Commun (2018) 9(1):1092. doi: 10.1038/s41467-018-03301-0
132. Chen S, Wainwright DA, Wu JD, Wan Y, Matei DE, Zhang Y, et al. CD73: An Emerging Checkpoint for Cancer Immunotherapy. Immunotherapy (2019) 11(11):983–97. doi: 10.2217/imt-2018-0200
133. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
134. Hasan SS, Ashraf GM, Banu N. Galectins - Potential Targets for Cancer Therapy. Cancer Lett (2007) 253(1):25–33. doi: 10.1016/j.canlet.2006.11.030
135. Xu YP, Dong ZN, Wang SW, Zheng YM, Zhang C, Zhou YQ, et al. Circhmgcs1-016 Reshapes Immune Environment by Sponging miR-1236-3p to Regulate CD73 and GAL-8 Expression in Intrahepatic Cholangiocarcinoma. J Exp Clin Cancer Res (2021) 40(1):290. doi: 10.1186/s13046-021-02095-2
136. Arnaout MK, Radomski KM, Srivastava DK, Tong X, Belt JR, Raimondi SC, et al. Treatment of Childhood Acute Myelogenous Leukemia With an Intensive Regimen (AML-87) That Individualizes Etoposide and Cytarabine Dosages: Short- and Long-Term Effects. Leukemia (2000) 14(10):1736–42. doi: 10.1038/sj.leu.2401906
137. Shipley JL, Butera JN. Acute Myelogenous Leukemia. Exp Hematol (2009) 37(6):649–58. doi: 10.1016/j.exphem.2009.04.002
138. De Kouchkovsky I, Abdul-Hay M. Acute Myeloid Leukemia: A Comprehensive Review and 2016 Update. Blood Cancer J (2016) 6(7):e441. doi: 10.1038/bcj.2016.50
139. Emamdoost F, Khanahmad H, Ganjalikhani-Hakemi M, Doosti A. The miR-125a-3p Inhibits TIM-3 Expression in AML Cell Line HL-60 In Vitro. Indian J Hematol Blood Transfus (2017) 33(3):342–7. doi: 10.1007/s12288-016-0733-4
140. Dianzani C, Minelli R, Mesturini R, Chiocchetti A, Barrera G, Boscolo S, et al. B7h Triggering Inhibits Umbilical Vascular Endothelial Cell Adhesiveness to Tumor Cell Lines and Polymorphonuclear Cells. J Immunol (2010) 185(7):3970–9. doi: 10.4049/jimmunol.0903269
141. Fan X, Wang J, Qin T, Zhang Y, Liu W, Jiang K, et al. Exosome miR-27a-3p Secreted From Adipocytes Targets ICOS to Promote Antitumor Immunity in Lung Adenocarcinoma. Thorac Cancer (2020) 11(6):1453–64. doi: 10.1111/1759-7714.13411
142. Lawrie CH. MicroRNAs and Lymphomagenesis: A Functional Review. Br J Haematol (2013) 160(5):571–81. doi: 10.1111/bjh.12157
143. Medina PP, Nolde M, Slack FJ. OncomiR Addiction in an In Vivo Model of microRNA-21-Induced Pre-B-Cell Lymphoma. Nature (2010) 467(7311):86–90. doi: 10.1038/nature09284
144. Zheng Z, Xu PP, Wang L, Zhao HJ, Weng XQ, Zhong HJ, et al. MiR21 Sensitized B-Lymphoma Cells to ABT-199 via ICOS/ICOSL-Mediated Interaction of Treg Cells With Endothelial Cells. J Exp Clin Cancer Res (2017) 36(1):82. doi: 10.1186/s13046-017-0551-z
145. Yoshinaga SK, Whoriskey JS, Khare SD, Sarmiento U, Guo J, Horan T, et al. T-Cell Co-Stimulation Through B7RP-1 and ICOS. Nature (1999) 402(6763):827–32. doi: 10.1038/45582
146. Shen Y, Wang Q, Zhao Q, Zhou J. Leptin Promotes the Immune Escape of Lung Cancer by Inducing Proinflammatory Cytokines and Resistance to Apoptosis. Mol Med Rep (2009) 2(2):295–9. doi: 10.3892/mmr_00000099
147. Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front Immunol (2018) 9:847. doi: 10.3389/fimmu.2018.00847
148. Flies DB, Chen L. The New B7s: Playing a Pivotal Role in Tumor Immunity. J Immunother (2007) 30(3):251–60. doi: 10.1097/CJI.0b013e31802e085a
149. Steinberger P, Majdic O, Derdak SV, Pfistershammer K, Kirchberger S, Klauser C, et al. Molecular Characterization of Human 4Ig-B7-H3, a Member of the B7 Family With Four Ig-Like Domains. J Immunol (2004) 172(4):2352–9. doi: 10.4049/jimmunol.172.4.2352
150. Gregorio A, Corrias MV, Castriconi R, Dondero A, Mosconi M, Gambini C, et al. Small Round Blue Cell Tumours: Diagnostic and Prognostic Usefulness of the Expression of B7-H3 Surface Molecule. Histopathology (2008) 53(1):73–80. doi: 10.1111/j.1365-2559.2008.03070.x
151. Whiteway SL, Harris PS, Venkataraman S, Alimova I, Birks DK, Donson AM, et al. Inhibition of Cyclin-Dependent Kinase 6 Suppresses Cell Proliferation and Enhances Radiation Sensitivity in Medulloblastoma Cells. J Neurooncol (2013) 111(2):113–21. doi: 10.1007/s11060-012-1000-7
152. Kanchan RK, Perumal N, Atri P, Chirravuri Venkata R, Thapa I, Klinkebiel DL, et al. MiR-1253 Exerts Tumor-Suppressive Effects in Medulloblastoma via Inhibition of CDK6 and CD276 (B7-H3). Brain Pathol (2020) 30(4):732–45. doi: 10.1111/bpa.12829
153. Ingebrigtsen VA, Boye K, Nesland JM, Nesbakken A, Flatmark K, Fodstad Ø. B7-H3 Expression in Colorectal Cancer: Associations With Clinicopathological Parameters and Patient Outcome. BMC Cancer (2014) 14:602. doi: 10.1186/1471-2407-14-602
154. Xu H, Cheung IY, Guo HF, Cheung NK. MicroRNA miR-29 Modulates Expression of Immunoinhibitory Molecule B7-H3: Potential Implications for Immune Based Therapy of Human Solid Tumors. Cancer Res (2009) 69(15):6275–81. doi: 10.1158/0008-5472.Can-08-4517
155. Zhao J, Lei T, Xu C, Li H, Ma W, Yang Y, et al. MicroRNA-187, Down-Regulated in Clear Cell Renal Cell Carcinoma and Associated With Lower Survival, Inhibits Cell Growth and Migration Though Targeting B7-H3. Biochem Biophys Res Commun (2013) 438(2):439–44. doi: 10.1016/j.bbrc.2013.07.095
156. Wang L, Zhang GC, Kang FB, Zhang L, Zhang YZ. Hsa_Circ0021347 as a Potential Target Regulated by B7-H3 in Modulating the Malignant Characteristics of Osteosarcoma. BioMed Res Int (2019) 2019:9301989. doi: 10.1155/2019/9301989
157. He L, Li Z. B7-H3 and Its Role in Bone Cancers. Pathol Res Pract (2019) 215(6):152420. doi: 10.1016/j.prp.2019.04.012
158. Ni L, Dong C. New B7 Family Checkpoints in Human Cancers. Mol Cancer Ther (2017) 16(7):1203–11. doi: 10.1158/1535-7163.Mct-16-0761
Keywords: miRNA, circRNA, lncRNA, cancer, immune checkpoints
Citation: Jiang Y, Zhao L, Wu Y, Deng S, Cao P, Lei X and Yang X (2022) The Role of NcRNAs to Regulate Immune Checkpoints in Cancer. Front. Immunol. 13:853480. doi: 10.3389/fimmu.2022.853480
Received: 12 January 2022; Accepted: 07 March 2022;
Published: 06 April 2022.
Edited by:
Haoyi Wang, Institute of Zoology (CAS), ChinaReviewed by:
Sandra Cascio, University of Pittsburgh, United StatesHaicheng Tang, Fudan University, China
Copyright © 2022 Jiang, Zhao, Wu, Deng, Cao, Lei and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyan Yang, yyangxiaoyan@163.com
 Yicun Jiang
Yicun Jiang Leilei Zhao1
Leilei Zhao1 Pu Cao
Pu Cao Xiaoyan Yang
Xiaoyan Yang