- 1Faculty of Biomedical Sciences, Epatocentro Ticino and Università della Svizzera Italiana, Lugano, Switzerland
- 2MowatLabs, Faculty of Life Sciences and Medicine, King’s College London, King’s College Hospital, London, United Kingdom
Genetic susceptibility to autoimmune liver diseases is conferred mainly by polymorphisms of genes encoding for the human leukocyte antigens (HLA). The strongest predisposition to autoimmune hepatitis type 1 (AIH-1) is linked to the allele DRB1*03:01, possession of which is associated with earlier disease onset and more severe course. In populations where this allele is very rare, such as in Asia, and in DRB1*03-negative patients, risk of AIH-1 is conferred by DRB1*04, which is associated with later disease onset and milder phenotype. AIH type 2 (AIH-2) is associated with DRB1*07. The pediatric condition referred to as autoimmune sclerosing cholangitis (ASC), is associated with the DRB1*13 in populations of Northern European ancestry. DRB1*1501 is protective from AIH-1, AIH-2 and ASC in Northern European populations. Possession of the DRB1*08 allele is associated with an increased risk of primary biliary cholangitis (PBC) across different populations. DRB1*03:01 and B*08:01 confer susceptibility to primary sclerosing cholangitis (PSC), as well as DRB1*13 and DRB1*15 in Europe. The hepatic blood supply is largely derived from the splanchnic circulation, suggesting a pathophysiological role of the gut microbiome. AIH appears to be associated with dysbiosis, increased gut permeability, and translocation of intestinal microbial products into the circulation; molecular mimicry between microbial and host antigens may trigger an autoaggressive response in genetically-predisposed individuals. In PBC an altered enteric microbiome may affect intestinal motility, immunological function and bile secretion. Patients with PSC have a gut microbial profile different from health as well as from patients with inflammatory bowel disease without PSC.
Introduction
Autoimmune liver diseases, including autoimmune hepatitis (AIH), primary biliary cholangitis (PBC) and primary sclerosing cholangitis (PSC) are believed to be triggered by as yet poorly characterized environmental factors that lead to loss of tolerance to self-antigens in genetically predisposed individuals. Among genetic factors, the most relevant lie within the human leukocyte antigen (HLA) region on chromosome 6. Among environmental factors, infections, drug/toxins and microbiome have been suggested to be involved. A recent animal model of AIH, using transgenic mice on the non-obese diabetic (NOD) background carrying human HLA-DR3 and immunized with a DNA plasmid coding for a fusion protein of human CYP2D6/FTCD – the autoantigens targeted in AIH type 2 -, showed
that not only did the human HLA-DR3 predispose the animals to AIH, but also influenced the microbiome composition compared to the HLA-DR3 negative controls (1), suggesting a direct link between HLA and microbiome. The present short review recapitulates current knowledge on HLA associations and microbiome changes in autoimmune liver disease.
The human leukocyte antigen
HLA are proteins expressed on the surface of a variety of cell types involved in antigen recognition by T cells. HLA molecules physiologically create a groove which embeds a short antigenic peptide to be presented to the T cell receptor. The genes encoding for the HLA proteins, located on the short arm of chromosome 6, are highly polymorphic, ensuring that different individuals are able to recognize and respond to a wide range of different antigenic peptides. Genetic predisposition to autoimmune diseases is often conferred by HLA alleles, and autoimmune liver diseases are no exception. Class I HLA proteins, expressed on all nucleated cells and recognized by CD8 T cells, display peptides derived from cytosolic proteins; class II HLA molecules, expressed on B cells, dendritic cells and macrophages, and recognized by CD4 T cells, display peptides derived from extracellular proteins, upon endocytosis. The HLA genes are characterized also by linkage disequilibrium extending over a large number of genes, meaning that certain alleles are inherited together with higher frequency than expected by random recombination during meiosis. This feature may make it difficult to assess the impact of a specific allele on the pathophysiology of diseases associated with HLA alleles. Interestingly, the so called 8.1 ancestral genotype is a very long haplotype containing alleles predisposing to AIH and PSC, as well as to a host of autoimmune diseases in Caucasian populations, including myositis syndromes, Sjögren syndrome, myasthenia gravis, and celiac disease (2, 3). The clinical observation of the frequent coexistence of hepatic and extrahepatic autoimmunity is possibly linked to a shared genetic predisposition involving different autoantigens.
Autoimmune hepatitis
AIH is a rare chronic inflammatory liver disease affecting all ages, characterized by positive autoantibodies, elevated immunoglobulin G (IgG) serum levels, interface hepatitis at liver histology, and swift response to immunosuppressive treatment (4). Some 10-20% of AIH patients respond unsatisfactorily to pharmacologic treatment and progress to liver failure, requiring liver transplantation (5). AIH is subdivided into type 1 (AIH-1) and type 2 (AIH-2) according to autoimmune serology, AIH-1 being associated with positive anti-nuclear (ANA) and/or anti-smooth muscle (SMA) antibodies, and AIH-2 being associated with anti-liver kidney microsomal (LKM) and/or anti-liver cytosol type 1 (LC-1) antibodies (5). While AIH-1 affects all ages, AIH-2 affects mainly children and adolescents. Autoimmune sclerosing cholangitis (ASC) is a pediatric condition defined by AIH, almost universally AIH-1, coexisting at diagnosis with abnormal cholangiogram (6). It reportedly affects 40-50% of AIH-1 pediatric patients, and has a more aggressive course, requiring liver transplantation more frequently than classical AIH (6, 7).
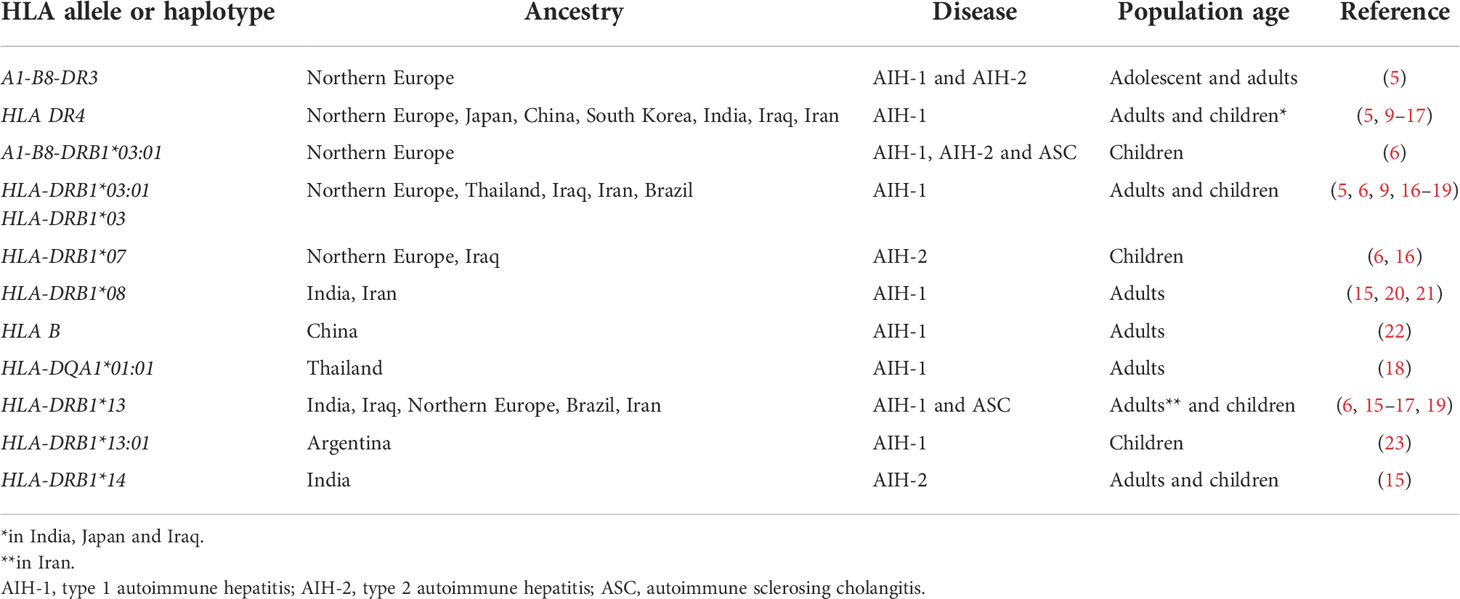
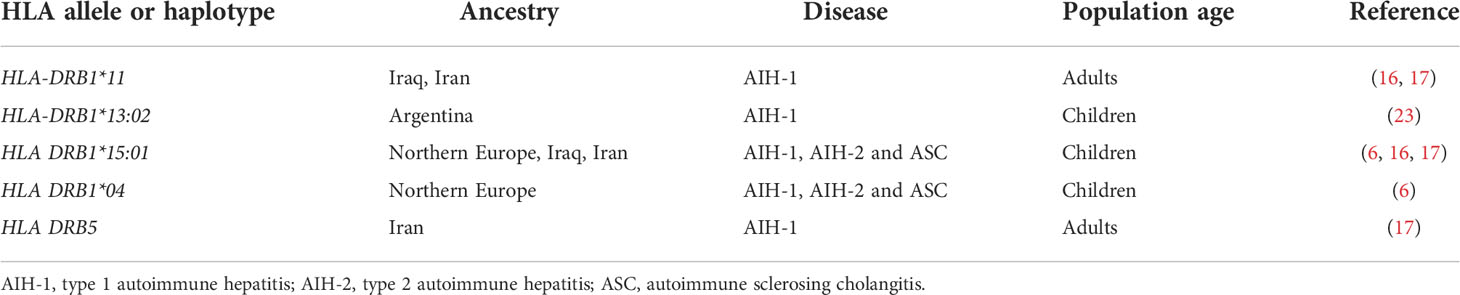
Although the aetiology of AIH and ASC remains unknown, genetic and environmental risk factors have been described, the latter including exposure to drugs and viruses (5). While non-HLA polymorphisms have been reported to increase the risk of AIH, the strongest genetic predisposition is conferred by possession of certain HLA alleles: the high diagnostic value of the predisposing HLA alleles is underscored by the inclusion of HLA in the revised AIH diagnostic criteria (5, 8). The HLA predisposing role to AIH, recognized already in 1977 by Opelz et al. and in 1980 by Mackay and Tait, was confirmed in 1991 in a landmark study from King’s College Hospital, London, UK, which identified a significantly higher frequency of the haplotype A1-B8-DR3 in a cohort including 96 Northern European AIH adolescent and adult patients (three anti-LKM-positive) as compared to healthy controls (9). Possession of this haplotype was also associated to younger age at diagnosis, more frequent relapse and need for liver transplantation (9) (Table 1). In DR3-negative subjects, AIH susceptibility was conferred by HLA DR4, which was associated with later disease onset and milder phenotype. Recently, a large paediatric study from the same centre confirmed the association of the A1-B8-DR3 haplotype with all types of paediatric autoimmune liver diseases, DR3 homozygosity conferring the highest risk, being also associated with fibrosis at disease onset (22). Possession of the HLA-DRB1*03:01 allele and of the A1-B8-DRB1*0301 haplotype is associated with treatment failure and increased risk of cirrhosis, suggesting a more aggressive phenotype in DRB1*03-positive patients (10). Interestingly, this haplotype is linked also to a polymorphism of the TNF-A gene, which leads to a higher production of tumour necrosis factor alpha (TNF-α) (10). HLA-DRB1*03 and DRB1*04 encode the amino acid sequences LLEQKR and LLEQRR at positions 67-72 of the HLA β chain (11). HLA-DRB1*1501, which is protective from AIH-1, AIH-2 and ASC in Northern European populations, encodes alanine at position 71, implying that the amino acid at this position is essential to confer susceptibility (22) (Table 2).
The strong association of AIH-1 with HLA polymorphisms has been confirmed by the two genome-wide association studies (GWAS) performed so far in AIH, both including only AIH-1 adult patients (12, 13). The first GWAS, carried out in a Northern European population, identified the class II HLA-DRB1*0301 as the allele with the strongest association with AIH-1, and HLA-DRB1*0401 as the second most strongly AIH-1- associated allele. The second GWAS study, carried out in a Chinese population, identified a single nucletotide polymorphism in the class I HLA-B locus as the strongest association with AIH-1. The study also identified two non-HLA loci associated with AIH-1, i.e. the CD28-CTLA4-ICOS and the SYNPR loci, the first coding for proteins involved in T cell activation, and the latter coding for a protein expressed on synaptic membranes (13).
There are geographical and ethnic differences in HLA allele frequencies, mirrored by differences in HLA associated to AIH in different populations. In Japan, China, South Korea and Taiwan, adult AIH-1 is associated with DRB1*04, DRB1*03 being very rare in the general population, thus potentially explaining the different AIH phenotype in East Asia, which is characterised by later onset and milder disease (14–16, 18, 23) (Table 1). However, in Thailand AIH-1 is associated with HLA-DRB1*03:01, and HLA-DQA1*01:01 (19). In India, a study including both children and adults, identified DRB1*04 and DRB1*08 as risk alleles for AIH-1, and HLA-DRB1*04 as risk factor for paediatric AIH, in contrast to its protective role in paediatric Northern European populations, and in line with Japanese data (16, 20). Similarly, a small paediatric study from Iraq, not distinguishing between AIH and ASC, reported DRB1*03, DRB1*04 and DRB1*13 being all associated to AIH-1 (17). DRB1*13 was also found to be associated with paediatric AIH-1 in the above-mentioned study from India, which also found an association of AIH-2 with HLA-DRB1*14 (20). Interestingly, a large study from Argentina found that DRB1*13:01 confers susceptibility for paediatric AIH-1, whereas DRB1*13:02, which differs only in one amino acid, confers protection (24) (Table 1).
Genetic studies in AIH-2 have been limited by the rarity of the disease. The largest cohort investigated so far is the above-mentioned study by the King’s College Hospital including only children of European ancestry, which reported an association of AIH-2 with HLA-DRB1*07, whereas HLA-DRB1*15 is protective, as it is for AIH-1 and ASC (22). These results have been confirmed by a smaller study from Iraq (17). The predisposing role of DRB1*07 for AIH-2 has been shown also in Brazil (25). The literature on genetic associations of ASC is also scant, since the differentiation of AIH-1 from ASC is not systematically done in paediatric studies. The association of ASC with DRB1*13 reported in the King’s College Hospital study was confirmed by a small study from Brazil (26); this allele was found to be associated to AIH-2 in Iran (27).
A Japanese study reported a role for class I HLA and killer cell immunoglobulin like receptors (KIR) in association in the predisposition to AIH-1 (15). Natural killer (NK) cells express KIR inhibitory receptors on their surface, which recognize self MHC class I molecules, expressed on all nucleated cells: engagement of these inhibitory receptors protects healthy cells from destruction by NK cells. The investigators, besides confirming that HLA-DRB1*04 confers an increased risk of AIH-1 in Japanese adults, found a strong protective role of the combination KIR3DL1/HLA-B Bw4-80Thr, suggesting a role for NK cells in the pathophysiology of AIH-1 (15).
Attempts have been made to investigate the link between HLA alleles and AIH-1 sub-phenotypes, besides the mentioned association of DRB3 with more severe disease. According to a retrospective Japanese study, the frequency of DRB1*04 is not different in adult AIH-1 patients presenting with acute severe disease as compared to those presenting with chronic AIH (14). Similar results have been reported by a small Greek study (28). A large German study reported that absence of HLA B8 and presence of HLA DR7 are associated with an increased risk of AIH-induced acute liver failure (29).
Primary biliary cholangitis
PBC, previously referred to as primary biliary cirrhosis, is an uncommon autoimmune liver disease characterized by inflammation and destruction of the small- and medium-sized intrahepatic bile ducts (21). It affects mainly middle-aged women and is associated with anti-mitochondrial antibody and/or PBC-specific ANA, i.e. ANA displaying a rim-like or a multiple dots nuclear staining pattern on HEp2 cells. Similarly to AIH, despite unknown aetiology, predisposing genetic background and environmental triggers have been reported (21, 30). The important predisposition conferred by genetics is reflected by the high PBC concordance rate of 63% in identical twins, being the highest among a host of autoimmune diseases (31). The unique genealogic database of Iceland offered the opportunity to investigate the familial risk of PBC: Ornolfsson et al. not only did confirm an increased risk in first-degree relatives, but also showed that the risk is increased up to fifth-degree relatives (32). Owing to the strong autoimmune features of PBC, HLA polymorphisms have been investigated, leading to the demonstration that DRB1*08 confers predisposition across different populations (33, 34). In a study including a population from the UK and one from Northern Italy, DRB1*08:01, was associated with increased PBC risk, whereas DRB1*13 was protective; DRB1*11 was protective only in the Italian population (35, 36). In China and Japan, the strongest predisposition has been linked to possession of the DRB1*08:03, which is only one amino acid different from DRB1*08:01 and may have a similar role in antigen presentation (37–39). DRB1*11 (in Italy and Japan but not in the UK) and DRB1*13 (in the UK, Northern Italy and Japan) are protective (35, 38, 40). In Japan, HLA-DQB1*06:04 and HLA-DQB1*03:01 have been reported to be protective, the latter being protective also in China, Europe and North America (37, 39, 41, 42). Due to linkage disequilibrium with HLA-DRB1*08:03, DQA1*01:03 and DQB1*06:01 have also been associated to PBC in Japan and China (38, 39). A distinctive HLA association with the haplotype DRB1*0301-DQB1*02:01 has been reported in Sardinia, known for its genetic isolation (43). Associations have been reported also for HLA DP alleles, including HLA DPB1*03:01 in a German study and DPB1*17:01 in a Chinese study (37, 44).
The first GWAS in PBC published in 2009 and performed in a Nort American population, identified HLA-DQB1 has having the strongest association (45). All subsequent GWAS confirmed the prominent association of HLA with PBC (42, 46–54).
Primary sclerosing cholangitis
PSC is a rare cholestatic liver disease leading to fibrosis and strictures of the bile ducts, associated with inflammatory bowel disease (IBD) in 80% of patients of Northern European origin, and showing a male preponderance (55). IBD comorbidity is lower in Southern European and Asiatic subjects (55). Affected patients are at increased risk of cholangiocarcinoma, colorectal cancer, gallbladder carcinoma and hepatocellular carcinoma (56).
Although the cause of PSC remains unknown, there is a genetic predisposition, mirrored by a nearly 10-times higher disease risk in first-degree relatives of affected patients (55). Similarly to AIH and PBC, the strongest genetic predisposition is conferred by HLA alleles, i.e. DRB1*03:01 and B*08:01, both being part of the above-mentioned 8.1 ancestral phenotype, which is associated with a host of autoimmune diseases, including AIH and ASC (2, 57–59). A European study, later confirmed by a study including European Americans, Hispanics and African Americans, identified also DRB1*13 as a risk allele, which is included in the haplotype HLA-DRB1*13:01-DQA1*01:03-DQB1*06:03 (60, 61). Since HLA-DRB1*03:01 is also associated to AIH and ASC, and HLA-DRB1*13 is also associated to ASC, these alleles predispose to AIH, ASC and PSC: whether PSC represents a late stage phenotype of ASC, remains an open question; of note, both diseases lack female preponderance (62). However, the HLA-DRB1*15:01, reported to represent a risk allele in PSC (60), was found to be protective to all forms of paediatric autoimmune diseases in an ethnically similar population, suggesting that HLA polymorphisms are only a component of the complex etiopathogenesis of AIH, ASC and PSC.
The PSC-associated haplotypes A*01-C*07-B*08-DRB1*0301-DQB1*0201 and A*03-C*07-B*07 -DRB1*1501-DQB1*0602 share class I antigens with the same binding properties to KIR: these haplotypes do not carry the HLA-Bw4 and HLA-C2 alleles, which are ligands for the inhibitory KIRs 3DL1 and 2DL1 (63). This observation suggests a role for NK cells in the pathophysiology of PSC (57).
The GWAS studies carried out so far in PSC confirm that HLA has the strongest association with PSC (55, 56). This observation supports the notion that PSC is an autoimmune disease, despite the paradox of lack of response to immunosuppression, a feature shared with the other cholestatic autoimmune liver disease, namely PBC.
Main HLA alleles and haplotypes predisposing to or protecting from PBC and PSC are summarized in Table 3.

Table 3 Main HLA alleles and haplotypes predisposing to or protecting from primary biliary cholangitis and primary sclerosing cholangitis.
Potential treatment implications
Though the predisposition conferred by HLA points towards a prominent pathophysiological role of the adaptive immune system in autoimmune liver diseases, the target antigens of the autoimmune attack in PSC, AIH-1 and ASC remain largely unknown. A precise knowledge of the antigenic peptides binding to the HLA proteins of an individual patient would pave the way to peptide-based therapies, by using modified peptides which block autoreactive T cells upon binding to the HLA groove, forming an HLA peptide inhibitory complex. Therefore, future research should focus on identifying autoantigenic proteins and their immunodominant epitopes.
Whether HLA can be included in an interactome-based approach to identify new drugs needs to be investigated (54, 64).
Gut microbiome
The human microbiome has been implicated in the occurrence of several immune-mediated diseases, ranging from rheumatoid arthritis, type 1 diabetes, IBD, and multiple sclerosis (65). Intestinal microbiome may be involved also in the pathogenesis of autoimmune liver disease (66, 67). The blood supply to the liver derives in large amount from the splanchnic circulation, making it exposed to bacteria and bacterial products from the intestinal microbiome. Alterations in the intestinal microbiome might lead to liver damage.
Autoimmune hepatitis
Changes in the intestinal microbiome have been documented in an experimental model of AIH (1) employing HLA-DR3 transgenic mice on the non-obese diabetic (NOD) background. AIH was induced by immunization with a DNA plasmid coding for a fusion protein of human CYP2D6/FTCD, the autoantigens targeted in AIH-2. HLA-DR3, as mentioned above, is strongly linked to AIH. Not only the HLA-DR3 positive mice that were immunized did develop chronic liver injury recapitulating AIH, but they also had a different microbiome composition compared to the HLA-DR3 negative mice. Mice that developed AIH displayed a reduced diversity of their microbiota. Another experimental model using Tet2ΔDVAV mice, which are deficient in the epigenetic regulator Tet methylcytosine dioxygenase 2 (TET2), shows a key role of microbiome and cytotoxic T lymphocytes in inducing a liver disease closely resembling AIH (68). Moreover, a compound of 15 probiotics has been reported to decrease hepatic inflammation, serum transaminase levels, Th1 and Th17 cells, and increase the number of regulatory T cells in a murine model of AIH, while protecting intestinal barrier integrity, blocking lipopolysaccharide (LPS) translocation, inhibiting the toll-like receptor 4/nuclear factor κB (TLR4/NF-κB) pathway activation and the production of inflammatory cytokines in both liver and ileum (69).
Also in humans, AIH appears to be associated with dysbiosis, increased gut permeability, and translocation of intestinal microbial products into the systemic circulation (70). Changes in the microbiome in association to a leakier mucosal barrier might permit translocation of bacteria and their products promoting an inflammatory response. Moreover, molecular mimicry between microbial and host antigens may trigger or intensify an autoaggressive response in genetically-predisposed individuals (71, 72).
Zona occludens 1 and occludin, structural proteins that function in the binding of intestinal epithelial cells, are decreased in patients with AIH compared to health (70), and this decrease worsens with progression of liver disease. Moreover, AIH patients display a reduced number of anaerobes (bifidobacterium and lactobacillus species) within the enteric microbiome. The concept of bacterial translocation is supported by the observed increase in plasma levels of lipo-polysaccharide (LPS) in AIH (70), which worsens with more advanced liver damage. Plasma LPS produced from bacterial translocation can reach the liver through the portal vein. LPS binds to toll-like receptors (TLR) on liver cells, including hepatocytes, stellate cells, Kupffer cells, and sinusoidal epithelial cells, leading to their activation and resulting in an inflammatory milieu predisposing to autoimmunity and fibrosis. The bacterial colonization may also favour the secretion of IL-10 by regulatory T cells (73, 74).
Breech of the intestinal mucosal barrier allows migration of gut-derived bacteria and bacterial products from the intestinal lumen to extra-intestinal sites including the liver. Once the translocated bacterial products, including LPS and unmethylated cytosine–phosphate–guanine (CpG), potent stimulators of the innate immune system, reach the liver they activate TLRs and non-obese diabetes (NOD)-like receptors (NLRs). Migration of activated T lymphocytes from the gut to the liver, including those expressing gut-specific homing receptor alpha4beta7-integrin, has also been implicated in the pathogenesis of autoimmune liver damage within a ‘gut/liver axis’ framework (75), though they can also be found in other forms of liver disease (76).
Primary biliary cholangitis
In PBC an altered enteric microbiome may affect intestinal motility, immunological function and bile secretion (77–79). PBC patients have fewer Acidobacteria, Lachnobacterium, Bacteroides, and Ruminococcus species than healthy individuals and a higher number of opportunistic microbes, such as Y-Proteobacteria, Enterobacteriaceae, Neisseriaceae, Spirochaetaceae, Veillonella, Streptococcus, Klebsiella, and Actinol species. In addition, PBC patients have elevated plasma levels of a variety of cytokines including IL-1β, IL-6,IL-8, IL-18, IL-16, IP-10, MIG, IL-2RA, TNF-α, and macrophage migration inhibitory factor; the lower the levels of microbes normally present in health the higher the levels of inflammatory cytokines (80).
These findings suggest that alterations within the microbiome in PBC have an impact on immune function. PBC has also been associated with abnormal accumulation of LPS in biliary epithelial cells (81, 82), which might activate host immune response. Polymorphisms in the gene coding for TLR 4, which binds to LPS, have been demonstrated in PBC and might favour LPS accumulation in biliary epithelial cells (BEC) triggering host immune responses, further supporting a link between microbial products, the immune system, and PBC (83–85). In addition to LPS, other intestinal microbial components, such as flagellin and cytosine-phosphorothioate-guanine oligonucleotide, concur in eliciting BEC-destructive immune responses (65).
Several studies have investigated the role of infectious agents as triggers of the immune attack on BEC in PBC (86). Anti-mitochondrial antibodies exhibit cross-reactivity with antigens of Escherichia coli and Novosphingobium aromaticivorans (87, 88). Escherichia coli or N. aromaticivorans infection might provoke, in genetically susceptible individuals, the production of anti-mitochondrial antibodies that target pyruvate dehydrogenase complex-E2 (PDC-E2) through a process of molecular mimicry (83, 87, 89–92). This theory is supported by an animal model where mice inoculated with N. Aromaticivorans develop PBC-like disease (93), and by the observation that PDC-E2 shares sequence homologies with intestinal Escherichia coli (83, 87, 89–92). Recently, it has been shown that exposure to Escherichia coli elicits specific antibody to Escherichia coli PDC-E2 that leads to epitope spreading and production of the classical human anti-PDC-E2 autoantibody, possibly being the first step to development of human PBC (30).
In addition to Escherichia coli, Klebsiella species have been implicated in the development of PBC through a mechanism of molecular mimicry (94).
Primary sclerosing cholangitis
The frequent association of PSC with IBD strengthen the notion of a link between the gut microbiome and the liver. Owing to intestinal inflammation allowing translocation of enteric pathogens through the mucosal barrier, enteric microbial components may reach the liver through the portal circulation and provoke damage to cholangiocytes (95). Alternatively, gut activated T cells may home on to the liver and trigger immune-mediated damage (75, 96). However, as mentioned above, presence of gut-derived activated T cells is not confined to PSC and other autoimmune liver diseases (76).
Patients with PSC have a gut microbial profile different from health as well as from patients with IBD without PSC (97–99). PSC patients have a reduction of intestinal bacterial diversity, with an increased presence of Veillonella, Rothia, Enterococcus, Streptococcus, Clostridium, Lactobacillus, Fusobacterium and Haemophilus species. In particular, Enterococcus, Lactobacillus, and Fusobacterium species are associated with PSC independently of IBD, probiotic use, cirrhosis, liver transplantation, ursodeoxycholic acid or antibiotic treatment (97–99).
The use of antibiotics, including vancomycin, metronidazole, tetracycline, sulfasalazine, azithromycin, and minocycline has been reported to improve laboratory indices in PSC, but not the progression of liver disease (100). The improvement observed with the use of antibiotics may derive from modifications in the microbiome that results in a decreased production of inflammatory molecules.
Faecal microbiota transplantation (FMT), which alters the host microbiome, has been attempted as a therapeutic measure for PSC in a small pilot study. Microbial diversity increased and the alkaline phosphatase levels decreased in treated patients, though it is unclear whether this was associated with a clinical benefit (101). In this pilot study FMT was applied through the colonic route, but a small bowel route might be more appropriate based on animal studies showing that small bowel, but not colonic, dysbiosis is associated with hepatobiliary inflammation, and that small bowel bacterial overgrowth results in a PSC-like disease (102). Larger clinical trials are needed to investigate the role of FMT in PSC.
Both PSC and, to a lesser extent, AIH are associated with positivity for the autoantibody referred to as atypical peripheral anti-neutrophil cytoplasmic antibody (pANCA), also known as peripheral anti-nuclear neutrophil antibody (pANNA). This antibody is reportedly directed to Beta-tubulin and cross-reacts with a bacterial antigen, FtsZ (103), an evolutionary precursor protein of Beta-tubulin present in almost all intestinal bacteria, suggesting that molecular mimicry to bacterial products may also play a role in the development of PSC and AIH.
Conclusions
The strongest genetic predisposition to all autoimmune liver diseases is conferred by HLA alleles, suggesting a preponderant role of adaptive immunity and particularly of T cells in their pathophysiology. Therefore, knowledge of the autoantigens recognized by T cells is key to advance our understanding of the immunopathogenesis of these diseases. Even in the diseases where the autoantigens are known (AIH-2 and PBC), little is known about the T cells autoantigenic peptides, knowledge of which could pave the way to novel therapeutic approaches. The role of the intestinal microbiome in shaping the phenotype of autoimmune liver disease is currently under investigation. Though most microbiome studies were conducted in wealthy countries in North America and Europe, skewing our understanding of human-microbe interactions, preliminary findings highlight the potential importance of its manipulation in therapeutic interventions.
Author contributions
BTB-P drafting the first part of the manuscript, critical review and approval of final version. GM-V, DV: drafting the second part of the manuscript, critical review and approval of final version. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Yuksel M, Wang Y, Tai N, Peng J, Guo J, Beland K, et al. A novel “humanized mouse” model for autoimmune hepatitis and the association of gut microbiota with liver inflammation. Hepatology (2015) 62:1536–50. doi: 10.1002/hep.27998
2. Candore G, Lio D, Colonna Romano G, Caruso C. Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: Effect of multiple gene interactions. Autoimmun Rev (2002) 1:29–35. doi: 10.1016/s1568-9972(01)00004-0
3. Gambino CM, Aiello A, Accardi G, Caruso C, Candore G. Autoimmune diseases and 8.1 ancestral haplotype: An update. HLA (2018) 92:137–43. doi: 10.1111/tan.13305
4. Terziroli Beretta-Piccoli B, Mieli-Vergani G, Vergani D. Autoimmmune hepatitis. Cell Mol Immunol (2021) 19(2): 158–76. doi: 10.1038/s41423-021-00768-8
5. Mieli-Vergani G, Vergani D, Czaja AJ, Manns MP, Krawitt EL, Vierling JM, et al. Autoimmune hepatitis. Nat Rev Dis Primers (2018) 4:18017. doi: 10.1038/nrdp.2018.17
6. Gregorio GV, Portmann B, Karani J, Harrison P, Donaldson PT, Vergani D, et al. Autoimmune hepatitis/sclerosing cholangitis overlap syndrome in childhood: a 16-year prospective study. Hepatology (2001) 33:544–53. doi: 10.1053/jhep.2001.22131
7. Di Giorgio A, Hadzic N, Dhawan A, Deheragoda M, Heneghan MA, Vergani D, et al. Seamless management of juvenile autoimmune liver disease: Long-term medical and social outcome. J Pediatr (2020) 218:121–129.e3. doi: 10.1016/j.jpeds.2019.11.028
8. Alvarez F, Berg PA, Bianchi FB, Bianchi L, Burroughs AK, Cancado EL, et al. International autoimmune hepatitis group report: Review of criteria for diagnosis of autoimmune hepatitis. J Hepatol (1999) 31:929–38. doi: 10.1016/S0168-8278(99)80297-9
9. Donaldson PT, Doherty DG, Hayllar KM, McFarlane IG, Johnson PJ, Williams R. Susceptibility to autoimmune chronic active hepatitis: Human leukocyte antigens DR4 and A1-B8-DR3 are independent risk factors. Hepatology (1991) 13:701–6. doi: 10.1002/hep.1840130415
10. Czaja AJ, Cookson S, Constantini PK, Clare M, Underhill JA, Donaldson PT. Cytokine polymorphisms associated with clinical features and treatment outcome in type 1 autoimmune hepatitis. Gastroenterology (1999) 117:645–52. doi: 10.1016/s0016-5085(99)70458-0
11. Doherty DG, Donaldson PT, Underhill JA, Farrant JM, Duthie A, Mieli-Vergani G, et al. Allelic sequence variation in the HLA class II genes and proteins in patients with autoimmune hepatitis. Hepatology (1994) 19:609–15. doi: 10.1002/hep.1840190311
12. de Boer YS, van Gerven NMF, Zwiers A, Verwer BJ, van Hoek B, van Erpecum KJ, et al. Genome-wide association study identifies variants associated with autoimmune hepatitis type 1. Gastroenterology (2014) 147:443–452.e5. doi: 10.1053/j.gastro.2014.04.022
13. Li Y, Sun Y, Liu Y, Wang B, Li J, Wang H, et al. Genome-wide meta-analysis identifies novel susceptibility loci for autoimmune hepatitis type 1. Hepatology (2022)76(3): 564–75. doi: 10.1002/hep.32417
14. Abe M, Onji M, Kawai–Ninomiya K, Michitaka K, Matsuura B, Hiasa Y, et al. Clinicopathologic features of the severe form of acute type 1 autoimmune hepatitis. Clin Gastroenterol Hepatol (2007) 5:255–8. doi: 10.1016/j.cgh.2006.10.011
15. Umemura T, Joshita S, Saito H, Yoshizawa K, Norman GL, Tanaka E, et al. KIR/HLA genotypes confer susceptibility and progression in patients with autoimmune hepatitis. JHEP Rep (2019) 1:353–60. doi: 10.1016/j.jhepr.2019.09.003
16. Furumoto Y, Asano T, Sugita T, Abe H, Chuganji Y, Fujiki K, et al. Evaluation of the role of HLA-DR antigens in Japanese type 1 autoimmune hepatitis. BMC Gastroenterol (2015) 15:144. doi: 10.1186/s12876-015-0360-9
17. Ibraheem MF, Ahmed SJ. The association between juvenile autoimmune hepatitis and HLA-DRB1 alleles: Iraqi tertiary center experience. Clin Exp Hepatol (2021) 7:178–82. doi: 10.5114/ceh.2021.106865
18. Katsumi T, Ueno Y. Epidemiology and surveillance of autoimmune hepatitis in Asia. Liver Int (2022)42(9): 2015–2022. doi: 10.1111/liv.15155
19. Tanwandee T, Wanichapol S, Vejbaesya S, Chainuvati S, Chotiyaputta W. Association between HLA class II alleles and autoimmune hepatitis type 1 in Thai patients. J Med Assoc Thai (2006) 89 Suppl 5:S73–78.
20. Kaur N, Minz RW, Anand S, Saikia B, Aggarwal R, Das A, et al. HLA DRB1 alleles discriminate the manifestation of autoimmune hepatitis as type 1 or type 2 in north Indian population. J Clin Exp Hepatol (2014) 4:14–8. doi: 10.1016/j.jceh.2013.12.002
21. Terziroli Beretta-Piccoli B, Mieli-Vergani G, Vergani D, Vierling JM, Adams D, Alpini G, et al. The challenges of primary biliary cholangitis: What is new and what needs to be done. J Autoimmun (2019), 105 102328. doi: 10.1016/j.jaut.2019.102328
22. Ma Y, Su H, Yuksel M, Longhi MS, McPhail MJ, Wang P, et al. Human leukocyte antigen profile predicts severity of autoimmune liver disease in children of european ancestry. Hepatology (2021) 74:2032–46. doi: 10.1002/hep.31893
23. Zhang H-P, Liu Y-M, Li Z, Ma Y-X, Li L-J, Zhao D-T, et al. Clinical characteristics and HLA genotypes in Chinese patients with anti-SLA/LP-positive autoimmune hepatitis. Ann Transl Med (2021) 9:153. doi: 10.21037/atm-20-8036
24. Pando M, Larriba J, Fernandez GC, Fainboim H, Ciocca M, Ramonet M, et al. Pediatric and adult forms of type I autoimmune hepatitis in Argentina: Evidence for differential genetic predisposition. Hepatology (1999) 30:1374–80. doi: 10.1002/hep.510300611
25. Bittencourt PL, Goldberg AC, Cançado EL, Porta G, Laudanna AA, Kalil J. Different HLA profiles confer susceptibility to autoimmune hepatitis type 1 and 2. Am J Gastroenterol (1998) 93:1394–5. doi: 10.1111/j.1572-0241.1998.1394a.x
26. Nunes MEG, Rosa DV, Fagundes EDT, Ferreira AR, de MDM, Ferri Liu PM. Hla-Drb1 gene polymorphisms in pediatric patients with type 1 autoimmune hepatitis and type 1 autoimmune hepatitis overlap syndrome with autoimmune cholangitis. Arq Gastroenterol (2019) 56:146–50. doi: 10.1590/S0004-2803.201900000-29
27. Baharlou R, Faghihi-Kashani A, Faraji F, Najafi-Samei M, Setareh M, Zamani F, et al. HLA-DRB1 alleles of susceptibility and protection in iranians with autoimmune hepatitis. Hum Immunol (2016) 77:330–5. doi: 10.1016/j.humimm.2016.01.007
28. Zachou K, Arvaniti P, Azariadis K, Lygoura V, Gatselis NK, Lyberopoulou A, et al. Prompt initiation of high-dose i.v. corticosteroids seems to prevent progression to liver failure in patients with original acute severe autoimmune hepatitis. Hepatol Res (2019) 49:96–104. doi: 10.1111/hepr.13252
29. Anastasiou OE, Dogan-Cavus B, Kucukoglu O, Baba H, Kahraman A, Gerken G, et al. Corticosteroid therapy improves the outcome of autoimmune hepatitis-induced acute liver failure. Digestion (2018) 98:104–11. doi: 10.1159/000487940
30. Yang Y, Choi J, Chen Y, Invernizzi P, Yang G, Zhang W, et al. E. coli and the etiology of human PBC: Antimitochondrial antibodies and spreading determinants. Hepatology (2022) 75:266–79. doi: 10.1002/hep.32172
31. Selmi C, Mayo MJ, Bach N, Ishibashi H, Invernizzi P, Gish RG, et al. Primary biliary cirrhosis in monozygotic and dizygotic twins: Genetics, epigenetics, and environment. Gastroenterology (2004) 127:485–92. doi: 10.1053/j.gastro.2004.05.005
32. Örnolfsson KT, Olafsson S, Bergmann OM, Gershwin ME, Björnsson ES. Using the icelandic genealogical database to define the familial risk of primary biliary cholangitis. Hepatology (2018) 68:166–71. doi: 10.1002/hep.29675
33. Gerussi A, Carbone M, Corpechot C, Schramm C, Asselta R, Invernizzi P. The genetic architecture of primary biliary cholangitis. Eur J Med Genet (2021) 64:104292. doi: 10.1016/j.ejmg.2021.104292
34. Invernizzi P, Selmi C, Poli F, Frison S, Floreani A, Alvaro D, et al. Human leukocyte antigen polymorphisms in italian primary biliary cirrhosis: A multicenter study of 664 patients and 1992 healthy controls. Hepatology (2008) 48:1906–12. doi: 10.1002/hep.22567
35. Donaldson PT, Baragiotta A, Heneghan MA, Floreani A, Venturi C, Underhill JA, et al. HLA class II alleles, genotypes, haplotypes, and amino acids in primary biliary cirrhosis: A large-scale study. Hepatology (2006) 44:667–74. doi: 10.1002/hep.21316
36. Invernizzi P, Ransom M, Raychaudhuri S, Kosoy R, Lleo A, Shigeta R, et al. Classical HLA-DRB1 and DPB1 alleles account for HLA associations with primary biliary cirrhosis. Genes Immun (2012) 13:461–8. doi: 10.1038/gene.2012.17
37. Wang C, Zheng X, Tang R, Han C, Jiang Y, Wu J, et al. Fine mapping of the MHC region identifies major independent variants associated with han Chinese primary biliary cholangitis. J Autoimmun (2020) 107:102372. doi: 10.1016/j.jaut.2019.102372
38. Onishi S, Sakamaki T, Maeda T, Iwamura S, Tomita A, Saibara T, et al. DNA Typing of HLA class II genes; DRB1*0803 increases the susceptibility of Japanese to primary biliary cirrhosis. J Hepatol (1994) 21:1053–60. doi: 10.1016/s0168-8278(05)80617-8
39. Zhao D-T, Liao H-Y, Zhang X, Liu Y-M, Zhao Y, Zhang H-P, et al. Human leucocyte antigen alleles and haplotypes and their associations with antinuclear antibodies features in Chinese patients with primary biliary cirrhosis. Liver Int (2014) 34:220–6. doi: 10.1111/liv.12236
40. Umemura T, Joshita S, Ichijo T, Yoshizawa K, Katsuyama Y, Tanaka E, et al. Human leukocyte antigen class II molecules confer both susceptibility and progression in Japanese patients with primary biliary cirrhosis. Hepatology (2012) 55:506–11. doi: 10.1002/hep.24705
41. Yasunami M, Nakamura H, Tokunaga K, Kawashima M, Nishida N, Hitomi Y, et al. Principal contribution of HLA-DQ alleles, DQB1*06:04 and DQB1*03:01, to disease resistance against primary biliary cholangitis in a Japanese population. Sci Rep (2017) 7:11093. doi: 10.1038/s41598-017-11148-6
42. Juran BD, Hirschfield GM, Invernizzi P, Atkinson EJ, Li Y, Xie G, et al. Immunochip analyses identify a novel risk locus for primary biliary cirrhosis at 13q14, multiple independent associations at four established risk loci and epistasis between 1p31 and 7q32 risk variants. Hum Mol Genet (2012) 21:5209–21. doi: 10.1093/hmg/dds359
43. Clemente MG, Frau F, Bernasconi M, Macis MD, Cicotto L, Pilleri G, et al. Distinctive HLA-II association with primary biliary cholangitis on the island of Sardinia. United Eur Gastroenterol J (2017) 5:527–31. doi: 10.1177/2050640616665030
44. Mella JG, Roschmann E, Maier KP, Volk BA. Association of primary biliary cirrhosis with the allele HLA-DPB1*0301 in a German population. Hepatology (1995) 21:398–402. doi: 10.1002/hep.1840210221
45. Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med (2009) 360:2544–55. doi: 10.1056/NEJMoa0810440
46. Liu X, Invernizzi P, Lu Y, Kosoy R, Lu Y, Bianchi I, et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet (2010) 42:658–60. doi: 10.1038/ng.627
47. Liu JZ, Almarri MA, Gaffney DJ, Mells GF, Jostins L, Cordell HJ, et al. Dense fine-mapping study identifies new susceptibility loci for primary biliary cirrhosis. Nat Genet (2012) 44:1137–41. doi: 10.1038/ng.2395
48. Nakamura M, Nishida N, Kawashima M, Aiba Y, Tanaka A, Yasunami M, et al. Genome-wide association study identifies TNFSF15 and POU2AF1 as susceptibility loci for primary biliary cirrhosis in the Japanese population. Am J Hum Genet (2012) 91:721–8. doi: 10.1016/j.ajhg.2012.08.010
49. Qiu F, Tang R, Zuo X, Shi X, Wei Y, Zheng X, et al. A genome-wide association study identifies six novel risk loci for primary biliary cholangitis. Nat Commun (2017) 8:14828. doi: 10.1038/ncomms14828
50. Mells GF, Floyd JAB, Morley KI, Cordell HJ, Franklin CS, Shin S-Y, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet (2011) 43:329–32. doi: 10.1038/ng.789
51. Cordell HJ, Han Y, Mells GF, Li Y, Hirschfield GM, Greene CS, et al. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun (2015) 6:8019. doi: 10.1038/ncomms9019
52. Paziewska A, Habior A, Rogowska A, Zych W, Goryca K, Karczmarski J, et al. A novel approach to genome-wide association analysis identifies genetic associations with primary biliary cholangitis and primary sclerosing cholangitis in polish patients. BMC Med Genomics (2017) 10:2. doi: 10.1186/s12920-016-0239-9
53. Kawashima M, Hitomi Y, Aiba Y, Nishida N, Kojima K, Kawai Y, et al. Genome-wide association studies identify PRKCB as a novel genetic susceptibility locus for primary biliary cholangitis in the Japanese population. Hum Mol Genet (2017) 26:650–9. doi: 10.1093/hmg/ddw406
54. Cordell HJ, Fryett JJ, Ueno K, Darlay R, Aiba Y, Hitomi Y, et al. An international genome-wide meta-analysis of primary biliary cholangitis: Novel risk loci and candidate drugs. J Hepatol (2021) 75:572–81. doi: 10.1016/j.jhep.2021.04.055
55. Karlsen TH, Folseraas T, Thorburn D, Vesterhus M. Primary sclerosing cholangitis - a comprehensive review. J Hepatol (2017) 67:1298–323. doi: 10.1016/j.jhep.2017.07.022
56. Jiang X, Karlsen TH. Genetics of primary sclerosing cholangitis and pathophysiological implications. Nat Rev Gastroenterol Hepatol (2017) 14:279–95. doi: 10.1038/nrgastro.2016.154
57. Karlsen TH. Genetic epidemiology of primary sclerosing cholangitis. WJG (2007) 13:5421. doi: 10.3748/wjg.v13.i41.5421
58. Schrumpf E, Fausa O, Førre Ø, Dobloug JH, Ritland S, Thorsby E, et al. And immunoregulatory t cells in ulcerative colitis associated with hepatobiliary disease. Scandinavian J Gastroenterol (1982) 17:187–91. doi: 10.3109/00365528209182038
59. Chapman RW, Varghese Z, Gaul R, Patel G, Kokinon N, Sherlock S. Association of primary sclerosing cholangitis with HLA-B8. Gut (1983) 24:38–41. doi: 10.1136/gut.24.1.38
60. Spurkland A, Saarinen S, Boberg KM, Mitchell S, Broome U, Caballeria L, et al. HLA class II haplotypes in primary sclerosing cholangitis patients from five European populations. Tissue Antigens (1999) 53:459–69. doi: 10.1034/j.1399-0039.1999.530502.x
61. Bowlus CL, Li C-S, Karlsen TH, Lie BA, Selmi C. Primary sclerosing cholangitis in genetically diverse populations listed for liver transplantation: Unique clinical and human leukocyte antigen associations. Liver Transpl (2010) 16:1324–30. doi: 10.1002/lt.22161
62. Terziroli Beretta-Piccoli B, Vergani D, Mieli-Vergani G. Autoimmune sclerosing cholangitis: Evidence and open questions. J Autoimmun (2018)95: 15–25. doi: 10.1016/j.jaut.2018.10.008
63. Karlsen TH, Boberg KM, Olsson M, Sun J-Y, Senitzer D, Bergquist A, et al. Particular genetic variants of ligands for natural killer cell receptors may contribute to the HLA associated risk of primary sclerosing cholangitis. J Hepatol (2007) 46:899–906. doi: 10.1016/j.jhep.2007.01.032
64. Guney E, Menche J, Vidal M, Barábasi A-L. Network-based in silico drug efficacy screening. Nat Commun (2016) 7:10331. doi: 10.1038/ncomms10331
65. Czaja AJ. Factoring the intestinal microbiome into the pathogenesis of autoimmune hepatitis. World J Gastroenterol (2016) 22:9257–78. doi: 10.3748/wjg.v22.i42.9257
66. Zheng Y, Ran Y, Zhang H, Wang B, Zhou L. The microbiome in autoimmune liver diseases: Metagenomic and metabolomic changes. Front Physiol (2021) 12:715852. doi: 10.3389/fphys.2021.715852
67. Qian Q, He W, Tang R, Ma X. Implications of gut microbiota in autoimmune liver diseases. Minerva Gastroenterol (Torino) (2021). doi: 10.23736/S2724-5985.21.02860-9
68. Pandey SP, Bender MJ, McPherson AC, Phelps CM, Sanchez LM, Rana M, et al. Tet2 deficiency drives liver microbiome dysbiosis triggering Tc1 cell autoimmune hepatitis. Cell Host Microbe (2022) 30(7): 1003–19. doi: 10.1016/j.chom.2022.05.006
69. Liu Q, Tian H, Kang Y, Tian Y, Li L, Kang X, et al. Probiotics alleviate autoimmune hepatitis in mice through modulation of gut microbiota and intestinal permeability. J Nutr Biochem (2021) 98:108863. doi: 10.1016/j.jnutbio.2021.108863
70. Lin R, Zhou L, Zhang J, Wang B. Abnormal intestinal permeability and microbiota in patients with autoimmune hepatitis. Int J Clin Exp Pathol (2015) 8:5153–60.
71. Sánchez B, Hevia A, González S, Margolles A. Interaction of intestinal microorganisms with the human host in the framework of autoimmune diseases. Front Immunol (2015) 6:594. doi: 10.3389/fimmu.2015.00594
72. Singh R, Bullard J, Kalra M, Assefa S, Kaul AK, Vonfeldt K, et al. Status of bacterial colonization, toll-like receptor expression and nuclear factor-kappa b activation in normal and diseased human livers. Clin Immunol (2011) 138:41–9. doi: 10.1016/j.clim.2010.09.006
73. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA (2010) 107:12204–9. doi: 10.1073/pnas.0909122107
74. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science (2011) 332:974–7. doi: 10.1126/science.1206095
75. Henriksen EKK, Jørgensen KK, Kaveh F, Holm K, Hamm D, Olweus J, et al. Gut and liver T-cells of common clonal origin in primary sclerosing cholangitis-inflammatory bowel disease. J Hepatol (2017) 66:116–22. doi: 10.1016/j.jhep.2016.09.002
76. Graham JJ, Mukherjee S, Yuksel M, Sanabria Mateos R, Si T, Huang Z, et al. Aberrant hepatic trafficking of gut-derived T cells is not specific to primary sclerosing cholangitis. Hepatology (2022) 75:518–30. doi: 10.1002/hep.32193
77. Dyson JK, Hirschfield GM, Adams DH, Beuers U, Mann DA, Lindor KD, et al. Novel therapeutic targets in primary biliary cirrhosis. Nat Rev Gastroenterol Hepatol (2015) 12:147–58. doi: 10.1038/nrgastro.2015.12
78. Selmi C, Bowlus CL, Gershwin ME, Coppel RL. Primary biliary cirrhosis. Lancet (2011) 377:1600–9. doi: 10.1016/S0140-6736(10)61965-4
79. Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol (2013) 58:949–55. doi: 10.1016/j.jhep.2013.01.003
80. Lv L-X, Fang D-Q, Shi D, Chen D-Y, Yan R, Zhu Y-X, et al. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ Microbiol (2016) 18:2272–86. doi: 10.1111/1462-2920.13401
81. Sasatomi K, Noguchi K, Sakisaka S, Sata M, Tanikawa K. Abnormal accumulation of endotoxin in biliary epithelial cells in primary biliary cirrhosis and primary sclerosing cholangitis. J Hepatol (1998) 29:409–16. doi: 10.1016/s0168-8278(98)80058-5
82. Maldonado RF, Sá-Correia I, Valvano MA. Lipopolysaccharide modification in gram-negative bacteria during chronic infection. FEMS Microbiol Rev (2016) 40:480–93. doi: 10.1093/femsre/fuw007
83. Wang A-P, Migita K, Ito M, Takii Y, Daikoku M, Yokoyama T, et al. Hepatic expression of toll-like receptor 4 in primary biliary cirrhosis. J Autoimmun (2005) 25:85–91. doi: 10.1016/j.jaut.2005.05.003
84. Benias PC, Gopal K, Bodenheimer H, Theise ND. Hepatic expression of toll-like receptors 3, 4, and 9 in primary biliary cirrhosis and chronic hepatitis c. Clin Res Hepatol Gastroenterol (2012) 36:448–54. doi: 10.1016/j.clinre.2012.07.001
85. Seki E, Brenner DA. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology (2008) 48:322–35. doi: 10.1002/hep.22306
86. Mattner J. Impact of microbes on the pathogenesis of primary biliary cirrhosis (pbc) and primary sclerosing cholangitis (PSC). Int J Mol Sci (2016) 17:E1864. doi: 10.3390/ijms17111864
87. Bogdanos D-P, Baum H, Grasso A, Okamoto M, Butler P, Ma Y, et al. Microbial mimics are major targets of crossreactivity with human pyruvate dehydrogenase in primary biliary cirrhosis. J Hepatol (2004) 40:31–9. doi: 10.1016/s0168-8278(03)00501-4
88. Olafsson S, Gudjonsson H, Selmi C, Amano K, Invernizzi P, Podda M, et al. Antimitochondrial antibodies and reactivity to n. aromaticivorans proteins in icelandic patients with primary biliary cirrhosis and their relatives. Am J Gastroenterol (2004) 99:2143–6. doi: 10.1111/j.1572-0241.2004.40397.x
89. Kikuchi K, Lian Z-X, Yang G-X, Ansari AA, Ikehara S, Kaplan M, et al. Bacterial CpG induces hyper-IgM production in CD27(+) memory b cells in primary biliary cirrhosis. Gastroenterology (2005) 128:304–12. doi: 10.1053/j.gastro.2004.11.005
90. Mao TK, Lian Z-X, Selmi C, Ichiki Y, Ashwood P, Ansari AA, et al. Altered monocyte responses to defined TLR ligands in patients with primary biliary cirrhosis. Hepatology (2005) 42:802–8. doi: 10.1002/hep.20859
91. Honda Y, Yamagiwa S, Matsuda Y, Takamura M, Ichida T, Aoyagi Y. Altered expression of TLR homolog RP105 on monocytes hypersensitive to LPS in patients with primary biliary cirrhosis. J Hepatol (2007) 47:404–11. doi: 10.1016/j.jhep.2007.03.012
92. Shimoda S, Harada K, Niiro H, Shirabe K, Taketomi A, Maehara Y, et al. Interaction between toll-like receptors and natural killer cells in the destruction of bile ducts in primary biliary cirrhosis. Hepatology (2011) 53:1270–81. doi: 10.1002/hep.24194
93. Mattner J, Savage PB, Leung P, Oertelt SS, Wang V, Trivedi O, et al. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe (2008) 3:304–15. doi: 10.1016/j.chom.2008.03.009
94. Smyk DS, Bogdanos DP, Kriese S, Billinis C, Burroughs AK, Rigopoulou EI. Urinary tract infection as a risk factor for autoimmune liver disease: From bench to bedside. Clin Res Hepatol Gastroenterol (2012) 36:110–21. doi: 10.1016/j.clinre.2011.07.013
95. Trivedi PJ, Adams DH. Gut-liver immunity. J Hepatol (2016) 64:1187–9. doi: 10.1016/j.jhep.2015.12.002
96. Lazaridis KN, LaRusso NF. Primary sclerosing cholangitis. N Engl J Med (2016) 375:1161–70. doi: 10.1056/NEJMra1506330
97. Kummen M, Holm K, Anmarkrud JA, Nygård S, Vesterhus M, Høivik ML, et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut (2017) 66:611–9. doi: 10.1136/gutjnl-2015-310500
98. Sabino J, Vieira-Silva S, Machiels K, Joossens M, Falony G, Ballet V, et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut (2016) 65:1681–9. doi: 10.1136/gutjnl-2015-311004
99. Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, Stehlikova Z, et al. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol (2017) 23:4548–58. doi: 10.3748/wjg.v23.i25.4548
100. Glassner K, Quigley EM, Franco L, Victor DW. Autoimmune liver disease and the enteric microbiome. AIMS Microbiol (2018) 4:334–46. doi: 10.3934/microbiol.2018.2.334
101. Allegretti JR, Kassam Z, Carrellas M, Mullish BH, Marchesi JR, Pechlivanis A, et al. Fecal microbiota transplantation in patients with primary sclerosing cholangitis: a pilot clinical trial. Am J Gastroenterol (2019) 114:1071–9. doi: 10.14309/ajg.0000000000000115
102. Verdier J, Luedde T, Sellge G. Biliary mucosal barrier and microbiome. Viszeralmedizin (2015) 31:156–61. doi: 10.1159/000431071
Keywords: autoimmune hepatitis, AIH, primary biliary cholangitis (PBC), primary sclerosing cholangites (PSC), gut microbiome, human leucocyte antigen (HLA)
Citation: Terziroli Beretta-Piccoli B, Mieli-Vergani G and Vergani D (2022) HLA, gut microbiome and hepatic autoimmunity. Front. Immunol. 13:980768. doi: 10.3389/fimmu.2022.980768
Received: 28 June 2022; Accepted: 25 July 2022;
Published: 18 August 2022.
Edited by:
Nanda Kerkar, University of Rochester, United StatesCopyright © 2022 Terziroli Beretta-Piccoli, Mieli-Vergani and Vergani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benedetta Terziroli Beretta-Piccoli, benedetta.terziroli@hin.ch
 Benedetta Terziroli Beretta-Piccoli
Benedetta Terziroli Beretta-Piccoli Giorgina Mieli-Vergani
Giorgina Mieli-Vergani Diego Vergani
Diego Vergani