- 1Soil Science, Faculty of Agricultural and Environmental Sciences, University of Rostock, Rostock, Germany
- 2Department of Microbiology, Faculty of Biology and Environmental Protection, Nicolaus Copernicus University in Toruń, Toruń, Poland
- 3Department of Crop Production Ecology, Swedish University of Agricultural Sciences, Uppsala, Sweden
- 4Uppsala BioCenter, Department of Forest Mycology and Plant Pathology, Swedish University of Agricultural Sciences, Uppsala, Sweden
The roots of Salix spp. can be colonized by two types of mycorrhizal fungi (ectomycorrhizal and arbuscular) and furthermore by dark-septate endophytes. The fungal root colonization is affected by the plant genotype, soil properties and their interactions. However, the impact of host diversity accomplished by mixing different Salix genotypes within the site on root-associated fungi and P-mobilization in the field is not known. It can be hypothesized that mixing of genotypes with strong eco-physiological differences changes the diversity and abundance of root-associated fungi and P-mobilization in the mycorrhizosphere based on different root characteristics. To test this hypothesis, we have studied the mixture of two fundamentally eco-physiologically different Salix genotypes (S. dasyclados cv. ‘Loden’ and S. schwerinii × S. viminalis cv. ‘Tora’) compared to plots with pure genotypes in a randomized block design in a field experiment in Northern Germany. We assessed the abundance of mycorrhizal colonization, fungal diversity, fine root density in the soil and activities of hydrolytic enzymes involved in P-mobilization in the mycorrhizosphere in autumn and following spring after three vegetation periods. Mycorrhizal and endophytic diversity was low under all Salix treatments with Laccaria tortilis being the dominating ectomyorrhizal fungal species, and Cadophora and Paraphaeosphaeria spp. being the most common endophytic fungi. Interspecific root competition increased richness and root colonization by endophytic fungi (four taxa in the mixture vs. one found in the pure host genotype cultures) more than by ectomycorrhizal fungi and increased the activities of hydrolytic soil enzymes involved in the P-mineralization (acid phosphatase and β-glucosidase) in mixed stands. The data suggest selective promotion of endophytic root colonization and changed competition for nutrients by mixture of Salix genotypes.
Introduction
Mycorrhizal fungi are central to soil fertility and can affect both crop productivity and cropping security (Rooney et al., 2009). Both arbuscular mycorrhizal (AM) and ectomycorrhizal (EM) fungi are known to increase the uptake of nutrients like phosphorus (P) and nitrogen (N) by the host plants, especially in infertile soils. However, their functions and benefits for the host plants might not be equivalent (Jones et al., 1998). For this reason, a change in the proportions of mycorrhizal colonization, or type of mycorrhizal association when the host plant can form dual mycorrhiza, might affect the mycorrhizal benefit of the host plant. The mycorrhizal colonization of plants can be affected by soil nutrient level and water content, site management as well as the diversity of the vegetation composition. An increased host plant diversity is known to influence the nutrient fluxes and the interactions with soil microorganisms (Courty et al., 2011). Endophytic fungi and mycorrhizal fungi can interact in their impact on the plant growth (Rillig et al., 2014). For example, AM fungi were revealed to be able to modulate the impact of endophytic fungi beneficially for the plant growth (Wężowicz et al., 2017).
Although the majority of higher plants form mycorrhizal associations, only a low number is dual mycorrhizal, forming both AM and EM associations (Smith and Read, 2008). Salix belongs to the dual mycorrhizal plant genera, and can also harbor diverse endophytic fungi, which may affect the hosts growth and ecophysiological traits (An et al., 2015). Although Salix spp. were mainly shown to be pre-dominantly EM (Püttsepp et al., 2004; Hrynkiewicz et al., 2010b), the opposite has also been shown with a dominance of AM colonization, e.g., in response to flooding (Lodge, 1989). The ability to change their mycorrhizal association in response to the environmental conditions makes them well-suitable model plants to investigate the changes in the mycorrhizosphere in response to a changed host diversity.
Fast growing Salix genotypes have been cultivated successfully in short rotation coppice (SRC) for a long time, mainly for biomass production for energy purposes in boreal climates (Weih, 2004). They are viewed as a sustainable source of biomass with a positive greenhouse gas balance due to their potential to fix and accumulate carbon (Cunniff et al., 2015). Furthermore, compared to annual systems grown on arable land, the management of perennial Salix stands in SRC leads to decreased mechanical disturbance of the soil and changed biochemical soil properties; along with changes in the abundance and diversity of soil organisms including mycorrhizal fungi (Baum et al., 2009a; Hrynkiewicz et al., 2010a; Fransson et al., 2013; Toju et al., 2013a; Goldmann et al., 2015). In addition, Salix genotype identity can significantly affect the soil enzyme activities at the same site (Baum and Hrynkiewicz, 2006), and Salix stands grown in a floodplain revealed higher activities of the soil enzyme β-glucosidase compared to perennial grassland (Zimmer et al., 2012). Changed soil enzyme activities are considered to be a direct expression of the soil community to metabolic requirements and available nutrients, and can therefore be used as indicators of soil functional diversity. Enzyme activities might be more indicative for ecosystem productivity and stability than the taxonomic diversity of soil microorganisms (Caldwell, 2005). Acid phosphatases and β-glucosidases are hydrolytic soil enzymes involved in P- and C-cycling of the soil and originate from plant roots and microorganisms. Mycorrhizal fungi can significantly increase the activities of these enzymes in the soil (Burke et al., 2011).
In SRC, Salix is usually planted with single genotypes, but this could possibly make them less resistant to pest organisms, diseases and abiotic stress. This is a major concern as a SRC must remain viable for up to 20 years to be profitable (Aravanopoulos et al., 1999). For this reason, an increased host plant diversity within Salix stands would be advantageous, but it will also change the below-ground plant–plant interactions within the stand. The soil ecological consequences of mixed Salix stands on former arable land are scarcely known so far (Hoeber et al., 2017).
We hypothesize that the mixture of genotypes with strong eco-physiological differences changes the mycorrhiza formation and activities in the mycorrhizosphere, based on different root characteristics of the genotypes involved. To test this hypothesis we have studied the mixture of two fundamental eco-physiological different Salix genotypes (S. schwerinii × S. viminalis cv. ‘Tora’ and S. dasyclados cv. ‘Loden’) grown in pure and mixed cultures in a field experiment in Northern Germany. Specifically, we expected (1) a higher diversity of root-associated fungi under mixed culture than in culture with the pure genotypes; and (2) changed fine root growth, abundance of root-associated fungi and enzymatic activities in the mycorrhizosphere under host genotype mixtures resulting from changed competitive conditions for the individual genotypes. Such insights into the overall structure of belowground plant–fungal associations will help to understand mechanisms that regulate the coexistence of eco-physiological diverse dual mycorrhizal plant species.
Materials and Methods
Study Site and Test Plants
The field site Rostock (54°04′12′′ N, 12°04′58′′ E) has been established in spring 2014 on former arable soil and is one out of three experimental field sites within the ECOLINK-Salix project (Hoeber et al., 2018), which is part of a global tree diversity network (TreeDivNet, Verheyen et al., 2016; Paquette et al., 2018). The plantation is a SRC stand with two willow genotypes [‘Tora,’ Svalöf-Weibull (SW) Cultivar No. 910007, S. schwerinii E. Wolf × S. viminalis L.; and ‘Loden’ SW 890129, S. dasyclados] grown in plots with one genotype being present (pure) and the combination (mixture) of the two. ‘Tora’ and ‘Loden’ in pure culture (‘Tora P’ and ‘Loden P’) were monocultures. In the mixture the two genotypes (‘Tora M’ and ‘Loden M’) were planted alternating one by one within the rows. Therefore, each genotype was directly associated with plants of the other genotype in the mixture.
Shoot biomass was harvested for the first time after 3 years of growth (in winter 2016/2017). The overall goal of the ECOLINK-Salix project is to assess the effects of genotype identity and diversity in willow SRC on various ecosystem functions. The experiment has a randomized block design, with three replicates (blocks, plot size: 92.16 m2). Planting density was c. 15600 plants ha-1. Each plot contains nine subplots (3.2 m × 3.2 m, 16 plants each).
The dominating soil type at the test site is a Stagnic Cambisol (FAO classification). The annual mean temperature is 8.5°C; the annual mean precipitation 592 mm. The texture class of the topsoil is predominantly loamy sand. General soil chemical properties (0–10 cm soil depth) were: pH (CaCl2) 6.2, Corg 6.89 g kg-1, Ntotal 0.86 g kg-1, C/N 8.01, Ptotal 0.11 g kg-1, and Stotal 0.09 g kg-1.
Soil and Fine Root Sample Collection
Soil and fine root sampling was done in November 2016 (at the end of the first rotation period after 3 years of growth) and in April 2017 (after the first shoot biomass harvest during the winter rest in February 2017). Nine soil cores (diameter 30 mm, depth 0–100 mm) per treatment were collected per genotype (2), sampling date (2), and planting layout (2: mono- [pure] and di-clonal [mix]) in the center of the plots for the determination of the fine root density, and of soil chemical and biochemical analyses (72 soil samples in total). The same number of samples and the same sampling design was used to collect the samples for the mycorrhizal analyses, but using 10 cm × 10 cm × 10 cm soil cubes taken with a sharp knife. The samples were collected from the uppermost 0–10 cm of soil, c. 20 cm from the stem base of willow plants and kept at 4°C until analyses. The majority of fine roots were found in 0–10 cm under diverse Salix species in SRC (Cunniff et al., 2015). For this reason the upper part of the topsoil seems to be most relevant for investigations of the mycorrhizosphere.
Soil Properties
The soil analyses were done using air-dried < 2 mm soil. Determination of soil pH was performed electrometrically using a glass electrode in 0.01 M CaCl2 with a soil:solution ratio of 1:2.5. Total carbon (Ctotal), total nitrogen (Ntotal), and total sulfur (Stotal) contents of the soils were determined with a Vario EL elemental analyzer (Elementar Analysensysteme GmbH, Hanau, Germany). All C was organic C (Corg) due to the acidic pH. The total phosphorus (Ptotal) content was extracted from 0.5 g dry soil material by microwave-assisted digestion with aqua regia solution (3:1 hydrochloric acid–nitric acid) (Chen and Ma, 2001). The P concentrations in the digests were measured by inductively coupled plasma optical emission spectroscopy (ICP-OES) (Optima 8300, PerkinElmer LAS GmbH, Rodgau, Germany).
Soil Enzyme Activities
The activities of acid phosphatases (EC 3.1.3.2) and β-glucosidases (EC 3.2.1.3) in the soil were determined colorimetrically according to Tabatabai (1982) and Eivazi and Tabatabai (1988). The enzyme activities were expressed as μg p-nitrophenol (pNP) g-1 soil h-1 released from the pre-given substrate solution (p-nitrophenyl-phosphate for acid phosphatases and p-nitrophenyl-β-D-glucosid for β-glucosidase) within 1 h of incubation.
Fine Root Density
Soil cores with plant roots were soaked in tap water for 1 h in a bowl and then carefully separated with forceps and washed with the help of a sieve. Roots were dried at 65°C for 48 h and the dry weights were calculated per soil volume and m2 within the upper 0–10 cm soil depth.
Fungal Colonization of Fine Roots
For analyses of AM and other endophytic colonization roots were rinsed and cut into 10 mm segments. The segments were cleared with 10% (w/v) KOH for 15 min at 90°C, bleached in 10% H2O2 for 1 h, acidified with 1% HCl (van der Heijden, 2001) and stained with 0.05% (w/v) chlorazol black E for 90 min at 80°C. The AM colonization was quantified microscopically using the intersection method of McGonigle et al. (1990), considering arbuscle formation as indicator of AM fungi vs. formation of sclerotia as indicator of dark septate endophytes (DSE). A minimum of 200 fine root intersections for AM colonization and a minimum of 200 fine root tips for DSE colonization were observed per plot.
For EM colonization and morphological characterization, 10 sub-samples of root fragments were randomly chosen on a grid for microscopical quantification of EM colonization. The numbers of living non-colonized root tips vs. obviously colonized EM root tips were counted using the method of Agerer (1991). In total c. 4800 root tips were scanned. A minimum of 200 root tips per genotype and treatment was investigated for each sampling date.
A minimum of two root tips per sub-sample was selected for subsequent molecular identification of the fungal partners. The morphological EM types were distinguished by macroscopical characteristics of the fungal mantle, such as color, surface appearance, presence of emanating hyphae and hyphal strands (Agerer, 1987–2002). Two to five root tips per sub-sample were separately frozen in Eppendorf-tubes and stored at -20°C for molecular analysis.
Molecular and Phylogenetic Identification of Root Colonizing Fungi
The fungal taxa from the root samples were identified using analysis of DNA sequences of the internal transcribed spacer (ITS) region. DNA was isolated from root tips using the Plant & Fungi Purification Kit (EurX, Poland) according to the manufacturer’s instructions. The ITS region within the ribosomal RNA genes was amplified using the primer pair ITS1F and ITS4 (White et al., 1990; Gardes and Bruns, 1993). Amplification of the ITS region was performed in a 25 μl final volume containing 50 ng of DNA, 0.125 μl of each primer (100 pM/μl), 10 μl of Master Mix (Qiagen) and 13 μl of nuclease-free water. The amplification protocol was as follows: 5 min at 94°C, 40 cycles of 30 s at 94°C, 1 min at 50°C, and 1 min at 72°C, and a final step at 72°C for 5 min. The PCR products obtained were purified using GeneMATRIX PCR/DNA Clean-Up Purification Kit protocol (EurX, Poland) and sequenced using primers ITS1F and ITS4. Sequence editing was performed using Sequencher TW Version 5.1 (Gene Codes, Ann Arbor, MI, United States). BLAST searching with ITS-sequences was performed on the GenBank (Altschul et al., 1990). If the sequence of the fungus showed 98% identities over the whole length of the sequence (about 600 to 700 bp) with a known fungus, this fungus was assumed to be the fungal root colonizing taxa. Contigs of ITS sequences were edited with EditSeq and aligned using Clustal W (DNASTAR®). DNA sequences were submitted to GenBank and accession numbers are presented in Table 1.
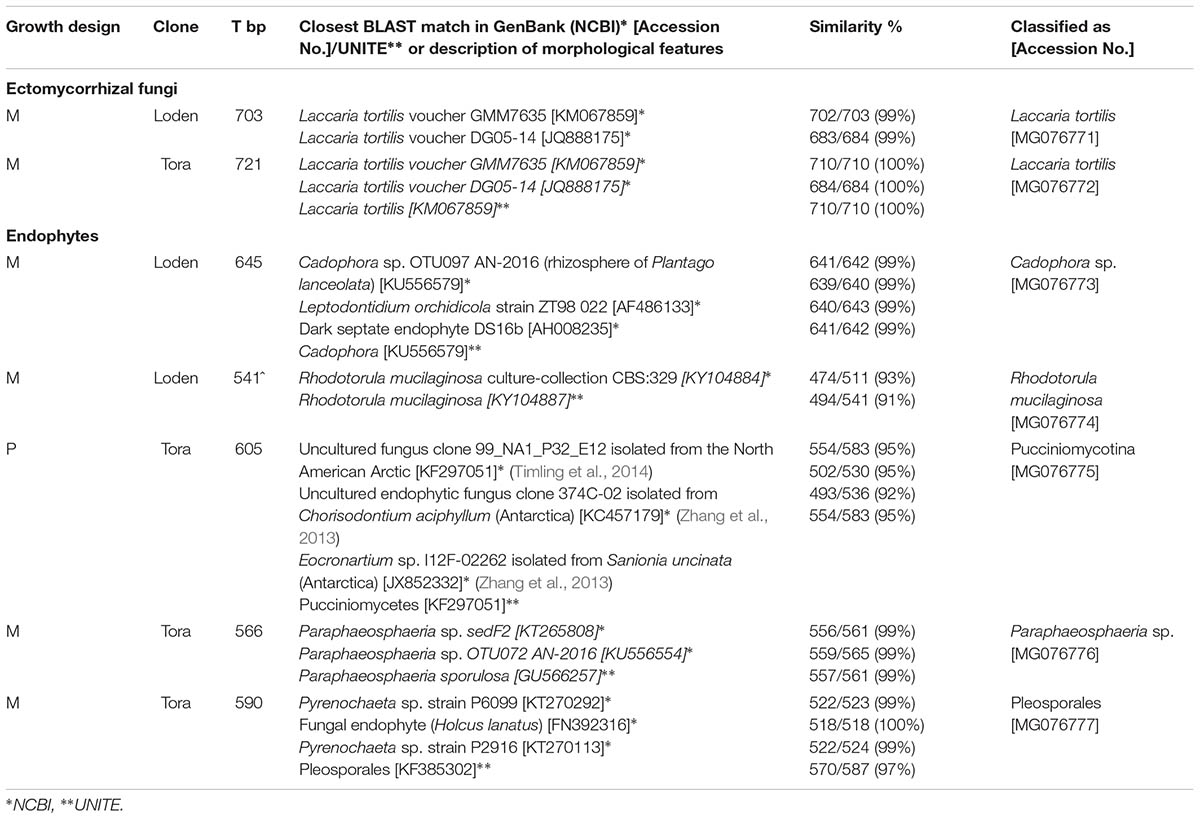
TABLE 1. Molecular identification of fungal partners in fine roots of Salix genotypes ‘Tora’ and ‘Loden’ in pure (P) and mixed (M) growth design in 0–10 cm soil depth at the short rotation coppice Rostock in autumn 2016 and spring 2017.
Statistical Analyses
The effect of the genotype-, the growth design (pure vs. mixed) and the interaction of both on biological and biochemical properties were analyzed by two-way ANOVA. The Detrended correspondence analysis (DCA) was used to disclose the effects of different host plant diversity (pure vs. mixed culture) on the root fungal colonization and soil enzyme activities in the mycorrhizosphere of Salix. Statistical analyses were computed using the software PAST (Hammer et al., 2001).
Results
Fine Root Biomass and Fungal Colonization
The fine root biomass was significantly affected by the interaction of genotype × growth design (pure vs. mixture) (Table 2). In pure culture ‘Loden P’ revealed higher fine root biomass than ‘Tora P,’ but under the mixture no significant differences between the two genotypes were observed (Figure 1). The means of the fine root biomass decreased in all treatments from autumn 2016 to spring 2017.
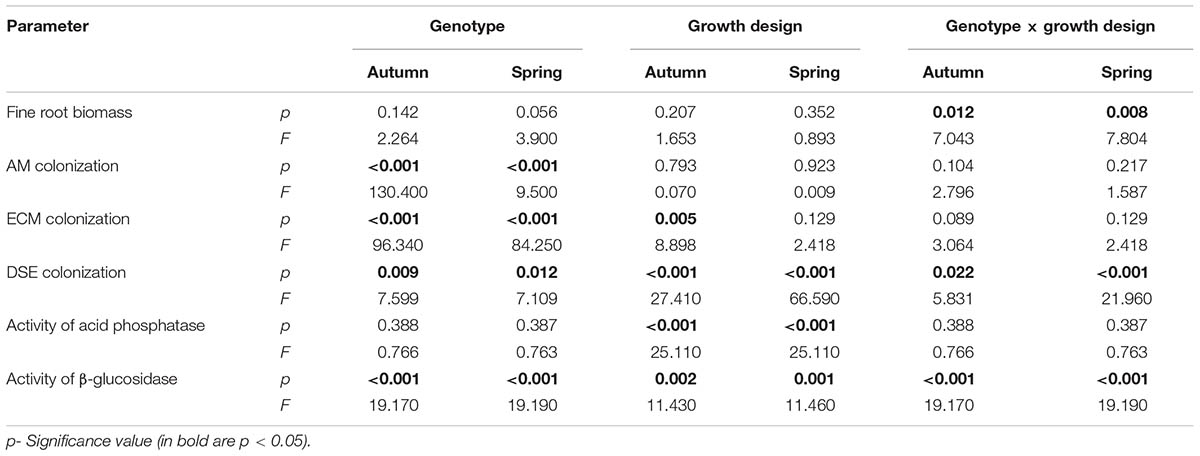
TABLE 2. Results of two-way analysis of variance (ANOVA) on the effect of the genotype, the growth design (with different host plant diversity; pure vs. mixture) and their interactions (genotype × growth design) on root and soil properties in the mycorrhizosphere of Salix from autumn 2016 (autumn) and spring 2017 (spring).
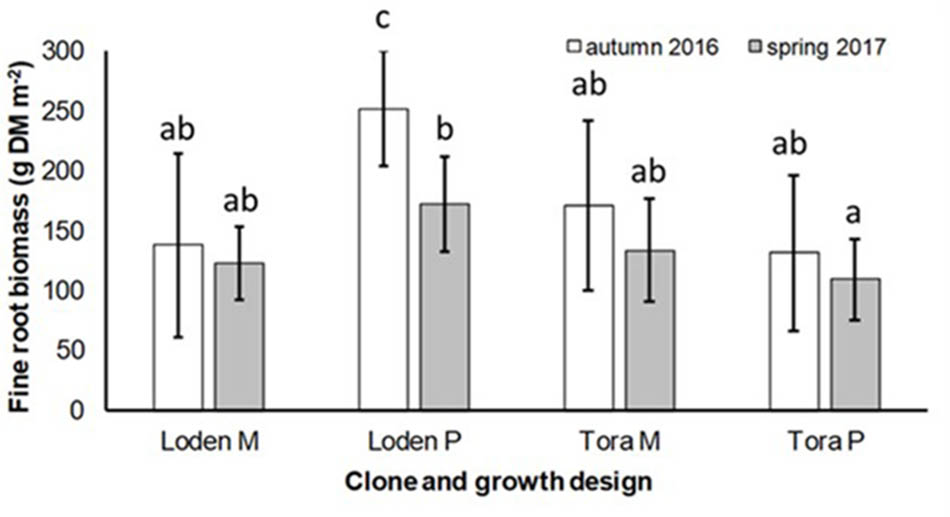
FIGURE 1. Fine root biomass (g DM per m2) under Salix genotypes ‘Tora’ and ‘Loden’ in pure (P) and mixed (M) growth in 0–10 cm soil depth at the short rotation coppice (SRC) Rostock in autumn 2016 and spring 2017 (means ± standard deviation, n = 9). Bars with different small letters (a, b, c) indicate significant differences among different treatments or sampling dates as defined by Duncan’s test (p < 0.05).
The AM colonization was relatively small (<12% of the total fine root length) in all treatments, but higher under ‘Tora M and P,’ than under ‘Loden M and P’ (Figure 2). The AM colonization of the fine roots was not significantly affected by the growth design (pure or mixed culture; Table 2). It was higher in spring than in autumn (Figure 2).
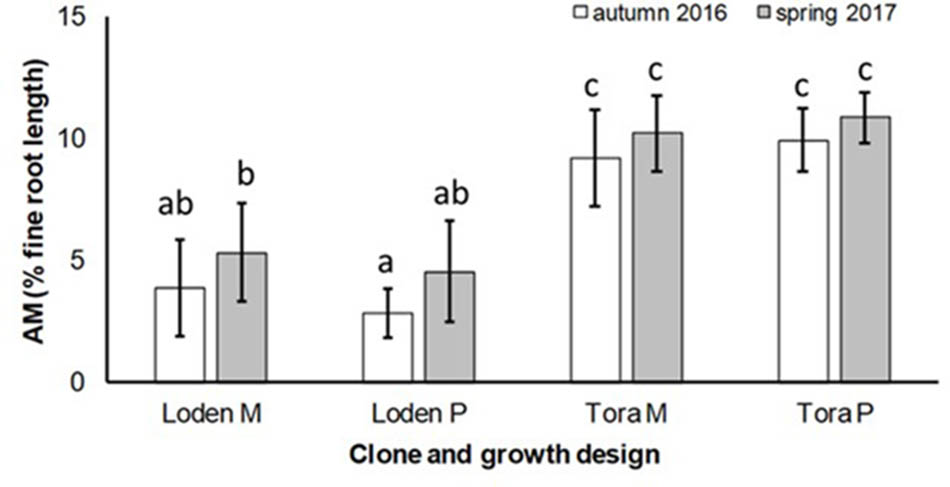
FIGURE 2. Arbuscular mycorrhizal colonization (% root length) and spore density (number of spores per 10 g DM soil) under Salix genotypes ‘Tora’ and ‘Loden’, in pure (P) and mixed (M) growth in 0–10 cm soil depth at the SRC Rostock in autumn 2016 and spring 2017 (means ± standard deviation, n = 9). Bars with different small letters (a, b, c) indicate significant differences among different treatments or sampling dates as defined by Duncan’s test (p < 0.05).
The EM colonization was significantly larger under ‘Loden M and P’ than under ‘Tora M and P,’ and significantly higher under the mixture in autumn 2016. In spring 2017 no significant impact of the growth design on the EM colonization was observed was (Figure 3A and Table 2). The EM colonization was in the mean slightly higher in autumn than in spring.
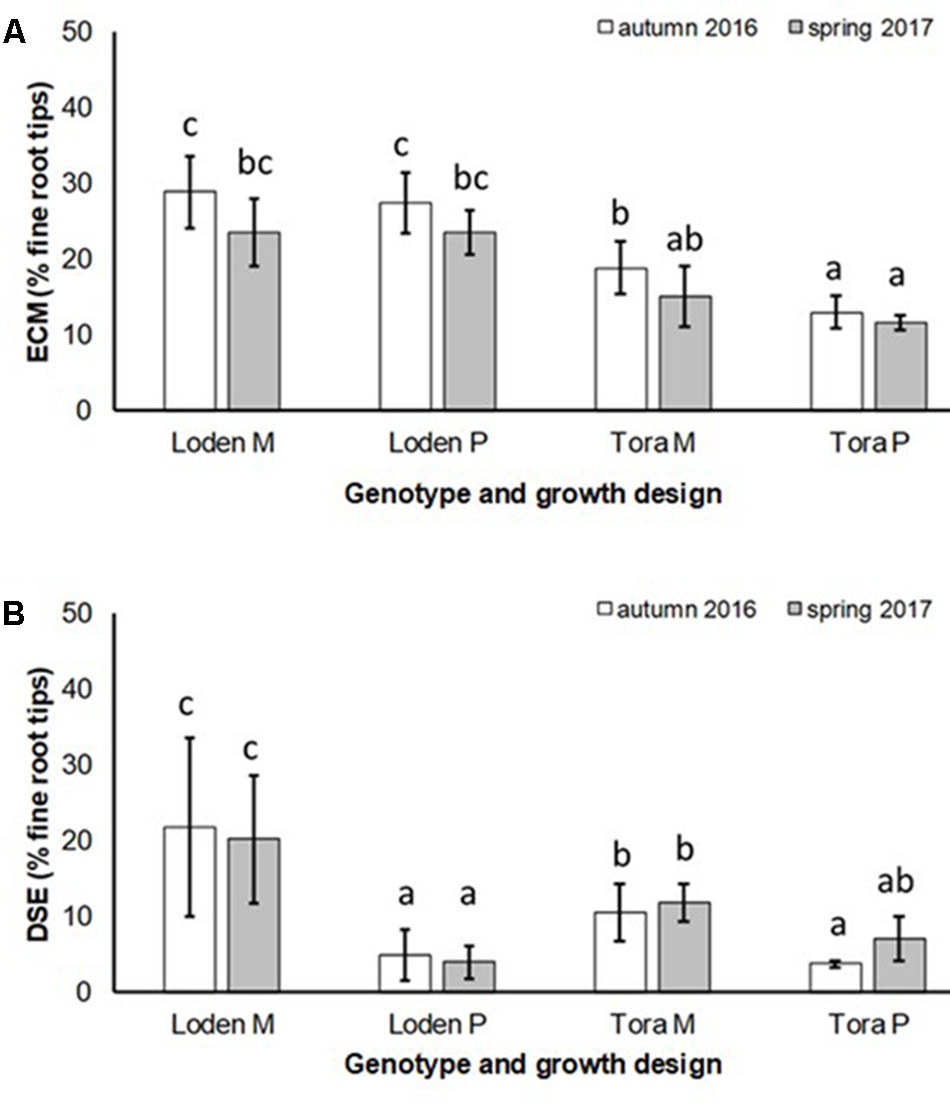
FIGURE 3. (A) Ectomycorrhizal colonization (ECM) and (B) dark septate endophytic colonization (DSE) of fine roots of Salix genotypes ‘Tora’ and ‘Loden in pure (P) and mixed (M) growth in 0–10 cm soil depth at the SRC Rostock in autumn 2016 and spring 2017 (means ± standard deviation, n = 9). Bars with different small letters (a, b, c) indicate significant differences among different treatments or sampling dates as defined by Duncan’s test (p < 0.05).
The DSE colonization was significantly larger in the mixture of genotypes (‘Loden M’ and ‘Tora M’) than in their pure cultures (‘Loden P’ and ‘Tora P’) (Figure 3B).
Fungal Diversity on the Fine Roots
With more than 95% of the total EM colonization (data not shown), Laccaria tortilis dominated as the single fungal partner in EM formation in all plots and was isolated from the roots of both genotypes (Table 1). Sporocarps of Laccaria tortilis were observed in all plots during the autumn sampling 2016. Other rarely occurring EM morphotypes observed were morphologically–anatomically determined to belong to Inocybe and Russula spp. and were found under both clones.
One endophytic fungal taxa was identified from the genotype ‘Tora’ in pure culture (Pucciniomycotina) and four endophytic fungal taxa from the genotypes in mixed culture (Cadophora sp., Paraphaeosphaeria sp., Rhodotorula mucilaginosa, Pleosporales) (Table 1 and Supplementary Figure 1).
Soil Enzyme Activities
The activity of acid phosphatase in the soil was significantly higher under the mixed growth design, but not significantly affected by the genotype and the interaction of genotype x growth design (Figure 4A and Table 2). The activities of β-glucosidase in the soil were higher under the genotype ‘Loden P’ in pure culture than under ‘Tora P’ in pure culture (Figure 4B and Table 2).
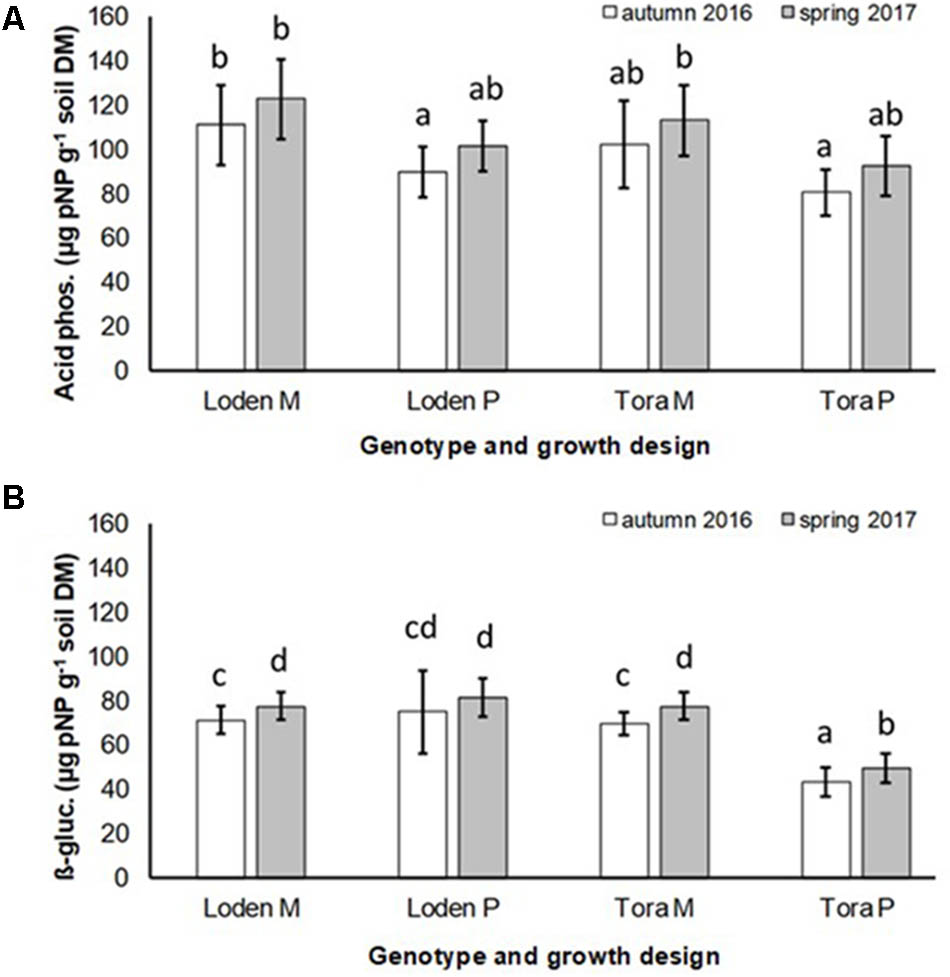
FIGURE 4. Soil enzyme activities of (A) acid phosphatase and (B) β-glucosidases of Salix genotypes ‘Tora’ and ‘Loden’ in pure (P) and mixed (M) growth in 0–10 cm soil depth at the SRC Rostock in autumn 2016 and spring 2017 (means ± standard deviation, n = 9). Bars with different small letters (a, b, c, d) indicate significant differences among different treatments or sampling dates as defined by Duncan’s test (p < 0.05).
Comparison Between Plots With Pure Host Genotypes and Genotype Mixtures
The mixed growth design of the genotypes increased significantly the colonization of fine roots by DSE, and the activities of acid phosphatases and β-glucosidases in the soil (Table 2).
The DCA of all fungal parameters and the two soil enzyme activities (acid phosphatase and β-glucosidase) clearly differentiated the genotypes (L for ‘Loden,’ T for ‘Tora’) and the host genotype mixture (M) from the two pure cultures (P) in autumn 2016 (Figure 5A) and spring 2017 (Figure 5B). The difference between the pure and the mixed culture was larger in spring 2017 than in autumn 2016, since the data differentiate the treatments along axis 2 (explaining about one quarter of the variation) in autumn, but along axis 1 in spring (explaining about half of the variation). The pure culture ‘Tora P’ was always closer to the mixture than the pure culture ‘Loden P.’
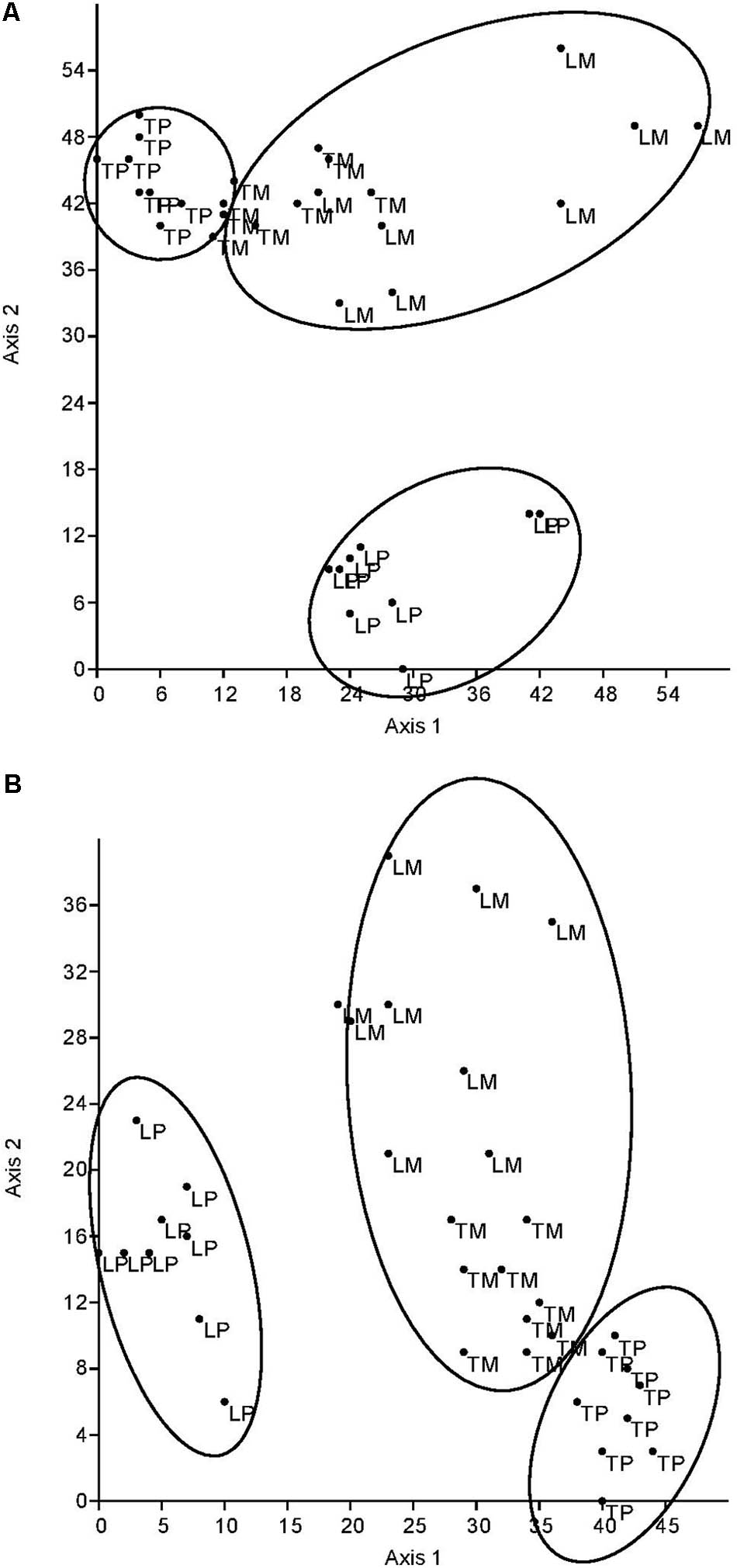
FIGURE 5. Detrended correspondence analysis (DCA) plot based on square root transformed, normalized data and Euclidean distance matrix. Comparison of the two planting designs [pure (P) and mixture (M)] of Salix [‘Loden’ (L) and ‘Tora’ (T)] by root and soil properties (A) in autumn 2016 (axis 1: Eigenvalue = 581.9, variance explained: 52.1%, axis 2: Eigenvalue = 281.9, variance explained: 25.2%) and (B) in spring 2017 (axis 1: Eigenvalue = 501.2, variance explained: 56.6%, axis 2: Eigenvalue = 230.8, variance explained: 26.1%).
Discussion
The present results confirm the significant impact of the Salix genotype on its phenotypical traits (Cunniff et al., 2015) including the mycorrhizal (see Figures 2, 3A) and endophytic (see Figure 3B) fungal colonization and host-specific differences in the fungal root colonization within a plant community as described by Toju et al. (2013b). The soil properties of the test site indicated P-deficiency (see section “Study Site and Test Plants”), which increases the importance of mycorrhizal fungi for the P-supply of the host plants (Smith et al., 2011). Assuming that plant neighbors with a faster growth rate can increase the competition for plants with slower growth, in the present mixture ‘Tora M’ had an advantage over ‘Loden M.’ Both, decreased fine root biomass under ‘Loden M’ grown in a mixture (see Figure 1) and greater similarity of the root traits and soil enzyme activities of the mixture compared with pure cultures ‘Tora P’ or ‘Loden P’ support this assumption (see Figure 5). These results are in line with our hypothesis that fine root growth, fungal abundance and activities are changed under host genotype mixtures, caused by changed competitive conditions for the individual genotypes and thereby changed interactions between them. However, the genotype-effect on the mycorrhizal colonization exceeds the impact of the plant design (pure or mixture of genotypes) on the mycorrhizal abundance (see Figures 2, 3A and Table 2).
We predicted that the increased diversity of host plants in the genotype mixture will increase the diversity of root associated fungi too, since aboveground and belowground diversity can be linked (De Deyn and Van der Putten, 2005). However, we found increased fungal diversity only in endophytic fungi, not in mycorrhizal ones. This increased diversity was associated with increased colonization of the roots with DSE. Generally, low mycorrhizal diversity on Salix found here by only one dominating EM fungal species (Laccaria tortilis; see Table 1) causes problems when interpreting the results. However, the low EM diversity is in line with the results on other genotypes of Salix (Püttsepp et al., 2004; Hrynkiewicz et al., 2012). It might be caused by the genotypic-specificity of this plant genus and additionally by the former arable site-conditions with lack of EM host plants. Therefore, we have focused rather on the ratios of root associations than on total mycorrhizal and endophytic diversity.
For the first time, we disclosed that increased host diversity of Salix genotypes can promote endophytic root colonization and increase soil enzyme activities involved in P-mobilization (see Figures 2–4). These results of a Continental SRC support the assumption of Pérez-Izquierdo et al. (2017) taken for Mediterranean forests, who highlighted the importance of structural shifts in fungal communities for their possible functional consequences. Functional consequences were revealed in an increased enzymatic P mobilization (see Figure 4) in the mycorrhizosphere with increased host plant diversity (mixture). These increased enzymatic activities can be caused directly by fungal impact, but also by the plant impact or most probably by a combination of both.
Dark septate endophytes were previously described to be the dominating root associated fungi under S. caprea on heavy metal contaminated sites (Likar and Regvar, 2013). In agreement with the results of these authors, we also found Cadophora spp. as a very common DSE in the roots of the two tested Salix genotypes at the non-contaminated arable site. This endophyte revealed plant growth promotion in heavy metal contaminated soils (Likar and Regvar, 2013). Also the second most common DSE in the present study (Paraphaeosphaeria sp., see Table 1), revealed plant growth promoting traits on another Salix sp. in heavy metal contaminated soil (An et al., 2015). The endophytic yeast Rhodotorula mucilaginosa, found under mixed Salix growth in the present study, was revealed to be plant growth promoting in Populus (Xin et al., 2009). However, from the present data no statement can be made on the growth response of Salix on the different fungal root colonization and the impact of DSE can vary from growth promotion to growth suppression of host plants (Aguilar-Trigueros and Rillig, 2016).
Increased colonization of the roots with DSE combined with consistent colonization with AM fungi in the mixture (see Figures 1, 3B) were accompanied by increased soil enzyme activities involved in P-mobilization (see Figure 4). Della Monica et al. (2015) assumed, that DSE can be more effective to mobilize organic P, than AM fungi. However, in the present study increased enzyme activities might be stronger affected directly by the changed nutrient competition of the host plants in the mixture. Increased abundance of DSE without suppression of mycorrhiza formation supports that these fungal groups can interact beneficially in a joint host plant (Wężowicz et al., 2017). This might be caused by exudates of DSE, which can even stimulate the growth of mycorrhizal fungi (Scervino et al., 2009). Oppositely to the promotion of DSE in the mixture of different Salix genotypes, Becklin et al. (2012) revealed promoted of EM fungi on Salix in increased plant diversity by combination with herbaceous plants in an alpine meadow. However, these results have in common that host plant diversity changes the fungal root colonization of Salix.
The present data on dual mycorrhizal colonization of AM and EM fungi on Salix clearly confirmed the assumption of Lodge and Wentworth (1990), that these types of mycorrhizal fungi colonize roots with an antagonistic behavior. Interestingly, Chen et al. (2000) found that antagonistic behavior of AM and EM fungi was most severe when the EM fungus Laccaria was present, which was the dominating EM fungus in the present study. Low fungal diversity was observed also in the sporocarp production in other SRCs (Baum et al., 2002).
The slow-growing genotype ‘Loden’ seems to be rather EM dominated, whereas the fast-growing genotype ‘Tora’ seems to promote AM formation (see Figures 2, 3). This might be affected by the different litter quality of these genotypes, since the litter quality affects mycorrhizal communities (Lambers et al., 1998). In a pot experiment, leaves of ‘Loden’ revealed higher phenolic concentrations in the later growth than ‘Tora’ under the same environmental conditions (Baum et al., 2009b). Leaf litter with higher phenolic concentrations was described to support EM formation, whereas low phenolic concentrations were found to promote AM colonization of the host plants (Lambers et al., 1998).
Interestingly, the colonization of the roots with DSE was stronger affected by the host plant diversity than by the genotype-specific differences, which was revealed by an increased colonization of the roots (see Figure 3B) combined with increased species richness (see Table 1) in the mixture. Increased stress tolerance of Salix spp. was previously linked to colonization by several DSE (An et al., 2015). However, root endophytes can also include potential pathogens and each plant species and combination with different fungal strains might respond differently in plant growth (Aguilar-Trigueros and Rillig, 2016). Furthermore, the colonization of roots with DSE is also affected by edaphic properties of the site (Xie et al., 2017). The impact of edaphic differences within the present test site was initially balanced between the treatments by the randomized plot design in the present study. However, the increased colonization by DSE might also lead back to changed nutrient mobilization caused by the changed root growth of at least one of the mixed genotypes (see Figure 1) and indicated by changed enzymatic P mobilization under the mixture (see Figure 4).
Based on our results, it can be concluded that increased genotype diversity in Salix cultivation can lead to changed root competition, fungal association and enzymatic P-mobilization in the mycorrhizosphere. Subsequent investigations should assess the generality of promotion of DSE and P-mobilization in increased host diversity.
Author Contributions
MW conceived and designed the field experiment within the frame of ECOLINK-Salix. CB managed the field site Rostock, collected the samples, did soil and root analyses and statistical analyses and wrote the first draft of the manuscript; did all soil chemical and biochemical analyses and wrote part of Section “Materials and Methods.” NV and KH isolated the ectomycorrhizal fungi, created figures and tables and did statistical analyses. KH and SS identified the fungal species. NV analyzed the colonization density of mycorrhizal fungi and analyzed these parts of the results. SH and PF evaluated and discussed the mycorrhizal and endophytic results. All authors edited and revised the manuscript and approved the publication.
Funding
The authors gratefully acknowledge the German Federal Ministry of Education and Research (BMBF) for funding the BonaRes project InnoSoilPhos (Project No. 031A558). The work of MW and PF was partly funded by the Swedish Energy Agency (Project Nos. 36654-1 and 36654-2). Partial support to SH was provided by the Swedish Research Council Formas (Project No. 2016-00998).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Ms. E. Heilmann (University of Rostock, Germany) for valuable technical assistance during the chemical soil analyses. This research was performed within the scope of the Leibniz ScienceCampus Phosphorus Research Rostock. We are grateful for the very valuable comments of the reviewers.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01012/full#supplementary-material
References
Agerer, R. (1991). “Characterization of ectomycorrhiza,” in Techniques for the Study of Mycorrhiza. Methods Microbiology, Vol. 23, eds J. R. Norris, D. J. Read, and A. K. Varma (London: Academic Press), 25–73.
Aguilar-Trigueros, C. A., and Rillig, M. C. (2016). Effect of different root endophytic fungi on plant community structure in experimental microcosms. Ecol. Evol. 6, 8149–8158.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
An, H., Liu, Y., Zhao, X., Huang, Q., Yuan, S., Yang, X., et al. (2015). Characterization of cadmium-resistant endophytic fungi from Salix variegata Franch. in Three Gorges Reservoir Region. China. Microbiol. Res. 176, 29–37. doi: 10.1016/j.micres.2015.03.013
Aravanopoulos, F. A., Kim, K. H., and Zsuffa, L. (1999). Genetic diversity of superior Salix clones selected for intensive forestry plantations. Biomass Bioenergy 16, 249–255.
Baum, C., and Hrynkiewicz, K. (2006). Clonal and seasonal shifts in communities of saprotrophic microfungi and soil enzyme activities in the mycorrhizosphere of Salix spp. J. Plant Nutr. Soil Sci. 169, 481–487.
Baum, C., Leinweber, P., Weih, M., Lamersdorf, N., and Dimitriou, I. (2009a). Effects of short rotation coppice with willows and poplar on soil ecology. Agric. Forest Res. 59, 183–196.
Baum, C., Toljander, Y. K., Eckhardt, K.-U., and Weih, M. (2009b). The significance of host-fungus combinations in ectomycorrhizal symbioses in the chemical quality of willow foliage. Plant Soil 323, 213–224.
Baum, C., Weih, M., Verwijst, T., and Makeschin, F. (2002). The effects of nitrogen fertilization and soil properties on mycorrhizal formation of Salix viminalis. For. Ecol. Manage. 160, 35–43.
Becklin, K. M., Pallo, M. L., and Galen, C. (2012). Willows indirectly reduce arbuscular mycorrhizal fungal colonization in understorey communities. J. Ecol. 100, 343–351. doi: 10.1111/j.1365-2745.2011.01903.x
Burke, D. J., Weintraub, M. N., Hewins, C. R., and Kalisz, S. (2011). Relationship between soil enzyme activities, nutrient cycling and soil fungal communities in a northern hardwood forest. Soil Biol. Biochem. 43, 795–803. doi: 10.1016/j.soilbio.2010.12.014
Caldwell, B. A. (2005). Enzyme activities as a component of soil biodiversity: a review. Pedobiologia 49, 637–644.
Chen, M., and Ma, L. Q. (2001). Comparison of three aqua-regia methods for twenty Florida soils. Soil Sci. Soc. Am. J. 65, 491–499.
Chen, Y. L., Brundrett, M. C., and Dell, B. (2000). Effects of ectomycorrhizas and vesicular-arbsucular mycorrhizas, alone and in competition, on root colonization and growth of Eucalyptus globules and E. urophylla. New Phytol. 146, 545–556.
Courty, P. E., Labbe, J., Kohler, A., Marçais, B., Bastien, C., Churin, J. L., et al. (2011). Effect of poplar genotypes on mycorrhizal infection and secreted enzyme activities in mycorrhizal and non-mycorrhizal roots. J. Exp. Bot. 62, 249–260. doi: 10.1093/jxb/erq274
Cunniff, J., Purdy, S. J., Barraclough, T. J., Castle, M., Maddison, A. L., Jones, L. E., et al. (2015). High yielded biomass genotypes of willow (Salix spp.) show differences in below ground biomass allocation. Biomass Bioenergy 80, 114–127.
De Deyn, G. B., and Van der Putten, W. H. (2005). Linking aboveground and belowground diversity. Trends Ecol. Evol. 20, 625–663.
Della Monica, I. F., Saparrat, M. C., Godeas, A. M., and Scervino, J. M. (2015). The co-existence between DSE and AMF symbionts affects plant P pools through P mineralization and solubilization processes. Fungal Ecol. 17, 10–17.
Eivazi, F., and Tabatabai, M. A. (1988). Glucosidases and galactosidases in soils. Soil. Biol. Biochem. 20, 601–606.
Fransson, P. M. A., Toljander, Y. K., Baum, C., and Weih, M. (2013). Plant genotype - ectomycorrhizal fungus combination drives resource allocation in willow - evidence for complex genotype-genotype interaction from a simple experiment. Ecoscience 20, 112–121.
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes—Application to identification of mycorhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Goldmann, K., Schöning, I., Buscot, F., and Wubet, T. (2015). Forest management type influences diversity and community composition of soil fungi across temperate forest ecosystems. Front. Microbiol. 6:1300. doi: 10.3389/fmicb.2015.01300
Hammer,Ø., Harper, D. A. T., and Ryan, P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9.
Hoeber, S., Arranz, C., Nordh, N.-E., Baum, C., Low, M., Nock, C., et al. (2018). Genotype identity has a more important influence than genotype diversity on shoot biomass productivity in willow short rotation coppices. GCB Bioenergy doi: 10.1111/gcbb.12521
Hoeber, S., Fransson, P., Prieto-Ruiz, I., Mazoni, S., and Weih, M. (2017). Two salix genotypes differ in productivity and nitrogen economy when grown in monoculture and mixture. Front. Plant Sci. 8:231. doi: 10.3389/fpls.2017.00231
Hrynkiewicz, K., Baum, C., Leinweber, P., Weih, M., and Dimitriou, I. (2010a). The significance of rotation periods for mycorrhiza formation in Short Rotation Coppice. For. Ecol. Manag. 260, 1943–1949.
Hrynkiewicz, K., Ciesielska, A., Haug, I., and Baum, C. (2010b). Ectomycorrhiza formation and willow growth promotion as affected by associated bacteria: role of microbial metabolites and use of C sources. Biol. Fertil. Soils 46, 139–150.
Hrynkiewicz, K., Toljander, Y., Baum, C., Fransson, P., Taylor, A., and Weih, M. (2012). Correspondence of ectomycorrhizal diversity and colonisation of willows (Salix spp.) grown in Short Rotation Coppice on arable sites and adjacent natural stands. Mycorrhiza 22, 603–613. doi: 10.1007/s00572-012-0437-z
Jones, M. D., Durall, D. M., and Tinker, P. B. (1998). A comparison of arbuscular and ectomycorrhizal Eucalyptus coccifera: growth response, phosphorus uptake efficiency and external hyphal production. New Phytol. 140, 125–134.
Lambers, H., Chapin, F. S., and Pons, T. L. (eds). (1998). “Role in ecosystem and global processes,” in Plant Physiological Ecology (New York, NY: Springer).
Likar, M., and Regvar, M. (2013). Isolates of dark septate endophytes reduce metal uptake and improve physiology of Salix caprea L. Plant Soil 370, 593–604.
Lodge, D. J. (1989). The influence of soil moisture and flooding on formation of VA- endo- and ectomycorrhizae in Populus and Salix. Plant Soil 117, 255–262.
Lodge, D. J., and Wentworth, T. R. (1990). Negative associations among VA-mycorrhizal fungi and some ectomycorrhizal fungi inhabiting the same root system. Oikos 57, 347–356.
McGonigle, T. P., Miller, M. H., Evans, D. G., Fairchild, G. L., and Swan, J. A. (1990). A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115, 495–501. doi: 10.1111/nph.1990.115.issue-3
Paquette, A., Hector, A., Castagneyrol, B., Vanhellemont, M., Koricheva, J., Scherer-Lorenzen, M., et al. (2018). A million and more trees for science. Nat. Ecol. Evolut. 2, 763–766. doi: 10.1038/s41559-018-0544-0
Pérez-Izquierdo, L., Zabal-Aguirre, M., Flores-Rentería, D., Gonzáles-Martínez, S. C., Buée, M., and Rincón, A. (2017). Functional outcomes of fungal community shifts driven by tree genotype and spatial-temporal factors in Mediterranean pine forests. Environ. Microbiol. 19, 1639–1652. doi: 10.1111/1462-2920.13690
Püttsepp,Ü., Rosling, A., and Taylor, A. F. S. (2004). Ectomycorrhizal fungal communities associated with Salix viminalis L. and S. dasyclados Wimm. Clones in a short-rotation forestry plantation. For. Ecol. Manag. 196, 413–424.
Rillig, M., Wendt, S., Antonovics, J., Hempel, S., Kohler, J., Wehner, J., et al. (2014). Interactive effects of root endophytes and arbuscular mycorrhizal fungi on an experimental plant community. Oecologia 174, 263–270. doi: 10.1007/s00442-013-2759-8
Rooney, D. C., Killham, K., Bending, G. D., Baggs, E., Weih, M., and Hodge, A. (2009). Mycorrhizas and biomass crops: opportunities for future sustainable development. Trends Plant Sci. 14, 542–549. doi: 10.1016/j.tplants.2009.08.004
Scervino, J. M., Gottlieb, A., Silvani, V. A., Pérgola, M., Fernández, L., and Godeas, A. M. (2009). Exudates of dark septate endophyte (DSE) modulate the development of the arbuscular mycorrhizal fungus (AMF) Gigaspora rosea. Soil Biol. Biochem. 41, 1753–1756. doi: 10.1016/j.soilbio.2009.04.021
Smith, S. E., Jakobsen, I., Gronlund, M., and Smith, F. A. (2011). Roles of arbuscular mycorrhizas in plant phosphorus nutrition; interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol. 156, 1050–1057.
Tabatabai, M. A. (1982). “Soil enzymes,” in Methods of Soil Analysis. Part 2, Agronomy 9, eds A. L. Page, R. H. Miller, and D. R. Keeney (Madison, WI: American Society of Agronomy), 903–947. doi: 10.1016/j.tplants.2009.08.004
Timling, I., Walker, D. A., Nusbaum, C., Lennon, N. J., and Taylor, D. L. (2014). Rich and cold: diversity, distribution and drivers of fungal communities in patterned-ground ecosystems of the North American Arctic. Mol. Ecol. 23, 3258–3272. doi: 10.1111/mec.12743
Toju, H., Sato, H., Yamamoto, S., Kohmei, K., Tanabe, A. S., Yazawa, S., et al. (2013a). How are plant and fungal communities linked to each other in belowground ecosystems? A massively parallel pyrosequencing analysis of the association specificity of root-associated fungi and their host plants. Ecol. Evol. 3, 3112–3124. doi: 10.1002/ece3.706
Toju, H., Yamamoto, S., Sato, H., and Tanabe, A. S. (2013b). Sharing of diverse mycorrhizal and root-endophytic fungi among plant species in an oak-dominated cool–temperate forest. PLoS One 8:e78248. doi: 10.1371/journal.pone.0078248
van der Heijden, E. (2001). Differential benefits of arbuscular mycorrhizal and ectomycorrhizal infection of Salix repens. Mycorrhiza 10, 185–193. doi: 10.1007/s005720000077
Verheyen, K., Vanhellemont, M., Auge, H., Baeten, L., Baraloto, C., Barsoum, N., et al. (2016). Contributions of a global network of tree diversity experiments to sustainable forest plantations. Ambio 45, 29–41. doi: 10.1007/s13280-015-0685-1
Weih, M. (2004). Intensive short rotation forestry in boreal climates: present and future perspectives. Can. J. For. Res. 34, 1369–1378.
Wężowicz, K., Rozpądek, P., and Turnau, K. (2017). Interactions of arbuscular mycorrhizal and endophytic fungi improve seedling survival and growth in post-mining waste. Mycorrhiza 27, 499–511. doi: 10.1007/s00572-017-0768-x
White, T. J., Bruns, T., Lee, S., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (New York, NY: Academic Press Inc), 315–322.
Xie, L., He, X., Wang, K., Hou, L., and Sun, Q. (2017). Spatial dynamics of dark septate endophytes in the roots and rhizospheres of Hedysarum scoparium in northwest China and the influence of edaphic variables. Fungal Ecol. 26, 135–143.
Xin, G., Glawe, D., and Doty, S. L. (2009). Characterization of three endophytic, indole-3-acetic acid producing yeasts occurring in Populus trees. Mycol. Res. 113, 973–980. doi: 10.1016/j.mycres.2009.06.001
Zhang, T., Xiang, H. B., Zhang, Y. Q., Liu, H. Y., Wei, Y. Z., Zhao, L. X., et al. (2013). Molecular analysis of fungal diversity associated with three bryophyte species in the Fildes Region, King George Island, maritime Antarctica. Extremophiles 17, 757–765. doi: 10.1016/j.mycres.2009.06.001
Keywords: arbuscular mycorrhizal fungi, ectomycorrhiza, soil enzymes, phosphorus, fine root density, short rotation coppice, willows
Citation: Baum C, Hrynkiewicz K, Szymańska S, Vitow N, Hoeber S, Fransson PMA and Weih M (2018) Mixture of Salix Genotypes Promotes Root Colonization With Dark Septate Endophytes and Changes P Cycling in the Mycorrhizosphere. Front. Microbiol. 9:1012. doi: 10.3389/fmicb.2018.01012
Received: 13 October 2017; Accepted: 30 April 2018;
Published: 18 May 2018.
Edited by:
Erika Kothe, Friedrich-Schiller-Universität Jena, GermanyReviewed by:
Fred Asiegbu, University of Helsinki, FinlandKatarzyna Turnau, Jagiellonian University, Poland
Johanna Witzell, Southern Swedish Forest Research Centre, Swedish University of Agricultural Sciences, Sweden
Copyright © 2018 Baum, Hrynkiewicz, Szymańska, Vitow, Hoeber, Fransson and Weih. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christel Baum, christel.baum@uni-rostock.de
 Christel Baum
Christel Baum Katarzyna Hrynkiewicz
Katarzyna Hrynkiewicz Sonia Szymańska
Sonia Szymańska Nora Vitow
Nora Vitow Stefanie Hoeber
Stefanie Hoeber Petra M. A. Fransson
Petra M. A. Fransson Martin Weih
Martin Weih