- 1Department of Parasitology, Xiangya School of Basic Medicine, Central South University, Changsha, China
- 2Department of Pharmacology, Robert Wood Johnson Medical School, Rutgers University, Piscataway, NJ, United States
- 3Greehey Children’s Cancer Research Institute, University of Texas Health San Antonio, San Antonio, TX, United States
- 4Department of Molecular Medicine, University of Texas Health San Antonio, San Antonio, TX, United States
Cells reprogram their transcriptome in response to stress, such as heat shock. In free-living bacteria, the transcriptomic reprogramming is mediated by increased DNA-binding activity of heat shock sigma factors and activation of genes normally repressed by heat-induced transcription factors. In this study, we performed transcriptomic analyses to investigate heat shock response in the obligate intracellular bacterium Chlamydia trachomatis, whose genome encodes only three sigma factors and a single heat-induced transcription factor. Nearly one-third of C. trachomatis genes showed statistically significant (≥1.5-fold) expression changes 30 min after shifting from 37 to 45°C. Notably, chromosomal genes encoding chaperones, energy metabolism enzymes, type III secretion proteins, as well as most plasmid-encoded genes, were differentially upregulated. In contrast, genes with functions in protein synthesis were disproportionately downregulated. These findings suggest that facilitating protein folding, increasing energy production, manipulating host activities, upregulating plasmid-encoded gene expression, and decreasing general protein synthesis helps facilitate C. trachomatis survival under stress. In addition to relieving negative regulation by the heat-inducible transcriptional repressor HrcA, heat shock upregulated the chlamydial primary sigma factor σ66 and an alternative sigma factor σ28. Interestingly, we show for the first time that heat shock downregulates the other alternative sigma factor σ54 in a bacterium. Downregulation of σ54 was accompanied by increased expression of the σ54 RNA polymerase activator AtoC, thus suggesting a unique regulatory mechanism for reestablishing normal expression of select σ54 target genes. Taken together, our findings reveal that C. trachomatis utilizes multiple novel survival strategies to cope with environmental stress and even to replicate. Future strategies that can specifically target and disrupt Chlamydia’s heat shock response will likely be of therapeutic value.
Introduction
In response to adverse environmental conditions, living cells activate certain genes and repress others in order to survive and thrive. For bacteria, a variety of environmental insults, such as temperature variations, osmotic changes, antibiotics, solvents, and host immune response, can all elicit a stress response. Among these, sudden temperature increase is the most widely used model for studying the impact of stress (for reviews see Yura et al., 1993; Wu, 1995; Hecker et al., 1996; Roncarati and Scarlato, 2017).
Research on stress response has mainly focused on the mechanisms of gene activation. In bacteria, heat shock genes can be activated through two mechanisms in response to stress signals (Yura et al., 1993; Wu, 1995; Hecker et al., 1996; Narberhaus, 1999; Roncarati and Scarlato, 2017). Positive transcriptional regulation is achieved by specific alternative sigma factors of the RNA polymerase (e.g., σ32 in the Gram-negative Escherichia coli and σB in the Gram-positive Bacillus subtilis; Neidhardt and VanBogelen, 1981; Yamamori and Yura, 1982; Grossman et al., 1984; Völker et al., 1994; Yura, 1996; Hughes and Mathee, 1998). These sigma factors (i.e., heat shock sigma factors) guide the polymerase to the promoters of heat shock genes (Haldenwang, 1995; Yura, 1996; Helmann, 1999; Mooney et al., 2005; Feklistov et al., 2014; Rodriguez Ayala et al., 2020). Alternatively, alleviation of negative regulation can occur following heat-induced dissociation of transcriptional repressors from promoters of heat shock genes (Schulz and Schumann, 1996; Baldini et al., 1998; Narberhaus, 1999; Wilson and Tan, 2002; Hu et al., 2007; Roncarati et al., 2019). The most widely distributed transcriptional repressor controlling heat shock response in bacteria is the heat-inducible HrcA (Schulz and Schumann, 1996; Baldini et al., 1998; Wilson and Tan, 2002; Hu et al., 2007; Roncarati et al., 2019). Some bacteria encode additional transcriptional repressors, such as HspR and/or CtsR (Derre et al., 1999a,b, 2000; Narberhaus, 1999; Nair et al., 2000; Stewart et al., 2002; Spohn et al., 2004; Holmes et al., 2010; Pepe et al., 2018; Roncarati and Scarlato, 2018). Both HrcA and HspR negatively regulate the expression of a limited set of molecular chaperones including the Hsp70 DnaK and its cochaperone GrpE, and the Hsp60 GroEL (aka chaperonin) and its cochaperone GroES (Yura et al., 1993; Wu, 1995; Hecker et al., 1996; Schulz and Schumann, 1996; Baldini et al., 1998; Narberhaus, 1999; Stewart et al., 2002; Wilson and Tan, 2002; Spohn et al., 2004; Hu et al., 2007; Holmes et al., 2010; Roncarati and Scarlato, 2017, 2018; Pepe et al., 2018; Roncarati et al., 2019). Chaperones maintain numerous proteins (including HrcA and HspR) in their functional conformation, whereas stress, particularly heat shock and solvents, cause protein denaturation. As a result of heat shock, protein chaperones are titrated away from HrcA and HspR, thus leading to their functional loss and consequent activation of their target genes (Yura et al., 1993; Wu, 1995; Hecker et al., 1996; Narberhaus, 1999; Roncarati and Scarlato, 2017). Protein chaperones may also target some cellular proteins, such as σ32 and σH, for degradation under normal growth conditions whereas heat shock frees the sigma factors to activate their target genes (Herman et al., 1995; Lund, 2001).
Chlamydia is a small Gram-negative bacterium that replicates strictly inside eukaryotic host cells. Chlamydia has a unique infectious cycle characterized by two cellular forms (Abdelrahman and Belland, 2005). The infectious but non-dividing elementary body (EB) invades host cells through receptor-mediated endocytosis. Within vacuoles termed inclusions, EBs convert into the proliferative but noninfectious reticulate bodies (RB). As RBs amass inside the inclusion, they differentiate back into EBs, which exit the cells (Hybiske and Stephens, 2007a,b).
The chlamydial developmental cycle leading to formation of progeny EBs can be disrupted by various stress conditions, such as interferon-γ, iron starvation, and antibiotic treatment (Belland et al., 2003; Huston et al., 2008; Xue et al., 2017; Brinkworth et al., 2018; Slade et al., 2019; Yang et al., 2020; Brockett and Liechti, 2021). As a result, chlamydiae enter a state referred to as persistence. During persistence, RBs continue to grow but fail to divide or differentiate into EBs, resulting in abnormally large cells referred to as aberrant bodies (Belland et al., 2003; Huston et al., 2008; Xue et al., 2017; Brinkworth et al., 2018; Slade et al., 2019; Yang et al., 2020; Brockett and Liechti, 2021; Shima et al., 2021). Upon return to favorable conditions, aberrant bodies convert to RBs, which resume the normal developmental cycle. Chlamydial persistence and recovery represent a significant clinical problem.
Given that chlamydial infections can cause fever in the host (Luger, 1948; Qvigstad et al., 1982; Dan et al., 1987; Wu et al., 2000; Reinhold et al., 2008, 2012; Stoner and Cohen, 2015; Clemmons et al., 2019), heat shock responses in Chlamydia cells have long been suspected. Indeed, Engel et al. (1990) first demonstrated increased mRNA and protein levels of GrpE and DnaK in Chlamydia muridarum cultures shortly after they were incubated at 45°C, whereas another work demonstrated heat shock-induced persistence in Chlamydia trachomatis (Huston et al., 2008). However, apart from recent studies focusing on transcriptional regulation by HrcA (Tan et al., 1996; Wilson and Tan, 2002, 2004; Wilson et al., 2005; Chen et al., 2011; Hanson and Tan, 2015), relatively little is known about the molecular mechanisms of heat shock response in Chlamydia.
We were interested in examining the full spectrum of the chlamydial response to heat shock, particularly in light of the fact that the pathogen encodes only three sigma factors (Stephens et al., 1998; Thomson et al., 2008). Its primary or housekeeping sigma factor σ66 is the counterpart of σ70 of E. coli (Engel and Ganem, 1990), while its two alternative sigma factors, σ28 and σ54, have counterparts with the same names in E. coli (Stephens et al., 1998; Thomson et al., 2008). Interestingly, we found that a 30 min incubation of C. trachomatis at 45°C upregulated the expression of both σ66 and σ28 and hrcA. The upregulations of these transcription regulators are likely responsible for the increased expression of at least 15.4% of C. trachomatis genes (judged by ≥1.5-fold increase, p < 0.005). For the first time, we document concurrent σ54 downregulation and atoC upregulation in a bacterium in response to heat shock. The discordant expression changes between σ54 and its activator atoC suggest a novel mechanism for fine-tuning the expression of σ54 target genes in response to stress.
Materials and Methods
Host Cells, Chlamydia, and Culture Conditions
Mouse L929 fibroblasts were used as host cells for C. trachomatis. Cells were grown as monolayer cultures at 37°C with air containing 5% CO2 using Dulbecco’s modified Eagle’s medium (DMEM) containing 4.5 g/L glucose and 0.11 g/L sodium pyruvate (Sigma Millipore) supplemented with fetal bovine serum (FBS; Omega Scientific) and gentamicin [final concentrations: 5% (vol/vol) and 10 μg/ml, respectively]. Chlamydia trachomatis L2 (strain 434/BU) was originally purchased from ATCC (Balakrishnan et al., 2006). CtL2/RFP was generated by transforming 434/BU with the pTRL2 (Δgfp) plasmid (Wurihan et al., 2020, 2021a), which carries a far-red fluorescence protein-encoding mKate gene downstream of a C. trachomatis promoter (Wickstrum et al., 2013).
Chlamydia trachomatis Heat Shock and Recovery
For heat shock experiments, EBs were purified via MD-76 gradient ultracentrifugation as described previously (Caldwell et al., 1981). Near-confluent L929 cells grown on 6-well plates were inoculated with EBs at a multiplicity of infection (MOI) of one inclusion-forming unit per cell. The plates were subjected to 20 min centrifugation at 900 × g at RT to synchronize the infection (Wurihan et al., 2021b). Following washes with Hank’s balanced salt saline, the infected cells were cultured in the above medium containing 1 μg/ml cycloheximide in a 37°C incubator. To determine effects of heat shock on the C. trachomatis transcriptome, at 15.5 h postinoculation, a plate with triplicate cultures of wild-type C. trachomatis was transferred into a 45°C incubator and incubated for 30 min, while the control plate was kept in the 37°C incubator. Cultures were terminated at 16 h postinoculation. To determine effects of heat shock on chlamydial growth, heat shock of wild-type C. trachomatis- or CtL2/RFP-infected cultures was initiated at 16 h postinoculation. After incubation at 45°C for 2 to 8 h or re-incubation at 37°C for an additional 6 h, CtL2/RFP cultures were imaged under an Olympus IX51 microscope. Inclusion areas and RFP intensities were quantified using the ImageJ software as previously described (Wurihan et al., 2021a,b). Wild-type C. trachomatis-infected cultures were terminated by removal of the culture medium. Cells were collected into a 1.0 ml of 0.85% NaCl. Following centrifugation, the cell pellet was dissolved with 100 μl of 25 mM NaOH. The lysate was incubated at 95°C for 15 min and subsequently neutralized with 100 μl of 40 mM Tris-HCl (pH 7.2). Samples of the neutralized lysate were used as PCR template using the Applied Biosystems PowerUp SYBR Green Master Mix. Thermo Fisher QS5 qPCR machine was used for qPCR analyses for quantifying the relative copy number of the C. trachomatis genome (Wurihan et al., 2021b).
Cellular RNA Isolation
Total host RNA and chlamydial RNA were isolated using TRI reagent (Millipore Sigma). DNA decontamination was achieved by using RNase-free DNase I (New England Biolabs). RNA concentration was determined using Qubit RNA HS assay kits (Thermo Fisher). Aliquots of the DNA-free RNA samples were stored at −80°C.
RNA Sequencing and Analyses
RNA-Seq was performed as described recently (Wurihan et al., 2021b). Briefly, total RNA integrity was determined using Fragment Analyzer (Agilent) prior to RNA-Seq library preparation. Illumina MRZE706 Ribo-Zero Gold Epidemiology rRNA Removal kit was used to remove mouse and chlamydial rRNAs. Oligo(dT) beads were used to remove mouse mRNA. RNA-Seq libraries were prepared using Illumina TruSeq stranded mRNA-Seq sample preparation protocol, subjected to quantification process, pooled for cBot amplification, and sequenced with Illumina HiSeq 3000 platform with 50 bp single-read sequencing module. Short read sequences were first aligned to the CtL2 chromosome (GenBank accession # NC_010287.1) using TopHat2 aligner and then quantified for gene expression by HTSeq to obtain raw read counts per gene, and then converted to FPKM (Fragment Per Kilobase of gene length per Million reads of the library; Anders and Huber, 2010; Trapnell et al., 2012; Anders et al., 2015). DESeq, an R package commonly used for analysis of data from RNA-Seq studies and test for differential expression, was used to normalize data and find group-pairwise differential gene expression based on three criteria: p < 0.05, average FPKM >1, and fold change ≥1.
Quantitative Reverse Transcription PCR
Quantitative reverse transcription PCR (qRT-PCR) was performed using the Luna Universal One-Step RT-qPCR kit (NEB, Cat. # E3005E) following manufacturer’s instructions. For each reaction with the exception of 23S rRNA qRT-PCR reactions, 10 ng of purified total host and bacterial RNA was used as initial template for cDNA synthesis. For quantifying 23 rRNA, 10 pg. of RNA was used for each reaction. All RT-qPCR reactions were performed in technical duplicate or triplicate. Thermo Fisher QS5 qPCR machine was used for qRT-PCR analyses.
Functional Classification of Genes
COG functional classification of the C. trachomatis proteome (Galperin et al., 2015) was performed with modifications described in previous publications (Wu et al., 2011; Wurihan et al., 2021b) or community-developed Chlamydia databases, namely, Chlambase (Putman et al., 2019) and Chlamdb (Pillonel et al., 2020) perform.
Controlling Inverted Repeat of Chaperone Expression Element and σ28 Promoter Search
The FIMO (find individual motif occurrences) program (Grant et al., 2011) was used to search for controlling inverted repeat of chaperone expression (CIRCE) elements and σ28 promoters in the C. trachomatis genome. The reference motifs were the modified CIRCE element sequence (TAGCA-N15-TGCTAA) identified by De Barsy et al. (2016) and the consensus σ28 promoter binding sequence identified by Yu et al. (TAAAGWWY-N11/12-RYCGAWRN). The search was restricted to 500 nucleotides upstream of the predicted translation start sites for all genes.
Transcriptional Regulatory Network Development
The heat shock transcriptional regulatory network (TRN) was developed for significantly differentially regulated genes (i.e., genes with a ≥1.5-fold change, p < 0.005). Previously identified physical and/or functional associations were automatically integrated into the heat shock TRN using STRING v11 (Szklarczyk et al., 2018). The STRING v11 network was exported to Gephi (Bastian et al., 2009) on which associations identified based on the literature and/or experimental findings from this study were manually developed.
Statistical Analysis
Inclusion area and RFP intensity, and qRT-PCR data were analyzed using t-tests in Excel of Microsoft Office. When applicable, values of p were adjusted for multiple comparisons by Benjamini-Hochberg procedure to control the false discovery rate (Benjamini and Hochberg, 1995).
Results
Heat Shock Induces Robust Transcriptomic Changes in Chlamydia trachomatis
To obtain a snapshot of C. trachomatis heat shock response at the transcriptomic level, we performed RNA sequencing (RNA-Seq) analyses and compared the transcriptome of a set of triplicate C. trachomatis cultures incubated at 45°C for 30 min with that of a set of control cultures maintained at the routine culture temperature 37°C (Figure 1). We chose these experimental settings to determine the full capacity of transcriptomic reprogramming in response to heat shock, even though C. trachomatis infection almost certainly does not increase the body temperature to such a degree. Previous heat shock studies with C. trachomatis and C. muridarum were performed under similar conditions (Engel et al., 1990; Karunakaran et al., 2003; Hanson and Tan, 2015). Importantly, C. trachomatis can recover from heat shock at 45°C. As shown in Figure 1A, red fluorescence protein-expressing C. trachomatis cultures incubated at 45°C for 2 h formed statistically significantly larger inclusions with more intense RFP signals following a 6 h recovery at 37°C, compared to cultures maintained at 45°C without the recovery (Figure 1A). Likewise, quantitative PCR analysis detected a higher level of the bacterial genome in wild-type C. trachomatis cultures exposed to a 2-h incubation at 45°C followed by a 6 h recovery at 37°C relative to heat-shocked cultures without the recovery (Figure 1B). Interestingly, it appears that C. trachomatis partially regained the capacity to replicate its genome when cultured at 45°C for an extended period following an initial complete halt of genome replication. The genome-doubling time at 45°C is estimated to be approximately 6 h, which is three times longer than the normal 2 h (Figure 1B).
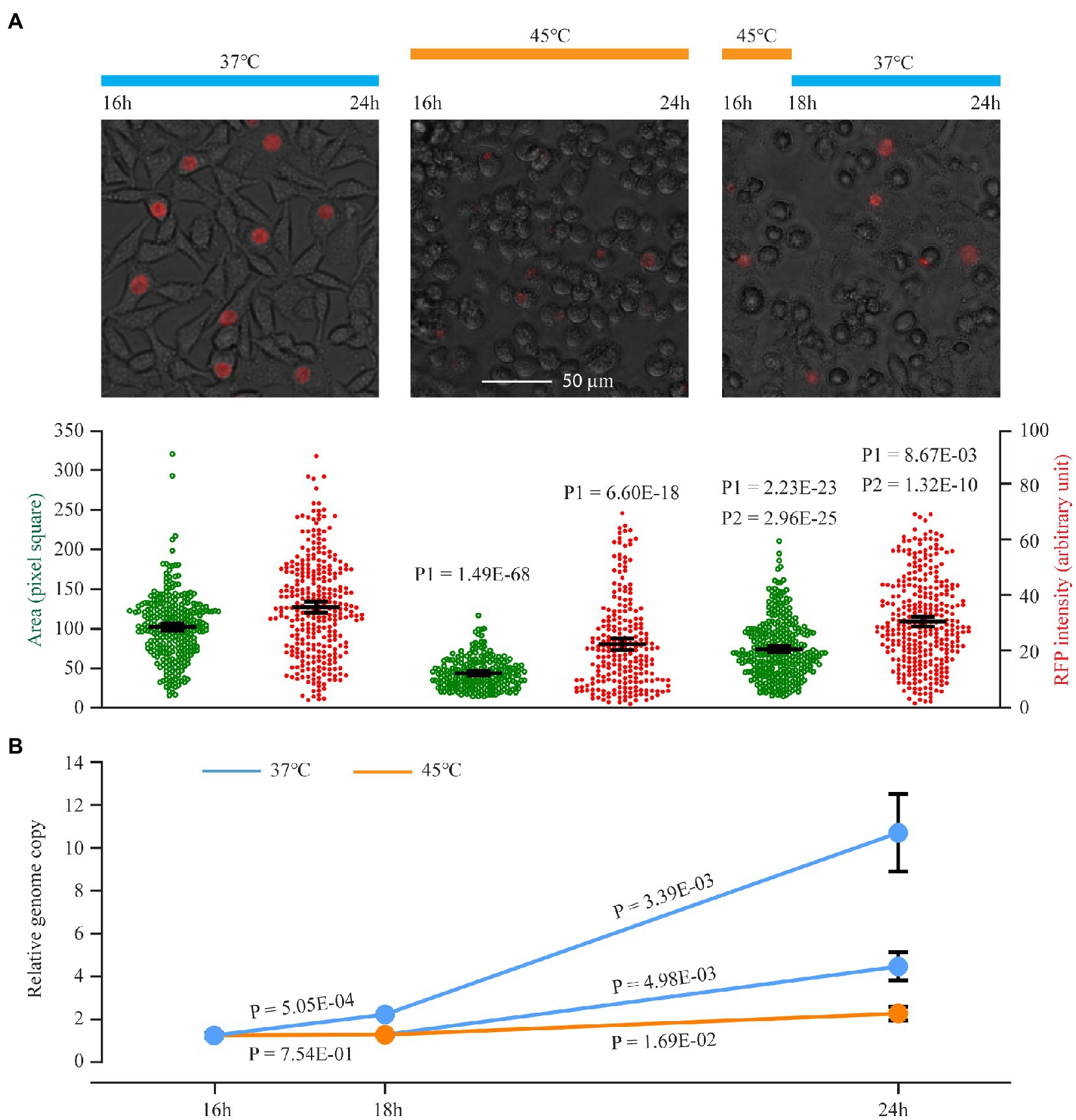
Figure 1. An initial C. trachomatis growth arrest following a switch to 45°C, adaptation upon continued incubation at 45°C, and growth recovery after return to 37°C. (A) Expansion of inclusion size and increased plasmid-expressed RFP intensity upon recovery from heat shock at 45°C. Chlamydia trachomatis CtL2/RFP-infected cells were cultured at 37°C. At 16 h postinoculation, cultures were left at 37°C (left) or subjected to heat shock at 45°C for 8 h (center) or 2 h followed by a 6 h recovery at 37°C (right). At 24 h, RFP and white light phase-contrast images were acquired. The culture temperatures during the experiments are shown on top, representative cell images in the middle, and quantitative inclusion areas and RFP intensities (averages ±95% confidential intervals) at the bottom. The scale bar is applicable to all three images. P1 indicates values of p of t-tests between heat-shocked cultures and control cultures. P2 indicates values of p of t-tests between cultures incubated at 45°C for 2 h followed by a 6 h recovery at 37°C and cultures incubated at 45°C for 8 h (i.e., no recovery). (B) Genome replication in C. trachomatis cultured at 45°C. Wild-type C. trachomatis were subjected to heat shock as in (A). Relative genome copy numbers were quantified using qPCR. Data represent averages ± standard deviations of biological triplicates.
Following incubation at 45°C for 30 min, 303 (30.9%) of the total 979 genes in the C. trachomatis genome underwent ≥1.50-fold expression changes with values of p < 0.005. Transcript copy numbers of 151 (15.4%) genes increased (Supplementary Table S1), while those of 152 (15.5%) genes decreased (Supplementary Table S2). These findings suggest that C. trachomatis is capable of mounting a robust heat shock response via transcriptomic reprogramming even though it lacks a typical heat shock sigma factor.
When the differentially regulated genes were placed into functional groups, it became apparent that certain functional gene groups are disproportionately regulated by heat shock (Figure 2A). While genes involved in energy metabolism, posttranslational modification, protein turnover, type III secretion, and plasmid-encoded genes are disproportionately upregulated (Figure 2A), genes involved in amino acid and peptide transport and metabolism and ribosomal structure and biogenesis are disproportionately downregulated (Figure 2B). As detailed below, quantitative reverse transcription PCR (qRT-PCR) analyses were performed to validate RNA-Seq changes, while pathway analyses were performed to reveal the effects of heat shock on distinct physiological functions and to elucidate the underlying regulatory mechanisms in C. trachomatis.
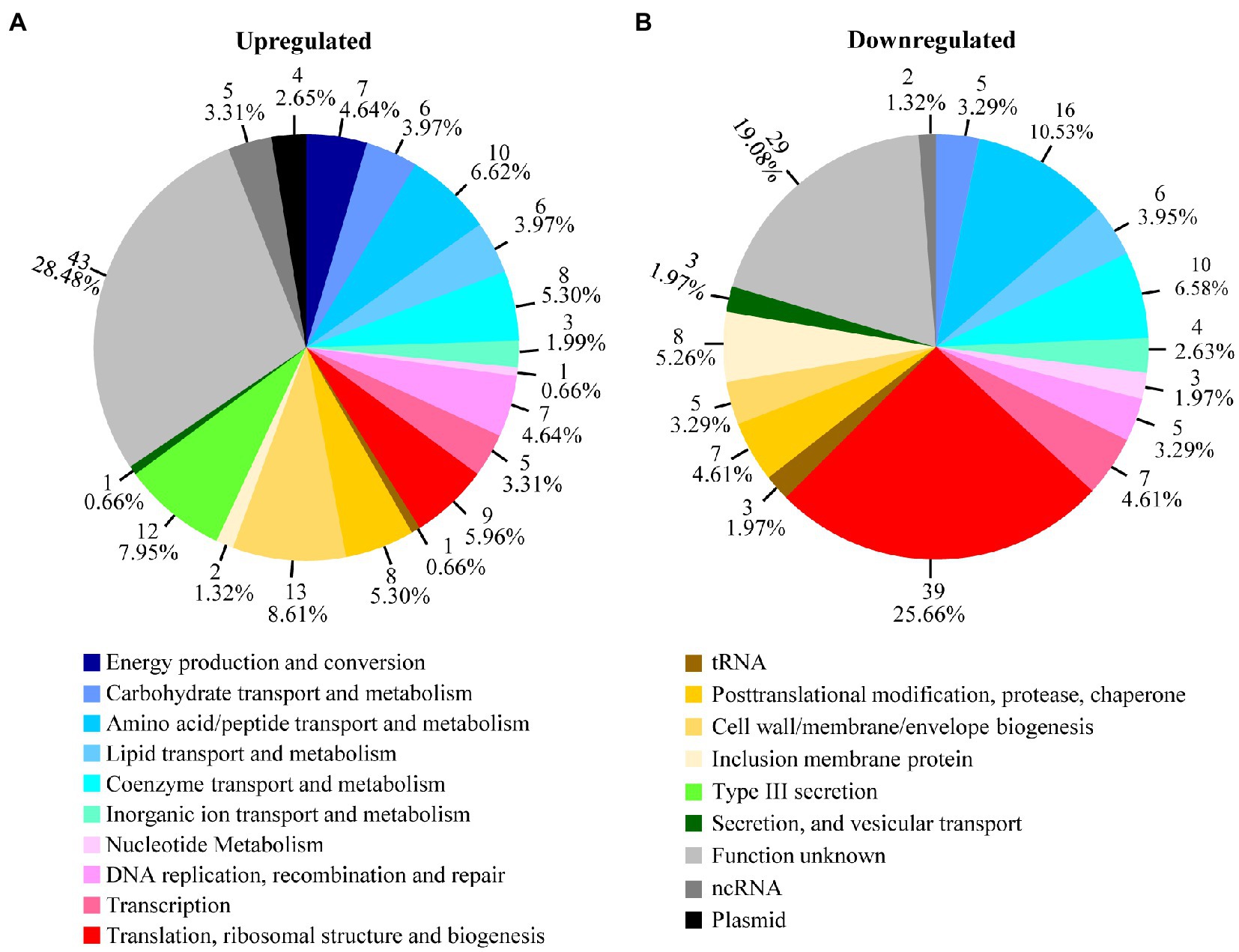
Figure 2. Heat shock-regulated genes in functional groups. Transcripts that were upregulated (A) or downregulated (B) by ≥1.5-fold (p < 0.005) as determined by RNA-Seq following culture at 45°C between 15.5 and 16.0 h postinoculation are organized in pie charts by their functional groups. Shown adjacently to each pie slice is the number of genes and the percentages of up- or downregulated genes in the group. See Supplementary Tables S1 and S2 for identities and functions of individual genes in the functional groups.
Heat Shock Upregulates Energy Production and Conversion
Seven (4.6%) of the 151 genes with increased expression, but none of the genes with decreased expression, were in the functional group of energy metabolism. Four of these seven genes are involved in NADH production. Of these four, pdhA encodes the alpha subunit of the E1 component of the pyruvate dehydrogenase that produces NADH while oxidizing pyruvate to acetyl-coenzyme A (CoA), whereas sucA (2-oxoglutarate dehydrogenase subunit E1) and mdhC (malate dehydrogenase) are involved in generating NADH via the incomplete tricarboxylic acid (TCA) cycle (Stephens et al., 1998; Iliffe-Lee and McClarty, 1999; Figure 3A). ctl0594 (2-oxoisovalerate dehydrogenase subunit alpha/beta) participates in the catalysis of NADH production in the branching amino acid catabolic pathway (Figure 3A). Among the three remaining upregulated genes involved in energy metabolism, ctl0482 (sodium:dicarboxylate symport protein) facilitates the uptake of host-derived glutamate that feeds into the TCA cycle (Figure 3A), while nqrA (Na+-translocating NADH-quinone reductase subunit A) and atpE (V-type ATP synthase subunit E) are involved in reactions in the oxidative phosphorylation chain that produces ATP (Figure 3A).
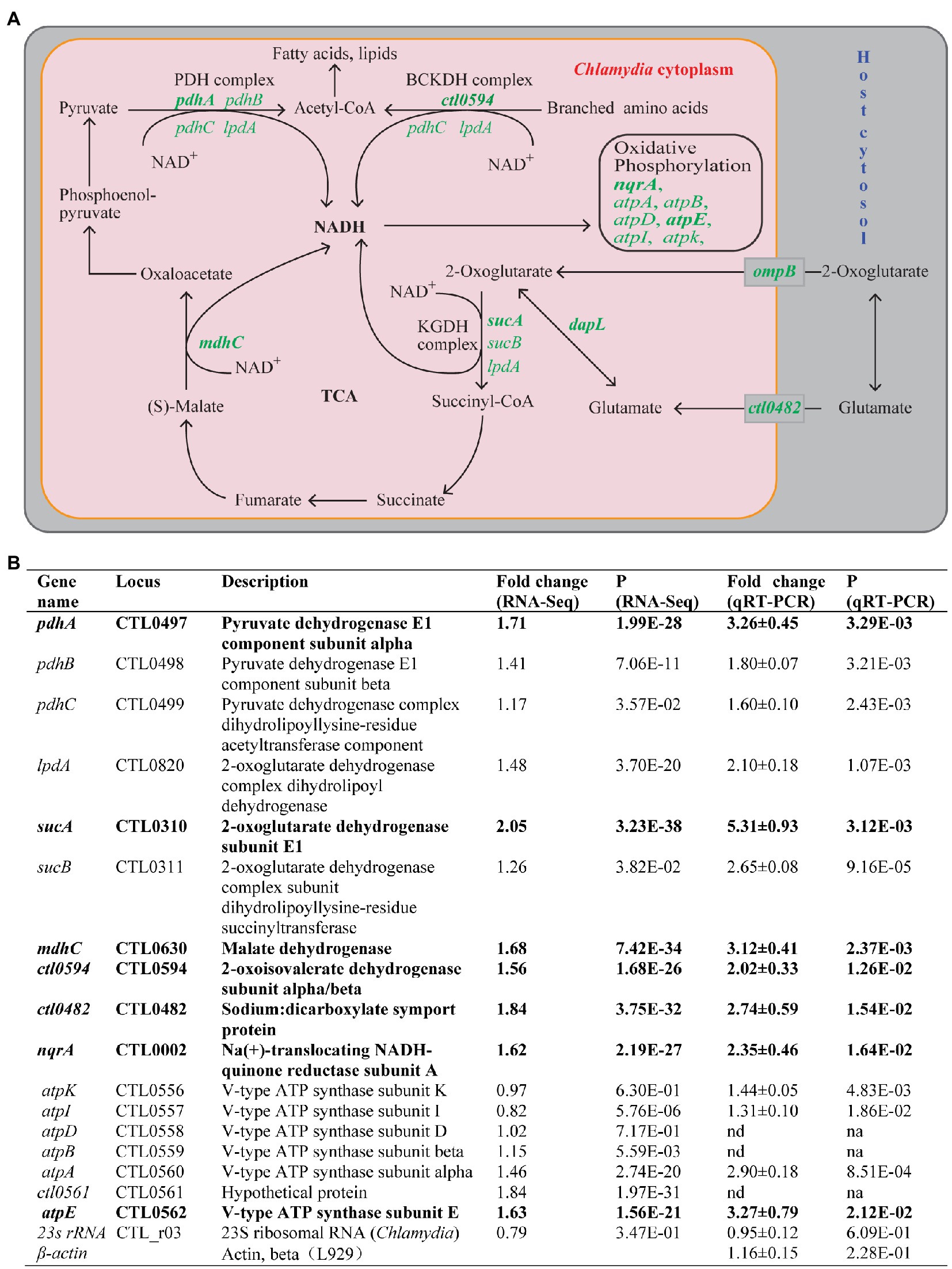
Figure 3. Upregulation of energy metabolism genes in response to heat shock. (A) Energy production and conversion pathways in C. trachomatis. Genes upregulated by heat shock are shown in green. Pink and gray rectangles depict chlamydial and host cytosols, respectively. (B) Functions and expression data of genes with demonstrated or presumptive expression increases. (A,B) Genes that displayed ≥1.5-fold upregulation (p < 0.005) in RNA-Seq are shown in bold text, while those that did not meet the upregulation criterion in RNA-Seq but displayed expression increases in qRT-PCR analysis or presumed to be increased are shown in normal text. atpB and atpD (as well as ctl0561) are presumed to be upregulated by heat shock (even though their expression was not analyzed by qRT-PCR) because they are located within the seven gene operon whose other four genes were significantly upregulated. (B) Mature 23S rRNA and host β-actin were analyzed as controls. Abbreviations: nd, not determined; na, not applicable.
We performed qRT-PCR analyses to validate the expression changes detected by RNA-Seq for the seven aforementioned upregulated energy metabolism-related genes. We included cotranscribed genes in the same operon of these genes in our qRT-PCR analysis. Importantly, we observed expression increases of all three subunits of the pyruvate dehydrogenase (pdhA, pdhB, and pdhC; Figure 3B), as well as both components of the 2-oxoglutarate dehydrogenase complex (sucA and sucB; Figure 3B). Additionally, three genes from the V-ATP synthase operon (atpA, atpI, and atpK) cotranscribed with the aforementioned atpE gene showed expression increases despite the fact that RNA-Seq detected only small or no expression increases for these genes. Although qRT-PCR was not performed for the genes atpD and atpB located in the middle of the V-ATPase operon, our findings predict the expression of these genes was likewise upregulated. Our qRT-PCR analysis also confirmed expression increases of four non-operon genes functionally involved in energy metabolism: mdhC, nqrA, ct0594, and ctl0482 (Figure 3B). In addition, qRT-PCR detected a 2.1-fold expression increase of lpdA whose expression increase in RNA-Seq was slightly below the 1.5-fold change threshold in RNA-Seq (Figure 3B; Supplementary Table S1). LpdA participates in multiple NADH-producing reactions (Figure 3A). As controls, we performed qRT-PCR for C. trachomatis 23S rRNA and host mouse actin RNA; neither was changed by heat shock (Figure 3B). In summary, at least 16 energy production and conversion genes were upregulated thus suggesting that in response to heat shock, C. trachomatis increases its energy production.
In addition to the above “core” energy metabolic genes, RNA-Seq results also showed a 2-fold increase in ompB in response to heat shock (Supplementary Table S1). ompB encodes a porin PorB, which mediates the acquisition of 2-oxoglutarate from the host cell (Kubo and Stephens, 2001; Figure 3A). Furthermore, RNA-Seq also showed a 2.1-fold increase of dapL whose gene product L, L-diaminopimelate aminotransferase converts glutamate to 2-oxoglutarate. Together, these gene upregulations provide C. trachomatis with increased 2-oxoglutarate for the production of NADH.
Heat Shock Upregulates Type III Secretion
The C. trachomatis type III secretion (T3S) system (T3SS) secretes effector proteins to establish and maintain its intracellular growth niche (Fields and Hackstadt, 2000; Subtil et al., 2000; Fields et al., 2003, 2005; Clifton et al., 2004; Hefty and Stephens, 2007; Jamison and Hackstadt, 2008; Hobolt-Pedersen et al., 2009; Markham et al., 2009; Spaeth et al., 2009; Lutter et al., 2010; Chen et al., 2014; Nans et al., 2015; Bugalhão and Mota, 2019). Similar to energy metabolism genes, T3S-related genes are overly represented in the heat shock-upregulated gene group (Figure 2A; Supplementary Table S1). Twelve (7.9%) of the total 151 upregulated genes, but none of the 152 downregulated, were related to T3S (Figure 2; Supplementary Tables S1 and S2). Six of these genes encode proteins that constitute the T3SS structural apparatus (Figure 4A). While flhA, sctC, sctJ, sctL, and sctQ encode components of the basal body, copB and copD encode components of the translocon located in the inclusion membrane. CopB and CopD are also considered effector proteins since they are delivered to the inclusion membrane through the T3SS (Figure 4A). Included among the other six upregulated T3SS-related genes detected by RNA-Seq were copN, ctl0338, ctl0399, ctl0884, and ctl0886, all of which encode T3SS effectors (Figure 4A), as well as ctl0003 which encodes a T3S chaperone that facilitates the secretion of effector proteins. While the functions of CTL0338 and CTL0399 have yet to be determined, CopN and CTL0884 interact with the host cytoskeleton protein tubulin (Archuleta et al., 2011; Nawrotek et al., 2014; Campanacci et al., 2019) and components of the endosomal sorting complexes required for transport (ESCRT; Vromman et al., 2016; Hamaoui et al., 2020), respectively. Furthermore, CTL0886 interacts with the host protein ATG16L1 and counteracts the restriction of ATG16L1 on chlamydial inclusion expansion (Hamaoui et al., 2020).
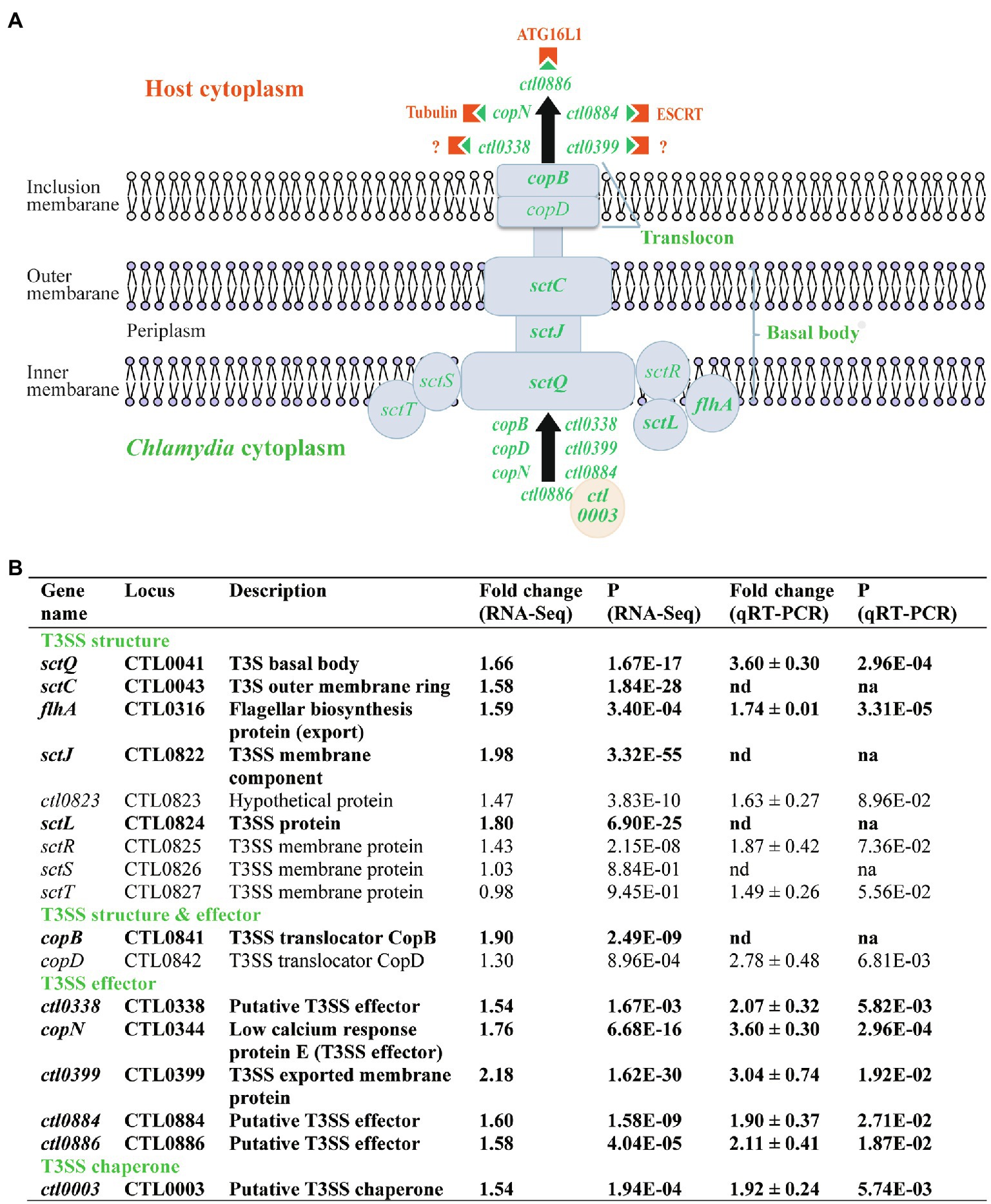
Figure 4. Upregulation of T3S genes in response to heat shock. (A) Schematic presentation of chlamydial T3SS, T3S effectors, and host components that interact with the T3S effectors. Only heat shock-upregulated T3SS and T3S effector genes are specified (shown in green). Host components targeted by the T3S effectors are shown in orange. (B) Functions and expression data of genes with expression increases. (A,B) Genes that displayed ≥1.5-fold upregulation (p < 0.005) in RNA-Seq are shown in bold text, while those that did not meet the upregulation criterion in RNA-Seq but displayed expression increases in qRT-PCR analysis or presumed to be increased are shown in normal text. Note sctS is presumed to be upregulated by heat shock (even though its expression was not analyzed by qRT-PCR) because it is cotranscribed in the same operon along with sctJ, ctl0823, sctL, sctR, and sctT, all of which showed increased transcripts by qRT-PCR and/or RNA-Seq at 45°C.
To validate the RNA-Seq findings, we performed qRT-PCR analyses for eight of the 12 upregulated T3SS genes and confirmed increases in their expression in response to heat shock (Figure 4B). We extended the qRT-PCR studies to include several T3SS operon genes that showed relatively small or even no expression increases by RNA-Seq despite the fact that their cotranscribed genes were upregulated. In the operon spanning from ctl0822 to ctl0827 (Hefty and Stephens, 2007), which encode mostly components of the basal body, RNA-Seq showed a 1.43-fold increase for sctR, whereas qRT-PCR showed a trending significant 1.87-fold increase (Figure 4B). qRT-PCR also showed a trending significant 1.49-fold increase in sctT expression even though RNA-Seq did not detect an expression change. Given these findings, it appears likely that the expression of sctS, the second to the last gene of the operon, is also upregulated in response to heat shock despite it not being included in this study (Figure 4B). Interestingly, we detected a statistically significant 2.78-fold increase in copD expression from the copBD operon, whereas the earlier RNA-Seq analysis had only measured a 1.30-fold increase (Figure 4B). In summary, at least 16 T3S-related genes were significantly upregulated in response to heat shock thus suggesting that C. trachomatis RBs secrete elevated levels of these distinct effector proteins to increase their chance of survival.
Heat Shock Upregulates Five of the Eight Plasmid-Encoded Genes
The C. trachomatis plasmid encodes eight proteins (Pgp1-8) whose functions include fitness improvement (pgp3; Ma et al., 2020), transcriptional regulation (pgp4 and possibly pgp5; Song et al., 2013; Liu et al., 2014a; Zhang et al., 2020), and plasmid maintenance (pgp1, pgp2, pgp6, and pgp8; Figure 5A). Interestingly, RNA-Seq analysis showed 2.4-, 1.67- 1.78-, and 1.60-fold upregulation for pgp1, pgp2, pgp3, and pgp6, respectively, following heat shock. qRT-PCR confirmed the upregulation of these genes (Figure 5B). In addition, qRT-PCR detected a 1.70-fold increase in pgp4 RNA in heat-shocked cultures, even though RNA-Seq did not detect an increase. Notably, the expression of pgp5, pgp7, and pgp8 were unaffected by heat shock either positively or negatively (Figure 5B).
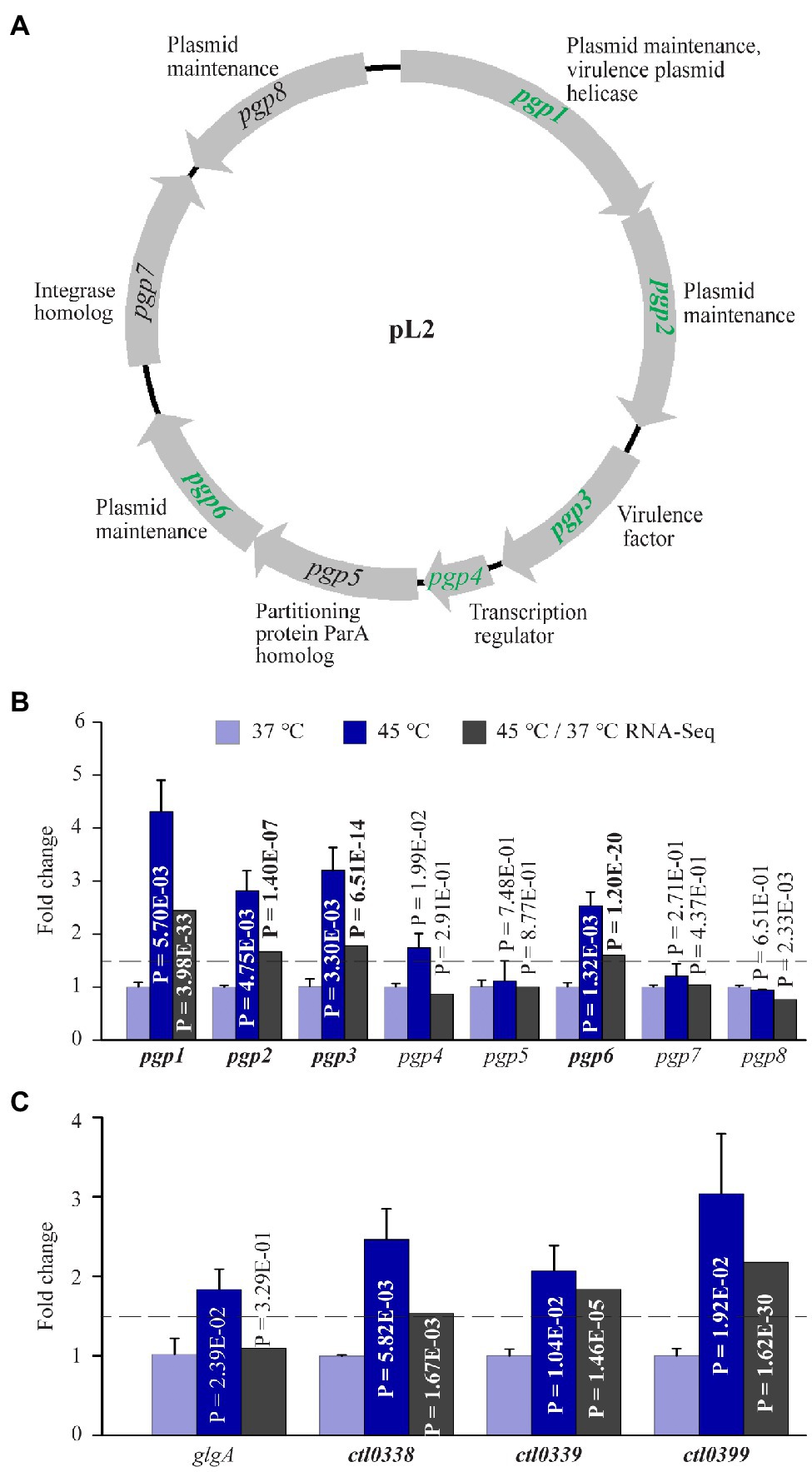
Figure 5. Upregulation of plasmid-encoded genes and Pgp4-regulated chromosomal genes in response to heat shock. (A) The pL2 plasmid map. Shown adjacently to each gene is its function. Genes upregulated by heat shock are shown in green. (B) qRT-PCR and RNA-Seq data for all eight plasmid-encoded genes. (C) Increased expression of all four Pgp4 target genes analyzed by qRT-PCR. (A–C) Genes that displayed ≥1.5-fold upregulation (p < 0.005) in RNA-Seq are shown in bold text, while those that did not meet the upregulation criterion in RNA-Seq but displayed expression increases in qRT-PCR analysis or presumed to be increased are shown in regular text.
The Pgp4 regulon is composed of pgp3 and numerous chromosomal genes (Song et al., 2013; Patton et al., 2018; Zhang et al., 2020). To examine whether the expression of other Pgp4 regulon genes was effected by heat shock, we performed qRT-PCR for four confirmed Pgp4 chromosomal target genes including glgA, ctl0338, ctl0339, and ctl0399 (Song et al., 2013; Zhang et al., 2020). RNA-Seq detected expression increases for ctl0338, ctl0339, and ctl0399, which was confirmed by qRT-PCR data (Figure 5C). Although RNA-Seq failed to detect an expression change in glgA following heat shock, qRT-PCR detected a 1.84-fold increase (Figure 5C). In addition, ctl0638, another Pgp4 target gene, was found by RNA-Seq analysis to be upregulated by 3.69-fold following heat shock (Supplementary Table S1). These findings are consistent with a role for Pgp4 in upregulation of its target genes in response to stress. Taken together, the increased expression of plasmid-encoded genes supports the notion that the C. trachomatis plasmid serves as an important virulence determinant that helps chlamydiae survive during heat shock.
Heat Shock Disproportionately Downregulates Genes With Functions in Protein Synthesis
In contrast to energy metabolism, T3S, and plasmid-encoded genes, RNA-Seq revealed that genes involved in protein translation, ribosomal structure and biogenesis are disproportionately downregulated in heat-shocked C. trachomatis cells. More than a quarter of the 152 downregulated genes (Figure 2B), but only 6% of the 151 upregulated genes (Figure 2A), encode proteins with functions in protein synthesis. The difference in the number of ribosomal protein genes in the two categories was particularly striking: 20 were downregulated, while only two were upregulated (Table 1). We performed qRT-PCR analyses for six of the downregulated genes with different functions in protein synthesis and confirmed their decreased expression (Table 1). Given that ribosomal proteins and factors involved in protein synthesis are the most abundant proteins in the bacterial cytosol, our findings suggest that RBs reduce protein synthesis in general to conserve energy and resources in response to heat shock.
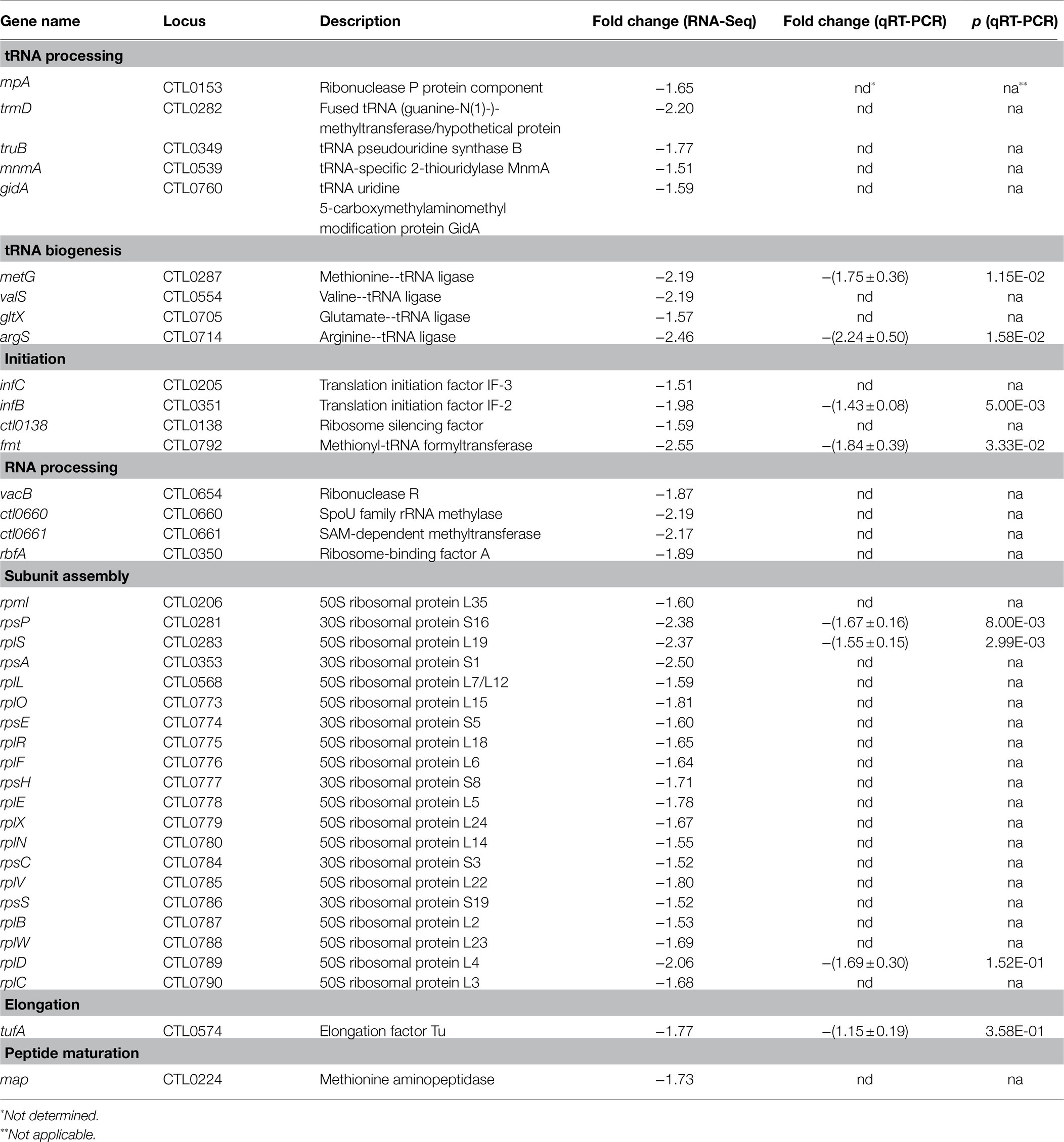
Table 1. Heat shock-downregulated genes associated with translation and ribosomal structure and biogenesis.
Heat Shock Upregulates Expression of hrcA and Its Known and Potential Target Genes
Having validated numerous heat shock-induced transcriptomic changes, we next searched for the underling regulatory mechanisms by analyzing the expression of genes known to regulate stress response and to control gene expression in general. The Chlamydia genome encodes a single heat-inducible transcriptional repressor HrcA (Stephens et al., 1998; Read et al., 2000, 2003; Thomson et al., 2005) whose previously characterized regulon includes two operons (Tan et al., 1996; Wilson and Tan, 2002, 2004; Hanson and Tan, 2015). One of the HrcA-regulated operons encodes HrcA itself, the molecular chaperone DnaK (aka Hsp70), and its cochaperone GrpE. The other operon encodes the molecular chaperone GroEL (also known as chaperonin) and its cochaperone GroES (Figure 6A). HrcA represses transcription of these operons by binding to CIRCE elements within or near their promoters at the physiological temperature (Schulz and Schumann, 1996; Baldini et al., 1998; Narberhaus, 1999; Lund, 2001; Wilson and Tan, 2002; Hu et al., 2007; Roncarati et al., 2019). At abnormally high temperatures or under other stress conditions, HrcA loses the capacity to bind CIRCE elements, leading to activation of its target genes (Schulz and Schumann, 1996; Baldini et al., 1998; Narberhaus, 1999; Lund, 2001; Wilson and Tan, 2002; Hu et al., 2007; Roncarati et al., 2019). As expected, RNA-Seq detected elevated RNA levels of hrcA, grpE, dnaK, groES, and groEL (Figure 6B). For each of these HrcA target genes, qRT-PCR detected an even higher level of increased expression than that measured by RNA-Seq (Figure 6B). By contrast, neither RNA-Seq nor qRT-PCR detected changes in the expression of groEL2 and groEL3 (Figure 6B), which also encode protein chaperones but are not regulated by HrcA due to the absence of CIRCE elements in their promoters (Hanson and Tan, 2015).
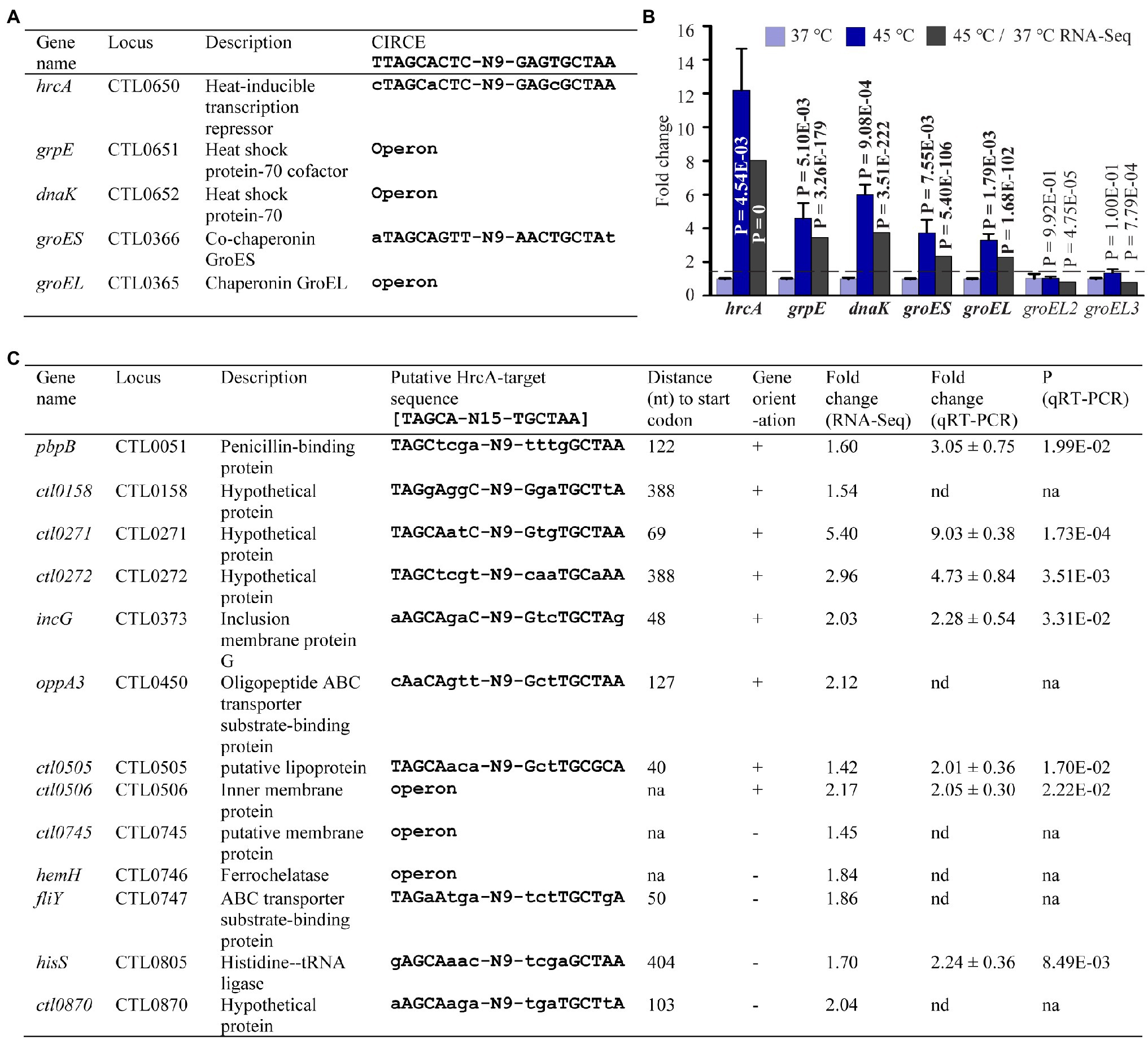
Figure 6. Upregulation of HrcA target and potential target genes in response to heat shock. (A) Name, function, and organization of five known HrcA target genes along with associated CIRCE element sequence previously shown to bind HrcA (see text for references). (B) qRT-PCR and RNA-Seq data for all the five known HrcA target genes. groEL2 and groEL3 are not target genes of HrcA. (C) Newly predicted potential novel HrcA target genes along with associated putative HrcA target binding sequence. Upregulated fold changes in gene expression as detected by RNA-Seq and/or qRT-PCR following heat shock are indicated.
Whereas the consensus sequence for a CIRCE element is 5′-TTAGCACTC-(N)9-GAGTGCTAA-3′ (Hecker et al., 1996; Narberhaus, 1999), De Barsy et al. (2016) recently proposed 5′-TAGCA-(N)15-TGCTAA-3′ as a bona fide HrcA-binding sequence based on studies with Waddlia chondrophila, a Chlamydia-like organism. Using this revised CIRCE consensus sequence, we performed motif search analyses in the C. trachomatis genome and identified eight non-operon and five operon genes carrying the upstream putative HrcA target sequence (Figure 6C). Interestingly, the expression of these genes increased by 1.42- to 9.03-fold as shown by RNA-Seq and/or qRT-PCR analyses (Figure 6C). Notably, these genes encode functionally diverse proteins including four hypothetical proteins (Figure 6C). Our findings thus suggest that transcriptional regulation by C. trachomatis HrcA not only includes chaperone-encoding genes, but genes with a variety of functions.
Heat Shock Induces Expression Changes in All Three Chlamydia trachomatis Sigma Factors and Other Transcriptional Regulators
HrcA-mediated transcriptional regulation accounts for only a small proportion of the heat shock-induced upregulated genes in C. trachomatis (Figure 6). In other bacteria, dedicated heat shock sigma factors of the RNA polymerase are responsible for the activation of numerous heat shock genes (Neidhardt and VanBogelen, 1981; Yamamori and Yura, 1982; Grossman et al., 1984; Völker et al., 1994; Yura, 1996; Hughes and Mathee, 1998). Interestingly, although C. trachomatis does not encode a clearly established heat shock sigma factor, our RNA-Seq data revealed a heat shock-induced 1.85-fold increase of fliA encoding σ28, and a concurrent 3.45-fold reduction of rpoN encoding σ54 (Supplementary Table S1). In addition, RNA-Seq also detected a statistically significant 1.45-fold increase in the RNA of rpoD encoding the housekeeping σ66. Consistent with the RNA-Seq analyses, qRT-PCR analysis detected 3.12- and 2.14-fold increases for fliA and rpoD, respectively, and a 1.56-fold decrease for rpoN (Figure 7A). These findings thus suggest that the relative up- or downregulation of all three sigma factors is an important determinant in facilitating the appropriate transcriptional response following heat shock.
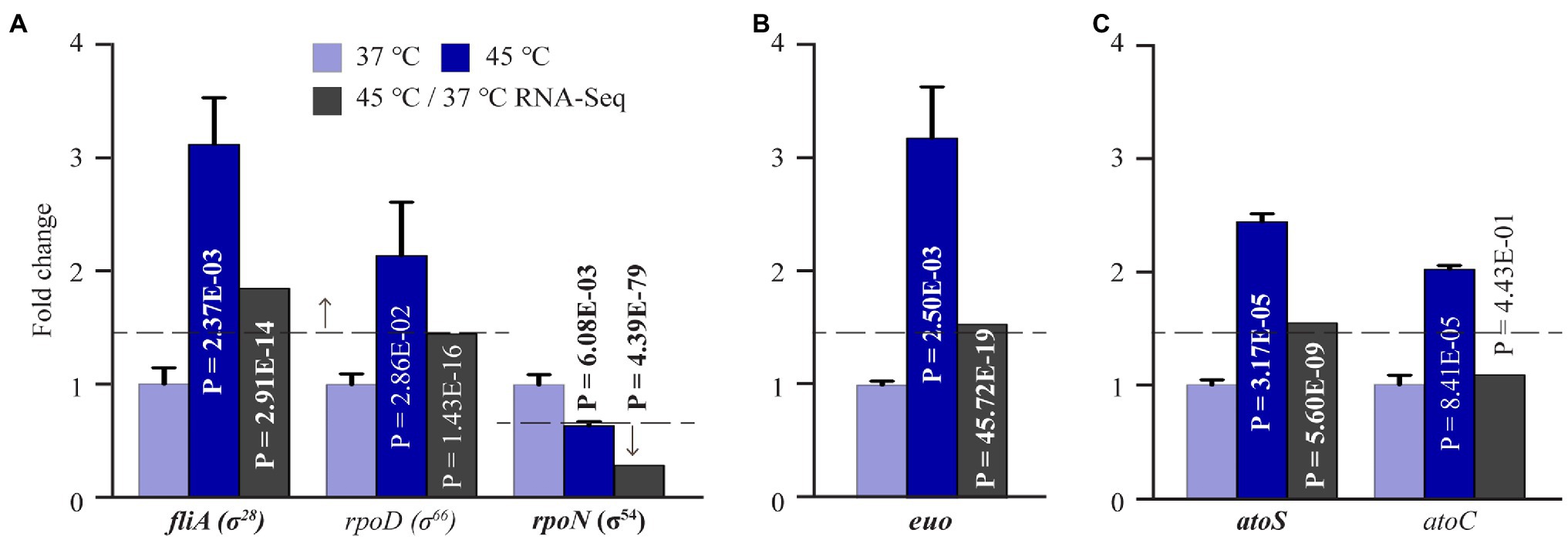
Figure 7. Expression changes of C. trachomatis transcription regulators in response to heat shock. (A) Increased fliA, rpoD expression and decreased rpoN expression as determined by both RNA-Seq and qRT-PCR. (B) Increased euo expression as determined by both RNA-Seq and qRT-PCR. (C) Increased atoS and atoC expression as detected by RNA-Seq and/or qRT-PCR.
In addition to hrcA, two other genes encoding transcription factors showed increased expression following heat shock. Transcripts of ctl0478, which encodes a Crp family transcriptional regulator, increased by 1.52-fold (Supplementary Table S1). This observation is consistent with the role of Crp as a transcriptional regulator of stress response in free-living bacteria (Kallipolitis and Valentin-Hansen, 1998; Ma et al., 2003; Shimizu, 2013). Moreover, transcripts of euo, which encodes a transcriptional repressor of late genes (Rosario and Tan, 2012; Rosario et al., 2014), showed a 1.53-fold expression increase in response to heat shock. Consistent with RNA-Seq, qRT-PCR detected a 3.18-fold increase in euo expression (Figure 7B). However, Euo target genes (e.g., omcA) did not show corresponding expression decreases following heat shock. The possible mechanism for these seemingly inconsistent observations will be discussed below.
RNA-Seq detected a significant 1.55-fold increase in the expression of atoS, a sensor histidine kinase, following heat shock that was confirmed by qRT-PCR (Supplementary Table S1). qRT-PCR additionally revealed a 2.1-fold increase in the expression of atoC, which encodes a σ54 RNAP transcriptional activator (Figure 7C). Together, AtoS and AtoC constitute a two-component system that regulates the σ54 RNA polymerase (Koo and Stephens, 2003; Soules et al., 2020). The significance of the upregulation of this transcriptional regulatory system and concurrent downregulated expression of rpoN (σ54) will also be discussed below.
Remarkably, RNA-Seq detected a heat shock-induced 2.5-fold decrease in nusA, encoding the transcription termination/antitermination protein NusA, and a 2.24-fold reduction in rho, encoding the transcription termination factor Rho (Supplementary Table S2). Furthermore, RNA-Seq detected significant downregulated expression of ctl0818 (ATP-dependent helicase) and ctl0463 (putative transcriptional regulator of the AlgH/UPF0301 family; Supplementary Table S2). Coupled with our detection of altered sigma factor-encoding gene expression, these findings suggest that alterations in transcription initiation, elongation, and termination all contribute to the heat shock-induced changes in the C. trachomatis transcriptome.
Heat Shock-Upregulated Genes Have σ28 Promoters
As mentioned above, both RNA-Seq and qRT-PCR showed a heat shock-induced increase in σ28 (Figure 7A; Supplementary Table S1). To further investigate the role of σ28 in heat shock response, we parsed the RNA-Seq data for previously identified σ28 target genes (Yu et al., 2006b). Interestingly, among the 17 known σ28 targets in C. trachomatis, 6 (pgk, trxB, dnaK, ctl0303, ctl0684, and ctl0815) were found to be upregulated following heat shock, whereas 2 (obgE and ctl0895) were downregulated. These results suggest that upregulation of σ28 in response to heat shock in turn mediates the expression of a subset of its known target genes.
We next performed our own σ28 promoter search within the C. trachomatis genome. Using the consensus σ28 promoter binding sequence identified by Yu et al. (2006a; TAAAGWWY-N11/12-RYCGAWRN; where W is A/T, R is a purine, Y is a pyrimidine, and N is any nucleotide), motif-based sequence analyses identified 33 potential σ28 binding sites within 500 bps upstream of the ATG initiation codon among the 151 heat shock-upregulated genes. qRT-PCR was carried out for 15 of the 33 genes and confirmed their expression was increased in C. trachomatis cells upon heat shock (Table 2). In sum, our findings are consistent with the notion that σ28 plays an important functional role in mediating heat shock-induced gene expression.
Most σ54 Target Genes Are Upregulated in the Presence of Downregulated rpoN and Upregulated atoC Following Heat Shock
The concurrent decreased expression of σ54 and increased expression of the σ54 RNAP activator AtoC (Figure 7; Supplementary Tables S1 and S2) suggests complexity in the regulated expression of σ54 target genes following heat shock. To investigate this apparent paradox more closely, we parsed the RNA-Seq data for the 13 known σ54 target genes recently identified by Soules et al. (2020). Interestingly, only three of the 13 σ54 target genes showed decreased RNA levels in C. trachomatis following heat shock, while the remaining 10 genes showed increased levels. We performed qRT-PCR analysis and confirmed the expression changes revealed by RNA-Seq (Table 3).
Among the 10 σ54 target genes upregulated by heat shock are hrcA and grpE, which are on the same operon with tandem σ66–σ54 promoters (Tan et al., 1996; Soules et al., 2020). As shown in Figures 6A,B, expression of hrcA and grpE from the σ66 promoter is negatively regulated by HrcA and activated by heat shock. omcA is another heat shock-upregulated σ54 target gene with tandem σ66–σ54 promoters (Soules et al., 2020). Thus, decreased σ54 expression would allow for increased transcription from the σ66 promoters of the hrcA-grpE operon and omcA (Soules et al., 2020). Increased expression level of AtoC is likely responsible for the upregulated expression of the remaining seven genes lacking either a σ66 or σ28 promoter in response to heat shock (Soules et al., 2020). Taken together, the data presented in Figures 7A,C, and Table 3 suggest that concurrent downregulation of σ54 and upregulation of AtoC have differential effects on σ54 target gene expression. A hypothetical underlying regulatory mechanism will be presented below.
Heat Shock Alters RNase Expression
RNA-Seq detected expression changes for several RNase-encoding genes following heat shock. Among them, cafE (RNase E) and vacB (RNase R) regulate mRNA decay in bacteria (Nilsson and Uhlin, 1991; Cheng and Deutscher, 2005; Mackie, 2013; Zhang and Gross, 2021). While cafE increased by 1.52-fold (Supplementary Table S1), vacB decreased by 1.87-fold (Supplementary Table S2). These data suggest that in addition to transcription, RNA decay also regulates C. trachomatis transcriptomic reprogramming in response to heat shock.
Chlamydia trachomatis Heat Shock Transcriptional Regulatory Network
To provide a more systemic view of the heat shock-induced transcriptional reprogramming in C. trachomatis, we generated a C. trachomatis TRN diagram, in which red and green nodules represent heat-downregulated and upregulated genes, respectively (Figure 8). We used STRING to produce the heat shock TRN based on the program’s ability to integrate previously identified functional and structural association networks (Szklarczyk et al., 2018). We manually developed edges (using blue lines) for the three sigma factors based on previously reported target genes (Ricci et al., 1993, 1995; Fahr et al., 1995; Tan et al., 1996; Mathews and Timms, 2000; Shen et al., 2000, 2006; Yu and Tan, 2003; Yu et al., 2006b; Hefty and Stephens, 2007; Soules et al., 2020), and from Euo and Pgp4 to their known target genes (Rosario and Tan, 2012; Song et al., 2013; Rosario et al., 2014; Zhang et al., 2020). We also manually developed edges (using red lines) from hrcA and σ28 to their newly identified putative novel target genes (Figure 6; Table 2).
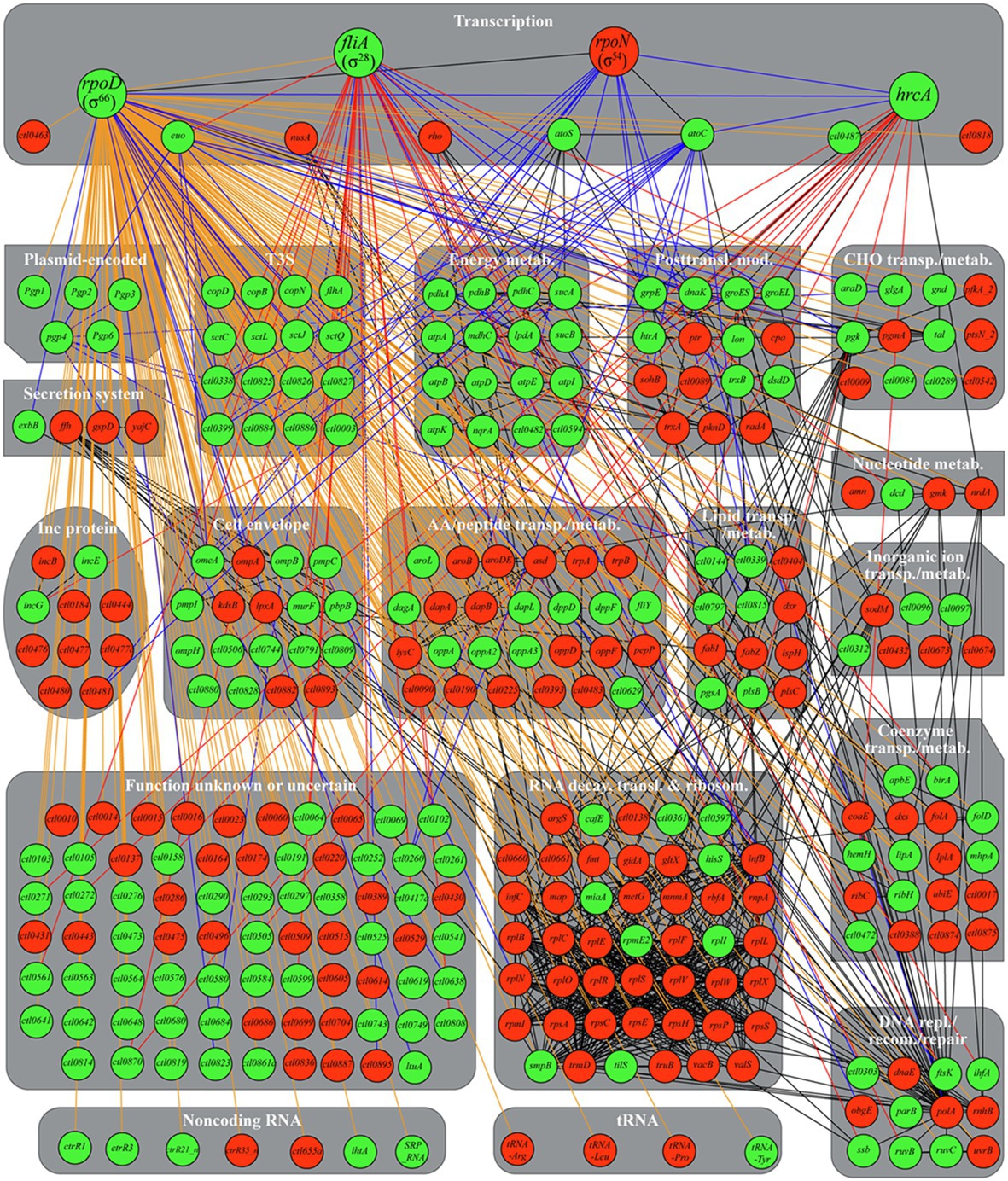
Figure 8. Chlamydia trachomatis heat shock transcriptional regulatory network. Green and red nodules signify heat-upregulated or downregulated genes, respectively, as determined by RNA-Seq and/or qRT-PCR (in cases of disagreements between the two analyses, qRT-PCR results were used to determine expression changes). Physical and/or functional associations identified by STRING are shown with black edges. σ66, σ28, σ54, HrcA, and Pgp4 target genes established in the literature but unrecognized by STRING are identified with blue edges. Putative σ28 and HrcA target genes identified in this study are identified with red edges. Genes without either a σ28 promoter or a σ54 promoter are treated as hypothetical σ66 targets and identified with orange edges. An interactive gephi file, which allows for clear viewing of individual associations, is presented as Supplementary Figure S1. Abbreviations: metab., metabolism; posttransl. mod., posttranslational modification; CHO, carbohydrate; transp., transport; Inc., inclusion membrane; AA, amino acid; transl., translation; ribosom., ribosomal structural and biogenesis; repl, replication; recom., recombination.
The addition of blue and red edges based on the literature and our own findings still left 251 heat shock-regulated genes without a sigma factor. We assume the remaining genes are σ66 targets given its “housekeeping” role in C. trachomatis (Engel and Ganem, 1990; Fahr et al., 1995; Douglas and Hatch, 2000) and manually developed edges to these genes using orange lines. An interactive Gephi Project file (Bastian et al., 2009) for this regulatory network can be accessed at the Figshare public data repository.1
The heat shock TRN shows that σ66 plays the most prominent role in reprogramming the transcriptome based on the number of its target genes (Figure 8). Notably, the transcription of the hrcA-grpE-dnaK operon is controlled not only by σ66 but additionally by a CIRCE element recognized by HrcA. During heat shock, HrcA is denatured, leading to greater induction of chaperone genes (connected to HrcA via black lines) which help maintain the structure and function of key bacterial proteins. Heat-induced HrcA denaturation also likely results in the derepression of a number of genes (connected to HrcA via red lines) with a variety of additional novel cellular functions (see Figure 6). Pgp4, which functions as a transcriptional coactivator (Song et al., 2013; Patton et al., 2018; Zhang et al., 2020), also positively reenforces the expression increases of at least four of σ66 target genes.
σ28 and the σ54/AtoC complex also play major roles in the heat-induced transcriptomic reprogramming as indicated by the numbers of genes connected to these transcriptional regulators (Figure 8). In addition to the numerous genes upregulated by elevated σ66 and σ28 levels, many σ66 target genes are downregulated (Figure 8) thus implicating functional roles for other transcriptional regulators. Interestingly, STRING identified eight genes regulated by the elongation factor and termination regulator NusA (infB, rbfA, rplL, and truB), the transcription terminator Rho (sohB and coaE), or both NusA and Rho (rpsA and polA). Similar to the observed nusA and rho downregulation, these eight genes were likewise downregulated, thus suggesting that NusA and Rho are responsible for their expression. However, how nusA and rho, which are presumed σ66 target genes, are downregulated is unclear. Several additional putative transcriptional regulators (e.g., CTL0463, CTL0487, and CTL0818) also undergo expression changes in response to heat shock, yet the function of these transcription factors in C. trachomatis remains unclear. The potential mechanisms of differential σ54 target gene expression in response to heat shock are discussed below.
Expression changes of a number of heat shock-regulated genes likely involve multiple transcriptional regulators and sigma factors. For example, while a major tandem σ66–σ54 promoter is upstream of the hrcA-grpE-dnaK operon (Soules et al., 2020), an additional σ28 promoter is upstream of the dnaK open reading frame (Yu et al., 2006b). Ctl0338 is also a target of both σ66 and σ54 (Soules et al., 2020) and is further regulated by both Euo and Pgp4 (Zhang et al., 2020).
cafE (RNase E) and vacB (RNase R), which regulate mRNA decay (Nilsson and Uhlin, 1991; Cheng and Deutscher, 2005; Mackie, 2013), are also expected to participate in the C. trachomatis transcriptome reprogramming during heat shock. In summary, the heat shock TRN incorporates novel transcriptional regulatory relations identified in this study as well as those reported in the literature. However, the roles of additional transcriptional regulators and RNases in the transcriptomic reprogramming in response to heat shock in C. trachomatis require further investigations.
Discussion
In response to infections, the human body raises its core temperature to inhibit the growth of pathogens including chlamydiae (Luger, 1948; Qvigstad et al., 1982; Dan et al., 1987; Wu et al., 2000; Reinhold et al., 2008, 2012; Stoner and Cohen, 2015; Clemmons et al., 2019). In this report, we have demonstrated that C. trachomatis is capable of mounting a very robust heat shock response. Our findings have important implications for chlamydial physiology and pathogenesis.
(Patho)Physiological Significance of Transcriptomic Response in Response to Heat Shock
The functions of the proteins up- and downregulated by heat shock support the notion that C. trachomatis reprograms its transcriptome to survive during fever. Among the most striking transcriptional reprogramming events in C. trachomatis following heat shock is the increased expression of energy production and conversion genes (Figures 2, 3; Supplementary Table S1) coupled with broad expression decreases in genes encoding ribosomal proteins and proteins with functions in ribosomal biogenesis (Figure 2; Table 1). Under normal growth conditions, the majority of ATP molecules in bacteria may be spent on protein synthesis (Tempest and Neijssel, 1984; Szaflarski and Nierhaus, 2007). Ribosomal proteins and proteins involved in ribosomal biogenesis represent the two most abundant groups of bacterial cytosolic proteins (Ishihama et al., 2008). Reduction in their synthesis would result in tremendous energy savings. This, coupled with increased ATP production, would help RBs meet the increased demands for ATP-dependent processes, including chaperone-mediated protein folding (Xu et al., 1997; Derre et al., 1999a; Ranson et al., 2006; Clare et al., 2012; Wu et al., 2012; Sarbeng et al., 2015), proteolytic degradation (Goldberg, 1990; Liu et al., 2015; Roncarati and Scarlato, 2017; Wood et al., 2019, 2020), and T3S (Lugert et al., 2004; Wilharm et al., 2007).
In general, the eukaryotic intracellular environment is considered hostile to microorganisms (Moulder, 1982). Secretion of chlamydial effector proteins, mostly through T3S, into host cells is thought to be a crucial mechanism for chlamydial adaption to an obligate intracellular life (Fields and Hackstadt, 2000; Subtil et al., 2000; Fields et al., 2003, 2005; Clifton et al., 2004; Hefty and Stephens, 2007; Jamison and Hackstadt, 2008; Hobolt-Pedersen et al., 2009; Markham et al., 2009; Spaeth et al., 2009; Lutter et al., 2010; Chen et al., 2014; Nans et al., 2015; Bugalhão and Mota, 2019). It is likely that heat shock makes the intracellular environment even less accommodating to chlamydiae. To survive, RBs need to secrete higher amounts of effectors to prevent it from becoming inhospitable. In response to heat shock, C. trachomatis upregulates genes encoding T3SS structural components and chaperones, as well as T3S effectors (Figures 2, 4; Supplementary Table S1). Among the specific T3S effectors upregulated by heat shock, the increased expression of CTL0884 and CTL0886 is particularly intriguing. CTL0886 relieves the inhibitory effect of host protein ATG16L1 on C. trachomatis inclusion expansion (79). However, the precise mechanisms by which CTL0884 contributes to invasion and growth remain unresolved. CTL0884 contains a DUF582 sequence which interacts with components of the ESCRT (Vromman et al., 2016). Some ESCRT complexes are required for the multivesicular body (MVB) pathway and cytokinesis (Babst, 2011; Schmidt and Teis, 2012). Others may be involved in C. trachomatis extrusion exit (Zuck and Hybiske, 2019). Although previous research showed that the host proteins interacting with DUF582 are dispensable for C. trachomatis infection and growth, it is possible that they play more prominent roles for chlamydial survival under stress conditions.
Studies performed on animals indicate that the chlamydial plasmid and its encoded Pgp3 are virulence determinants (Liu et al., 2014b; Zhong, 2017). In this study, we found that heat shock upregulates pgp3 and pgp4, as well as three other plasmid-encoded genes required for the plasmid maintenance, suggesting that Pgp3 and Pgp4 are important for chlamydial survival under stress conditions. Consistent with a role in stress response, it was previously shown that Pgp3 facilitates acid tolerance as evidenced by the fact that, unlike the wild-type bacterium, pgp3-deficient C. muridarum are incapable of colonizing the murine gastrointestinal tract following oral inoculation (Zhong, 2017; Shao et al., 2018; Zhang et al., 2019). Although a role for pgp4 in pathogenicity has not been directly tested in animals, C. muridarum with mutations in glgA, a Pgp4 target gene that is also upregulated by heat shock, is attenuated in pathogenicity in the upper genital tract (Hu et al., 2020). The findings here thus reveal a correlation between proteins previously implicated in chlamydial pathogenicity with Chlamydia survival under stress conditions.
Mechanisms of Heat Shock-Induced Transcriptomic Reprogramming
Heat shock sigma factors, found in most free-living bacteria but not Chlamydia, are responsible for the positive transcriptional regulation of heat shock genes (Neidhardt and VanBogelen, 1981; Yamamori and Yura, 1982; Grossman et al., 1984; Völker et al., 1994; Yura, 1996; Hughes and Mathee, 1998). The observation of increased rpoD and fliA expression in C. trachomatis during heat shock suggests that σ66 and σ28 activate numerous genes in response to the stress. However, it is somewhat counterintuitive that transcript copies of numerous presumptive σ66 target genes as well as some σ28 target genes were downregulated in response to heat shock. Notably, in addition to HrcA, several other transcription factor-encoding genes also showed expression changes following heat shock. Those transcription factors likely fine-tunes the expression of σ66 and σ28 target genes. Furthermore, the increased RNase E expression plausibly mediates the downregulation of some σ66 and σ28 target genes.
rpoN, which encodes σ54, was named for its role in response to nitrogen starvation in E. coli (Hunt and Magasanik, 1985). Previous studies have reported increased σ54 RNAP activity in response to different types of stress including heat shock in some bacteria (Schmid and Lidstrom, 2002). Surprisingly, we found that heat shock significantly downregulates rpoN expression in C. trachomatis. To the best of our knowledge, this is the first documentation of rpoN downregulation in response to environmental stress in bacteria. Unlike the σ66 and σ28 RNAP holoenzymes, the σ54 RNAP holoenzyme requires transcriptional activators, such as AtoC, in C. trachomatis for its functional activity (Merrick, 1993; Ghosh et al., 2010; Soules et al., 2020). Interestingly, we show here that σ54 downregulation is accompanied by an upregulation of the atoS-atoC operon, leading to upregulation of most, but not all, σ54 target genes. We hypothesize that the concurrent downregulation of σ54 and upregulation of AtoC differentially regulate the expression of genes with σ54 promoters depending on the inherent promoter affinity for the σ54 RNAP and whether or not the genes additionally carry σ66 or σ28 promoters. For example, when σ54 is downregulated, “simple” low-affinity σ54 promoters would lose access to the σ54 RNAP leading to reduced transcription. Moreover, σ54 RNAP might also serve as a de facto transcriptional repressor for genes with tandem σ66–σ54 or σ28–σ54 promoters when the AtoC levels are rate-limiting. In this case, downregulation of σ54 would allow σ66 RNAP or σ28 RNAP to transcribe such genes. On the other hand, activation of “high-affinity” σ54 promoters would be more dependent on elevated AtoC levels provided that σ54 levels do not become so low, such as to prevent any access to the promoter.
AtoS, which functions as a sensor kinase, and AtoC constitute a two-component system that regulates transcription of σ54 target genes (Koo and Stephens, 2003; Soules et al., 2020). The functional role of AtoS in facilitating this process has yet to be defined. Although we have not compared the kinase activity of AtoS at normal culture temperatures vs. heat shock conditions, we speculate that AtoS might serve as a sensor of temperature (or possibly of a metabolite whose levels have risen or fallen) during heat shock.
HrcA is the most widely distributed stress-inducible transcriptional repressor in bacteria and has long been recognized to bind and regulate target genes with CIRCE elements in their promoters (Baldini et al., 1998; Narberhaus, 1999; Stewart et al., 2002; Wilson and Tan, 2002, 2004; Spohn et al., 2004; Wilson et al., 2005; Holmes et al., 2010; Hanson and Tan, 2015; Pepe et al., 2018; Roncarati and Scarlato, 2018; Roncarati et al., 2019). By performing chromatin immunoprecipitation, De Barsy et al. (2016) detected HrcA binding to promoters lacking inverted repeat sequences. In this study, we identified DNA sequences that match the revised CIRCE element upstream of 13 C. trachomatis genes. Importantly, all 13 genes were upregulated in response to heat shock, supporting the notion that they serve as novel HrcA regulatory targets. Work is currently underway in our lab to investigate their functional roles in C. trachomatis in response to environmental stress.
Euo functions as a repressor of late genes (Rosario and Tan, 2012; Rosario et al., 2014). Despite increased euo expression, transcript copies of Euo target genes were also increased in response to heat shock. It is possible that the increased euo mRNA did not translate to increased Euo protein. Alternatively, the transcripts of Euo target genes might be stabilized during heat shock. Incidentally, we observed heat shock-downregulated vacB expression. It is conceivable that RNase R, the protein product of vacB, controls the decay of the RNAs of Euo target genes in C. trachomatis, based on previous findings made from other bacteria (Cheng and Deutscher, 2005; Zhang and Gross, 2021).
In summary, we have shown that C. trachomatis, which encodes only three sigma factors and a limited number of transcription factors, is able to mount a robust heat shock response that involves an extensive reprogramming of its transcriptome. This reprogramming meets the cell’s demand for: (a) increased energy production, (b) increased communication with host cells, (c) conservation of resources for protein synthesis, and (d) retention of the fitness and virulence determinants of its plasmid. The reprogramming also enables the RB to replicate its genome at a reduced rate. We show that the reprogramming is accomplished through the actions of all three sigma factors, the heat-inducible transcriptional repressor HrcA, the plasmid-encoded transcription regulator Pgp4, the σ54 RNAP transcriptional activator AtoC, and additional yet-to-be-defined transcription regulators. Clearly, and despite its small size, Chlamydia is a sophisticated organism. Such sophistication likely has made Chlamydia a highly successful parasite. Future strategies that can specifically target and disrupt Chlamydia’s heat shock response will likely be of therapeutic value.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YH, XW, and HF conceived and designed the project. YH, WW, BL, YW, and KW carried out the experiments. YH, WW, YZ, JDF, ZL, XW, and HF analyzed the data. YH, JDF, and HF wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by grants from the National Institutes of Health (grant nos. AI140167 and AI154305 to HF) and National Natural Sciences Foundation of China (grant nos. 82072306 and 81371834 to XW). Genome Sequencing Facility at UTHSA is supported by NIH-NCI P30 CA054174 (Cancer Center at UT Health San Antonio), NIH Shared Instrument grant OD021805 (S10 grant), and CPRIT Core Facility Award (RP160732). YH was supported by a scholarship (award no. 201806370171) from the China Scholarship Council (CSC) from September 2018 to September 2020.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.812448/full#supplementary-material
Footnotes
References
Abdelrahman, Y. M., and Belland, R. J. (2005). The chlamydial developmental cycle. FEMS Microbiol. Rev. 29, 949–959. doi: 10.1016/j.femsre.2005.03.002
Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. doi: 10.1186/gb-2010-11-10-r106
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Archuleta, T. L., Du, Y., English, C. A., Lory, S., Lesser, C., Ohi, M. D., et al. (2011). The Chlamydia effector chlamydial outer protein N (CopN) sequesters tubulin and prevents microtubule assembly. J. Biol. Chem. 286, 33992–33998. doi: 10.1074/jbc.M111.258426
Babst, M. (2011). MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol. 23, 452–457. doi: 10.1016/j.ceb.2011.04.008
Balakrishnan, A., Patel, B., Sieber, S. A., Chen, D., Pachikara, N., Zhong, G., et al. (2006). Metalloprotease inhibitors GM6001 and TAPI-0 inhibit the obligate intracellular human pathogen Chlamydia trachomatis by targeting peptide deformylase of the bacterium. J. Biol. Chem. 281, 16691–16699. doi: 10.1074/jbc.M513648200
Baldini, R. L., Avedissian, M., and Gomes, S. L. (1998). The CIRCE element and its putative repressor control cell cycle expression of the Caulobacter crescentus groESL operon. J. Bacteriol. 180, 1632–1641. doi: 10.1128/JB.180.7.1632-1641.1998
Bastian, M., Heymann, S., and Jacomy, M. (2009). Gephi: an open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 3, 361–362.
Belland, R. J., Nelson, D. E., Virok, D., Crane, D. D., Hogan, D., Sturdevant, D., et al. (2003). Transcriptome analysis of chlamydial growth during IFN-γ-mediated persistence and reactivation. Proc. Natl. Acad. Sci. 100, 15971–15976. doi: 10.1073/pnas.2535394100
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300.
Brinkworth, A. J., Wildung, M. R., and Carabeo, R. A. (2018). Genomewide transcriptional responses of iron-starved Chlamydia trachomatis reveal prioritization of metabolic precursor synthesis over protein translation. mSystems 3:e00184-00117. doi: 10.1128/mSystems.00184-17
Brockett, M. R., and Liechti, G. W. (2021). Persistence alters the interaction between Chlamydia trachomatis and its host cell. Infect. Immun. 89:e0068520. doi: 10.1128/IAI.00685-20
Bugalhão, J. N., and Mota, L. J. (2019). The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the iceberg. Microbial Cell 6, 414–449. doi: 10.15698/mic2019.09.691
Caldwell, H. D., Kromhout, J., and Schachter, J. (1981). Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31, 1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981
Campanacci, V., Urvoas, A., Cantos-Fernandes, S., Aumont-Nicaise, M., Arteni, A. A., Velours, C., et al. (2019). Insight into microtubule nucleation from tubulin-capping proteins. Proc. Natl. Acad. Sci. U. S. A. 116, 9859–9864. doi: 10.1073/pnas.1813559116
Chen, Y. S., Bastidas, R. J., Saka, H. A., Carpenter, V. K., Richards, K. L., Plano, G. V., et al. (2014). The chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog. 10:e1003954. doi: 10.1371/journal.ppat.1003954
Chen, A. L., Wilson, A. C., and Tan, M. (2011). A Chlamydia-specific C-terminal region of the stress response regulator HrcA modulates its repressor activity. J. Bacteriol. 193, 6733–6741. doi: 10.1128/JB.05792-11
Cheng, Z. F., and Deutscher, M. P. (2005). An important role for RNase R in mRNA decay. Mol. Cell 17, 313–318. doi: 10.1016/j.molcel.2004.11.048
Clare, D. K., Vasishtan, D., Stagg, S., Quispe, J., Farr, G. W., Topf, M., et al. (2012). ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin. Cell 149, 113–123. doi: 10.1016/j.cell.2012.02.047
Clemmons, N. S., Jordan, N. N., Brown, A. D., Kough, E. M., Pacha, L. A., Varner, S. M., et al. (2019). Outbreak of Chlamydia pneumoniae infections and X-ray-confirmed pneumonia in army trainees at Fort Leonard Wood, Missouri, 2014. Mil. Med. 184, e196–e199. doi: 10.1093/milmed/usy402
Clifton, D. R., Fields, K. A., Grieshaber, S. S., Dooley, C. A., Fischer, E. R., Mead, D. J., et al. (2004). A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 101, 10166–10171. doi: 10.1073/pnas.0402829101
Dan, M., Tyrrell, L. D., and Goldsand, G. (1987). Isolation of chlamydia trachomatis from the liver of a patient with prolonged fever. Gut 28, 1514–1516. doi: 10.1136/gut.28.11.1514
De Barsy, M., Frandi, A., Panis, G., Théraulaz, L., Pillonel, T., Greub, G., et al. (2016). Regulatory (pan-)genome of an obligate intracellular pathogen in the PVC superphylum. ISME J. 10, 2129–2144. doi: 10.1038/ismej.2016.23
Derre, I., Rapoport, G., Devine, K., Rose, M., and Msadek, T. (1999a). ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32, 581–593. doi: 10.1046/j.1365-2958.1999.01374.x
Derre, I., Rapoport, G., and Msadek, T. (1999b). CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol. Microbiol. 31, 117–131. doi: 10.1046/j.1365-2958.1999.01152.x
Derre, I., Rapoport, G., and Msadek, T. (2000). The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37 degrees C. Mol. Microbiol. 38, 335–347. doi: 10.1046/j.1365-2958.2000.02124.x
Douglas, A. L., and Hatch, T. P. (2000). Expression of the transcripts of the sigma factors and putative sigma factor regulators of Chlamydia trachomatis L2. Gene 247, 209–214. doi: 10.1016/S0378-1119(00)00094-9
Engel, J. N., and Ganem, D. (1990). A polymerase chain reaction-based approach to cloning sigma factors from eubacteria and its application to the isolation of a sigma-70 homolog from Chlamydia trachomatis. J. Bacteriol. 172, 2447–2455. doi: 10.1128/jb.172.5.2447-2455.1990
Engel, J. N., Pollack, J., Perara, E., and Ganem, D. (1990). Heat shock response of murine Chlamydia trachomatis. J. Bacteriol. 172, 6959–6972.
Fahr, M. J., Douglas, A. L., Xia, W., and Hatch, T. P. (1995). Characterization of late gene promoters of Chlamydia trachomatis. J. Bacteriol. 177, 4252–4260. doi: 10.1128/jb.177.15.4252-4260.1995
Feklistov, A., Sharon, B. D., Darst, S. A., and Gross, C. A. (2014). Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376. doi: 10.1146/annurev-micro-092412-155737
Fields, K. A., Fischer, E. R., Mead, D. J., and Hackstadt, T. (2005). Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J. Bacteriol. 187, 6466–6478. doi: 10.1128/JB.187.18.6466-6478.2005
Fields, K. A., and Hackstadt, T. (2000). Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol. Microbiol. 38, 1048–1060. doi: 10.1046/j.1365-2958.2000.02212.x
Fields, K. A., Mead, D. J., Dooley, C. A., and Hackstadt, T. (2003). Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48, 671–683. doi: 10.1046/j.1365-2958.2003.03462.x
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/nar/gku1223
Ghosh, T., Bose, D., and Zhang, X. (2010). Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol. Rev. 34, 611–627. doi: 10.1111/j.1574-6976.2010.00239.x
Goldberg, A. L. (1990). ATP-dependent proteases in prokaryotic and eukaryotic cells. Semin. Cell Biol. 1, 423–432.
Grant, C. E., Bailey, T. L., and Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Grossman, A. D., Erickson, J. W., and Gross, C. A. (1984). The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell 38, 383–390. doi: 10.1016/0092-8674(84)90493-8
Haldenwang, W. G. (1995). The sigma factors of Bacillus subtilis. Microbiol. Rev. 59, 1–30. doi: 10.1128/mr.59.1.1-30.1995
Hamaoui, D., Cosse, M. M., Mohan, J., Lystad, A. H., Wollert, T., and Subtil, A. (2020). The Chlamydia effector CT622/TaiP targets a nonautophagy related function of ATG16L1. Proc. Natl. Acad. Sci. U. S. A. 117, 26784–26794. doi: 10.1073/pnas.2005389117
Hanson, B. R., and Tan, M. (2015). Transcriptional regulation of the Chlamydia heat shock stress response in an intracellular infection. Mol. Microbiol. 97, 1158–1167. doi: 10.1111/mmi.13093
Hecker, M., Schumann, W., and Volker, U. (1996). Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19, 417–428. doi: 10.1046/j.1365-2958.1996.396932.x
Hefty, P. S., and Stephens, R. S. (2007). Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J. Bacteriol. 189, 198–206. doi: 10.1128/JB.01034-06
Helmann, J. D. (1999). Anti-sigma factors. Curr. Opin. Microbiol. 2, 135–141. doi: 10.1016/S1369-5274(99)80024-1
Herman, C., Thévenet, D., D’ari, R., and Bouloc, P. (1995). Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. U. S. A. 92, 3516–3520.
Hobolt-Pedersen, A. S., Christiansen, G., Timmerman, E., Gevaert, K., and Birkelund, S. (2009). Identification of Chlamydia trachomatis CT621, a protein delivered through the type III secretion system to the host cell cytoplasm and nucleus. FEMS Immunol. Med. Microbiol. 57, 46–58. doi: 10.1111/j.1574-695X.2009.00581.x
Holmes, C. W., Penn, C. W., and Lund, P. A. (2010). The hrcA and hspR regulons of Campylobacter jejuni. Microbiology 156, 158–166. doi: 10.1099/mic.0.031708-0
Hu, Y., Oliver, H. F., Raengpradub, S., Palmer, M. E., Orsi, R. H., Wiedmann, M., et al. (2007). Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and σB in Listeria monocytogenes. Appl. Environ. Microbiol. 73, 7981–7991. doi: 10.1128/AEM.01281-07
Hu, C., Wu, H., Sun, Y., Kong, J., Shao, L., Chen, X., et al. (2020). GlgA plays an important role in the induction of hydrosalpinx by Chlamydia muridarum. Pathog. Dis. 78. doi: 10.1093/femspd/ftaa027
Hughes, K. T., and Mathee, K. (1998). The anti-sigma factors. Annu. Rev. Microbiol. 52, 231–286. doi: 10.1146/annurev.micro.52.1.231
Hunt, T. P., and Magasanik, B. (1985). Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc. Natl. Acad. Sci. U. S. A. 82, 8453–8457. doi: 10.1073/pnas.82.24.8453
Huston, W. M., Theodoropoulos, C., Mathews, S. A., and Timms, P. (2008). Chlamydia trachomatis responds to heat shock, penicillin induced persistence, and IFN-gamma persistence by altering levels of the extracytoplasmic stress response protease HtrA. BMC Microbiol. 8:190. doi: 10.1186/1471-2180-8-190
Hybiske, K., and Stephens, R. S. (2007a). Mechanisms of Chlamydia trachomatis entry into nonphagocytic cells. Infect. Immun. 75, 3925–3934. doi: 10.1128/IAI.00106-07
Hybiske, K., and Stephens, R. S. (2007b). Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U. S. A. 104, 11430–11435. doi: 10.1073/pnas.0703218104
Iliffe-Lee, E. R., and Mcclarty, G. (1999). Glucose metabolism in Chlamydia trachomatis: the ‘energy parasite’ hypothesis revisited. Mol. Microbiol. 33, 177–187. doi: 10.1046/j.1365-2958.1999.01464.x
Ishihama, Y., Schmidt, T., Rappsilber, J., Mann, M., Hartl, F. U., Kerner, M. J., et al. (2008). Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9:102. doi: 10.1186/1471-2164-9-102
Jamison, W. P., and Hackstadt, T. (2008). Induction of type III secretion by cell-free chlamydia trachomatis elementary bodies. Microb. Pathog. 45, 435–440. doi: 10.1016/j.micpath.2008.10.002
Kallipolitis, B. H., and Valentin-Hansen, P. (1998). Transcription of rpoH, encoding the Escherichia coli heat-shock regulator sigma32, is negatively controlled by the cAMP-CRP/CytR nucleoprotein complex. Mol. Microbiol. 29, 1091–1099.
Karunakaran, K. P., Noguchi, Y., Read, T. D., Cherkasov, A., Kwee, J., Shen, C., et al. (2003). Molecular analysis of the multiple GroEL proteins of Chlamydiae. J. Bacteriol. 185, 1958–1966. doi: 10.1128/JB.185.6.1958-1966.2003
Koo, I. C., and Stephens, R. S. (2003). A developmentally regulated two-component signal transduction system in Chlamydia. J. Biol. Chem. 278, 17314–17319. doi: 10.1074/jbc.M212170200
Kubo, A., and Stephens, R. S. (2001). Substrate-specific diffusion of select dicarboxylates through Chlamydia trachomatis PorB. Microbiology 147, 3135–3140. doi: 10.1099/00221287-147-11-3135
Liu, Y., Chen, C., Gong, S., Hou, S., Qi, M., Liu, Q., et al. (2014a). Transformation of Chlamydia muridarum reveals a role for Pgp5 in suppression of plasmid-dependent gene expression. J. Bacteriol. 196, 989–998. doi: 10.1128/JB.01161-13
Liu, Y., Huang, Y., Yang, Z., Sun, Y., Gong, S., Hou, S., et al. (2014b). Plasmid-encoded Pgp3 is a major virulence factor for Chlamydia muridarum to induce hydrosalpinx in mice. Infect. Immun. 82, 5327–5335. doi: 10.1128/IAI.02576-14
Liu, C., Niu, Y., Zhou, X., Zheng, X., Wang, S., Guo, Q., et al. (2015). Streptococcus mutans copes with heat stress by multiple transcriptional regulons modulating virulence and energy metabolism. Sci. Rep. 5:12929. doi: 10.1038/srep12929
Luger, N. M. (1948). “Fever of unknown origin” due to lymphogranuloma venereum. N. Engl. J. Med. 238, 44–47. doi: 10.1056/NEJM194801082380202
Lugert, R., Kuhns, M., Polch, T., and Gross, U. (2004). Expression and localization of type III secretion-related proteins of Chlamydia pneumoniae. Med. Microbiol. Immunol. 193, 163–171. doi: 10.1007/s00430-003-0206-x
Lund, P. A. (2001). Microbial molecular chaperones. Adv. Microb. Physiol. 44, 93–140. doi: 10.1016/S0065-2911(01)44012-4
Lutter, E. I., Bonner, C., Holland, M. J., Suchland, R. J., Stamm, W. E., Jewett, T. J., et al. (2010). Phylogenetic analysis of Chlamydia trachomatis tarp and correlation with clinical phenotype. Infect. Immun. 78, 3678–3688. doi: 10.1128/IAI.00515-10
Ma, J., He, C., Huo, Z., Xu, Y., Arulanandam, B., Liu, Q., et al. (2020). The cryptic plasmid improves Chlamydia fitness in different regions of the gastrointestinal tract. Infect. Immun. 88:e00860-00819. doi: 10.1128/IAI.00860-19
Ma, Z., Richard, H., and Foster, J. W. (2003). pH-dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of the GadX regulator and Escherichia coli acid resistance. J. Bacteriol. 185, 6852–6859. doi: 10.1128/JB.185.23.6852-6859.2003
Mackie, G. A. (2013). RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 11, 45–57. doi: 10.1038/nrmicro2930
Markham, A. P., Jaafar, Z. A., Kemege, K. E., Middaugh, C. R., and Hefty, P. S. (2009). Biophysical characterization of Chlamydia trachomatis CT584 supports its potential role as a type III secretion needle tip protein. Biochemistry 48, 10353–10361. doi: 10.1021/bi901200y
Mathews, S. A., and Timms, P. (2000). Identification and mapping of sigma-54 promoters in Chlamydia trachomatis. J. Bacteriol. 182, 6239–6242. doi: 10.1128/JB.182.21.6239-6242.2000
Merrick, M. J. (1993). In a class of its own--the RNA polymerase sigma factor sigma 54 (sigma N). Mol. Microbiol. 10, 903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x
Mooney, R. A., Darst, S. A., and Landick, R. (2005). Sigma and RNA polymerase: an on-again, off-again relationship? Mol. Cell 20, 335–345. doi: 10.1016/j.molcel.2005.10.015
Moulder, J. W. (1982). The relation of basic biology to pathogenic potential in the genus Chlamydia. Infection 10(Suppl. 1), S10–S18. doi: 10.1007/BF01640709
Nair, S., Derre, I., Msadek, T., Gaillot, O., and Berche, P. (2000). CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35, 800–811. doi: 10.1046/j.1365-2958.2000.01752.x
Nans, A., Kudryashev, M., Saibil, H. R., and Hayward, R. D. (2015). Structure of a bacterial type III secretion system in contact with a host membrane in situ. Nat. Commun. 6:10114. doi: 10.1038/ncomms10114
Narberhaus, F. (1999). Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31, 1–8. doi: 10.1046/j.1365-2958.1999.01166.x
Nawrotek, A., Guimaraes, B. G., Velours, C., Subtil, A., Knossow, M., and Gigant, B. (2014). Biochemical and structural insights into microtubule perturbation by CopN from Chlamydia pneumoniae. J. Biol. Chem. 289, 25199–25210. doi: 10.1074/jbc.M114.568436
Neidhardt, F. C., and Vanbogelen, R. A. (1981). Positive regulatory gene for temperature-controlled proteins in Escherichia coli. Biochem. Biophys. Res. Commun. 100, 894–900. doi: 10.1016/S0006-291X(81)80257-4
Nilsson, P., and Uhlin, B. E. (1991). Differential decay of a polycistronic Escherichia coli transcript is initiated by RNaseE-dependent endonucleolytic processing. Mol. Microbiol. 5, 1791–1799. doi: 10.1111/j.1365-2958.1991.tb01928.x
Patton, M. J., Chen, C. Y., Yang, C., Mccorrister, S., Grant, C., Westmacott, G., et al. (2018). Plasmid negative regulation of CPAF expression is Pgp4 independent and restricted to invasive Chlamydia trachomatis biovars. MBio 9. doi: 10.1128/mBio.02164-17
Pepe, S., Pinatel, E., Fiore, E., Puccio, S., Peano, C., Brignoli, T., et al. (2018). The Helicobacter pylori heat-shock repressor HspR: definition of its direct regulon and characterization of the cooperative DNA-binding mechanism on its own promoter. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01887
Pillonel, T., Tagini, F., Bertelli, C., and Greub, G. (2020). ChlamDB: a comparative genomics database of the phylum Chlamydiae and other members of the Planctomycetes-Verrucomicrobiae-Chlamydiae superphylum. Nucleic Acids Res. 48, D526–D534. doi: 10.1093/nar/gkz924
Putman, T., Hybiske, K., Jow, D., Afrasiabi, C., Lelong, S., Cano, M. A., et al. (2019). ChlamBase: a curated model organism database for the chlamydia research community. Database 2019. doi: 10.1093/database/baz041
Qvigstad, E., Skaug, K., Jerve, F., Vik, I. S., and Ulstrup, J. C. (1982). Therapeutic abortion and chlamydia trachomatis infection. Br. J. Vener. Dis. 58, 182–183. doi: 10.1136/sti.58.3.182
Ranson, N. A., Clare, D. K., Farr, G. W., Houldershaw, D., Horwich, A. L., and Saibil, H. R. (2006). Allosteric signaling of ATP hydrolysis in GroEL-GroES complexes. Nat. Struct. Mol. Biol. 13, 147–152. doi: 10.1038/nsmb1046
Read, T. D., Brunham, R. C., Shen, C., Gill, S. R., Heidelberg, J. F., White, O., et al. (2000). Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28, 1397–1406. doi: 10.1093/nar/28.6.1397
Read, T. D., Myers, G. S., Brunham, R. C., Nelson, W. C., Paulsen, I. T., Heidelberg, J., et al. (2003). Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31, 2134–2147. doi: 10.1093/nar/gkg321
Reinhold, P., Kirschvink, N., Theegarten, D., and Berndt, A. (2008). An experimentally induced Chlamydia suis infection in pigs results in severe lung function disorders and pulmonary inflammation. Vet. Res. 39:35. doi: 10.1051/vetres:2008012
Reinhold, P., Ostermann, C., Liebler-Tenorio, E., Berndt, A., Vogel, A., Lambertz, J., et al. (2012). A bovine model of respiratory Chlamydia psittaci infection: challenge dose titration. PLoS One 7:e30125. doi: 10.1371/journal.pone.0030125
Ricci, S., Cevenini, R., Cosco, E., Comanducci, M., Ratti, G., and Scarlato, V. (1993). Transcriptional analysis of the Chlamydia trachomatis plasmid pCT identifies temporally regulated transcripts, anti-sense RNA and sigma 70-selected promoters. Mol. Gen. Genet. 237, 318–326. doi: 10.1007/BF00279434
Ricci, S., Ratti, G., and Scarlato, V. (1995). Transcriptional regulation in the Chlamydia trachomatis pCT plasmid. Gene 154, 93–98. doi: 10.1016/0378-1119(94)00825-D
Rodriguez Ayala, F., Bartolini, M., and Grau, R. (2020). The stress-responsive alternative sigma factor SigB of Bacillus subtilis and its relatives: an old friend with new functions. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01761
Roncarati, D., Pinatel, E., Fiore, E., Peano, C., Loibman, S., and Scarlato, V. (2019). Helicobacter pylori stress-response: definition of the HrcA regulon. Microorganisms 7. doi: 10.3390/microorganisms7100436
Roncarati, D., and Scarlato, V. (2017). Regulation of heat-shock genes in bacteria: from signal sensing to gene expression output. FEMS Microbiol. Rev. 41, 549–574. doi: 10.1093/femsre/fux015
Roncarati, D., and Scarlato, V. (2018). The interplay between two transcriptional repressors and chaperones orchestrates Helicobacter pylori heat-shock response. Int. J. Mol. Sci. 19. doi: 10.3390/ijms19061702
Rosario, C. J., Hanson, B. R., and Tan, M. (2014). The transcriptional repressor EUO regulates both subsets of Chlamydia late genes. Mol. Microbiol. 94, 888–897. doi: 10.1111/mmi.12804
Rosario, C. J., and Tan, M. (2012). The early gene product EUO is a transcriptional repressor that selectively regulates promoters of Chlamydia late genes. Mol. Microbiol. 84, 1097–1107. doi: 10.1111/j.1365-2958.2012.08077.x
Sarbeng, E. B., Liu, Q., Tian, X., Yang, J., Li, H., Wong, J. L., et al. (2015). A functional DnaK dimer is essential for the efficient interaction with Hsp40 heat shock protein*. J. Biol. Chem. 290, 8849–8862. doi: 10.1074/jbc.M114.596288
Schmid, A. K., and Lidstrom, M. E. (2002). Involvement of two putative alternative sigma factors in stress response of the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 184, 6182–6189. doi: 10.1128/JB.184.22.6182-6189.2002
Schmidt, O., and Teis, D. (2012). The ESCRT machinery. Curr. Biol. 22, R116–R120. doi: 10.1016/j.cub.2012.01.028
Schulz, A., and Schumann, W. (1996). hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178, 1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996
Shao, L., Zhang, T., Melero, J., Huang, Y., Liu, Y., Liu, Q., et al. (2018). The genital tract virulence factor pGP3 is essential for Chlamydia muridarum colonization in the gastrointestinal tract. Infect. Immun. 86. doi: 10.1128/IAI.00429-17
Shen, L., Feng, X., Yuan, Y., Luo, X., Hatch, T. P., Hughes, K. T., et al. (2006). Selective promoter recognition by chlamydial sigma28 holoenzyme. J. Bacteriol. 188, 7364–7377. doi: 10.1128/JB.01014-06
Shen, L., Shi, Y., Douglas, A. L., Hatch, T. P., O’Connell, C. M., Chen, J. M., et al. (2000). Identification and characterization of promoters regulating tuf expression in Chlamydia trachomatis serovar F. Arch. Biochem. Biophys. 379, 46–56. doi: 10.1006/abbi.2000.1854
Shima, K., Kaufhold, I., Eder, T., Kading, N., Schmidt, N., Ogunsulire, I. M., et al. (2021). Regulation of the mitochondrion-fatty acid axis for the metabolic reprogramming of Chlamydia trachomatis during treatment with beta-lactam antimicrobials. MBio 12. doi: 10.1128/mBio.00023-21
Shimizu, K. (2013). Regulation Systems of Bacteria such as Escherichia coli in response to nutrient limitation and environmental stresses. Meta 4, 1–35. doi: 10.3390/metabo4010001
Slade, J. A., Brockett, M., Singh, R., Liechti, G. W., and Maurelli, A. T. (2019). Fosmidomycin, an inhibitor of isoprenoid synthesis, induces persistence in Chlamydia by inhibiting peptidoglycan assembly. PLoS Pathog. 15:e1008078. doi: 10.1371/journal.ppat.1008078
Song, L., Carlson, J. H., Whitmire, W. M., Kari, L., Virtaneva, K., Sturdevant, D. E., et al. (2013). Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect. Immun. 81, 636–644. doi: 10.1128/IAI.01305-12
Soules, K. R., Labrie, S. D., May, B. H., and Hefty, P. S. (2020). Sigma 54-regulated transcription is associated with membrane reorganization and type III secretion effectors during conversion to infectious forms of Chlamydia trachomatis. MBio 11. doi: 10.1128/mBio.01725-20
Spaeth, K. E., Chen, Y. S., and Valdivia, R. H. (2009). The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 5:e1000579. doi: 10.1371/journal.ppat.1000579
Spohn, G., Danielli, A., Roncarati, D., Delany, I., Rappuoli, R., and Scarlato, V. (2004). Dual control of Helicobacter pylori heat shock gene transcription by HspR and HrcA. J. Bacteriol. 186, 2956–2965. doi: 10.1128/JB.186.10.2956-2965.2004
Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., et al. (1998). Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282, 754–759. doi: 10.1126/science.282.5389.754
Stewart, G. R., Wernisch, L., Stabler, R., Mangan, J. A., Hinds, J., Laing, K. G., et al. (2002). Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarraysaaA list of the 100 ORFs most highly induced by heat shock is provided as supplementary data with the online version of this paper (http://mic.sgmjournals.org). Microbiology 148, 3129–3138. doi: 10.1099/00221287-148-10-3129
Stoner, B. P., and Cohen, S. E. (2015). Lymphogranuloma venereum 2015: clinical presentation, diagnosis, and treatment. Clin. Infect. Dis. 61, S865–S873. doi: 10.1093/cid/civ756
Subtil, A., Blocker, A., and Dautry-Varsat, A. (2000). Type III secretion system in Chlamydia species: identified members and candidates. Microbes Infect. 2, 367–369. doi: 10.1016/S1286-4579(00)00335-X
Szaflarski, W., and Nierhaus, K. H. (2007). Question 7: optimized energy consumption for protein synthesis. Orig. Life Evol. Biosph. 37, 423–428. doi: 10.1007/s11084-007-9091-4
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2018). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Tan, M., Wong, B., and Engel, J. N. (1996). Transcriptional organization and regulation of the dnaK and groE operons of Chlamydia trachomatis. J. Bacteriol. 178, 6983–6990. doi: 10.1128/jb.178.23.6983-6990.1996
Tempest, D. W., and Neijssel, O. M. (1984). The status of YATP and maintenance energy as biologically interpretable phenomena. Annu. Rev. Microbiol. 38, 459–513. doi: 10.1146/annurev.mi.38.100184.002331
Thomson, N. R., Holden, M. T. G., Carder, C., Lennard, N., Lockey, S. J., Marsh, P., et al. (2008). Chlamydia trachomatis: genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 18, 161–171. doi: 10.1101/gr.7020108
Thomson, N. R., Yeats, C., Bell, K., Holden, M. T., Bentley, S. D., Livingstone, M., et al. (2005). The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15, 629–640. doi: 10.1101/gr.3684805
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Völker, U., Engelmann, S., Maul, B., Riethdorf, S., Völker, A., Schmid, R., et al. (1994). Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140, 741–752. doi: 10.1099/00221287-140-4-741
Vromman, F., Perrinet, S., Gehre, L., and Subtil, A. (2016). The DUF582 proteins of Chlamydia trachomatis bind to components of the ESCRT machinery, which is dispensable for bacterial growth in vitro. Front. Cell. Infect. Microbiol. 6:123. doi: 10.3389/fcimb.2016.00123
Wickstrum, J., Sammons, L. R., Restivo, K. N., and Hefty, P. S. (2013). Conditional gene expression in Chlamydia trachomatis using the tet system. PLoS One 8:e76743. doi: 10.1371/journal.pone.0076743
Wilharm, G., Dittmann, S., Schmid, A., and Heesemann, J. (2007). On the role of specific chaperones, the specific ATPase, and the proton motive force in type III secretion. Int. J. Med. Microbiol. 297, 27–36. doi: 10.1016/j.ijmm.2006.10.003
Wilson, A. C., and Tan, M. (2002). Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J. Bacteriol. 184, 6566–6571. doi: 10.1128/JB.184.23.6566-6571.2002
Wilson, A. C., and Tan, M. (2004). Stress response gene regulation in Chlamydia is dependent on HrcA-CIRCE interactions. J. Bacteriol. 186, 3384–3391. doi: 10.1128/JB.186.11.3384-3391.2004
Wilson, A. C., Wu, C. C., Yates, J. R. 3rd, and Tan, M. (2005). Chlamydial GroEL autoregulates its own expression through direct interactions with the HrcA repressor protein. J. Bacteriol. 187, 7535–7542. doi: 10.1128/JB.187.21.7535-7542.2005
Wood, N. A., Blocker, A. M., Seleem, M. A., Conda-Sheridan, M., Fisher, D. J., and Ouellette, S. P. (2020). The ClpX and ClpP2 orthologs of Chlamydia trachomatis perform discrete and essential functions in organism growth and development. MBio 11. doi: 10.1128/mBio.02016-20
Wood, N. A., Chung, K. Y., Blocker, A. M., Rodrigues De Almeida, N., Conda-Sheridan, M., Fisher, D. J., et al. (2019). Initial characterization of the two ClpP paralogs of Chlamydia trachomatis suggests unique functionality for each. J. Bacteriol. 201:e00635-00618. doi: 10.1128/JB.00635-18
Wu, C. (1995). Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 11, 441–469. doi: 10.1146/annurev.cb.11.110195.002301
Wu, J. S., Lin, J. C., and Chang, F. Y. (2000). Chlamydia pneumoniae infection in community-acquired pneumonia in Taiwan. J. Microbiol. Immunol. Infect. 33, 34–38.
Wu, C. C., Naveen, V., Chien, C. H., Chang, Y. W., and Hsiao, C. D. (2012). Crystal structure of DnaK protein complexed with nucleotide exchange factor GrpE in DnaK chaperone system: insight into intermolecular communication. J. Biol. Chem. 287, 21461–21470. doi: 10.1074/jbc.M112.344358
Wu, S., Zhu, Z., Fu, L., Niu, B., and Li, W. (2011). WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics 12:444. doi: 10.1186/1471-2164-12-444
Wurihan, W., Huang, Y., Weber, A. M., Wu, X., and Fan, H. (2020). Nonspecific toxicities of Streptococcus pyogenes and Staphylococcus aureus dCas9 in Chlamydia trachomatis. Pathog. Dis. 77:ftaa005. doi: 10.1093/femspd/ftaa005
Wurihan, W., Weber, A. M., Gong, Z., Lou, Z., Sun, S., Zhou, J., et al. (2021a). GrgA overexpression inhibits Chlamydia trachomatis growth through sigma66- and sigma28-dependent mechanisms. Microb. Pathog. 156:104917. doi: 10.1016/j.micpath.2021.104917
Wurihan, W., Zou, Y., Weber, A. M., Weldon, K., Huang, Y., Bao, X., et al. (2021b). Identification of a GrgA-Euo-HrcA transcriptional regulatory network in chlamydia. mSystems 6:e0073821. doi: 10.1128/mSystems.00738-21
Xu, Z., Horwich, A. L., and Sigler, P. B. (1997). The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature 388, 741–750.
Xue, Y., Zheng, H., Mai, Z., Qin, X., Chen, W., Huang, T., et al. (2017). An in vitro model of azithromycin-induced persistent Chlamydia trachomatis infection. FEMS Microbiol. Lett. 364. doi: 10.1093/femsle/fnx145
Yamamori, T., and Yura, T. (1982). Genetic control of heat-shock protein synthesis and its bearing on growth and thermal resistance in Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 79, 860–864.
Yang, C., Kari, L., Lei, L., Carlson, J. H., Ma, L., Couch, C. E., et al. (2020). Chlamydia trachomatis plasmid gene protein 3 is essential for the establishment of persistent infection and associated immunopathology. MBio 11. doi: 10.1128/mBio.01902-20
Yu, H. H., Di Russo, E. G., Rounds, M. A., and Tan, M. (2006a). Mutational analysis of the promoter recognized by Chlamydia and Escherichia coli sigma(28) RNA polymerase. J. Bacteriol. 188, 5524–5531. doi: 10.1128/JB.00480-06
Yu, H. H., Kibler, D., and Tan, M. (2006b). In silico prediction and functional validation of sigma28-regulated genes in Chlamydia and Escherichia coli. J. Bacteriol. 188, 8206–8212. doi: 10.1128/JB.01082-06
Yu, H. H. Y., and Tan, M. (2003). Sigma28 RNA polymerase regulates hctB, a late developmental gene in Chlamydia. Mol. Microbiol. 50, 577–584. doi: 10.1046/j.1365-2958.2003.03708.x
Yura, T. (1996). Regulation and conservation of the heat-shock transcription factor sigma32. Genes Cells 1, 277–284. doi: 10.1046/j.1365-2443.1996.28028.x
Yura, T., Nagai, H., and Mori, H. (1993). Regulation of the heat-shock response in bacteria. Annu. Rev. Microbiol. 47, 321–350. doi: 10.1146/annurev.mi.47.100193.001541
Zhang, Y., and Gross, C. A. (2021). Cold shock response in bacteria. Annu. Rev. Genet. 55, 377–400. doi: 10.1146/annurev-genet-071819-031654
Zhang, T., Huo, Z., Ma, J., He, C., and Zhong, G. (2019). The plasmid-encoded pGP3 promotes Chlamydia evasion of acidic barriers in both stomach and vagina. Infect. Immun. 87. doi: 10.1128/IAI.00844-18
Zhang, Q., Rosario, C. J., Sheehan, L. M., Rizvi, S. M., Brothwell, J. A., He, C., et al. (2020). The repressor function of the Chlamydia late regulator EUO is enhanced by the plasmid-encoded protein Pgp4. J. Bacteriol. 202:e00793-00719. doi: 10.1128/JB.00793-19
Zhong, G. (2017). CHLAMYDIAL plasmid-dependent pathogenicity. Trends Microbiol. 25, 141–152. doi: 10.1016/j.tim.2016.09.006
Keywords: Chlamydia, heat shock response, stress response, transcriptome, transcriptional regulatory network, sigma factor, heat-induced transcription repressor, HrcA
Citation: Huang Y, Wurihan W, Lu B, Zou Y, Wang Y, Weldon K, Fondell JD, Lai Z, Wu X and Fan H (2022) Robust Heat Shock Response in Chlamydia Lacking a Typical Heat Shock Sigma Factor. Front. Microbiol. 12:812448. doi: 10.3389/fmicb.2021.812448
Edited by:
Harry Beller, U.S. Environmental Protection Agency (EPA), Region 9, United StatesReviewed by:
Brett Hanson, Roche (United States), United StatesKatelyn Soules, Merck Animal Health, United States
Copyright © 2022 Huang, Wurihan, Lu, Zou, Wang, Weldon, Fondell, Lai, Wu and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Wu, wxspring@126.com; Huizhou Fan, fanhu@rwjms.rutgers.edu
†ORCID: Yehong Huang orcid.org/0000-0001-8186-1088
Wurihan Wurihan orcid.org/0000-0001-7853-0130
Bin Lu orcid.org/0000-0002-3193-3941
Yuxuan Wang orcid.org/0000-0003-0771-1078
Joseph D. Fondell orcid.org/0000-0002-7046-9854
Xiang Wu orcid.org/0000-0001-9013-742X
Huizhou Fan orcid.org/0000-0002-4903-926X
 Yehong Huang
Yehong Huang Wurihan Wurihan
Wurihan Wurihan Bin Lu1,2†
Bin Lu1,2† Yuxuan Wang
Yuxuan Wang Zhao Lai
Zhao Lai Xiang Wu
Xiang Wu Huizhou Fan
Huizhou Fan