- 1Berks College, Penn State University, Reading, PA, United States
- 2Centre for Crop and Food Innovation, College of Science, Health, Engineering and Education, Food Futures Institute, Murdoch University, Murdoch, WA, Australia
- 3Murdoch University Associate, Murdoch, WA, Australia
- 4Sustainability and Biosecurity, Department of Primary Industries and Regional Development, South Perth, WA, Australia
- 5Soybean Genomics and Improvement Laboratory, United States Department of Agriculture, Beltsville, MD, United States
- 6Curtin University Sustainability Policy Institute, Curtin University, Bentley, WA, Australia
- 7Department of Energy (DOE) Joint Genome Institute, Berkeley, CA, United States
- 8Centre for Water Energy and Waste, Harry Butler Institute, Murdoch University, Murdoch, WA, Australia
Although Medicago sativa forms highly effective symbioses with the comparatively acid-sensitive genus Ensifer, its introduction into acid soils appears to have selected for symbiotic interactions with acid-tolerant R. favelukesii strains. Rhizobium favelukesii has the unusual ability of being able to nodulate and fix nitrogen, albeit sub-optimally, not only with M. sativa but also with the promiscuous host Phaseolus vulgaris. Here we describe the genome of R. favelukesii OR191 and genomic features important for the symbiotic interaction with both of these hosts. The OR191 draft genome contained acid adaptation loci, including the highly acid-inducible lpiA/acvB operon and olsC, required for production of lysine- and ornithine-containing membrane lipids, respectively. The olsC gene was also present in other acid-tolerant Rhizobium strains but absent from the more acid-sensitive Ensifer microsymbionts. The OR191 symbiotic genes were in general more closely related to those found in Medicago microsymbionts. OR191 contained the nodA, nodEF, nodHPQ, and nodL genes for synthesis of polyunsaturated, sulfated and acetylated Nod factors that are important for symbiosis with Medicago, but contained a truncated nodG, which may decrease nodulation efficiency with M. sativa. OR191 contained an E. meliloti type BacA, which has been shown to specifically protect Ensifer microsymbionts from Medicago nodule-specific cysteine-rich peptides. The nitrogen fixation genes nifQWZS were present in OR191 and P. vulgaris microsymbionts but absent from E. meliloti-Medicago microsymbionts. The ability of OR191 to nodulate and fix nitrogen symbiotically with P. vulgaris indicates that this host has less stringent requirements for nodulation than M. sativa but may need rhizobial strains that possess nifQWZS for N2-fixation to occur. OR191 possessed the exo genes required for the biosynthesis of succinoglycan, which is required for the Ensifer-Medicago symbiosis. However, 1H-NMR spectra revealed that, in the conditions tested, OR191 exopolysaccharide did not contain a succinyl substituent but instead contained a 3-hydroxybutyrate moiety, which may affect its symbiotic performance with Medicago hosts. These findings provide a foundation for the genetic basis of nodulation requirements and symbiotic effectiveness with different hosts.
Introduction
The symbiosis between legumes and root nodule bacteria (collectively known as rhizobia) is of global importance in both natural and agricultural ecosystems. It is established following a molecular dialogue between the legume and its microsymbiont that leads to infection, nodule organogenesis and the eventual endocytosis-like release of the bacteria into membrane-bound compartments in the nodule cells of the root cortex. Following this, the rhizobia differentiate into N2-fixing bacteroids and supply essential fixed N to the plant host.
Most current knowledge on the evolution of N2-fixing rhizobia is based on the comparative study of a relatively small number of symbiotically effective strains. Although this bias is understandable from an agronomic perspective, relatively little attention has been devoted to the study of strains that are poorly effective for N2-fixation, which may constitute the majority of rhizobial populations in soils (Bottomley and Jenkins, 1983; Segovia et al., 1991; Sullivan et al., 1996). These poorly effective strains, which are often well adapted to the soil environment, are a constraint to maximizing the productivity of the legume-rhizobia symbiosis in sustainable agricultural systems (Howieson et al., 2008). Sequencing the genomes of such strains could not only provide greater knowledge of their evolution, biogeography and symbiotic relationships, but could also contribute to an understanding of the molecular mechanisms that govern N2-fixation effectiveness.
The Rhizobium favelukesii strain OR191 was first isolated from a nodule of the perennial pasture legume alfalfa (Medicago sativa L.) growing in moderately acid (pH 5.5–5.7) soil at Corvallis, Oregon, in 1982, and was shown to be poorly effective on this host (Eardly et al., 1985). OR191, along with similar isolates from the same site, possessed a unique symbiotic host range; specifically, they were able to fix nitrogen with both alfalfa (Medicago sativa L.) and the common bean [Phaseolus vulgaris (L) Savi], albeit at levels much lower than the usual microsymbionts of these hosts. M. sativa is usually nodulated by strains of Ensifer (ex Sinorhizobium) meliloti (Béna et al., 2005), but this bacterial species is known to be acid-sensitive and unable to survive in low pH soils (Garau et al., 2005; Reeve et al., 2006). P. vulgaris microsymbionts are most commonly species of Rhizobium (Eardly et al., 1985; reviewed in Graham, 2008). Because these two hosts were thought to be nodulated by distinctly different rhizobial genera, and the genes determining symbiotic host range in Rhizobium spp. were known to reside on plasmids (Rogel et al., 2011), it was thought that perhaps these Oregon isolates might represent recombinants of one species that had somehow acquired the genetic host-range determinants of the other. However, comparative sequence analysis of a partial segment of the 16S rRNA gene of OR191 revealed that this strain belonged to a distinct and previously unrecognized taxon (Eardly et al., 1992). In the same study it was also observed that OR191 was able to grow on strongly acidic agar media (pH 5.2), as did most of the P. vulgaris rhizobial symbionts examined. In contrast, M. sativa symbionts were unable to grow at pH 5.2 or below.
Strains sharing the unique host range, limited symbiotic efficiency, and acid-tolerance characteristics of OR191 were subsequently isolated from alfalfa growing at two other locations. In 1992, several “OR191-like” strains, including the strain T1155, were isolated from weakly acid soils in Ontario, Canada (Bromfield et al., 2001, 2010). Subsequently other OR191-like strains were isolated from moderately acid soils in Buenos Aires, Argentina (Del Papa et al., 1999). One of these strains, LPU83T, has now been proposed as the type strain for the new species Rhizobium favelukesii (Torres Tejerizo et al., 2016). The genome of LPU83T (NZ_HG916852) has been established and consists of five replicons: the chromosome (4,195,305 bp); the chromid pLPU83d (1,932,030 bp); the accessory plasmids pLPU83a (151,687 bp) and pLPU83c (759,787 bp) and the symbiotic plasmid pLPU83b (ca. 531,535 bp) (Wibberg et al., 2014). pLPU83b carries all the nodulation and nitrogen fixation genes required to establish and maintain a nitrogen-fixing symbiosis (Wibberg et al., 2014).
OR191 and these other R. favelukesii strains thus represent a divergent lineage of acid-adapted strains within the genus Rhizobium that are able to fix N2 with both alfalfa and common bean, legume hosts that are each normally nodulated by different genera within the family Rhizobiaceae. Here we have sequenced the genome of R. favelukesii OR191, as part of the JGI 2010 GEBA-RNB project (Reeve et al., 2015; Seshadri et al., 2015). In this paper we provide an analysis of this genome and perform a comparison with the genomes of strains that are microsymbionts of alfalfa, or common bean, or of both. The genome properties of OR191 should provide insights into the diversification of the genus Rhizobium, the mechanisms involved in rhizobial acid-stress tolerance, and the roles that various genetic determinants play in the development of symbiotic relationships with two widely divergent legume hosts. In addition, due to the importance of exopolysaccharide (EPS) both for rhizobial symbiosis with legumes and for acid stress tolerance (Hawkins et al., 2017), we quantified the amount of EPS and substituents produced under neutral and acidic pH growth conditions, and determined its composition by NMR spectroscopy.
Materials and Methods
Bacterial Strains and Growth Conditions
Cultures of R. faveluksii OR191 were routinely sub-cultured on TYC solid medium (Howieson and Dilworth, 2016) incubated at 28°C for 3–4 days. For long-term maintenance, bacterial strains were grown in TYC broth and preserved in 20% glycerol at –80°C.
Genomic DNA Preparation
R. favelukesii OR191 was streaked onto TYC solid medium and grown at 28°C for 3 days to obtain well grown, well separated colonies. A single colony was selected and used to inoculate 5 ml TYC broth medium. The culture was grown for 48 h on a gyratory shaker (200 rpm) at 28°C. Subsequently 1 ml was used to inoculate 60 ml TYC broth medium and grown on a gyratory shaker (200 rpm) at 28°C until an OD600nm of 0.6 was reached. DNA was isolated from 60 ml of cells using a CTAB bacterial genomic DNA isolation method (Joint Genome Institute, 2022). Final concentration of the DNA was set to 0.5 mg ml–1.
Genome Sequencing and Assembly
The draft genome of Rhizobium favelukesii OR191 was generated at the DOE Joint Genome Institute (JGI) using Illumina data (Bennett, 2004). An Illumina standard shotgun library was constructed and sequenced using the Illumina HiSeq 2000 platform which generated 17,712,488 reads totaling 2,657 Mbp. All raw Illumina sequence data were passed through DUK, a filtering program developed at JGI, which removes known Illumina sequencing and library preparation artifacts (Mingkun, L., Copeland, A. and Han, J., unpublished). The following steps were then performed for assembly: (1) filtered Illumina reads were assembled using Velvet (version 1.1.04) (Zerbino, 2010), (2) 1–3 Kbp simulated paired end reads were created from Velvet contigs using wgsim (Github and GitHub, 2011), (3) Illumina reads were assembled with simulated read pairs using Allpaths–LG (version r39750) (Gnerre et al., 2011). Parameters for the assembly steps were 1) Velvet: –v –s 51 –e 71 –i 2 –t 1 –f “-shortPaired -fastq $FASTQ” –o “-ins_length 250 -min_contig_lgth 500” for Velvet and 2) wgsim: -e 0 -1 76 -2 76 -r 0 -R 0 -X 0. The final draft assembly contained 240 contigs in 240 scaffolds. The total size of the assembly is 7.4 Mbp with an average of 350 × coverage of the genome.
Genome Annotation
Genes were identified using Prodigal (Hyatt et al., 2010), as part of the DOE-JGI genome annotation pipeline (Mavromatis et al., 2009; Chen et al., 2013). The predicted CDSs were translated and used to search the National Center for Biotechnology Information (NCBI) non-redundant database, UniProt, TIGRFam, Pfam, KEGG, COG, and InterPro databases. The tRNAScanSE tool (Lowe and Eddy, 1997) was used to find tRNA genes, whereas ribosomal RNA genes were found by searches against models of the ribosomal RNA genes built from SILVA (Pruesse et al., 2007). Other non–coding RNAs such as the RNA components of the protein secretion complex and the RNase P were identified by searching the genome for the corresponding Rfam profiles using INFERNAL (Nawrocki and Eddy, 2013). Additional gene prediction analysis and manual functional annotation was performed within the IMG-Expert Review system (Markowitz et al., 2009) developed by the Joint Genome Institute, Berkeley, CA, United States.
Phylogenetic Analysis
We assessed the phylogenetic position of R. favelukesii OR191, relative to other Rhizobium type and non-type strains, using a 1,296 bp internal region of the 16S rRNA gene. The Azorhizobium caulinodans ORS 571T sequence was used as an outgroup. Phylogenetic analyses were performed using MEGA X (Kumar et al., 2018). The tree was built using the maximum likelihood method with the Tamura 3- parameter model (Tamura, 1992). A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+ G, parameter = 0.3233)]. The rate variation model allowed for some sites to be evolutionarily invariable [( + I), 42.2407% sites]. Bootstrap analysis (Felsenstein, 1985) with 500 replicates was performed to assess the support of the clusters.
For phylogenetic analysis of the NodA proteins, where more than one nodA allele was present in the genome, we used the translated amino acid sequence of the nodA gene that was immediately upstream of nodBC. The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura 3-parameter model. The tree with the highest log likelihood (4204.9163) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+ G, parameter: 3.2381)]. The rate variation model, used for nodA, allowed for some sites to be evolutionarily invariable [( + I), 18.6224% sites]. The trees were drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 41 nucleotide sequences for nodA. All positions containing gaps and missing data were eliminated. In the final dataset there were a total of 588. Evolutionary analyses were conducted in MEGA X (113).
Phylogenetic analysis and evolutionary history of the NifH proteins was inferred by using the Maximum Likelihood method and JTT matrix-based model (Jones et al., 1992). The tree with the highest log likelihood (–8402.67) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+ G, parameter = 2.4848)]. The rate variation model allowed for some sites to be evolutionarily invariable [( + I), 14.51% sites]. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 38 amino acid sequences; Rhizobium mesoamericanum STM3655 and Ensifer meliloti AK53 were excluded from the analysis due to a lack of sequencing information. There were a total of 572 positions in the final dataset. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018).
Genome Average Nucleotide Identity Pairwise Comparisons
gANI comparisons were performed using the IMG gANI analysis tool (Chen et al., 2021).
Exopolysaccharide Production
Cells of OR191 and Ensifer medicae WSM419 were grown to mid-exponential phase at pH 7.0 in JMM minimal salts media (Howieson and Dilworth, 2016), washed in JMM at pH 7.0 and then resuspended in JMM at either pH 7 or 5.8 to an OD600 nm of approximately 0.01 for pH 7.0 media (100 ml) and 0.25 for pH 5.8 media (100 ml). Cells were incubated at 28°C with shaking at 200 rpm for 4 days. Cells were then removed by centrifugation (10 min at 7,000 g) and EPS was precipitated from the supernatant by adding 0.1 vol. of a 2% hexadecyltrimethyl-ammonium bromide solution stored at 28°C. EPS was pelleted (10 min at 7,000 g) and redissolved in 10% (w/v) NaCl. The EPS was precipitated by adding 2 volumes of acetone, pelleted (10 min at 7,000 g), redissolved in sterile water and dialyzed in water. EPS was lyophilized, resuspended in D2O (99.96%.), lyophilized again, and dissolved in D2O (99.96%.) to a concentration of 15 mg ml–1. 1H-NMR spectra were recorded on a Varian 400-MR spectrometer at 80°C. A 30 μl aliquot of a 2% (w/v) solution of the succinoglycan-binding dye Calcofluor (Fluorescent brightener 28, Sigma) was added to 1 ml samples containing 7.5 mg EPS in saline [0.89% (w/v)] to visualize fluorescence at a wavelength of 365 nm.
Results and Discussion
OR191 Genome Characteristics, Properties and Phylogenetic Placement
OR191 is a rod shaped isolate that is fast growing and forms typically mucoid colonies on conventional media used to isolate rhizobia (Howieson and Dilworth, 2016; Supplementary Figure 1). Flagella are not present in the transmission electron micrograph of OR191 (Supplementary Figure 1B), which is consistent with the description of the R. favelukesii type strain LPU83T as non-motile (Torres Tejerizo et al., 2016). The classification, general features and genome sequencing project information for OR191 are provided in Supplementary Table 1, in accordance with the minimum information about a genome sequence (MIGS) recommendations (Field et al., 2008) published by the Genomic Standards Consortium (Field et al., 2011).
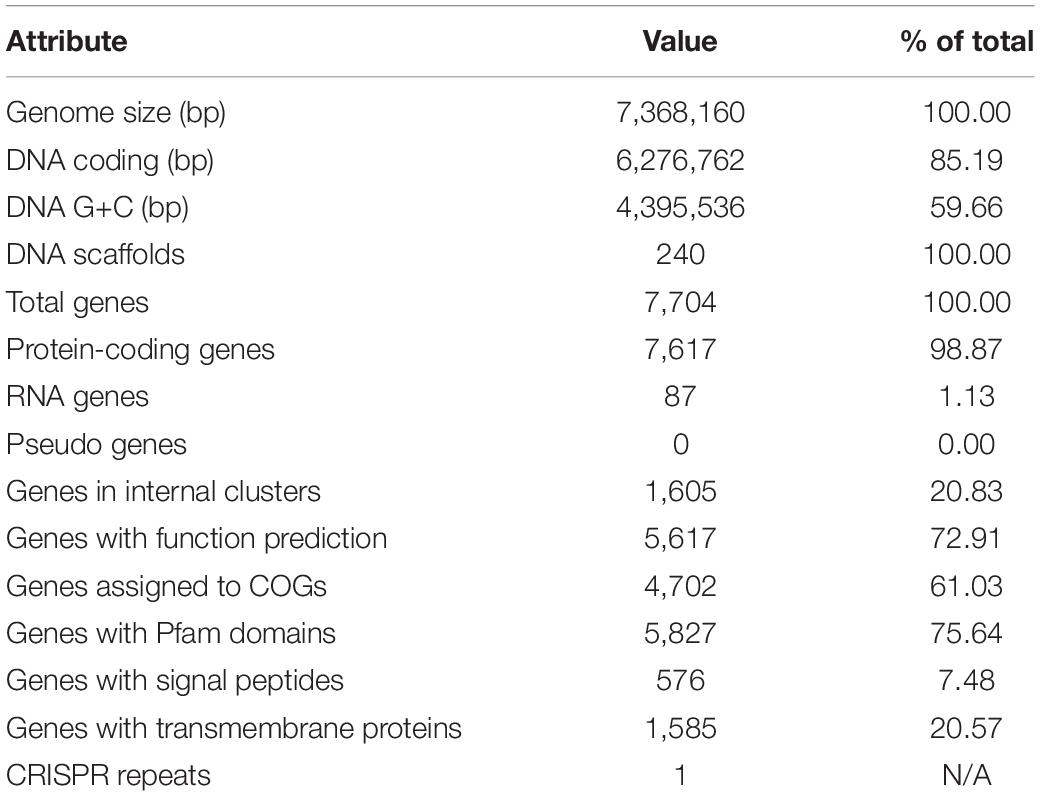
The draft genome of strain OR191 was generated at the DOE Joint Genome Institute (JGI) using Illumina data (see “Materials and Methods” section). A summary of the genome project features for OR191 is shown in Table 1. The genome is 7,368,160 bp with 59.66% GC content and is comprised of 240 scaffolds. From a total of 7,704 genes, 7,617 were protein encoding and 87 RNA-only encoding genes. The majority of genes (72.12%) were assigned a putative function whilst the remaining genes were annotated as hypothetical. The distribution of genes into COGs functional categories is presented in Supplementary Table 2.
Many of the features of OR191 are indistinguishable from those of R. favelukesii LPU83T and T1155, including host range, nod and nif genes, insertion-sequence hybridization profiles, PCR-based genomic fingerprints, and plasmid profiles (Eardly et al., 1985, 1992; Del Papa et al., 1999; Segundo et al., 1999; Wegener et al., 2001; Hou et al., 2009; Bromfield et al., 2010; Wibberg et al., 2014; Table 2). Consistent with the plasmid profiles, we identified genes encoding homologs of the Rep proteins (required for the replication and stable maintenance of plasmid replicons) in the OR191 genome that shared 100% identity with those present in the R. favelukesii pLPU83a, pLPU83b, pLPU83c, and pLPU83d plasmids, suggesting that both R. favelukesii strains possess the same number of replicons. Furthermore, the OR191 genome contains two separate regions that encode proteins required for DNA transfer replication and mating pair formation, which share very high identity with homologs in the characterized plasmids pLPU83a (which is conjugative) and pLPU83b (which is mobilizable) (Torres Tejerizo et al., 2010, 2014). We also found genes encoding a separate conjugative system in OR191 that shares very high identity with a conjugative system that is present on the pLPU83d chromid. LPU83T shares its lipopolysaccharide profile with OR191 (Wegener et al., 2001), while T1155 shares its phage-resistance type with OR191 (Bromfield et al., 2010). Furthermore, the DNA-DNA hybridization values between strains LPU83T and OR191 were greater than 84%, indicating that these strains are members of the same species (Torres Tejerizo et al., 2016). DNA alignments of OR191 essential “housekeeping” gene sequences (including 16S rRNA, 23S rRNA, recA, and atpD genes) to the corresponding genes of LPU83T, T1155 and the closely related Rhizobium tibeticum CGMCC 1.7071T gave 100% sequence identity of OR191 genes to those of LPU83T and from 98.83 to 100% sequence identity to those of T1155 or CGMCC 1.7071T (Table 2). In the 16S rRNA phylogenetic tree, OR191 is in a clade with R. grahamii CCGE 502T, R. favelukesii strains and R. tibeticum CGMCC 1.7071T (Supplementary Figure 2).
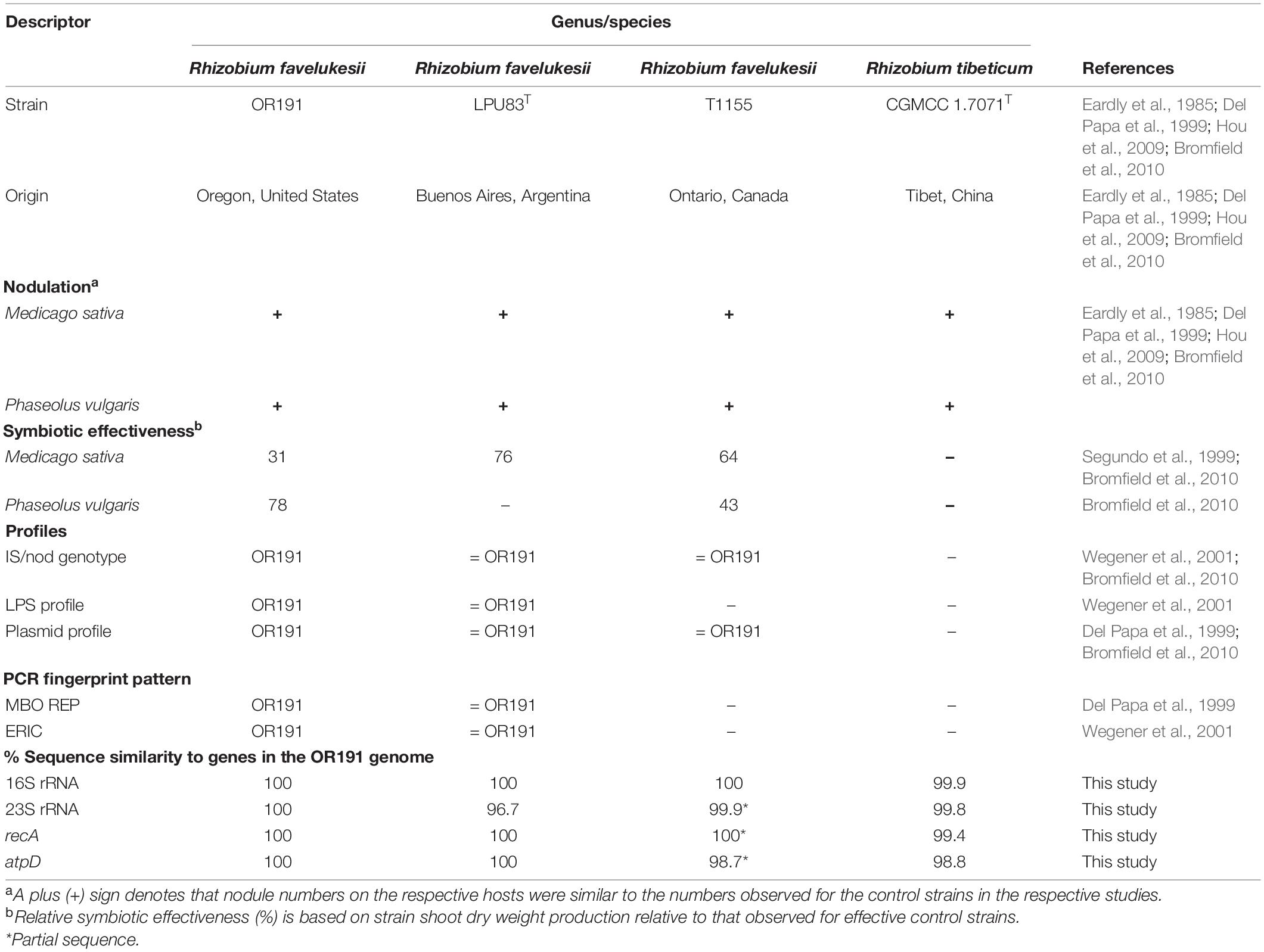
Table 2. A descriptive summary of the phenotypic and genotypic characteristics of Rhizobium favelukesii OR191 compared to three other closely related Rhizobium strains that have the same extended host range as OR191.
gANI pairwise comparisons were also calculated for Rhizobium genomes deposited in IMG that were most closely related to OR191 in the 16S rRNA phylogenetic tree. The genome of R. favelukesii T1155 could not be used in this analysis as it has not yet been sequenced. gANI values for the most closely related strains are shown in Table 3; scores over the defined species affiliation cut-off value of > 96.5 gANI (Varghese et al., 2015) are shown in bold font. The gANI pairwise comparisons, along with other previously presented phenotypic and genotypic data, confirmed that OR191 is conspecific with R. favelukesii LPU83T. The gANI results also suggest that R. tibeticum CGMCC 1.7071T belongs to the same species as LPU83T and OR191. However, the DND-DNA hybridization values obtained by Torres Tejerizo et al. (2016) indicate that R. favelukesii and R. tibeticum strains are separate species; moreover, R. tibeticum strain CGMCC 1.7071T shared lower sequence identities of the recA, atpD, and rpoB genes than the R. favelukesii strains and differed in its plasmid profile, fatty acid profile and ability to metabolize several sole carbon substrates.
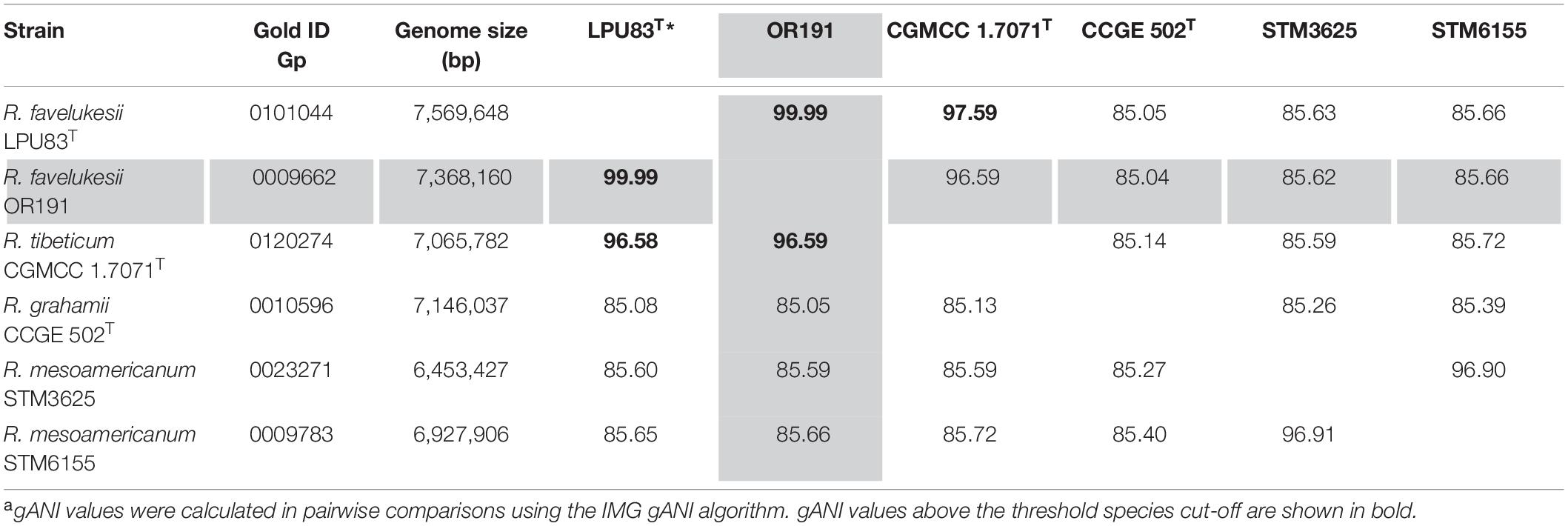
Table 3. Pairwise comparisons of gANI valuesa of Rhizobium favelukesii OR191 (shaded) to selected Rhizobium strains in IMG.
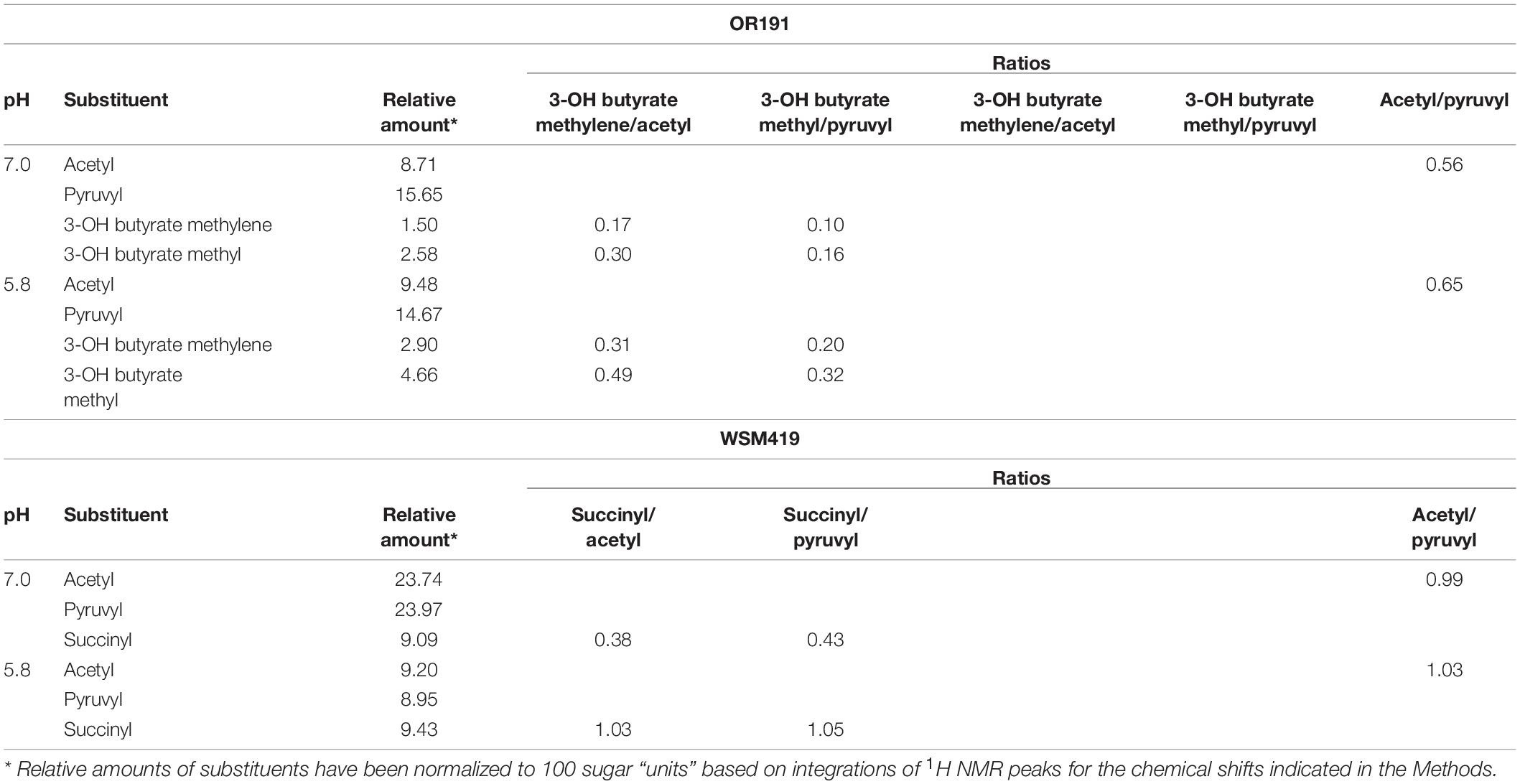
Table 4. Relative amounts of substitution on EPS from Rhizobium favelukesii OR191 and Ensifer medicae WSM419 grown at pH 7.0 and 5.8.
Whole Genome Comparisons of Rhizobium favelukesii Strains
The genes of the sequenced R. favelukesii strains OR191 and LPU83T were compared, using the default parameters of 30% protein identity and E-value of 1e-5 in the JGI IMG phylogenetic profiler. Of a total of 7,704 protein coding genes in OR191, 7,449 genes were found to be in common with LPU83T. The remaining 255 genes were unique to OR191, with nearly half annotated as hypothetical (128 genes representing 50.20%). These hypothetical genes included extensive prophage loci clustered on scaffold 24.25. By using the prophage predicting tool PHASTER (Arndt et al., 2016) these loci were shown to encode a complete resident prophage. Whole genome analysis of OR191 found four additional resident prophages present on scaffolds 4.5, 7.8, 84.85, and 91.92 (Supplementary Figure 3), in addition to the unique prophage on scaffold 24.25.
Identification and Comparison of Genes Involved in Acid Tolerance in OR191
Strain OR191 was isolated from alfalfa growing in moderately acidic soil (Eardly et al., 1985) and can grow in laboratory culture at pH 5.2 (Eardly et al., 1992). Most P. vulgaris Rhizobium microsymbionts can also grow in laboratory culture at pH 5.2 (Eardly et al., 1992). In contrast, the E. meliloti strains that are the usual microsymbionts of M. sativa are known to be particularly acid-sensitive in comparison to other rhizobial species, and in laboratory culture fail to grow below pH 5.6 (Graham and Parker, 1964; Eardly et al., 1992; Garau et al., 2005). In order to gain insights into the acid tolerance of OR191, we examined the genome for the presence of loci known to be important for the pH adaptation of very acid tolerant strains (such as Rhizobium tropici CIAT899T) and to less acid tolerant but well characterized strains (such as Ensifer meliloti 1021 and E. medicae WSM419). Although less acid-tolerant than CIAT899T, WSM419 is the dominant nodule occupant of Medicago species growing in moderately acidic soils in Sardinia and Greece (Garau et al., 2005).
Most of the genes found to be important for acid tolerance in Rhizobium strains had orthologs in WSM419 and Ensifer meliloti 1021 (Supplementary Table 3), including the most acid-activated genes discovered so far, lpiA and acvB (Reeve et al., 2006), encoding lysyl-phosphatidylglycerol synthase and lysyl-phosphatidylglycerol hydrolase, respectively, and required for modulation of lysyl-phosphatidylglycerol homeostasis (Sohlenkamp et al., 2007). However, the fsrR, tcrA, and tcsA regulatory system, which is implicated in the strong acid induction of the lpiA-acvB operon in WSM419, is not present in OR191 or in species other than E. medicae, indicating that an unknown regulatory system governs acid induction of these genes in other rhizobia (Reeve et al., 2006; Tian et al., 2017). In OR191 and several other Rhizobium strains, the lpiA-acvB operon is immediately downstream of a gene encoding a small-conductance mechanosensitive channel, a membrane protein that is important for bacterial survival (reviewed in Naismith and Booth, 2012). The olsC gene, which has been identified as being important for the production of hydroxylated ornithine lipid species that increase stress tolerance in CIAT899T (Rojas-Jiménez et al., 2005; Vences-Guzmán et al., 2011), was present in both the highly acid tolerant strain R. tropici CIAT899T and in R. favelukesii strains but was absent from E. medicae WSM419 and E. meliloti 1021. This accords with proteomic and transcriptomic studies of R. favelukesii LPU83T that revealed changes in proteins and genes associated with lipid metabolism involving the cell envelope in response to acid stress, however, these studies also suggested that some acid-responsive regulatory systems differ in LPU83T compared with Ensifer strains (Nilsson et al., 2019, 2020).
Comparisons of the OR191 Genome With Other Microsymbionts of Medicago and/or Phaseolus vulgaris
R. favelukesii OR191 nodulates and fixes nitrogen with both M. sativa and P. vulgaris. To identify features within the OR191 genome that enable it to be a microsymbiont of both these hosts, the IMG online database was used to compare relevant sequenced genomes of microsymbionts of Medicago species, or of common bean, or of both (strains listed in Supplementary Table 4).
We examined these genomes for genes with known roles in rhizobial symbiotic interactions. In addition to meeting or exceeding the cut-off values of > 30% protein identity over > 75% protein coverage, OR191 nod, nif and fix (cco) homologs were validated as orthologous if they were present within the neighborhood of symbiotic gene clusters. A summary of the identified OR191 genes with characterized roles in symbiotic interactions and their phylogenetic profile in microsymbionts of Medicago and P. vulgaris is provided in Supplementary Table 5.
Overview of the Nod Genes in OR191
Symbiosis between legumes and rhizobia is initiated by a molecular dialogue, in which the host secretes flavonoid signals from the root cells, triggering expression of the rhizobial nodulation (nod, noe, and nol) genes and production of lipochitoligosaccharide Nod factors, which are required for infection of the host and nodule organogenesis. Expression of the nodulation genes is controlled by the regulatory protein NodD. In addition to the Nod factor core biosynthetic genes (nodABC) and export genes (nodIJ), rhizobial genomes contain accessory nodulation genes, which encode moieties that decorate the basic Nod factor and act as specific host determinants (Perret et al., 2000).
The core and accessory nodulation genes of OR191 are distributed on four scaffolds (Supplementary Figure 4) and share 99–100% sequence identity with those of R. favelukesii LPU83T. While all rhizobial genomes contained the core nodulation genes nodABCDIJ, there were differences in the accessory nodulation genes present within Medicago and P. vulgaris microsymbionts.
OR191 Nod Genes That Are Common to Medicago Microsymbionts
In addition to the core nodABCIJ genes, all Medicago microsymbiont genomes (E. meliloti strains + OR191, LPU83T, R. tibeticum CGMCC 1.7071T and R. mongolense USDA 1844T) contained a specific set of accessory nod genes. All Medicago microsymbiont genomes harbored three nodD alleles, which in E. meliloti appear to be important in optimizing symbiotic interactions with Medicago hosts (Honma et al., 1990; Van Berkum et al., 1998). The OR191 nodD genes are most closely related to orthologs in LPU83T and the Medicago-nodulating Rhizobium strains R. tibeticum CGMCC 1.7071T and R. mongolense USDA 1844T. All Medicago microsymbiont genomes also contained the nodL and nodHPQ genes that code for synthesis of acetylated and sulfated Nod factors, respectively, which in Ensifer strains are required for symbiotic interaction with Medicago hosts (Lerouge et al., 1990). The Nod factors of R. favelukesii LPU83T include species with trimeric, tetrameric, and pentameric chitin backbones that contain unsaturated acyl chains and may be sulfated or methylated, however, sulfated Nod factors do not appear to be required for this strain to nodulate alfalfa, as mutation of nodH delayed, but did not abolish, nodulation of this host (Del Papa et al., 2007; Torres Tejerizo et al., 2011). Both LPU83T and OR191 contained the flavonoid-inducible nodL, noeA, and noeB operon, which is required for effective nodulation of Medicago sp. (Ardourel et al., 1995). NoeA and NoeB are hypothesized to modify the Nod factor for accurate recognition by Medicago spp., allowing the microsymbiont to successfully infect the host.
The nodEF genes, which are required for production of Nod factors with α, β-unsaturated acyl chains (Dénarié et al., 1996), were also present in all Medicago microsymbiont genomes. NodE has homology to β-ketoacyl synthases, while NodF is homologous to acyl carrier proteins (Demont et al., 1993). The nodG gene, encoding a 3-oxoacyl-acyl carrier protein reductase, was identified in all Medicago-nodulating E. meliloti genomes. NodG has a postulated role in fatty acid synthesis in the M. sativa microsymbiont strain E. meliloti 1021 (Mao et al., 2016), but is not required for synthesis of the characteristic α, β-unsaturated acyl chains of the Nod factors (Debellé et al., 2001). Although nodG is not essential for nodulation of M. sativa, it does increase nodulation efficiency (Mao et al., 2016). In OR191, the gene immediately upstream of nodP has sequence similarity to the characterized nodG of E. meliloti but encodes a truncated protein of 55 amino acids instead of the 245 amino acids found for E. meliloti NodG. The lack of a functional NodG might partially explain OR191’s less efficient nodulation performance on M. sativa, in comparison to 1021. R. favelukesii LPU83T also contains a truncated version of nodG (Torres Tejerizo et al., 2011). We identified similarly truncated nodG genes in the same gene neighborhoods within the genomes of R. tibeticum CGMCC 1.7071T and R. mongolense USDA 1844T, indicating that nodG pseudogenes may be a feature of these strains. The nolFG genes, encoding an RND-type efflux pump, are also characteristic of Medicago-nodulating Ensifer strains (Baev et al., 1991) and disruption of this region leads to delayed nodulation of alfalfa (Baev et al., 1991).
The OR191 nodABC, nodEF, nodL, nodHPQ, and nolFG genes are most closely related to those of LPU83T, CGMCC 1.7071T and USDA 1844T, and then to orthologs in Ensifer strains that nodulate Medicago. On the basis of its nod genes and symbiotic phenotype, Rogel et al. (2011) have included OR191 in the symbiovar orientale (Silva et al., 2005; Rogel et al., 2011), along with R. favelukesii LPU83T, R. gallicum CCBAU 01083, R. mongolense USDA 1844T, and R. tibeticum CGMCC 1.7071T (Van Berkum et al., 1998; Zhao et al., 2008; Hou et al., 2009; Torres Tejerizo et al., 2016).
OR191 Nod Genes Common to Phaseolus vulgaris Microsymbionts
A comparison of OR191 to other P. vulgaris microsymbionts revealed that the only nod genes they have in common are the core nodABCDIJ genes. Both the types of nod genes and the gene arrangements varied considerably in the P. vulgaris microsymbionts; for example, from two to six nodD alleles and from one to three nodA alleles were found and nodHPQ and nodEF clusters were present in some genomes but not others. Similarly, nodU, nodZ, noeI, noeT, and nolL genes were variably present or absent. However, all the P. vulgaris microsymbionts except OR191, LPU83T, R. tibeticum CGMCC 1.7071T and R. mongolense USDA 1844T contained nodS, which encodes an S-adenosyl-L-methionine (SAM)-dependent N-methyltransferase. Notably, nodS is absent from the genomes of all Medicago microsymbionts. A nodS-dependent N-methylation on the non-reducing end of the Nod Factor has been shown to be essential for the nodulation of P. vulgaris by the Rhizobium tropici strain CIAT 899T (Jabbouri et al., 1995). Although the Medicago microsymbionts (including the R. favelukesii, R. tibeticum, and R. mongolense strains) lack nodS, they contain the nodL gene required for production of O-acetylated Nod factors. NodL acetylation of the Nod factor functionally prevents the NodS-dependent transfer of the N-methyl group substituent, however, it has been proposed that NodL-dependent O-acetylation of the Nod factor can compensate for the lack of this N-methyl group (Waelkens et al., 1995; Lopez-Lara et al., 2001). Nodulation of P. vulgaris would appear to require either an acetylated or a methylated Nod factor, which can be accomplished by NodL or NodS, respectively.
The nodEF genes were found in R. favelukesii, R. tibeticum, and R. mongolense, Ensifer fredii GR64, E. meliloti GVPV12, Ensifer sp. strains 4H41 and BR816 and strains closely related to R. tropici CIAT 899T, but not in the remaining P. vulgaris-nodulating strains. Although the characterized strain R. tropici CIAT 899T contains nodEF, it does not produce Nod factors with α,β-unsaturated acyl chains (Ormeño-Orrillo et al., 2012). This can be related to the inability of R. tropici NodA to transfer unsaturated acyl chains to the Nod factor, whereas NodA of E. meliloti Medicago microsymbionts specifies the N-acylation of the Nod factor by an unsaturated or hydroxylated fatty acid (Debellé et al., 1996). However, the CIAT 899T hsnT, nodF, and nodE genes, which are in an operon with nodA2 in this strain, are important for the biosynthesis and specific decoration of the Nod Factors, with resulting impact on host specificity and symbiotic performance in some legume species (Gomes et al., 2019).
Because the transfer of unsaturated fatty acid chains to the Nod factor backbone requires a specific NodA protein (Debellé et al., 1996), we performed a comparative sequence analysis of the NodA proteins of the Medicago and P. vulgaris microsymbionts. The resulting phylogenetic tree grouped NodA into eight clusters (Group A—G, Figure 1) and showed that Cluster B (NodA of Medicago-nodulating R. favelukesii, R tibeticum, and R mongolense strains) formed a sister group to Cluster A of the Medicago-nodulating E. meliloti strains. This illustrates that even though E. meliloti 4H41 and GVPV12, Ensifer sp. BR816 and strains closely related to R. tropici CIAT 899T contain nodEF and therefore are potentially able to produce α, β-unsaturated acyl chains, their NodA is phylogenetically distinct from the NodA of Medicago-nodulating strains that can transfer unsaturated acyl chains to the Nod factor.
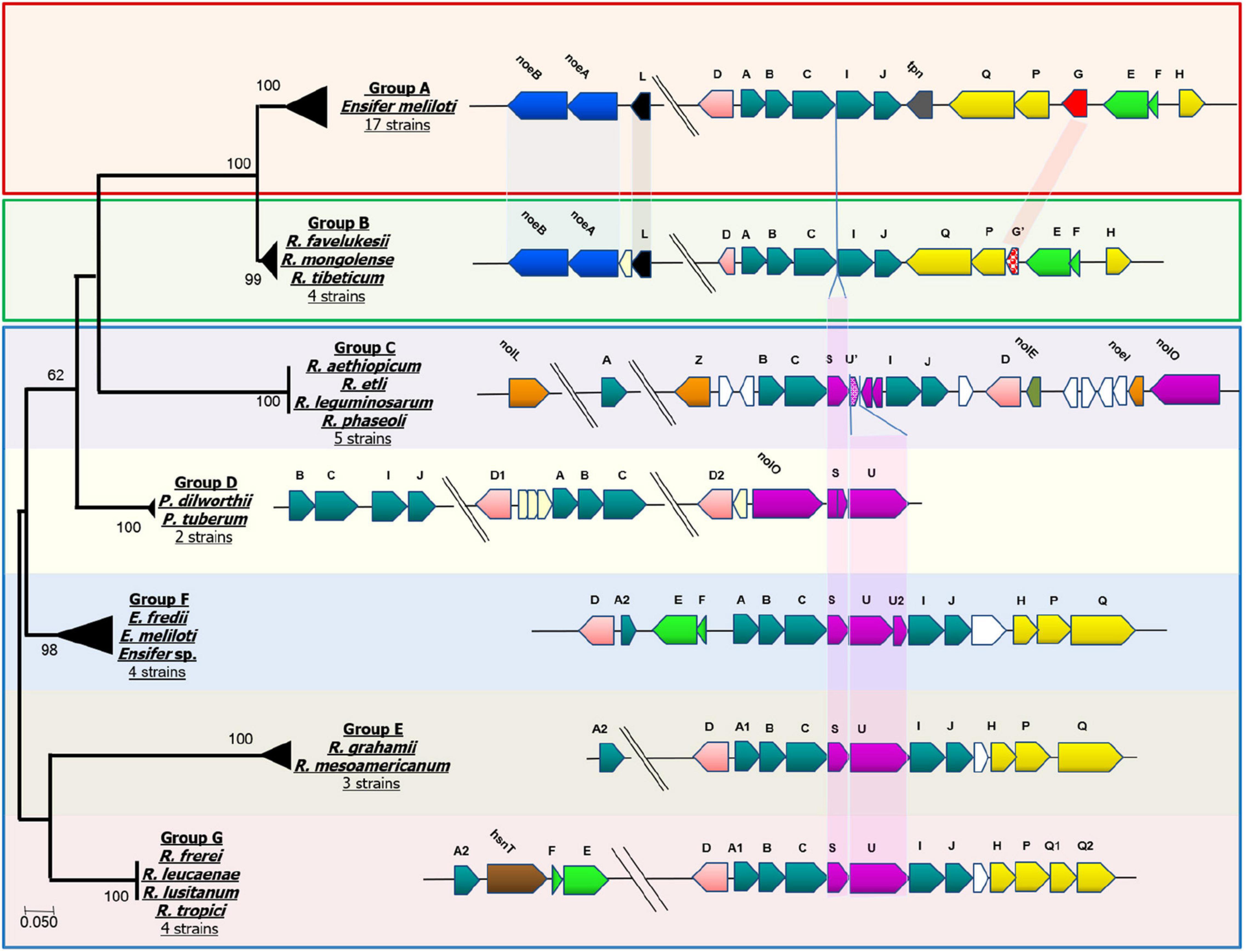
Figure 1. NodA phylogenetic distribution and comparison of gene neighborhoods. Comprehensive list of strains in each group shown and not shown can be found in Supplementary Table 4. Group A: Ensifer meliloti 1021, Ensifer meliloti 2011, Ensifer meliloti 5A14, Ensifer meliloti AE608H, Ensifer meliloti AK58, Ensifer meliloti AK83, Ensifer meliloti BL225C, Ensifer meliloti BO21CC, Ensifer meliloti CCNWSX0020, Ensifer meliloti CIAM1775, Ensifer meliloti GR4, Ensifer meliloti Mlalz-1, Ensifer meliloti MVII-I, Ensifer meliloti Rm41, Ensifer meliloti RRI128, Ensifer meliloti SM11 and Ensifer meliloti WSM1022. Group B: Rhizobium favelukesii OR191, Rhizobium favelukesii LPU83, Rhizobium mongolense USDA 1844, Rhizobium tibeticum CGMCC 1.7071. Group C: Rhizobium aethiopicum HBR26, Rhizobium etli CFN 42, Rhizobium leguminosarum s.s. 4292, Rhizobium leguminosarum genospecies K FA23, Rhizobium phaseoli CIAT 652. Group D: Paraburkholderia dilworthii WSM3556 and Paraburkholderia tuberum WSM4176. Group E: Rhizobium mesoamericanum STM 3625 and Rhizobium mesoamericanum STM6155. Group F: Ensifer fredii GR64, Ensifer sp. 4H41 and Ensifer meliloti GVPV12. Group G: Rhizobium leucaenae USDA 9039, Rhizobium lusitanum P1-7 and Rhizobium tropici CIAT899.
By mapping nod gene neighborhoods to the NodA phylogenetic tree, we identified that particular arrangements of nod genes are associated with each NodA group (Figure 1). The figure illustrates the high degree of synteny between the nod genes of the Medicago-nodulating Ensifer strains (Group A) and the R. favelukesii, R tibeticum and R mongolense strains (Group B). R. tibeticum CGMCC 1.7071T is the microsymbiont of Medicago ruthenica, while R. mongolense USDA 1844T is the microsymbiont of Medicago archiducis-nicolai (Van Berkum et al., 1998; Hou et al., 2009). Both these Medicago species belong to the basal section Platycarpae and are distributed in Siberia, Mongolia, Tibet and northern China, which is far to the northeast of alfalfa’s center of origin (Small, 2010; Steele et al., 2010), suggesting that there may have been a change of microsymbiont in Medicago species, due to either biogeographical factors or a change in host preference. As noted previously, all P. vulgaris-nodulating strains (Group B—G) have either nodS or nodL in their genomes. In the strains belonging to Group D to G, nodSU is consistently located between nodC and nodIJ. The truncated nodU seen in the P. vulgaris-nodulating strains of Group C would appear to have arisen from an ancestral nodSU form. In the Medicago-nodulating strains belonging to Group A and B, the deletion of nodSU would account for nodABCIJ being adjacent to each other.
Comparison of Nif and Fix Genes in Medicago and Phaseolus vulgaris Microsymbionts
The rhizobial nif, fix and fdx genes are required for the stepwise reduction of dinitrogen gas and incorporation of fixed N (Fischer, 1994). Whereas the characterized rhizobial diazotroph Azorhizobium caulinodans ORS 571T has 15 nif genes, most rhizobia are unable to fix nitrogen ex planta and their genomes contain fewer than 15 nif genes (Masson-Boivin et al., 2009). We identified 13 nif genes in OR191 (Supplementary Figure 4 and Supplementary Table 5). The genes and the associated processes that they are involved in are: nifA (regulation); nifH (dinitrogenase reductase); nifDK (α and β subunits of dinitrogenase); nifB (synthesis of the iron-sulfur-containing precursor of the FeMo-co); nifEN (assembly of the FeMo-co); nifQSX (FeMo-co biosynthesis), nifZ (nitrogenase maturation) and fixU/nifTnifW (nitrogen fixation accessory and stabilization proteins, respectively). OR191 lacked genes encoding NifU (iron-sulfur cluster scaffolding protein) and NifV (homocitrate synthase). The OR191 genome also contained genes required for production of ferredoxins (fdxBN), putative electron transfer proteins (fixABCX), the specific high-affinity cbb3-type cytochrome c oxidase (fixNOQP), a Cu2+-exporting ATPase (fixI), electron transfer components (fixGHS) and a putative FNR/CRP [fumarate and nitrate reductase regulator protein and cyclic AMP (cAMP) receptor protein] transcriptional regulator (fixK) (Supplementary Table 5). The OR191 nif and fix genes are most closely related to those of LPU83T, CGMCC 1.7071T and USDA 1844T and then to those of Rhizobium and Ensifer strains that nodulate P. vulgaris, rather than to the nif and fix genes of Medicago-nodulating Ensifer strain. Phylogenetic analysis of NifH and NifA shows that OR191, LPU83T, CGMCC 1.7071T, and USDA 1844T form a clade in comparison to other microsymbiont strains analyzed in this study (Figure 2A).
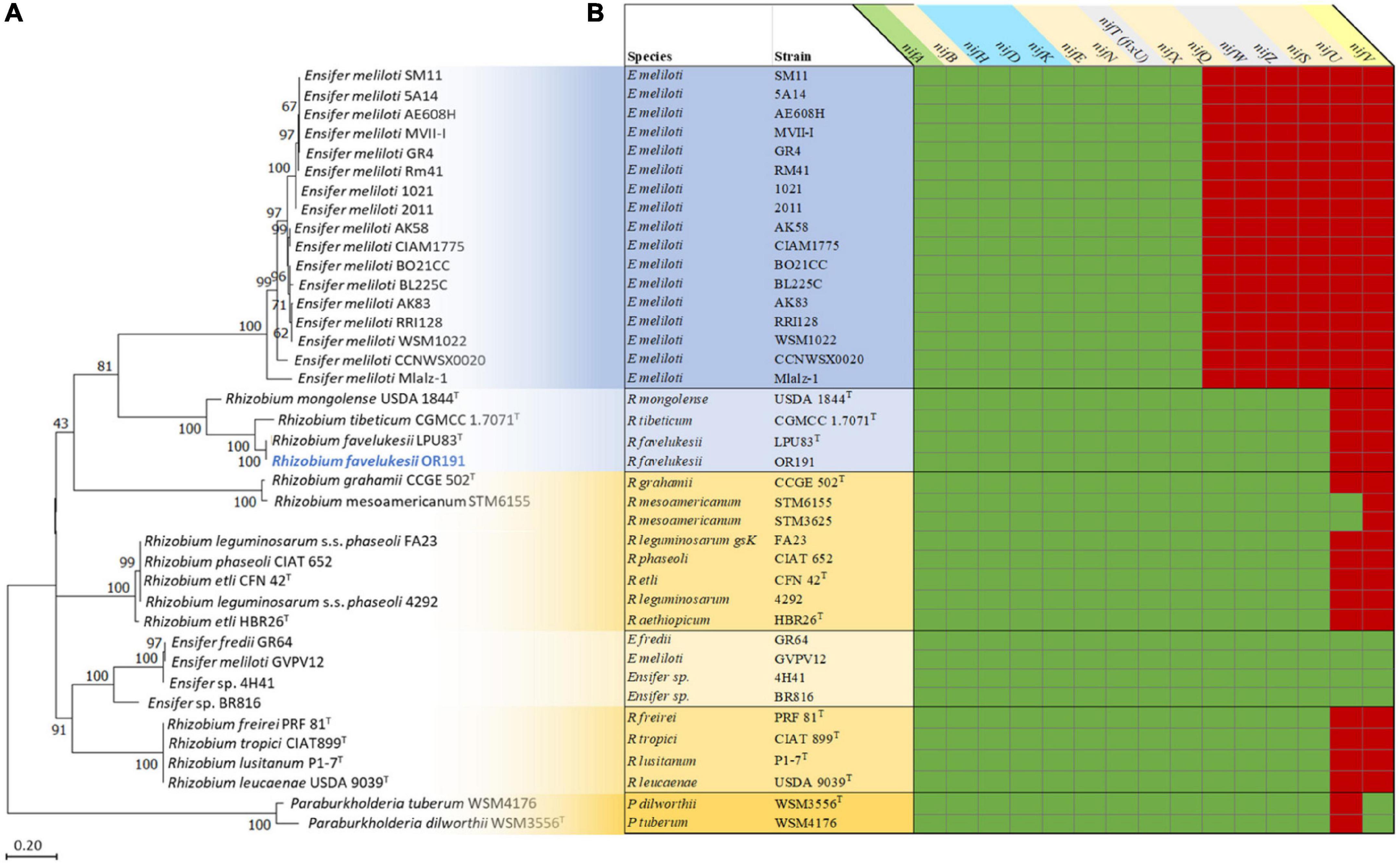
Figure 2. Analysis of the nitrogen fixation genes and proteins in Medicago and Phaseolus vulgaris microsymbionts (Supplementary Table 4). Phylogenetic analysis of the NifH proteins (A) and a map of nif genes present (green) or absent (red) in the genomes of the microsymbionts (B). All microsymbionts shaded in blue nodulate and fix with Medicago sp. Strains shaded in yellow nodulate and fix with Phaseolus vulgaris. Rhizobium favelukesii LPU83, Rhizobium mongolense USDA 1844, Rhizobium tibeticum CGMCC 1.7071 can nodulate both Medicago spp. and Phaseolus vulgaris.
Furthermore, a comparison of the nif genes present in the microsymbionts of Medicago spp. and P. vulgaris showed that all genomes contained the nitrogenase genes nifABHDKENX and fixU/nifT or nifT-like genes (encoding the specific NifT pfam06988), fdxBN and fixABCX genes (Figure 2B). All strains contained fixGHIS and fixNOQP, apart from the Paraburkholderia microsymbionts (previously reported in De Meyer et al., 2016). The most striking differences between the suite of nif genes found in the Medicago and P. vulgaris rhizobia was that Ensifer strains that are Medicago microsymbionts all lacked nifQWZSUV, whereas strains that nodulate and fix with P. vulgaris all contained nifQWZS. All the Rhizobium strains contained nifS but lacked nifU and nifV. Ensifer strains that were microsymbionts of P. vulgaris (E. fredii GR64, E meliloti GVPV12 and Ensifer sp. 4H41 and BR816) all contained nifU and nifV, in addition to the 13 nif genes identified in OR191. The nifU gene was absent from the Paraburkholderia strains, but as rhizobial Paraburkholderia strains are able to fix N ex planta (Elliott et al., 2007), nifU may be replaced by an unidentified gene encoding a functionally equivalent protein.
Comparisons of Other Symbiotically Relevant Genes in Medicago and Phaseolus vulgaris Microsymbionts
Succinoglycan Biosynthesis Genes
Other known rhizobial genes that are required for symbiotic interactions with legumes include those responsible for production of exopolysaccharides, which play an important role in the primary stage of infection by suppressing the plant defense response, as well as contributing to acidic pH tolerance (Skorupska et al., 2006; Hawkins et al., 2017). In the alfalfa-E. meliloti symbiosis, the specific exopolysaccharide succinoglycan (EPS I) is required for the formation of infection threads in Medicago root hairs and is necessary to prevent the expression of plant defense response genes (Jones and Walker, 2008), although the lack of EPS I may be overcome to some extent by the production of galactoglucan (EPS II) or capsular polysaccharide (Pellock et al., 2000). In the characterized strain E. meliloti 1021, the exo genes required for succinoglycan biosynthesis (exoBZQFYXUVWTIHKLAMONP) form a cluster on the pSymB of this strain (Finan et al., 2001; Janczarek, 2011).
Within the OR191 genome, we identified homologs of all these exo genes (Supplementary Table 5) except for exoI (the OR191 exoI-like gene, encoding a periplasmic endonuclease, shared greater sequence identity with paralogs of exoI-like genes within the 1021 genome). The exoB and exoN genes, encoding nucleotide sugar precursors, were located on scaffolds 2.3 and 77.78, respectively. The expR gene, encoding a LuxR family transcriptional regulator that regulates production of both EPS I and EPS II, was also located around 60 kbp downstream of exoB on scaffold 2.3. The remaining exo genes were in two separate clusters, with the nucleotide sequence of each cluster being highly conserved and nearly identical (>99% over the entire length) to corresponding exo clusters in R. favelukesii LPU83T. The exo genes of LPU83T have been characterized; one cluster is on the chromosome and one cluster is located on the plasmid pLPU83a between two inverted repeat regions, suggesting an integration of DNA from another organism (Castellani et al., 2019, 2021). In OR191, Cluster I on scaffold 50.51 contained exoZQFYXUWHKAMOP and shared DNA sequence similarity of 99–100% with the exo cluster on the LPU83T chromosome (NCBI accession number HG916852). The R. favelukesii Cluster I exo genes shared greatest synteny and sequence identity with exo genes of the Rhizobium strains R. tibeticum CGMCC 1.7071T, R. grahamii CCGE 502T and R. mesoamericanum, all closely related to R. favelukesii strains. Cluster II, on scaffold 118.119, contained exoPOO’MLKHTWV and an exoA pseudogene and shared 99–100% sequence similarity with the LPU83T exo cluster harbored on the accessory plasmid LPU83a (NCBI accession number HG916853). In contrast to Cluster I, the R. favelukesii Cluster II exo genes shared greatest synteny and sequence identity with the E. meliloti exo gene cluster, rather than with other Rhizobium strains.
All Medicago microsymbiont genomes contained the full set of exopolysaccharide biosynthetic genes required to synthesize succinoglycan. The E. meliloti strains, including GVPV12 that nodulates P. vulgaris, as well as the closely related Ensifer sp. 4H41, contained a full set of the required succinoglycan biosynthetic genes, whereas E. fredii GR64 and Ensifer sp. BR816 did not. Only some P. vulgaris microsymbionts, including all those within symbiovar orientale and additionally R. grahamii CCGE 502T, R. lusitanum P1-7T, R. mesoamericanum strains and R. freirei PRF 81T, contained the exoH gene that is specifically required for the succinylation of succinoglycan (reviewed in Skorupska et al., 2006).
Interestingly, although succinoglycan is required for the effective symbiotic interaction of alfalfa with E. meliloti, it does not seem to be necessary for the alfalfa-R. favelukesii symbiosis. According to Castellani et al. (2021), LPU83T produces an identical EPS I to the one produced by E. meliloti and does not produce EPS II or capsular polysaccharide, yet LPU83T mutants devoid of the genes required for succinoglycan biosynthesis are able to infect and nodulate with alfalfa and are not impaired for symbiosis, compared with the wild type.
Characterization of Exopolysaccharide Produced by OR191
Following bioinformatics analysis of EPS biosynthetic genes in OR191, we quantified the amount of EPS and substituents produced under neutral and acidic pH growth conditions (Table 4), and determined its composition by NMR spectroscopy. OR191 produced 0.1 mg ml–1 and 0.5 mg ml–1 EPS at pH 7.0 and 5.8, respectively, representing a fivefold increase in EPS production in acidic conditions. This differs from previous reports of EPS production in R. favelukesii LPU83T, which found no significant difference in the amount of EPS obtained from cells grown under neutral and acidic conditions (Nilsson et al., 2020). In contrast, the E. medicae strain WSM419 produced 0.05 mg ml–1 and 4.65 mg ml–1 EPS at pH 7.0 and 5.8, respectively, representing a 93-fold increase in EPS production in acidic conditions. This accords with the up-regulation of EPS production at low pH that was previously observed for the succinoglycan-producing strain WSM419 (Dilworth et al., 1999). Purified EPS from WSM419 was highly viscous whereas OR191 EPS was less viscous. In the presence of calcofluor, WSM419 EPS showed an intense greenish fluorescence whereas OR191 EPS gave a distinct blueish fluorescence (Figure 3), indicating structural differences in the EPS.
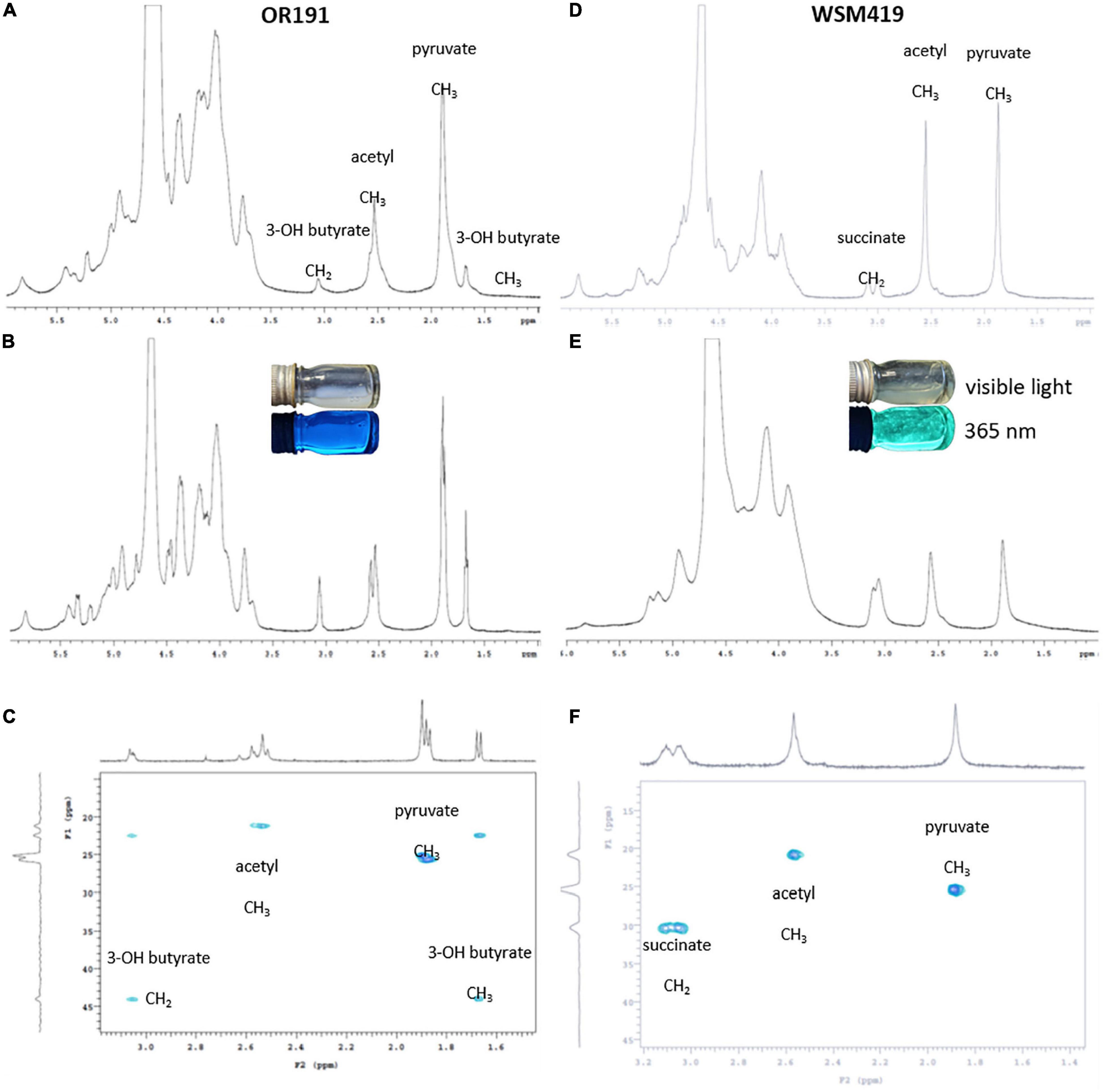
Figure 3. NMR spectra for EPS purified from Rhizobium favelukesii OR191 (A–C) and Ensifer medicae WSM419 (D,E). (A,D) 1H spectrum at pH 7.0. (B,E) 1H spectra at pH 5.8. (C,F) HSQC-TOCSY. EPS samples containing Calcofuor (B,E) were imaged using visible light and at a wavelength of 365 nm.
The 1H-NMR pattern for the isolated EPS from OR191 (Figures 3A,B) was consistent with that described by Reeve et al. (1997) for Rhizobium leguminosarum. Signals at δH 1.85 and 2.55 were assigned to pyruvyl and acetyl methyl groups, respectively. Absolute values of the chemical shifts are different from those reported in similar EPS (McNeil et al., 1986; Reeve et al., 1997) due to different methods for calibrating the chemical shifts. Signals at δH 3.05 and 1.65 were identified as methylene and methyl, respectively, from a 3-hydroxybutyrate moiety. These assignments were confirmed by correlations between carbons at δC 22 and 24 with the 1H signals at 2.05 and 1.65 in the HSQC-TOCSY spectrum (Figure 3C). These correlations confirm that the protons for these two signals must be in the same spin system. The HSQC-TOCSY data also confirmed that assigned pyruvyl and acetyl 1H signals were correlated to only a single carbon, consistent with the isolated spin system for each of those methyl groups. The presence of 3-hydroxybutyrate substitution in the OR191 EPS is consistent with the finding that hydroxybutanoyl substituents have been found in the EPS isolated from a variety of Rhizobium strains (O’neill et al., 1991).
NMR spectra for the EPS isolated from WMS419 (Figures 3D–F) indicated that this was succinoglycan, with the pattern consistent with that reported previously for this strain (Dilworth et al., 1999) and recently from LPU83T (Castellani et al., 2021). Signals at δH 1.88 and 2.56 were assigned as pyruvyl and acetyl methyls, respectively, and this is supported by correlations of those signals to carbons at δC 25.4 and 20.9 in the HSQC-TOCSY. The signals at δH 3.05 and 3.10 are consistent with those for succinyl methylene as previously reported (Dilworth et al., 1999). An exclusive correlation in the HSQC-TOCSY between a carbon at δC 30.36 and the 1H signals at δH 3.05 and 3.10 are consistent with those of a succinic methylene. The broadness of the 1H-NMR signals δH 3.0–3.1 in the spectra for the EPS isolated from OR191 and WMS419 suggests that these represent the same structural moiety in the two strains. However, the clear and distinct correlations in the HSQC-TOCSY data clearly indicate that there is no succinate present in OR191 EPS and there is no evidence to indicate the presence of a 3-hydroxybutyl moiety in the EPS isolated from WMS419.
A possible explanation for the difference in EPS produced by OR191 and LPU83T may be that the strains were grown in different media. Osmolarity, ammonium and phosphate availability, and the type of carbon source and age of the culture have been shown to modify the amount and composition of EPS produced by rhizobial strains (Janczarek, 2011). In E. meliloti, succinoglycan is essential for establishment of an effective symbiosis with species of Medicago, and overexpression of exoY in E. meliloti 1021, resulting in an increase in succinoglycan production, promoted a more effective symbiosis with Medicago truncatula (Jones, 2012). Our results suggest that OR191 does not produce succinoglycan under the given experimental conditions. Future work could look at the genetic regulation of EPS production in OR191 in response to different environmental conditions and different hosts. Creating strains that express succinoglycan could determine whether this EPS improves effectiveness of the OR191 symbiosis with Medicago spp.
The Role of the bacA Gene
Medicago hosts belong to a group of legumes within the IRLC that target NCR peptides to their microsymbionts, resulting in endoreduplication, pleomorphism, and terminal differentiation of the bacteroids (reviewed in Alunni and Gourion, 2016). Both the number of NCR peptides and the bacteroid morphotype vary according to the IRLC legume species (Montiel et al., 2017). In contrast to IRLC legumes, phaseoloid legumes such as P. vulgaris do not produce NCR peptides, and their bacteroids are not endoreduplicated and remain viable (Mergaert et al., 2006; Haag et al., 2013). The survival of rhizobia within nodules that produce NCR peptides depends on the presence of the rhizobial transporter protein BacA (Alunni and Gourion, 2016). Moreover, the different bacterial orthologs of bacA are not functionally interchangeable and the requirement for a specific type of BacA varies according to the particular legume species (Dicenzo et al., 2017). We identified orthologs of bacA in all microsymbiont genomes except the two Paraburkholderia strains. The OR191 BacA grouped with the E. meliloti -type BacA clade (Dicenzo et al., 2017), along with BacA of R. favelukesii LPU83T, R. tibeticum CGMCC 1.7071T, R. grahamii CCGE 502T, R. mesoamericanum STM6155 and STM3625, R. lusitanum P1-7T and all E. meliloti strains. This accords with results obtained by Dicenzo et al. (2017), who found that E. medicae and E. meliloti BacA had evolved to accommodate a specific interaction with Medicago and were divergent from BacA of other rhizobia. Although the R. favelukesii BacA is similar to the E. meliloti BacA, bacteroids of OR191 and LPU83T show a lack of differentiation inside the nodules (Eardly et al., 1992; Wegener et al., 2001), which has been suggested as an explanation for the low rate of N2 fixation in the alfalfa-R. favelukesii symbiosis (Eardly et al., 1992; Wegener et al., 2001; Castellani et al., 2021). This also suggests that while the R. favelukesii BacA is able to maintain the viability of bacteria inside alfalfa nodule cells, there are differences in the effects of alfalfa NCR peptides on E. meliloti bacteroids compared with R. favelukesii bacteroids.
Conclusion
The cultivation of M. sativa outside its natural range, in moderately acid agricultural soils, has resulted in this host being nodulated by strains such as R. favelukesii OR191, LPU83T and T1155 that are well adapted to these edaphic conditions (Eardly et al., 1985; Del Papa et al., 1999; Bromfield et al., 2010). These R. favelukesii strains represent a divergent lineage of acid-adapted strains within the genus Rhizobium that are partially effective for N2 fixation with both alfalfa and common bean, two hosts that are usually nodulated by distinctly different rhizobial genera.
We identified orthologs of rhizobial acid-adaptation genes in OR191, including olsC, which is involved in the production of hydroxylated ornithine lipid species in the acid-tolerant strain R. tropici CIAT899T (Rojas-Jiménez et al., 2005; Vences-Guzmán et al., 2011). This gene is absent from the comparatively acid-sensitive E. medicae WSM419 and E. meliloti 1021 strains. Additionally, R. favelukesii contains the highly acid-induced lpiA and acvB genes, but lacks the associated regulatory system described for this operon in Ensifer (Reeve et al., 2006), indicating that an unidentified pH responsive regulatory system is present in R. favelukesii. Several mechanisms for acid tolerance in R. favelukesii LPU83T have been suggested by Nilsson et al. (2019, 2020).
The R. favelukesii nod genes are highly syntenic to the nod genes of E. meliloti Medicago microsymbionts. The nodA gene is closely related to nodA of E. meliloti Medicago microsymbionts, which is specifically required for the N-acylation of the Nod factor by an unsaturated fatty acid (Debellé et al., 1996). The presence of nodEF, nodHPQ and nodL suggests that OR191 produces the sulfated, acetylated Nod factors with α, β-unsaturated acyl chains that are required for symbiotic interactions with Medicago hosts (Honma et al., 1990; Lerouge et al., 1990; Debellé et al., 2001). However, the Nod factors produced by the closely related strain LPU83T are sulfated and methylated with unsaturated acyl chains (Torres Tejerizo et al., 2011). The nodG gene is truncated at the C-terminus in R. favelukesii strains, which could decrease Medicago nodulation efficiency compared to the E. meliloti Medicago microsymbionts. R. favelukesii genomes contain additional loci thought to be required for symbiosis with Medicago, including genes required for the biosynthesis of succinoglycan. This rhizobial exopolysaccharide is specifically required to prevent the expression of plant defense response genes and allow the formation of infection threads in Medicago hosts (Skorupska et al., 2006; Jones and Walker, 2008). We have shown that OR191 does not produce succinoglycan in the conditions tested, which differs from the results found for LPU83T (Castellani et al., 2021); instead, OR191 EPS contains a 3-hydroxybutyrate substituent. The lack of succinoglycan in OR191 may reduce its symbiotic performance with Medicago hosts. Additionally, the R. favelukesii bacA gene groups with bacA genes within the E. meliloti clade. E. meliloti BacA specifically protects the microsymbiont from the toxic effects of Medicago NCR peptides (Dicenzo et al., 2017). These Medicago-specific symbiotic determinants in R. favelukesii do not prevent P. vulgaris nodulation, however, NodL-acetylation or NodS-methylation of the Nod Factor appears to be required for symbiosis with this host. The presence of the nitrogenase genes nifQWZS in the genomes of R. favelukesii and other P. vulgaris microsymbionts, but their absence from the E. meliloti Medicago microsymbionts suggests that these genes are important for N2 fixation with P. vulgaris. Although the R. favelukesii strains nodulate and fix with M. sativa and P. vulgaris, they are poorly effective for N2 fixation compared with the usual microsymbionts of these hosts. Future studies will be aimed at identifying genetic determinants required not only for symbiotic interaction with legume hosts but also those that enable effective nitrogen fixation.
Data Availability Statement
The OR191 genome project is deposited in the Genomes On-Line Database (Reddy et al., 2015) and a high-quality permanent draft genome sequence is deposited in IMG (Markowitz et al., 2014). The data associated with this project is available at https://gold.jgi.doe.gov/project?id=9662 under the project number Gp0009662. Sequencing, finishing, and annotation were performed by the JGI.
Author Contributions
BE and PB supplied the strain, DNA and background information for this project. BE, WM, JA, JZ, and WR drafted the manuscript. WM, JA, JZ, and WR performed the bioinformatics analyses. DM, MG, PE, RS, TR, NI, AP, TW, and NK involved in sequencing the genome and/or editing the final manuscript. WR and JA extracted EPS from cell cultures. ML and DL performed the NMR spectra analysis of the EPS. All authors read and approved the final manuscript.
Funding
This work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, was supported under Contract No. DE-AC02-05CH11231. We gratefully acknowledge the funding received from the Curtin University Sustainability Policy Institute and from Murdoch University’s Small Research Grants Scheme in 2016.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Gordon Thompson and Rui Tian (Murdoch University) for the preparation of SEM and TEM photos.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.735911/full#supplementary-material
Supplementary Figure 3 | Resident prophages present in Rhizobium favelukesii OR191 imaged using PHASTER (Arndt et al., 2016). Scaffolds 4.5 and 84.85, as predicted by PHASTER, contain two putative prophages (A,C), whereas scaffolds 7.8 and 91.92 contain two questionable prophages (B,D), all of which are incomplete.
Abbreviations
EPS, Exopolysaccharide; FeMo-co, Nitrogenase iron–molybdenum cofactor; GEBA-RNB, Genomic Encyclopedia for Bacteria and Archaea-Root Nodule Bacteria; gANI, Genome Average Nucleotide Identity; GOLD, Genomes Online Database; IMG, Integrated Microbial Genomes; IRLC, Inverted Repeat Lacking Clade; JGI, Joint Genome Institute; NCR peptide, Nodule-specific Cysteine-Rich peptide; TYC, Tryptone yeast extract agar; YEM, Yeast extract mannitol.
References
Alunni, B., and Gourion, B. (2016). Terminal bacteroid differentiation in the legume-rhizobium symbiosis: nodule-specific cysteine-rich peptides and beyond. New Phytol. 211, 411–417. doi: 10.1111/nph.14025
Ardourel, M., Lortet, G., Maillet, F., Roche, P., Truchet, G., Promé, J.-C., et al. (1995). In Rhizobium meliloti, the operon associated with the nod box n5 comprises nodL, noeA and noeB, three host-range genes specifically required for the nodulation of particular Medicago species. Mol. Microbiol. 17, 687–699. doi: 10.1111/j.1365-2958.1995.mmi_17040687.x
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Baev, N., Endre, G., Petrovics, G., Banfalvi, Z., and Kondorosi, A. (1991). Six nodulation genes of nod box locus 4 in Rhizobium meliloti are involved in nodulation signal production: nodM codes for D-glucosamine synthetase. Mol. Gen. Genet. 228, 113–124. doi: 10.1007/BF00282455
Béna, G., Lyet, A., Huguet, T., and Olivieri, I. (2005). Medicago-Sinorhizobium symbiotic specificity evolution and the geographic expansion of Medicago. J. Evol. Biol. 18, 1547–1558. doi: 10.1111/j.1420-9101.2005.00952.x
Bottomley, P. J., and Jenkins, M. B. (1983). Some characteristics of Rhizobium meliloti isolates from alfalfa fields in Oregon. Soil Sci. Soc. Am. J. 47, 1153–1157. doi: 10.2136/sssaj1983.03615995004700060019x
Bromfield, E. S. P., Butler, G., and Barran, L. R. (2001). Temporal effects on the composition of a population of Sinorhizobium meliloti associated with Medicago sativa and Melilotus alba. Can. J. Microbiol. 47, 567–573. doi: 10.1139/w01-034
Bromfield, E. S. P., Tambong, J. T., Cloutier, S., Prevost, D., Laguerre, G., Van Berkum, P., et al. (2010). Ensifer. Phyllobacterium and Rhizobium species occupy nodules of Medicago sativa (alfalfa) and Melilotus alba (sweet clover) grown at a Canadian site without a history of cultivation. Microbiology 156, 505–520. doi: 10.1099/mic.0.034058-0
Castellani, L. G., Luchetti, A., Nilsson, J. F., Pérez-Giménez, J., Wegener, C., Schlüter, A., et al. (2021). Exopolysaccharide characterization of Rhizobium favelukesii LPU83 and its role in the symbiosis with alfalfa. Front. Plant Sci. 12:642576 doi: 10.3389/fpls.2021.642576
Castellani, L. G., Nilsson, J. F., Wibberg, D., Schlüter, A., Pühler, A., Brom, S., et al. (2019). Insight into the structure, function and conjugative transfer of pLPU83a, an accessory plasmid of Rhizobium favelukesii LPU83. Plasmid 103, 9–16. doi: 10.1016/j.plasmid.2019.03.004
Chen, I. M. A., Markowitz, V. M., Chu, K., Anderson, I., Mavromatis, K., Kyrpides, N. C., et al. (2013). Improving microbial genome annotations in an integrated database context. PLoS One 8:e54859. doi: 10.1371/journal.pone.0054859
Chen, I.-M. A., Chu, K., Palaniappan, K., Ratner, A., Huang, J., Huntemann, M., et al. (2021). The IMG/M data management and analysis system v. 6.0: new tools and advanced capabilities. Nucleic Acids Res. 49, D751–D763. doi: 10.1093/nar/gkaa939
De Meyer, S. E., Briscoe, L., Martínez-Hidalgo, P., Agapakis, C. M., De Los Santos, P. E., Seshadri, R., et al. (2016). Symbiotic Burkholderia species show diverse arrangements of nif/fix and nod genes and lack typical high-affinity cytochrome cbb3 oxidase genes. Mol. Plant Microbe Inter. 29, 609–619. doi: 10.1094/MPMI-05-16-0091-R
Debellé, F., Moulin, L., Mangin, B., Denarié, J., and Boivin, C. (2001). Nod genes and Nod signals and the evolution of the rhizobium legume symbiosis. Acta Biochim. Pol. 48, 359–365. doi: 10.18388/abp.2001_3921
Debellé, F., Plazanet, C., Roche, P., Pujol, C., Savagnac, A., Rosenberg, C., et al. (1996). The NodA proteins of Rhizobium meliloti and Rhizobium tropici specify the N-acylation of Nod factors by different fatty acids. Mol. Microbiol. 22, 303–314. doi: 10.1046/j.1365-2958.1996.00069.x
Del Papa, M. F., Balagué, L. J., Sowinski, S. C., Wegener, C., Segundo, E., Abarca, F. M., et al. (1999). Isolation and characterization of alfalfa-nodulating rhizobia present in acidic soils of Central Argentina and Uruguay. Appl. Environ. Microbiol. 65, 1420–1427. doi: 10.1128/AEM.65.4.1420-1427.1999
Del Papa, M. F., Pistorio, M., Draghi, W. O., Lozano, M. J., Giusti, M. A., Medina, C., et al. (2007). Identification and characterization of a nodH ortholog from the alfalfa-nodulating Or191-Like rhizobia. Mol. Plant Microbe Interact. 20, 138–145. doi: 10.1094/MPMI-20-2-0138
Demont, N., Debellé, F., Aurelle, H., Dénarié, J., and Promé, J. C. (1993). Role of the Rhizobium meliloti nodF and nodE genes in the biosynthesis of lipo-oligosaccharidic nodulation factors. J. Biol. Chem. 268, 20134–20142. doi: 10.1016/s0021-9258(20)80704-2
Dénarié, J., Debellé, F., and Promé, J. C. (1996). Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65, 503–535. doi: 10.1146/annurev.bi.65.070196.002443
Dicenzo, G. C., Zamani, M., Ludwig, H. N., and Finan, T. M. (2017). Heterologous complementation reveals a specialized activity for BacA in the Medicago–Sinorhizobium meliloti symbiosis. Mol. Plant Microbe Interact. 30, 312–324. doi: 10.1094/MPMI-02-17-0030-R
Dilworth, M. J., Rynne, F. G., Castelli, J. M., Vivas-Marfisi, A. I., and Glenn, A. R. (1999). Survival and exopolysaccharide production in Sinorhizobium meliloti WSM419 are affected by calcium and low pH. Microbiology 145, 1585–1593. doi: 10.1099/13500872-145-7-1585
Eardly, B. D., Hannaway, D. B., and Bottomley, P. J. (1985). Characterization of rhizobia from ineffective alfalfa nodules: ability to nodulate bean plants Phaseolus vulgaris (L) Savi]. Appl. Environ. Microbiol. 50, 1422–1427. doi: 10.1128/aem.50.6.1422-1427.1985
Eardly, B. D., Young, J. P. W., and Selander, R. K. (1992). Phylogenetic position of Rhizobium sp. strain OR191, a symbiont of both Medicago sativa and Phaseolus vulgaris, based on partial sequences of the 16s ribosomal RNA and nifH genes. Appl. Environ. Microbiol. 58, 1809–1815. doi: 10.1128/aem.58.6.1809-1815.1992
Elliott, G. N., Chen, W. M., Chou, J. H., Wang, H. C., Sheu, S. Y., Perin, L., et al. (2007). Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 173, 168–180. doi: 10.1111/j.1469-8137.2006.01894.x
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Field, D., Amaral-Zettler, L., Cochrane, G., Cole, J. R., Dawyndt, P., Garrity, G. M., et al. (2011). The genomic standards consortium. PLoS Biol. 9:e1001088. doi: 10.1371/journal.pbio.1001088
Field, D., Garrity, G., Gray, T., Morrison, N., Selengut, J., Sterk, P., et al. (2008). Towards a richer description of our complete collection of genomes and metagenomes “Minimum Information about a Genome Sequence” (MIGS) specification. Nat. Biotechnol. 26, 541–547.
Finan, T. M., Weidner, S., Wong, K., Buhrmester, J., Chain, P., Vorhölter, F. J., et al. (2001). The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. U.S.A. 98, 9889–9894. doi: 10.1073/pnas.161294698
Fischer, H. M. (1994). Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58, 352–386. doi: 10.1128/mr.58.3.352-386.1994
Garau, G., Reeve, W. G., Brau, L., Deiana, P., Yates, R. J., James, D., et al. (2005). The symbiotic requirements of different Medicago spp. suggest the evolution of Sinorhizobium meliloti and S. medicae with hosts differentially adapted to soil pH. Plant Soil 276, 263–277. doi: 10.1007/s11104-005-0374-0
Github and GitHub. (2011). GitHub - lh3/wgsim: Reads Simulator [Online]. Available online at: https://github.com/lh3/wgsim (accessed January 28, 2022).
Gnerre, S., Maccallum, I., Przybylski, D., Ribeiro, F. J., Burton, J. N., Walker, B. J., et al. (2011). High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. U.S.A. 108, 1513–1518. doi: 10.1073/pnas.1017351108
Gomes, D. F., Tullio, L. D., Del Cerro, P., Nakatani, A. S., Rolla-Santos, A. P., Gil-Serrano, A., et al. (2019). Regulation of hsnT, nodF and nodE genes in Rhizobium tropici CIAT 899 and their roles in the synthesis of Nod factors and in the symbiosis. Microbiology 165, 990–1000. doi: 10.1099/mic.0.000824
Graham, P. H. (2008). “Ecology of the root-nodule bacteria of legumes,” in Nitrogen-fixing Leguminous Symbioses, eds M. J. Dilworth, E. K. James, J. I. Sprent, and W. E. Newton (Dordrecht: Springer Netherlands), 23–58. doi: 10.1007/978-1-4020-3548-7_2
Graham, P. H., and Parker, C. A. (1964). Diagnostic features in the characterization of the root nodule bacteria of legumes. Plant Soil 20, 383–396. doi: 10.1007/bf01373828
Haag, A. F., Arnold, M. F. F., Myka, K. K., Kerscher, B., Dall’angelo, S., Zanda, M., et al. (2013). Molecular insights into bacteroid development during Rhizobium–legume symbiosis. FEMS Microbiol. Rev. 37, 364–383. doi: 10.1111/1574-6976.12003
Hawkins, J. P., Geddes, B. A., and Oresnik, I. J. (2017). Succinoglycan production contributes to acidic pH tolerance in Sinorhizobium meliloti Rm1021. Mol. Plant Microbe Interact. 30, 1009–1019. doi: 10.1094/MPMI-07-17-0176-R
Honma, M. A., Asomaning, M., and Ausubel, F. M. (1990). Rhizobium meliloti nodD genes mediate host-specific activation of nodABC. J. Bacteriol. 172, 901–911. doi: 10.1128/jb.172.2.901-911.1990
Hou, B. C., Wang, E. T., Li, Y., Jia, R. Z., Chen, W. F., Gao, Y., et al. (2009). Rhizobium tibeticum sp. nov., a symbiotic bacterium isolated from Trigonella archiducis-nicolai (Širj.) Vassilcz. Int. J. Syst. Evol. Microbiol. 59, 3051–3057. doi: 10.1099/ijs.0.009647-0
Howieson, J. G., and Dilworth, M. J. (2016). Working with Rhizobia. Canberra, ACT: Australian Centre for International Agricultural Research (ACIAR).
Howieson, J. G., Yates, R. J., Foster, K. J., Real, D., and Besier, R. B. (2008). “Prospects for the future use of legumes,” in Nitrogen-fixing Leguminous Symbioses, eds M. J. Dilworth, E. K. James, J. I. Sprent, and W. E. Newton (Dordrecht: Springer Netherlands), 363–394. doi: 10.1007/978-1-4020-3548-7_12
Hyatt, D., Chen, G. L., Locascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Jabbouri, S., Fellay, R., Talmont, F., Kamalaprija, P., Burger, U., Relić, B., et al. (1995). Involvement of nodS in N-methylation and nodU in 6-O-carbamoylation of Rhizobium sp. NGR234 Nod Factors. J. Biol. Chem. 270, 22968–22973.
Janczarek, M. (2011). Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int. J. Mol. Sci. 12, 7898–7933. doi: 10.3390/ijms12117898
Joint Genome Institute (2022). Joint Genome Institute Protocols [Online]. Available online at: http://jgi.doe.gov/user-program-info/pmo-overview/protocols-sample-preparation-information/ (accessed January 28, 2022).
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Jones, K. M. (2012). Increased production of the exopolysaccharide succinoglycan enhances Sinorhizobium meliloti 1021 symbiosis with the host plant Medicago truncatula. J. Bacteriol. 194, 4322–4331. doi: 10.1128/JB.00751-12
Jones, K. M., and Walker, G. C. (2008). Responses of the model legume Medicago truncatula to the rhizobial exopolysaccharide succinoglycan. Plant Signal Behav. 3, 888–890. doi: 10.4161/psb.3.10.6512
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lerouge, P., Roche, P., Faucher, C., Maillet, F., Truchet, G., Promé, J. C., et al. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulfated and acylated glucosamine oligosaccharide signal. Nature 344, 781–784. doi: 10.1038/344781a0
Lopez-Lara, I. M., Kafetzopoulos, D., Spaink, H. P., and Thomas-Oates, J. E. (2001). Rhizobial NodL O-acetyl transferase and NodS N-methyl transferase functionally interfere in production of modified Nod factors. J. Bacteriol. 183, 3408–3416. doi: 10.1128/JB.183.11.3408-3416.2001
Lowe, T. M., and Eddy, S. R. (1997). TRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Mao, Y.-H., Li, F., Ma, J.-C., Hu, Z., and Wang, H.-H. (2016). Sinorhizobium meliloti functionally replaces 3-oxoacyl-acyl carrier protein reductase (FabG) by overexpressing NodG during fatty acid synthesis. Mol. Plant Microbe Interact. 29, 458–467. doi: 10.1094/MPMI-07-15-0148-R
Markowitz, V. M., Chen, I. A., Palaniappan, K., Chu, K., Szeto, E., Pillay, M., et al. (2014). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42, D560–D567. doi: 10.1093/nar/gkt963
Markowitz, V. M., Mavromatis, K., Ivanova, N. N., Chen, I. M., Chu, K., and Kyrpides, N. C. (2009). IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25, 2271–2278. doi: 10.1093/bioinformatics/btp393
Masson-Boivin, C., Giraud, E., Perret, X., and Batut, J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends in Microbiol. 17, 458–466. doi: 10.1016/j.tim.2009.07.004
Mavromatis, K., Ivanova, N. N., Chen, I. M., Szeto, E., Markowitz, V. M., and Kyrpides, N. C. (2009). The DOE-JGI standard operating procedure for the annotations of microbial genomes. Stand. Genom. Sci. 1, 63–67. doi: 10.4056/sigs.632
McNeil, M., Darvill, J., Darvill, A. G., Albersheim, P., Van Veen, R., Hooykaas, P., et al. (1986). The discernible, structural features of the acidic polysaccharides secreted by different Rhizobium species are the same. Carbohydrate Res. 146, 307–326. doi: 10.1016/0008-6215(86)85048-0
Mergaert, P., Uchiumi, T., Alunni, B., Evanno, G., Cheron, A., Catrice, O., et al. (2006). Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium–legume symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 5230–5235. doi: 10.1073/pnas.0600912103
Montiel, J., Downie, J. A., Farkas, A., Bihari, P., Herczeg, R., Bálint, B., et al. (2017). Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc. Natl. Acad. Sci. U.S.A. 114, 5041–5046. doi: 10.1073/pnas.1704217114
Mukherjee, S., Stamatis, D., Bertsch, J., Ovchinnikova, G., Sundaramurthi, J. C., Lee, J., et al. (2021). Genomes OnLine Database (GOLD) v.8: overview and updates. Nucleic Acids Res. 49, D723–D733. doi: 10.1093/nar/gkaa983
Naismith, J. H., and Booth, I. R. (2012). Bacterial mechanosensitive channels—MscS: evolution’s solution to creating sensitivity in function. Annu. Rev. Biophys. 41, 157–177. doi: 10.1146/annurev-biophys-101211-113227
Nawrocki, E. P., and Eddy, S. R. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935. doi: 10.1093/bioinformatics/btt509
Nilsson, J. F., Castellani, L. G., Draghi, W. O., Mogro, E. G., Wibberg, D., Winkler, A., et al. (2020). Global transcriptome analysis of Rhizobium favelukesii LPU83 in response to acid stress. FEMS Microbiol. Ecol. 97:fiaa235
Nilsson, J. F., Castellani, L. G., Draghi, W. O., Pérez-Giménez, J., Torres Tejerizo, G. A., and Pistorio, M. (2019). Proteomic analysis of Rhizobium favelukesii LPU83 in response to acid stress. J. Proteome Res. 18, 3615–3629. doi: 10.1021/acs.jproteome.9b00275
O’neill, M. A., Darvill, A. G., and Albersheim, P. (1991). The degree of esterification and points of substitution by O-acetyl and O-(3-hydroxybutanoyl) groups in the acidic extracellular polysaccharides secreted by Rhizobium leguminosarum biovars viciae, trifolii, and phaseoli are not related to host range. J. Biol. Chem. 266, 9549–9555. doi: 10.1016/s0021-9258(18)92855-3
Ormeño-Orrillo, E., Menna, P., Almeida, L. G. P., Ollero, F. J., Nicolás, M. F., Pains Rodrigues, E., et al. (2012). Genomic basis of broad host range and environmental adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 which are used in inoculants for common bean (Phaseolus vulgaris L.). BMC Genomics 13:735. doi: 10.1186/1471-2164-13-735
Pellock, B. J., Cheng, H.-P., and Walker, G. C. (2000). Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182:4310. doi: 10.1128/JB.182.15.4310-4318.2000
Perret, X., Staehelin, C., and Broughton, W. J. (2000). Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64, 180–201. doi: 10.1128/MMBR.64.1.180-201.2000
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. D. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Reddy, T. B. K., Thomas, A. D., Stamatis, D., Bertsch, J., Isbandi, M., Jansson, J., et al. (2015). The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 43, D1099–D1106. doi: 10.1093/nar/gku950
Reeve, W. G., Bräu, L., Castelli, J., Garau, G., Sohlenkamp, C., Geiger, O., et al. (2006). The Sinorhizobium medicae WSM419 lpiA gene is transcriptionally activated by FsrR and required to enhance survival in lethal acid conditions. Microbiology 152, 3049–3059. doi: 10.1099/mic.0.28764-0
Reeve, W. G., Dilworth, M. J., Tiwari, R. P., and Glenn, A. R. (1997). Regulation of exopolysaccharide production in Rhizobium leguminosarum biovar viciae WSM710 involves exoR. Microbiology 143, 1951–1958. doi: 10.1099/00221287-143-6-1951
Rogel, M. A., Ormeno-Orrillo, E., and Romero, E. M. (2011). Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst. Appl. Microbiol. 34, 96–104. doi: 10.1016/j.syapm.2010.11.015
Rojas-Jiménez, K., Sohlenkamp, C., Geiger, O., Martínez-Romero, E., Werner, D., and Vinuesa, P. (2005). A ClC chloride channel homolog and ornithine-containing membrane lipids of Rhizobium tropici CIAT899 are involved in symbiotic efficiency and acid tolerance. Mol. Plant Microbe Interact. 18, 1175–1185. doi: 10.1094/MPMI-18-1175
Segovia, L., Pinero, D., Palacios, R., and Martinezromero, E. (1991). Genetic structure of a soil population of nonsymbiotic Rhizobium leguminosarum. Appl. Environ. Microbiol. 57, 426–433. doi: 10.1128/aem.57.2.426-433.1991
Segundo, E., Martinez-Abarca, F., Van Dillewijn, P., Fernández-López, M., Lageres, A., Martinez-Drets, G., et al. (1999). Characterisation of symbiotically efficient alfalfa-nodulating rhizobia isolated from acid soils of Argentina and Uruguay. FEMS Microbiol. Ecol. 28, 169–176. doi: 10.1111/j.1574-6941.1999.tb00572.x
Seshadri, R., Reeve, W. G., Ardley, J. K., Tennessen, K., Woyke, T., Kyrpides, N. C., et al. (2015). Discovery of novel plant interaction determinants from the genomes of 163 Root Nodule Bacteria. Sci. Rep. 5:16825. doi: 10.1038/srep16825
Silva, C., Vinuesa, P., Eguiarte, L. E., Souza, V., and Martínez-Romero, E. (2005). Evolutionary genetics and biogeographic structure of Rhizobium gallicum sensu lato, a widely distributed bacterial symbiont of diverse legumes. Mol. Ecol. 14, 4033–4050. doi: 10.1111/j.1365-294X.2005.02721.x
Skorupska, A., Janczarek, M., Marczak, M., Mazur, A., and Krol, J. (2006). Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb. Cell Fact. 5:7. doi: 10.1186/1475-2859-5-7
Small, E. (2010). Alfalfa And Relatives: Evolution And Classification Of Medicago. Ottawa, ON: NRC Research Press.
Sohlenkamp, C., Galindo-Lagunas, K. A., Guan, Z., Vinuesa, P., Robinson, S., Thomas-Oates, J., et al. (2007). The lipid lysyl-phosphatidylglycerol is present in membranes of Rhizobium tropici CIAT899 and confers increased resistance to Polymyxin B under acidic growth conditions. Mol. Plant Microbe Interact. 20, 1421–1430. doi: 10.1094/MPMI-20-11-1421
Steele, K. P., Ickert-Bond, S. M., Zarre, S., and Wojciechowski, M. F. (2010). Phylogeny and character evolution in Medicago (Leguminosae): evidence from analyses of plastid trnK/matK and nuclear GA3ox1 sequences. Am. J. Bot. 97, 1142–1155. doi: 10.3732/ajb.1000009
Sullivan, J. T., Eardly, B. D., Vanberkum, P., and Ronson, C. W. (1996). Four unnamed species of nonsymbiotic rhizobia isolated from the rhizosphere of Lotus corniculatus. Appl. Environ. Microbiol. 62, 2818–2825. doi: 10.1128/aem.62.8.2818-2825.1996
Tamura, K. (1992). Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 9, 678–687. doi: 10.1093/oxfordjournals.molbev.a040752
Tian, R., Heiden, S., Osman, W. A. M., Ardley, J. K., James, E. K., Gollagher, M. M., et al. (2017). Evolution of a multi-step phosphorelay signal transduction system in Ensifer: recruitment of the sigma factor RpoN and a novel enhancer binding protein triggers acid-activated gene expression. Mol. Microbiol. 103, 829–844. doi: 10.1111/mmi.13592
Torres Tejerizo, G., Del Papa, M. F., Soria-Diaz, M. E., Draghi, W., Lozano, M., Giusti, M. D. L., et al. (2011). The nodulation of alfalfa by the acid-tolerant Rhizobium sp. strain LPU83 does not require sulfated forms of lipochitooligosaccharide nodulation signals. J. Bacteriol. 193, 30–39. doi: 10.1128/JB.01009-10
Torres Tejerizo, G., Florencia Del, Papa, M., De Los Ángeles Giusti, M., Draghi, W., Lozano, M., et al. (2010). Characterization of extrachromosomal replicons present in the extended host range Rhizobium sp. LPU83. Plasmid 64, 177–185. doi: 10.1016/j.plasmid.2010.07.004
Torres Tejerizo, G., Pistorio, M., Althabegoiti, M. J., Cervantes, L., Wibberg, D., Schlüter, A., et al. (2014). Rhizobial plasmid pLPU83a is able to switch between different transfer machineries depending on its genomic background. FEMS Microbiol. Ecol. 88, 565–578. doi: 10.1111/1574-6941.12325
Torres Tejerizo, G., Rogel, M. A., Ormeño-Orrillo, E., Althabegoiti, M. J., Nilsson, J. F., Niehaus, K., et al. (2016). Rhizobium favelukesii sp. nov., isolated from the root nodules of alfalfa (Medicago sativa L). Int. J. Syst. Evol. Microbiol. 66, 4451–4457. doi: 10.1099/ijsem.0.001373
Van Berkum, P., Beyene, D., Bao, G., Campbell, T. A., and Eardly, B. D. (1998). Rhizobium mongolense sp. nov. is one of three rhizobial genotypes identified which nodulate and form nitrogen-fixing symbioses with Medicago ruthenica [(L.) Ledebour]. Int. J. Syst. Bacteriol. 48, 13–22. doi: 10.1099/00207713-48-1-13
Varghese, N. J., Mukherjee, S., Ivanova, N., Konstantinidis, K. T., Mavrommatis, K., Kyrpides, N. C., et al. (2015). Microbial species delineation using whole genome sequences. Nucleic Acids Res. 43, 6761–6771. doi: 10.1093/nar/gkv657
Vences-Guzmán, M. Á, Guan, Z., Ormeño-Orrillo, E., González-Silva, N., López-Lara, I. M., Martínez-Romero, E., et al. (2011). Hydroxylated ornithine lipids increase stress tolerance in Rhizobium tropici CIAT899. Mol. Microbiol. 79, 1496–1514. doi: 10.1111/j.1365-2958.2011.07535.x
Waelkens, F., Voets, T., Vlassak, K., Vanderleyden, J., and Van Rhijn, P. (1995). The nodS gene of Rhizobium tropici strain CIAT899 is necessary for nodulation on Phaseolus vulgaris and on Leucaena leucocephala. Mol. Plant Microbe Interact. 8, 147–154. doi: 10.1094/mpmi-8-0147
Wegener, C., Schroder, S., Kapp, D., Pühler, A., Lopez, E. S., Martinez-Abarca, F., et al. (2001). Genetic uniformity and symbiotic properties of acid-tolerant alfalfa-nodulating rhizobia isolated from dispersed locations throughout Argentina. Symbiosis 30, 141–162.
Wibberg, D., Tejerizo, G. T., Del Papa, M. F., Martini, C., Puhler, A., Lagares, A., et al. (2014). Genome sequence of the acid-tolerant strain Rhizobium sp LPU83. J. Biotechnol. 176, 40–41. doi: 10.1016/j.jbiotec.2014.02.008
Zerbino, D. R. (2010). Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinformatics 11:15. doi: 10.1002/0471250953.bi1105s31
Keywords: symbiotic nitrogen fixation (SNF), host—bacteria interaction, acid soils, Medicago sativa, Phaseolus vulgaris
Citation: Eardly B, Meor Osman WA, Ardley J, Zandberg J, Gollagher M, van Berkum P, Elia P, Marinova D, Seshadri R, Reddy TBK, Ivanova N, Pati A, Woyke T, Kyrpides N, Loedolff M, Laird DW and Reeve W (2022) The Genome of the Acid Soil-Adapted Strain Rhizobium favelukesii OR191 Encodes Determinants for Effective Symbiotic Interaction With Both an Inverted Repeat Lacking Clade and a Phaseoloid Legume Host. Front. Microbiol. 13:735911. doi: 10.3389/fmicb.2022.735911
Received: 03 July 2021; Accepted: 10 February 2022;
Published: 13 April 2022.
Edited by:
Nikolai Ravin, Institute of Bioengineering, Research Center of Biotechnology of the Russian Academy of Sciences, RussiaReviewed by:
Gonzalo Arturo Torres Tejerizo, CONICET Instituto de Biotecnologia y Biologia Molecular (IBBM), ArgentinaKarin E. Groten, Max Planck Institute for Chemical Ecology, Germany
Copyright © 2022 Eardly, Meor Osman, Ardley, Zandberg, Gollagher, van Berkum, Elia, Marinova, Seshadri, Reddy, Ivanova, Pati, Woyke, Kyrpides, Loedolff, Laird and Reeve. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wayne Reeve, W.Reeve@murdoch.edu.au
 Bertrand Eardly
Bertrand Eardly Wan Adnawani Meor Osman2
Wan Adnawani Meor Osman2 Julie Ardley
Julie Ardley Jaco Zandberg
Jaco Zandberg Margaret Gollagher
Margaret Gollagher Patrick Elia
Patrick Elia Dora Marinova
Dora Marinova Rekha Seshadri
Rekha Seshadri Natalia Ivanova
Natalia Ivanova Tanja Woyke
Tanja Woyke Nikos Kyrpides
Nikos Kyrpides Matthys Loedolff
Matthys Loedolff Damian W. Laird
Damian W. Laird Wayne Reeve
Wayne Reeve