- Institute of Biochemistry and Biophysics Polish Academy of Sciences, Warsaw, Poland
Here, we describe functional characterization of an early gene (gp46) product of a virulent Lactococcus lactis sk1-like phage, vB_Llc_bIBBF13 (abbr. F13). The GP46F13 protein carries a catalytically active RecA-like domain belonging to the P-loop NTPase superfamily. It also retains features characteristic for ATPases forming oligomers. In order to elucidate its detailed molecular function, we cloned and overexpressed the gp46 gene in Escherichia coli. Purified GP46F13 protein binds to DNA and exhibits DNA unwinding activity on branched substrates in the presence of adenosine triphosphate (ATP). Size exclusion chromatography with multi-angle light scattering (SEC-MALS) experiments demonstrate that GP46F13 forms oligomers, and further pull-down assays show that GP46F13 interacts with host proteins involved in replication (i.e., DnaK, DnaJ, topoisomerase I, and single-strand binding protein). Taking together the localization of the gene and the obtained results, GP46F13 is the first protein encoded in the early-expressed gene region with helicase activity that has been identified among lytic L. lactis phages up to date.
Introduction
Bacteriophages infecting the Lactococcus lactis species pose a great threat to industrial settings by perturbing or arresting milk fermentation processes (Garneau and Moineau, 2011). To elucidate the persistence of especially lytic lactococcal dairy phages, they have been quite extensively studied in terms of genome content and physio-morphological features (Deveau et al., 2006; Muhammed et al., 2017; Oliveira et al., 2018; Marcelli et al., 2020). Yet still, certain aspects of their biology, including genome replication, demand detailed characterization. The knowledge on the replication of lytic lactococcal phages and proteins participating in the process is of great value and may lead to developing new phage resistance mechanisms in lactococcal starter strains for controlling phage multiplication in dairy environments.
DNA replication is one of the pivotal processes in bacteriophage development. Distinct phages follow various replication pathways and exploit different sets of own and/or host proteins for multiplication of their genomic content (Weigel and Seitz, 2006 and references within). For example, the Gram(−) phage T4 and the Gram(+) phage phi29 possess all the necessary components of their replication machinery (i.e., initiator protein, primase, helicase loader, helicase, and DNA polymerase) (Meijer et al., 2001; Kulczyk and Richardson, 2016). In turn, other phages, such as Escherichia coli phage λ or Bacillus subtilis phage SPP1 encode their own replication initiation proteins, but depend on host proteins for further stages of the process. Specifically, proteins GP38, GP39, and GP40 of B. subtilis phage SPP1 execute the function of an origin initiation protein, helicase loader, and DNA helicase, respectively, and phage λ encodes only two proteins participating in replication–an origin-binding protein (protein O) and a helicase loader (protein P) (Mensa-Wilmot et al., 1989; Seco et al., 2013). Due to the generally restricted genome sizes, phage replication initiation functions quite often are coupled in one multi-activity protein, e.g., a primase-helicase like the GP4 protein of T7 and protein α of phage P4 (Bernstein and Richardson, 1988; Ziegelin et al., 1993), or the primase-polymerase of the deep-sea vent phage NrS-1 (Guo et al., 2019) and the corynephage BFK20 replication protein GP43 (Halgasova et al., 2012).
Studies on the replication of lactococcal phages and proteins engaged in the process have been limited essentially to temperate P335-group phages (McGrath et al., 1999; Ostergaard et al., 2001; Labrie et al., 2008). Phage TP901-1 ORF13 and phage Tuc2009 ORF16 (or Rep2009) were identified as the replisome organizer proteins that bind to regions rich in direct repeats located within their cognate genes and determined to serve as the origin region of phage replication (ori). Both elements, ori region and replisome organizer proteins, were shown to be directly engaged in phage replication and confer a PER (phage-encoded resistance) phenotype when cloned in trans. Sequence similarity searches performed for another P335-type phage, r1t, identified ORF11, and ORF12 as a homolog of the Rep2009 replisome organizer and an E. coli DnaC-like helicase loader, respectively (Zúñiga et al., 2002). The replication initiator (organizer) alone or together with the helicase loader is also found in the genomes of other P335-group phages often accompanied by gene encoding a single-strand DNA binding (SSB) protein (Weigel and Seitz, 2006).
The genes encoding putative replication functions of lytic L. lactis phages belonging to sk1-like and c2-like groups are located essentially in analogous genetic locations within the early gene expression region, upstream of the recombination genes, ssa (single-strand annealing) and ssb (single-strand binding). Our in silico genome analyses of sk1-like phages that were previously isolated from the Polish dairy environment (Chmielewska-Jeznach et al., 2018) indicated in this region the presence of a single gene encoding a P-loop NTPase domain. Yet, like for the majority of the early genes, the exact role of this product in DNA replication of sk1-like phages has not been experimentally studied.
P-loop NTPases hydrolyze nucleoside triphosphates (NTPs) and transfer this energy to drive various processes, such as sliding along the DNA and its unwinding (Longo et al., 2020). The structural core of P-loop NTPases is formed by a central 5-strand β-sheet surrounded by α-helices on both sides (Berger, 2008). On one rim of the sheet are localized residues taking part in NTP hydrolysis, organized in several sequence motifs characteristic for AAA+ and RecA-like proteins (for a brief summary see Berger, 2008). For instance, Walker A (also known as P-loop) and Walker B motifs together with Sensor I in AAA+ ATPases provide charged residues for NTP hydrolysis. Arginine finger completes the catalytic site in trans and is especially important for NTPases functioning in oligomeric assemblies, e.g., hexameric rings (Iyer et al., 2004; Miller and Enemark, 2016; Zhao et al., 2016). In the process of DNA replication, P-loop NTPases, with either AAA+ or RecA fold, play the role of clamp loaders, replication initiator proteins, helicase loaders, and DNA helicases (Davey et al., 2002; Ye et al., 2004; Miller and Enemark, 2016).
DNA helicases have key functions in DNA replication, recombination, transcription, and repair (Huttner and Hickson, 2013). The current classification divides helicases into six superfamilies (SF1-6), each characterized by unique structural and biochemical properties (Singleton et al., 2007). The distinctive features of helicases include nucleic acid-specificity (DNA or RNA), oligomerization forms (monomeric or dimeric SF1-2 and hexameric SF3-6 helicases), and unwinding directionality (5′-3′ or 3′-5′). One of the best studied models of DNA helicases is the SF4 bifunctional primase-helicase (gp4) of coliphage T7 (Richardson, 1983). It retains all sequence motifs characteristic for helicases (I, Ia, II, III, IV) and structural studies explain the detailed role of each of them for the function of this hexameric protein (Patel and Hingorani, 1993; Egelman et al., 1995; Sawaya et al., 1999; Singleton et al., 2007). Other similar helicases include bacteriophage T4 gene 41 protein (T4 gp41) and the RepA protein encoded by plasmid RSF1010–both hexameric and both preserving respective sequence motifs (Dong et al., 1995; Scherzinger et al., 1997).
In this study, we show that the product of gene 46 (GP46) of the L. lactis phage vB_Llc_bIBBF13 (phage F13) binds DNA as a dimer, and most likely as a higher order oligomer, and demonstrates adenosine triphosphate (ATP)-dependent 5′-3′ DNA unwinding activity. Through a series of functional experiments we show that the protein most efficiently displaces DNA strands from branched substrates, which imitate replication forks, with a 5′ extended arm. We predict that GP46F13 contains a catalytically active RecA-like protein domain homologous to RepA, RecA, DnaB, and AAA_25, among the others. Based on its sequence similarity to other helicases of known structure, e.g., RepA from E. coli RSF1010 plasmid and phage T7 gp4, we anticipated that GP46F13 would function as a hexameric, ring-shaped helicase. Size exclusion chromatography with multi-angle light scattering (SEC-MALS) assay indicated strong oligomerization potential of GP46, but the issue concerning its multimeric structure remains unresolved. Moreover, we identify among others an interaction of GP46F13 with host’s DnaK and DnaJ proteins as well as with single strand binding protein (SSB) and topoisomerase I (topoI). Altogether, these data suggest that GP46F13 is a DNA helicase engaged likely in replication. As orfs in the direct surrounding of gp46 do not encode for known replication proteins or possess domains typical for such activity, GP46F13 is the first functionally characterized early gene product among L. lactis sk1-like phages that defines the putative phage replication module and for which a role as a helicase is proposed. Our findings bring us closer to unraveling the role of specific protein players in replication of virulent L. lactis phages in general.
Materials and Methods
Bacteriophage, Bacteria, and Growth Conditions
Escherichia coli Rosetta™ (DE3) strain (Novagen) and TG1 strain (Gibson, 1984) were grown in Luria-Bertani (LB) medium at 37°C with shaking or on LB plates supplemented with 1.5% agar. L. lactis IL1403 strain (Chopin et al., 1984) was grown at 30°C in M17 broth (Oxoid) supplemented with 0.5% glucose (GM17) without shaking or on GM17-agar plates. Phage F13 (GenBank accession no. MG253653) propagation was performed on L. lactis strain IL1403 in liquid GM17 medium supplemented with 10 mM calcium chloride (GM17-CaCl2) as previously described (Chmielewska-Jeznach et al., 2018).
Cloning and Purification of GP46F13
The overproduction of the L. lactis phage F13 GP46 protein (GenBak accession no. ATW69819.1) was primarily attempted in E. coli using the IMPACT™ expression system (NEB). Yet, with this approach we were able to obtain only very low protein recovery levels, irrespectively of conditions assayed (induction temperature, IPTG and salt concentration, glycerol addition, etc.). To circumvent these problems, we cloned gene 46 (gp46) in the pET28a-SUMO-PIN vector (courtesy of M. Pastor). For this, gp46 was amplified from bacteriophage F13 lysate by PCR reaction using primers SF13for/SF13rev listed in Table 1. The resultant PCR product was digested with BamHI and XhoI (Fermentas) enzymes and ligated into BamHI and XhoI sites of the pET28a-SUMO-PIN vector creating the N-terminal fusion plasmid for the overproduction of the GP46F13 protein (pET28:SUMO:gp46). Ligated molecules were transformed into the E. coli TG1 cells. Transformants were selected on LB-agar medium containing 50 μg/ml kanamycin at 37°C. The presence of the cloned fragment was verified by PCR using SUMOF13_F and SUMOF13_R primers (Table 1) and sequencing.
The recombinant plasmid pET28:SUMO:gp46 was introduced into E. coli Rosetta™ (DE3) cells. Freshly grown colonies were inoculated into 1 L Luria broth (LB) medium containing 50 μg/ml kanamycin and 10 μg/ml chloramphenicol at 28°C until optical density at 600 nm (OD600) reached 0.7. Expression of the target protein was induced by adding isopropyl-b-D-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM and the culture was incubated further on an orbital shaker at 150 rpm for 24 h at 16°C. Next, cells were harvested by centrifugation at 5,000 rpm for 15 min at 4°C. All subsequent purification steps were carried out at 4°C. Bacterial pellet was resuspended in 30 ml of optimized suspension buffer (50 mM Tris–HCl pH 8, 500 mM NaCl, 20 mM imidazole, 2% Triton-X100) and lysed by sonication on ultrasonic cell disruptor XL (MISONIX) 10 times for 15 s with 15 s intervals on ice. Without addition of Triton-X100 the protein after sonication was in the membrane fraction. With low salt in the suspension buffer, the protein recovery was very low. After sonication, the cell debris was then removed by centrifugation at 11,000 rpm for 1 h at 4°C. The supernatant was applied on a HisTrap FF crude column pre-packed with the Ni-Sepharose 6 Fast Flow affinity medium (GE Healthcare) and equilibrated with column buffer (50 mM Tris–HCl pH 8, 500 mM NaCl, 20 mM imidazole). The resin-bound protein was rinsed with 50 ml of column buffer and washed out with elution buffer (50 mM Tris–HCl pH 8, 500 mM NaCl, 300 mM imidazole). The eluate was digested by SUMO protease at 4°C overnight to remove SUMO-tag and then analyzed for the presence of GP46F13 by SDS-PAGE. Protein concentration was determined using the Bradford assay on a UV spectrophotometer (Shimadzu).
Ion Exchange Chromatography
Further protein purification was performed by FPLC (Fast Protein Liquid Chromatography) on the ÄKTA Purifier using the HiTrap® Heparin HP column (GE Healthcare) equilibrated with 50 mM Tris-HCl buffer (pH 8). Before applying on the column, the elution buffer was exchanged to a buffer containing 50 mM Tris–HCl pH 8, 100 mM NaCl, by dialysis. The GP46F13 protein sample was loaded on the column at a flow rate of 0.4 ml/min. Protein elution was carried out using a growing gradient concentration of NaCl (from 100 mM to 1 M) in 50 mM Tris–HCl pH 8 buffer. The peak fractions were analyzed by SDS-PAGE and stored after adding 50% glycerol (v/v) at −20°C for further experiments.
DNA Binding Assays
The DNA-binding activity of GP46F13 was evaluated by electrophoretic mobility-shift assay (EMSA) with fluorescently labeled oligonucleotides (Cy5 on 5′ end) differing in size (22, 45, 68, 110 nt). The dsDNA 110 nt substrate was prepared by PCR reaction using L. lactis c2 phage genome as a template and a pair of complementary primers (ori110_F/ori110_R). The binding reactions were carried out for 30 min at 30°C in binding buffer (20 mM Tris–HCl pH 8, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.1 mg/ml of BSA, 2.5% glycerol) in 20 μl reaction volumes containing 1 nM of appropriate DNA substrate and the indicated amounts of analyzed protein (0.1–0.75 μM). Reaction samples were loaded onto 7% non-denaturing polyacrylamide gels and resolved for 40 min at 100 V in 0.5 × TBE buffer at 4°C with 1-h pre-run. Gels were developed using FluorChemQ MultiImageIII ChemiImager (Alpha Innotech, San Leandro, CA, United States) and the images were captured using Alpha View software (Alpha Innotech, San Leandro, CA, United States).
Helicase Activity Assay
Helicase activity of the GP46F13 protein was assessed on a variety of DNA substrates (Table 1). Asymmetric or forked DNA duplexes were prepared by annealing a 5′ Cy5-labeled oligonucleotides to a series of partially complementary unlabeled ssDNAs (as described by Curti et al., 2007). The helicase activity was assayed by mixing the protein with 1.5 nM of Cy5-labeled DNA substrate in a standard reaction buffer containing (if not stated otherwise) 50 mM Tris-HCl, pH 8, 50 mM NaCl, 5 mM MgCl2 at 20 μl of final volume. Reactions were initiated by the addition of 5 mM ATP (or other NTP/dNTP), and the mixtures were incubated at 30°C for 30 min (if not otherwise stated). Reactions were terminated by the addition of 6 μl of stop solution (10% glycerol, 0.4% SDS, 50 mM EDTA). To minimize reannealing of the unwound oligonucleotides, a 10-fold molar excess of unlabeled DNA trap, corresponding to the Cy5-labeled strand was added after 10 min of the reaction. As a negative control, the substrates were incubated in the reaction mix in the absence of the protein. Samples were resolved in a 15% polyacrylamide gel in 0.5 × TBE buffer for 40 min at 18°C with a 30-min pre-run. For data analysis, gels were developed in FluorChemQ MultiImageIII ChemiImager (Alpha Innotech, San Leandro, CA, United States) and the images were captured using Alpha View software (Alpha Innotech, San Leandro, CA, United States). The unwound DNA was quantified using ImageJ (Image Processing and Analyzing In Java).
Size Exclusion Chromatography With Multi-Angle Light Scattering Analysis
Size exclusion chromatography (SEC) coupled with multi-angle light-scattering (MALS) was done using a high-performance liquid chromatography (HPLC) equipment (1260 Infinity LC, Agilent Technologies Inc., Santa Clara, CA, United States) connected to a UV detector, a MALS detector (DAWN HELEOS II, Wyatt Technology Santa Barbara, CA, United States), and a differential refractometer (Optilab T-rEX, Wyatt Technology, Santa Barbara, CA, United States). The GP46F13 sample (100 μl) purified as described earlier was injected at 1.5 mg/ml final concentration onto a Superdex 200 Increase 10/300 column (GE Healthcare, Milwaukee, WI, United States) and run at room temperature and a 0.5 ml/min flow rate in buffer containing 50 mM Tris–HCl pH 8, 500 mM NaCl, 20 mM imidazole. Output data were analyzed using ASTRA v. 6 software (Wyatt Technology, Santa Barbara, CA, United States).
Pull-Down Assay
Preparation of Phage-Infected Bacterial Culture Extracts
The L. lactis IL1403 host strain culture (160 ml) was grown to OD600 0.4. After that a portion of the culture (40 ml) was taken, centrifuged (6,000 rpm, 4°C, 2 min) and frozen in liquid nitrogen (negative control). The remaining part was supplemented with 10 mM CaCl2 and infected with L. lactis phage F13 at MOI of 5. The infected cell culture was further incubated at 30°C and 40 ml portions were taken at times 5, 10, and 15 min after infection, centrifuged (6,000 rpm, 4°C, 2 min) and frozen in liquid nitrogen to stop the phage cycle and stored at −80°C until further use. At the time of analysis, the phage-infected culture samples were thawed. The pellet was resuspended in phosphate buffered saline (pH 7.4), bacterial cells were broken using glass beads (0.10–0.11 mm) and centrifuged (13,000 rpm, 4°C, 30 min). The supernatant samples containing phage-infected bacterial cell extracts were used in further analyses.
Immobilization of the GP46F13 Protein on the Ni2+ Resin
The GP46F13-His tagged protein from 250 ml culture was immobilized in 1 ml of HisPur™ Ni-NTA Superflow Agarose (Thermo Scientific™) applied to 5-ml PP columns (GE Healthcare) and rinsed with column buffer (50 mM Tris–HCl pH 8, 500 mM NaCl, 20 mM imidazole). Then, crude cell lysates of IL1403 host cells at 0, 5, 10, 15 min after infection with phage F13 were added and incubated overnight at 4°C. After excessive washing with 100 volumes of column buffer, the protein content bound to the resin was analyzed using mass spectrometry in the Mass Spectrometry Lab at IBB PAS.
Mass Spectrometry Analysis
The assay was performed with the use of the nanoHPLC nanoACQUITY system and the C18 pre-column and nanoAcquity BEH C18 column. Mobile phase consisted of water supplemented with 0.1% formic acid (FA) and acetonitrile with gradient elution. The column out-put was directly connected with the Q Exactive spectrophotometer. Fragmentation was done using the HCD method. Preprocessing and protein identification was performed respectively by employing the Mascot Distiller (version 2.6) and Server (version 2.4.1) and the L. lactis IL1403 (GenBank accession no.: AE005176.1) and phage F13 (GenBank accession no.: MG253653.1) protein sets. In order to distinguish non-specific binding to the resin, the obtained hit results were normalized against control affinity resin without tagged GP46F13.
Bioinformatic Analyses
Homologs of the GP46F13 protein were collected using PSI-BLAST (Altschul et al., 1997) search (three iterations, inclusion threshold 0.05) against non-redundant (nr) database with GP46F13 protein sequence (GenBank accession no. ATW69819.1) as a query. Sequence similarity to proteins with known structures and other Pfam families was scanned with HHpred at MPI Bioinformatics Toolkit (Zimmermann et al., 2018). All multiple sequence alignments were calculated using Mafft (L-INS-i) (Katoh et al., 2002). GP46F13 structure modeling was done using AlphaFold2 (Jumper et al., 2021). Multiple structure alignments between identified homologs of known structure and GP46F13 model were prepared manually.
Sequences of proteins belonging to other helicase families identified by HHpred were collected using PSI-BLAST searches starting with one representative sequence for each family. All the sequences, including GP46F13 homologs, were altogether clustered to 65% of sequence identity using CD-HIT (Li et al., 2001) to reduce redundancy. The dataset was further clustered using CLANS (Frickey and Lupas, 2004).
Results
In silico Analyses of GP46F13 Amino Acid Sequence and 3-D Fold
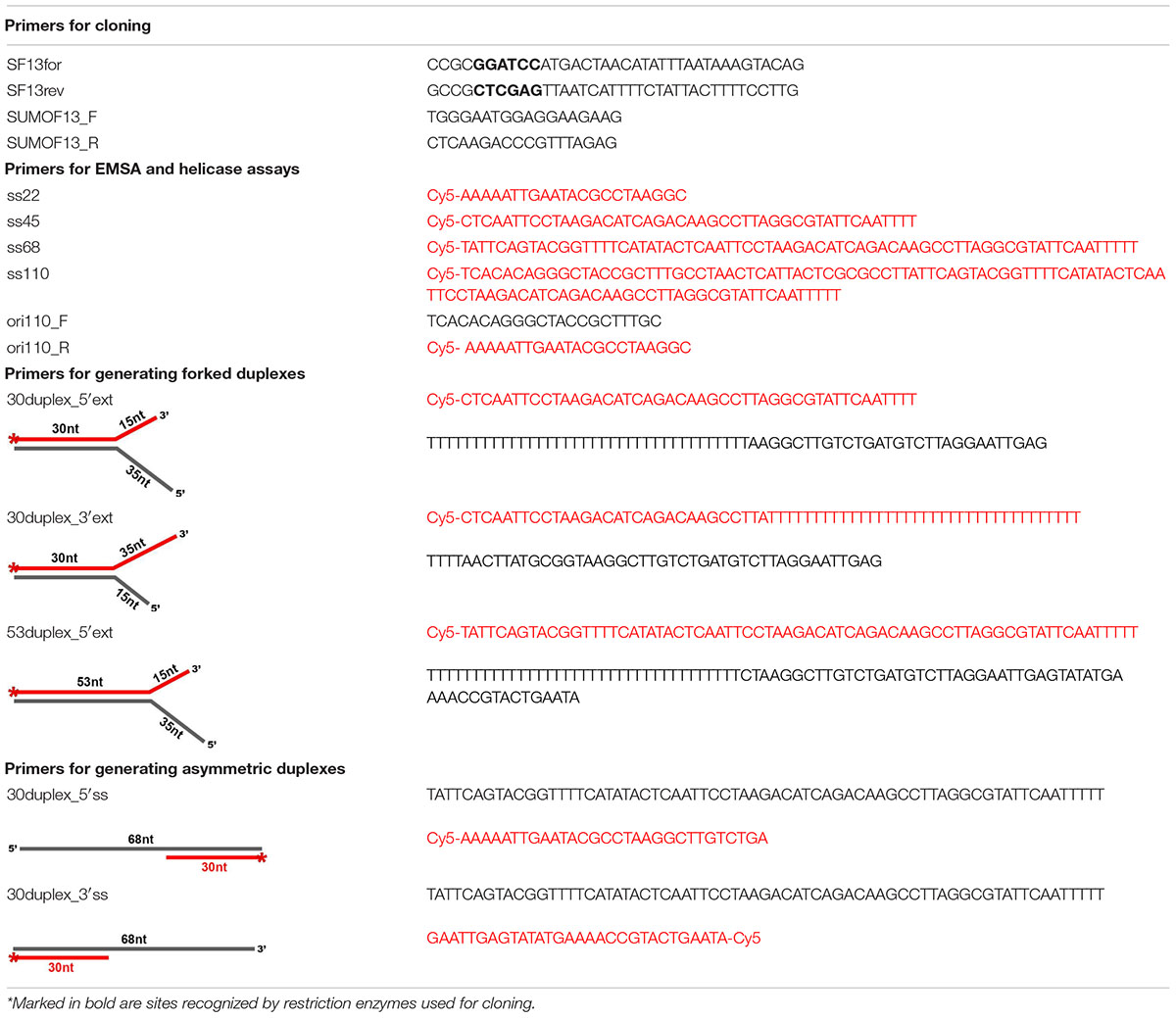
The GP46 protein of L. lactis phage F13 (GP46F13) according to HHpred mappings displays high sequence similarity (all hits with scores > 90 and estimated probability of being true positive > 99%) to several P-loop NTPase protein families, including RepA, RadA, RecA, GvpD, DnaB, RAD51, KaiC, and AAA_25 (Figure 1 and Supplementary Table 1). RepA and DnaB are replicative helicases (Niedenzu et al., 2001; Bailey et al., 2007), RadA is a DnaB-type helicase interacting with RecA in homologous recombination (Marie et al., 2017), RAD51 and RecA are recombinases forming helical filaments seeking for homology in homologous recombination (Chen et al., 2008; Short et al., 2016), GvpD regulates gas vesicle formation in Archaea, KaiC is an ATPase controlling cyanobacterial circadian clock (Abe et al., 2015) and AAA_25 proteins have no known function described yet. Due to the functional diversity of homologous protein families and taking into consideration sequence and structural similarities between them we performed a more detailed sequence clustering in order to provide a more reliable classification and annotation. Obtained results suggest that GP46F13 is closely related to the RepA family. It retains all sequence motifs characteristic for NTP hydrolysis, including residues Lys49 from Walker A (P-loop) motif, Glu82 (motif 1a), Asp136 (Walker B), and His177 (motif 3) (Figure 2A). It also has an arginine finger (Arg233) which might suggest a similar catalytic mechanism. Interestingly, the AAA_25 family identified among GP46F13 homologs also displays RecA-like features and will probably not function as an AAA ATPase (Figure 2A).

Figure 1. Sequence clustering of GP46F13 and the closest protein families. Each point represents one protein sequence. Protein sequences used as queries for collecting family members are marked with green dots and sequences of proteins of known structures with blue ones. Lines representing mappings with P-values lower and higher than 1E-15 are drawn in gray and teal, respectively.
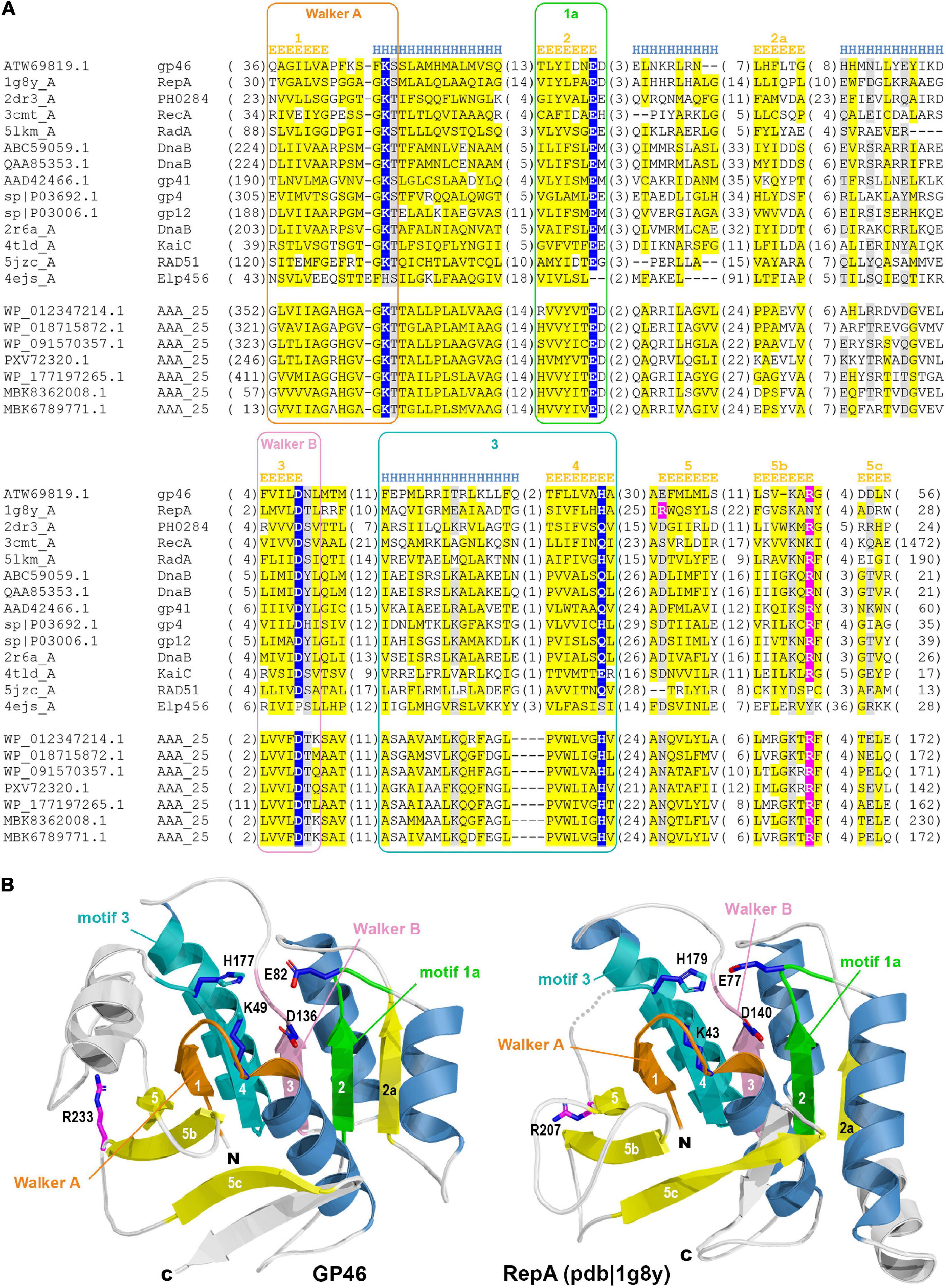
Figure 2. (A) Multiple sequence alignment of GP46F13 and the representatives of related families. Sequences are labeled with GenBank accession number, Uniprot ID, or PDB code. Sequence conservation is denoted with yellow and gray highlighting for non-polar and charged or polar residues, respectively. The numbers of residues omitted in the alignment are provided in parentheses. Residues involved in NTP hydrolysis are highlighted with blue, and the arginine from arginine finger–in pink. Sequence motifs are outlined with rectangles. Secondary structure elements are labeled according to James M Berger’s summary on helicase structures (Berger, 2008). (B) Comparison between 3D model of GP46F13 (left) and structure of the RepA (pdb| 1g8y). Non-core structural elements not present in the alignment (A) are rendered in white. Structural elements corresponding to defined motifs are colored in respective colors.
Molecular modeling of GP46F13 confirmed conservation of all structural elements essential for helicase function. The model computed by AlphaFold2 (pLDDT score 83.9; scores above 70 are regarded as confident) demonstrates no major differences compared to the RepA structure (Figure 2B) which suggest that GP46F13 may also function as hexameric helicase. Despite the similarity of GP46F13 to RepA, it retained the arginine finger in its canonical position at strand 5b, while in RepA it is located on the proximal strand 5 from where it executes its catalytic function.
Cloning and Purification the GP46F13 Protein
To obtain sufficient quantities of GP46F13 protein for in vitro studies the pET28a:SUMO-PIN expression system was used. For this, the phage F13 gp46 open reading frame encoding the 298-AA product was amplified by PCR and fused at its N-terminal end with 6xHis tag and SUMO protein-encoding sequences of the pET28a:SUMO-PIN vector in E. coli. The GP46F13 6xHis- and SUMO-tagged protein was purified by nickel-affinity chromatography, and after removal of the SUMO tag, further purification was done using the heparin-sepharose column (Supplementary Figure 1). SDS-PAGE analysis of the final purification protein revealed a major protein band with a molecular weight of approximately 34 kDa, which corresponds to the molecular weight calculated from the amino acid sequence (Supplementary Figure 1).
GP46F13 Binding to DNA Substrates
In order to elucidate the binding of GP46F13 to different DNA substrates in vitro EMSA assays were performed. For this purpose 1 nM of 5′ fluorescently labeled DNA substrates (Table 1) were incubated with varying GP46F13 protein concentrations.
As shown in Figure 3, GP46F13 interacts with ssDNA substrates of different length (22, 45, 68, and 110 nt) with nanomolar affinity and the relative amounts of unshifted ssDNA decrease proportionally to protein concentrations added. Clear retardation complexes could be seen for the 110-nt ssDNA (ss110). For shorter (ss22, ss45, ss68) substrates, a formation of discrete protein-DNA aggregates in the wells of the gel were observed. Although we see a fraction of the ssDNA substrate stuck in the well, most probably associated with the tendency of the protein to aggregate in vitro, our results indicate that GP46F13 can interact with DNA. At the same time, GP46F13 did not bind to dsDNA; no nucleoprotein complexes were observed with the 110-bp dsDNA (ds110) vs. ss110 substrate (Figure 3). To examine whether GP46F13 has affinity for substrates mimicking the replication forks, we performed EMSA assays using a branched 30-bp duplex with a short 15-nt 3′ tail and an extended, 35-nt 5′ ssDNA overhang (30duplex_5′ext). We show that the GP46F13 gives clear protein-DNA complexes with 30duplex_5’ext, suggesting that GP46F13 can bind also to heteroduplex structures.
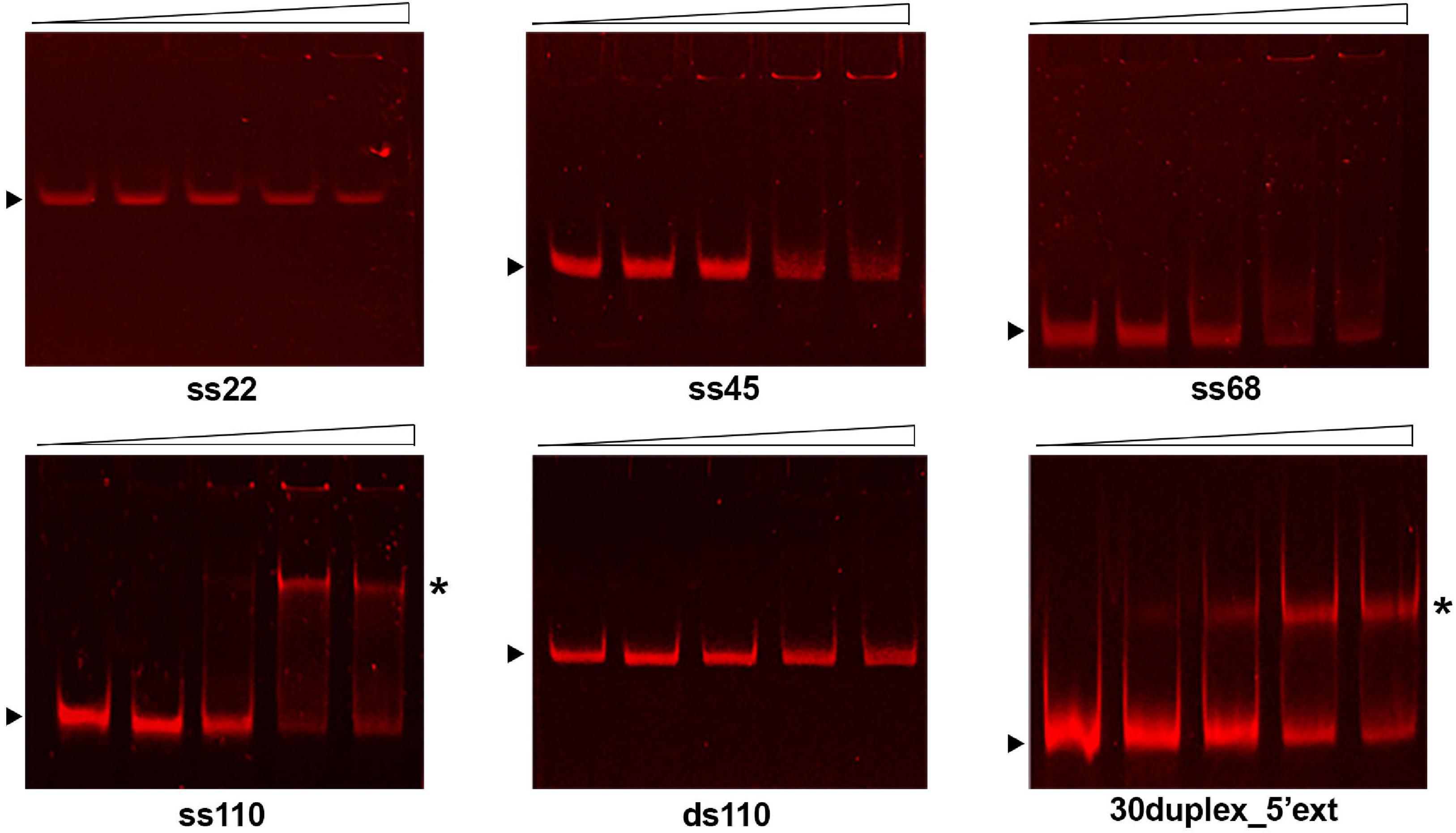
Figure 3. Binding of the GP46F13 protein to DNA substrates. Rising concentrations of GP46F13 (0.1, 0.25, 0.5, and 0.75 μM) were incubated with 1 nM of ssDNA (ss22, ss45, ss68, ss110), dsDNA (110ds) or forked substrate with elongated 5′ tail (30duplex_5′ext) for 30 min at 30°C in binding buffer (20 mM Tris–HCl pH 8, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.1 mg/ml of BSA, 2.5% glycerol) in 20 μl reaction volume. Reaction samples were analyzed on 7% native polyacrylamide gels. Nucleoprotein complexes are indicated by asterisk and unbound DNA substrates by black arrow.
DNA Unwinding by GP46F13
Results of our comparative analyses in silico suggested that GP46F13 is conserved in key amino acids and has similar fold structure to DNA helicases, including RepA from plasmid RSF1010 and phage T7 gp4 protein. To evaluate whether GP46F13 exhibits also a similar DNA unwinding ability, we performed helicase activity assays under various conditions. For these we used a series of asymmetric substrates with an ssDNA overhang from either 5′ or 3′ end or forked substrates with two free ssDNA ends of different length (Table 1). We determined that the GP46F13 protein was not active on blunt-ended dsDNA substrates (results not shown). Instead, we observed a preference for an asymmetric DNA substrate with 5′ tail (30duplex_5′ss), but not with a 3′ overhang (30duplex_3′ss), indicative of a 5′–3′ directionality of the protein’s unwinding activity (Figure 4).
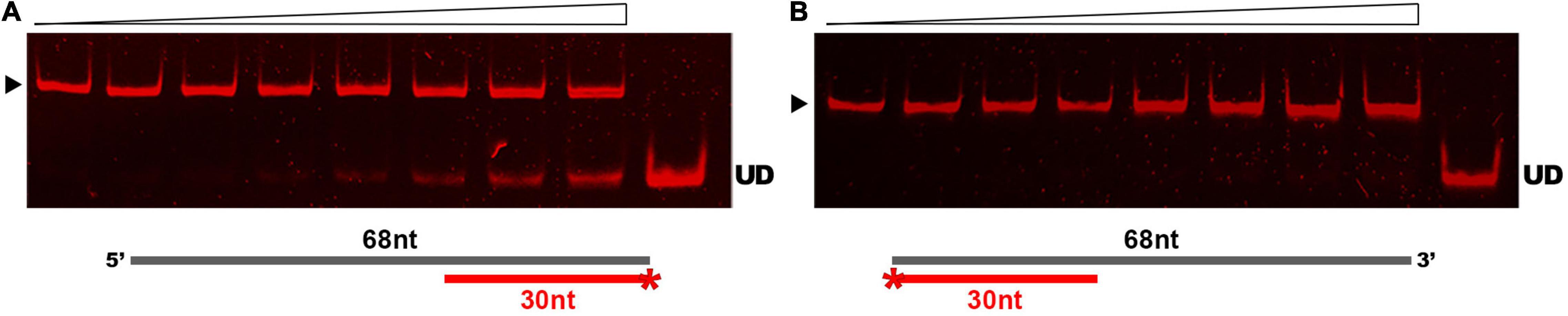
Figure 4. Substrate-specific unwinding by GP46F13. Reactions with asymmetric DNA substrate (A) 30duplex_5’ss or (B) 30duplex_3’ss. Rising concentrations of the GP46F13 protein (0, 0.1, 0.25, 0.5, 0.75, 1.0, 1.5, 2.0 μM) was incubated with DNA substrate (1.5 nM) for 30 min at 30°C in reaction buffer (50 mM Tris-HCl, pH 8, 50 mM NaCl, 5 mM MgCl2) and 5 mM ATP. Reaction samples were analyzed on native 15% polyacrylamide gels. ▶ indicates the expected migration of the substrate; UD indicates the expected migration of the unwound DNA (Cy5-labeled oligonucleotide) marked on the scheme in red and with an asterisk.
As DNA helicases can act at replication forks by breaking down the hydrogen bonds and separating the strands of the duplex DNA, we examined whether GP46F13 is capable of unwinding branched duplexes that mimic such structure. For this we used three types of substrates varying in the length of the dsDNA region and the free 3′ and 5′ ss ends (Table 1). Results showed clearly that extension (from 15 nt to 35 nt) of the 5′ end increases the rate of the separation of duplex DNA strands (Figures 5A–C). A 15-nt long 5′ ssDNA overhang was sufficient enough for GP46F13 unwinding of the forked substrate. We also showed that the protein is capable of displacing longer (53 bp) stretches of duplex DNA with 5′ tail (Figure 5C). Yet, liberation of the labeled oligonucleotide product occurred at slightly higher protein:DNA molar ratio for the 53duplex_5′ext vs. 30duplex_5′ext substrate (Figures 5A,C). To this end, the 30duplex_5′ext was recognized as the most preferable substrate and was used in further assays.
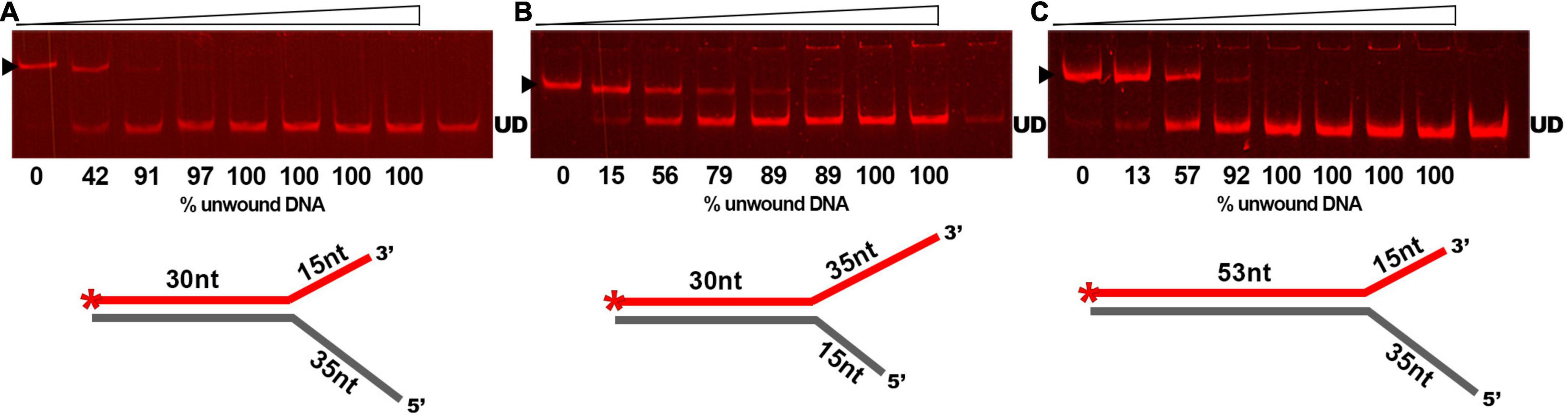
Figure 5. Substrate specific unwinding by GP46F13. Reactions with forked DNA substrates: (A) 30duplex_5′ext; (B) 30duplex_3′ext; (C) 53duplex_5′ext. Rising concentrations of the GP46F13 protein (0, 0.1, 0.25, 0.5, 0.75, 1,0, 1.5, 2.0 μM) were incubated with specific branched substrate (1.5 nM) for 30 min at 30°C in reaction buffer (50 mM Tris-HCl, pH 8, 50 mM NaCl, 5 mM MgCl2) and 5 mM ATP. Reaction samples were analyzed on native 15% polyacrylamide gels. ▶ indicates the expected migration of the substrate; UD indicates the expected migration of the unwound DNA (Cy5-labeled oligonucleotide) marked on the scheme in red and with an asterisk.
To determine the efficiency of the reaction of GP46F13 on the substrate DNA, we conducted a time-dependent DNA unwinding assay (Figure 6A). Results showed that the extent of displaced oligonucleotide increased in time and after 30 min it was almost completely liberated from the duplex DNA. Optimal temperature and pH conditions for DNA unwinding were determined to be at 30°C and pH 8 (Figure 6B). Under these conditions the strands of the duplex DNA were completely separated. As the unwinding activity of DNA helicase proteins is known to be fueled by various nucleotide substrates, we tested the specific requirement of GP46F13 for NTPs or dNTPs (Figure 6C). Results of the assay demonstrated efficient separation of duplex strands in a 30-min reaction in the presence of 5 mM ATP. Moderate displacement of the labeled oligonucleotide was detected for dATP and dTTP at the same concentrations, while no unwinding activity was observed for the remaining nucleotide substrates. We thus concluded that the unwinding activity of GP46F13 is ATP-dependent, while the binding to DNA can occur in the absence of ATP. Next, we examined the influence of rising ATP concentrations on the unwinding activity of GP46F13 (Figure 6D). Complete unwinding of the forked DNA substrate was observed at 5 mM ATP under the tested conditions, indicating that this is the optimal concentration of the supplied nucleotides. Lower and higher ATP concentrations allowed only for partial separation of duplex DNA strands. The activity of ATP-dependent helicases relies strongly on the presence of metal cofactors, particularly Mg2+ ions. The ratio of ATP to Mg2+ was documented previously to influence the efficiency of helicase activity. Therefore, we also investigated the effects of different concentrations of Mg2+ under constant ATP level on the unwinding activity of GP46F13. The most efficient separation of duplex DNA was determined when Mg2+ and ATP were supplied at an equimolar (5 mM) concentration (Figure 6E).
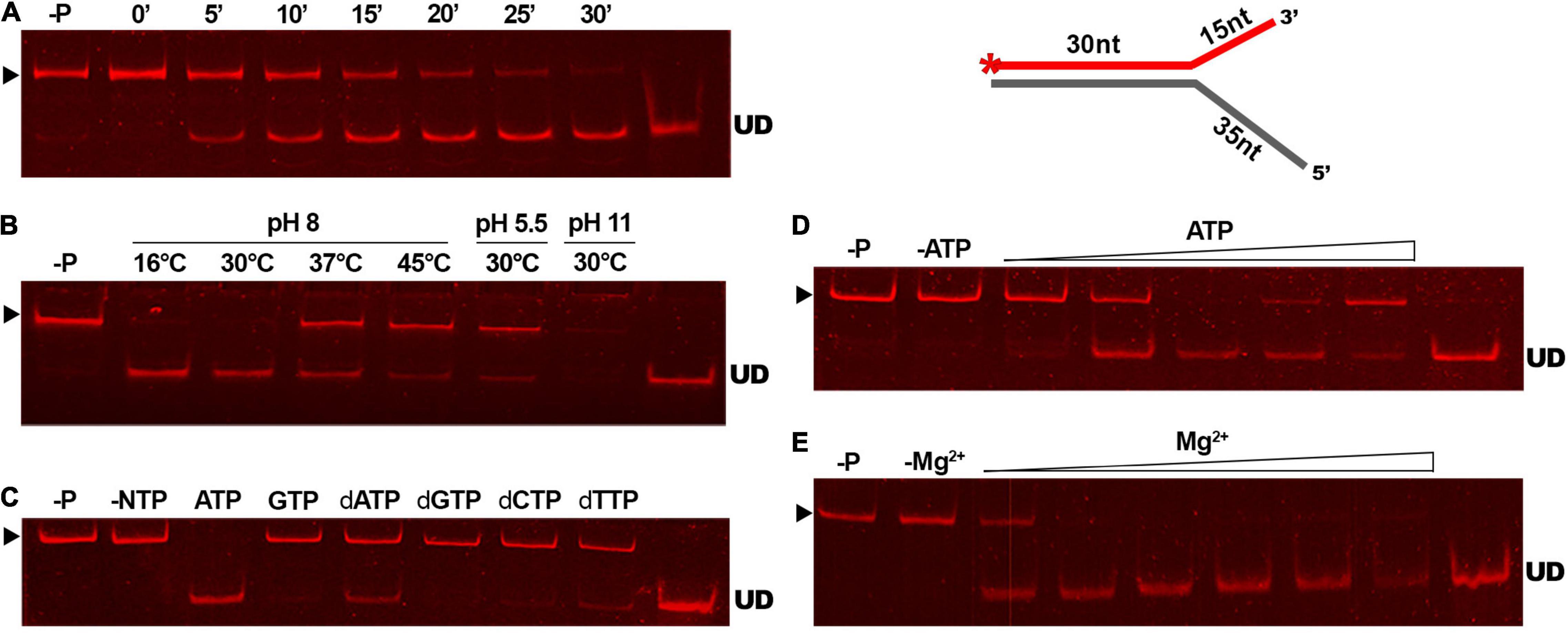
Figure 6. The unwinding activity of GP46F13 under different reaction conditions. The GP46F13 protein (0.5 μM) and Cy5-labeled branched substrate (1.5 nM) with extended 5′ tail (30duplex_5′ext) were incubated under varying conditions: time (0, 5, 10, 15, 20, 25, 30 min) (A); temperature (16°C, 30°C, 37°C, 45°C) and pH (5.5, 8.0, 11) (B); NTPs or dNTPs (5 mM) (C); ATP (0, 1, 2, 5, 10, 15 mM) (D); and MgCl2 (0, 1, 2, 5, 10, 15, 20 mM) (E). Reaction samples were analyzed on native 15% polyacrylamide gels. –P; control reaction without protein. –NTP/–ATP; control reaction without NTPs/ATP. –Mg2+; control reactions without Mg2+. ▶ indicates the expected migration of the substrate; UD indicates the expected migration of the unwound DNA (Cy5-labeled oligonucleotide) marked on the scheme in red and with an asterisk.
GP46F13 Multimerizes in Solution
DNA helicases generally function as multimeric forms (dimers or hexamers) (West, 1996). In order to infer the oligomerization status of GP46F13, we performed SEC-MALS analysis. For purification of GP46F13, we used in all the buffers high salt concentration (500 mM NaCl) to increase solubility and stability of the protein (see Section “Materials and Methods”). In turn, formation of oligomeric forms is generally promoted at low salt concentrations (50–200 mM). Thus, we initially performed several SEC runs at low salt conditions (50 and 200 mM). Unfortunately, in these cases the protein was eluted as a large multimeric form in the void volume of the SEC column (Supplementary Figure 2). To stabilize the protein, we increased the salt concentration to 500 mM and obtained a major single peak. By DLS assays we checked that addition of cofactors (DNA, ATP, and Mg2+) does not influence GP46F13 oligomerization (data not shown). In the light of these data, SEC-MALS analysis was performed at restored higher salt conditions without cofactors and revealed several GP46F13 populations–a monomer and two oligomeric forms (Figure 7). The main fraction had a mass value of 65 kDa, the second peak corresponded to 39 kDa fraction, while the smallest peak was dispersed with a mean mass of 410 kDa; yet, the signal detecting the largest peak was too weak for accurate size determination. The values detected were similar to the theoretical molecular weight (MW) of a dimer (2 × 34.25 = 68.5 kDa), a monomer (34.25 kDa), and possibly a 12-mer (12 × 34.25 = 411 kDa). Results of the assay indicate that GP46F13 can self-associate in solution. The presence of the monomeric fraction indicates that under the tested conditions the multimeric protein structure is not stable since high salt concentrations prompt possible dissociation. On the other hand, it is surprising to observe multimeric forms in a buffer containing 500 mM NaCl. This proves a strong GP46F13 tendency for oligomerization and suggests assembly of GP46F13 into higher multimeric forms.
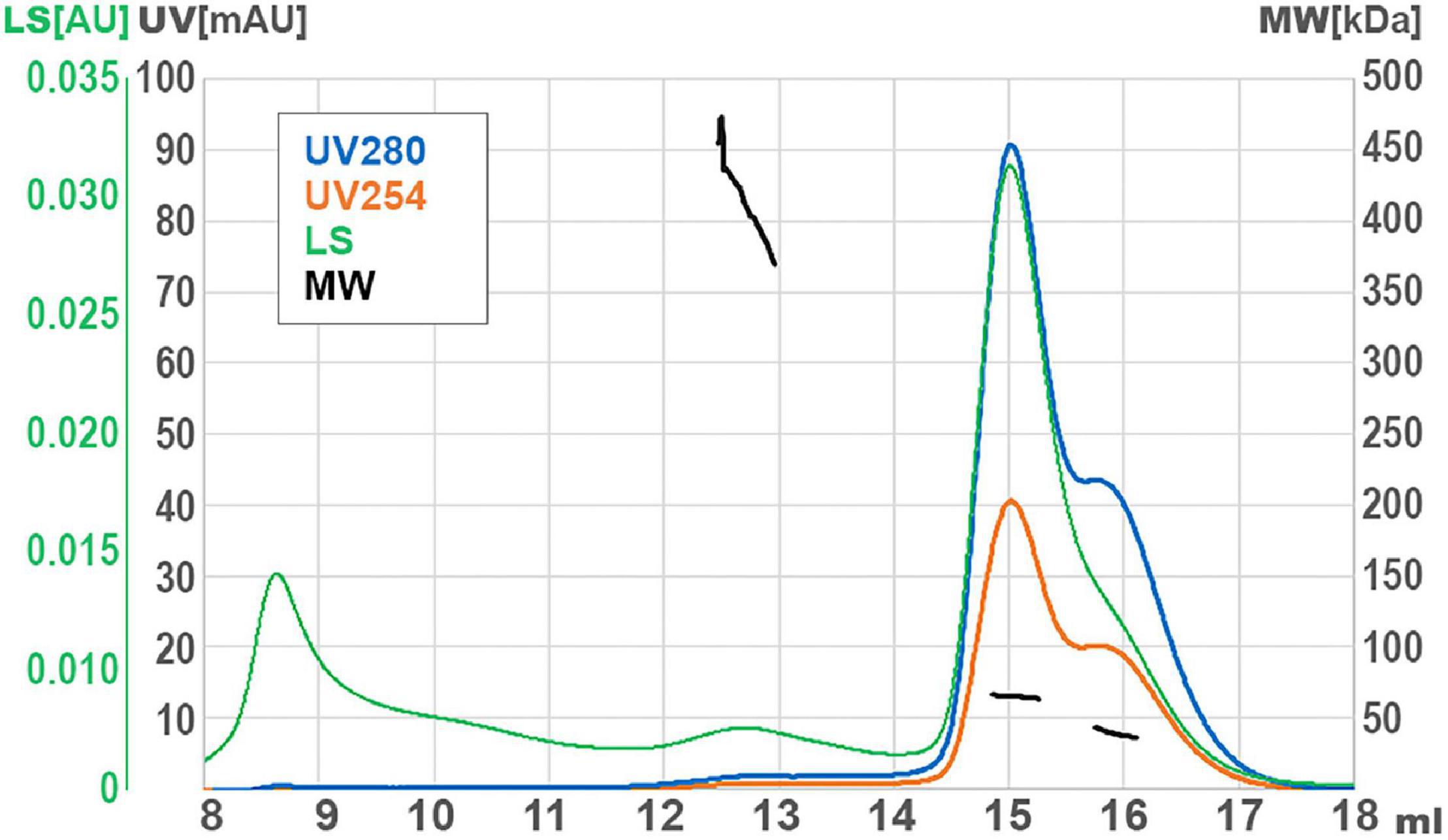
Figure 7. Size exclusion chromatography (SEC) with multi-angle light scattering (MALS) analysis for GP46F13. UV absorption and light scattering (LS) are shown on the left axis, molecular weight of protein (MW) are on the right axis. Short black lines mark the detected molecular mass.
Interaction of GP46F13 With Proteins From Phage F13 and the Lactococcus lactis IL1403 Host Strain
To probe for potential phage or host protein partners, we performed pull-down assays with tagged GP46F13 as bait, followed by LC-MS and computational analysis. We detected high distribution of peptides corresponding to GP46F13 and to three other non-structural phage proteins, i.e., GP23 (GenBank accession no.: ATW69796.1), GP28 (GenBank accession no.: ATW69801.1), and GP32 (GenBank accession no.: ATW69805.1), suggesting their interaction with the affinity-bound GP46F13 (Supplementary Table 2). By BlastP and HHPred searches we found that GP28 carries a Zn2+ binding domain, GP32 contains a conserved DUF3310 (domain of unknown function), and GP23 has no significant hits. Thus, the explicit function of all three detected proteins and their role in phage replication remains hypothetical. By LC-MS we also detected significant amounts of peptides representing host L. lactis IL1403 proteins—DnaK (GenBank accession no.: AAK05052.1), DnaJ (GenBank accession no.: AAK06322.1), topoisomerase I (GenBank accession no.: AAK05328.1), and SSB (GenBank accession no.: AAK06288.1). All of these proteins have previously been shown to be engaged in DNA replication. Affinity interactions of these proteins with the tagged GP46F13 in our pull down assay are a strong suggestion that they are partners in a common biological process. Their specific interaction with GP46F13 demands further experimental confirmation.
Discussion
It is generally considered that in virulent L. lactis sk1- and c2-like phages functions involved in phage DNA replication, recombination and translation are encoded in the early region. Yet, the majority of the genes therein remain uncharacterized. This hinders our understanding of the early stage of infection by L. lactis phages. We examined the activity of an early gene product, GP46, encoded by a virulent sk1-like L. lactis phage F13. By gene order, gp46 is located as the third open reading frame in the phage early-transcribed gene region. Its immediate genomic neighbors encode proteins of hypothetical function and the precise role of this region in phage development remains unknown. We demonstrate that GP46F13 belongs to the RepA cluster of P-loop NTPases, which are involved in managing diverse DNA processing pathways, such as DNA replication, repair, and recombination (Iyer et al., 2004; Miller and Enemark, 2016). It possesses all features characteristic for known hexameric helicases. For instance, it retains a complete catalytic site (Lys49, Glu82, Asp136, and His177) complemented with an arginine finger (Arg233), typical for oligomeric helicases.
GP46F13 is also highly similar to proteins annotated as hypothetical DNA polymerases encoded in homologous genome regions in other lytic phages. Yet, none of them seems to be functionally characterized. Several studies describe the identification of the origin of replication of sk1-like phage genomes (Chandry et al., 1997; Crutz-Le Coq et al., 2002), but no specific protein functions associated with phage DNA replication have been identified at that time. By employing a series of experimental approaches, including protein structure analysis, DNA binding and unwinding assays, and investigation of protein-protein interactions, we examined the activity of GP46F13. Our study is the first to functionally characterize a DNA helicase function encoded in the early gene region of a virulent L. lactis phage and propose its role in phage replication.
The closest structurally characterized homolog of GP46F13, RepA, from a broad-host range plasmid RFS1010, is an ATP-dependent hexameric 5′-3′ replicative helicase (Scherzinger et al., 1997). Our results demonstrate that in many aspects GP46F13 remains similar to RepARSF1010, including DNA binding, dissection of forked substrates in the 5′-3′ direction in the presence of ATP and the ability to form oligomers. Since substrate recognition by helicases engaged in DNA replication is rather structure- than sequence-specific (Anand and Khan, 2004; George et al., 2009), the effective assembly on branched duplex DNAs mimicking the replication forks implies the preference of the protein to unwind these types of substrates. RepARSF1010 is known to have the highest affinity to forked substrates but also to asymmetric DNA duplexes with free 5′ ends (Scherzinger et al., 1997). GP46F13 demonstrates a similar preference–unwinding of the DNA duplexes (both branched and asymmetric) was promoted given a free 5′ tail was provided. Interestingly, the GP46F13 showed more robust unwinding activity on the replication fork-mimicking substrate with extended 5′ end and a short 3′ flap than on DNA with only one free (5′) tail. These observations further support our hypothesis that GP46F13 plays a role in DNA processing pathway(s).
The DNA unwinding activity of helicases is propelled by the presence of nucleoside triphosphates (NTPs) and ion cofactors (Tuteja and Tuteja, 2006). Energy deriving from the hydrolysis of specific NTPs (or dNTPs) is the general driving force of helicase-based separation of DNA strands. Other studies showed that the activity of different DNA helicases is based on specific nucleotide requirements (Lohman and Bjornson, 1996). In our experiments the unwinding activity of GP46F13 was most potent in the presence of ATP and Mg2+ at equimolar ratio. For other NTPs/dNTPs and in the absence of Mg2+ lower rates or no strand displacement was observed. In this respect GP46F13 also resembles the RepARSF1010 which exhibited its highest activity in the presence of ATP and Mg2+ (Scherzinger et al., 1997).
Replicative DNA helicases act as ring oligomers that catalyze unwinding of dsDNA at replication forks (Patel and Picha, 2000; Trakselis, 2016). In general, various P-loop NTPases exhibiting helicase activity were determined to form oligomeric structures, including the bovine papillomavirus E1 and the Large-T-antigen of the simian virus 40 (Hickman and Dyda, 2005). Results of our SEC-MALS experiments showed that GP46F13 forms oligomers in solution, indicating that it most probably operates in such form, rather than as a monomer. Helicases work to unwind duplex DNA to generate ssDNA stretches that serve as intermediates in various DNA metabolic processes, including replication, recombination, or repair. In the cell, DNA helicases work as part of complex machinery systems which they form with other proteins (Patel and Picha, 2000). Moreover, the loading of helicases onto DNA is facilitated by the interaction with specific proteins at sequence-specific sites (Konieczny, 2003). For example, the RepA of plasmid RSF1010 is loaded at the oriV (origin of vegetative DNA replication) of plasmid RSF1010 initially melted by the plasmid-encoded RepC protein (Scherzinger et al., 1991). Replication of RSF1010 in vitro was shown to require chromosomally encoded proteins, including E. coli DNA gyrase, DnaZ protein (gamma subunit of PolIII holoenzyme), and SSB. For phage T7 gp4 primase-helicase, interactions were detected with phage-encoded gp2.5 SSB and gp5 DNA polymerase protein as well as with the host (E. coli) processivity factor (i.e., thioredoxin) (Zhang et al., 2011). The G40P helicase of B. subtilis phage SPP1 efficiently assembles on the DNA in the presence of phage G38P and G39P proteins (Ayora et al., 1999). To determine what are the protein partners of GP46F13 from both the phage and host side, we performed LC-MS analysis. Results of the assay revealed, among others, self-interaction of GP46F13 as well as interaction with host proteins, DnaK/DnaJ, SSB, and topoI. DnaK/DnaJ proteins set up a known chaperone protein system, both in Gram(−) and Gram(+) bacteria. They have previously been shown to be involved in replication of plasmids R1, P1, and F (Żylicz et al., 1989; Wickner et al., 1991; Giraldo-Suárez et al., 1993). In phage λ, DnaK/DnaJ were found to destabilize the oriλ:O:P:DnaB complex and release of the DnaB helicase from λP, which concomitantly led to the stimulation of its unwinding activity. Interaction of DnaB-like DNA helicases with SSB was also reported for several bacterial species. In E. coli, the native SSB protein (but also other heterologous SSBs) was recognized to stimulate the activity of the cognate DnaB helicase, albeit direct interaction of these two proteins is under discussion (Biswas et al., 2002). In turn, the SSB protein of phage T7, gp2.5, also physically interacts with its cognate helicase-primase (gp4) and by this modulates its DNA-unwinding activity (He and Richardson, 2004; Marintcheva et al., 2006). Finally, multiple evidence of the role of topoI in replication cannot preclude the possible contact with DNA helicases. Studies in E. coli have shown that the negative supercoiling relaxation by topoI at oriC (origin of replication) sites makes this region accessible for the replication machinery to initiate replication (Kraemer et al., 2019; Brochu et al., 2020). It has also been proposed that topoI may slow down DnaB helicase progression and halt replication by preventing excessive negative supercoiling at chromosomally encoded termination regions (Ters) (Valjavec-Gratian et al., 2005). Yet, confirmation of the direct interaction of all above-mentioned proteins with GP46F13 necessitates further studies.
Our study provides experimental evidence that GP46F13 is a 5′-3′ helicase. So far it is the only identified and functionally characterized protein with DNA unwinding activity encoded in the early gene region among sk1-like lytic L. lactis phages. The genomic localization and identified activity of GP46F13 implies its role in DNA replication and we suspect it defines the putative replication module. Weigel and Seitz (2006) have recognized a range of distinct replication modules of phage infecting various hosts. Yet, the study describes only the replication modules of L. lactis temperate P335-like phages. Results of this study brings us closer to deciphering the “dark matter” of the early gene expression region in sk1-like phages and other lytic lactococcal phages in general as well as the protein players engaged in the replication process of this group of phages.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
MC-J performed the experimental investigations, prepared the tables, and edited the manuscript. KS performed the bioinformatics analyses, prepared figures and tables, wrote and edited sections of the manuscript. KK performed the SEC-MALS analysis and edited parts of the manuscript. JB co-supervised the work. AS co-supervised the work, responsible for the experimental concept, critically reviewed and analyzed all data, and wrote the main text of the manuscript. All authors have read, revised, and approved the manuscript.
Funding
This research was funded by the Miniatura 3 grant no. 2019/03/X/NZ1/00467 and SONATA grant no. 2019/35/D/NZ2/03411 of the National Science Center and 79/E-35/SPUB/SP/2019 and POL-OPENSCREEN (DIR/Wk/2018/06) projects. The equipment used for mass spectrometry and FPLC analysis was sponsored in part by the Center for Preclinical Research and Technology (CePT), a project co-sponsored by European Regional Development Fund and Innovative Economy, The National Cohesion Strategy of Poland.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Michał Pastor from the Laboratory of Genome Engineering for his kind gift of the pET28a:SUMO-PIN plasmid and technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.840219/full#supplementary-material
Supplementary Figure 1 | GP46F13 protein overproduction and purification steps.
Supplementary Figure 2 | SEC analysis of GP46F13 protein fractions at different salt concentrations.
Supplementary Table 1 | Protein data set of GP46F13 homologs and related protein families.
Supplementary Table 2 | Mass spectrometry results presented as specific peptide hits against Lactococcus lactis IL1403 (GenBank accession no.: AE005176.1) and phage F13 (GenBank accession no.: MG253653.1) protein sets.
References
Abe, J., Hiyama, T. B., Mukaiyama, A., Son, S., Mori, T., Saito, S., et al. (2015). Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science 349, 312–316. doi: 10.1126/science.1261040
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. NAR 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anand, S. P., and Khan, S. A. (2004). Structure-specific DNA binding and bipolar helicase activities of PcrA. NAR 32, 3190–3197. doi: 10.1093/nar/gkh641
Ayora, S., Stasiak, A., and Alonso, J. C. (1999). The Bacillus subtilis bacteriophage SPP1 G39P delivers and activates the G40P DNA helicase upon interacting with the G38P-bound replication origin. J. Mol. Biol. 288, 71–85. doi: 10.1006/jmbi.1999.2662
Bailey, S., Eliason, W. K., and Steitz, T. A. (2007). Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318, 459–463. doi: 10.1126/science.1147353
Berger, J. M. (2008). SnapShot: nucleic acid helicases and translocases. Cell 134, 888–888.e1. doi: 10.1016/j.cell.2008.08.027
Bernstein, J. A., and Richardson, C. C. (1988). A 7-kDa region of the bacteriophage T7 gene 4 protein is required for primase but not for helicase activity. Proc. Natl. Acad. Sci. U.S.A. 85, 396–400. doi: 10.1073/pnas.85.2.396
Biswas, E. E., Chen, P. H., and Biswas, S. B. (2002). Modulation of enzymatic activities of Escherichia coli DnaB helicase by single-stranded DNA-binding proteins. NAR 30, 2809–2816. doi: 10.1093/nar/gkf384
Brochu, J., Breton, É. V., and Drolet, M. (2020). Supercoiling, R-loops, replication and the functions of bacterial type 1A topoisomerases. Genes 11:249. doi: 10.3390/genes11030249
Chandry, P. S., Moore, S. C., Boyce, J. D., Davidson, B. E., and Hillier, A. J. (1997). Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26, 49–64. doi: 10.1046/j.1365-2958.1997.5491926.x
Chen, Z., Yang, H., and Pavletich, N. P. (2008). Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–494. doi: 10.1038/nature06971
Chmielewska-Jeznach, M., Bardowski, J. K., and Szczepankowska, A. K. (2018). Molecular, physiological and phylogenetic traits of Lactococcus 936-type phages from distinct dairy environments. Sci. Rep. 8:12540. doi: 10.1038/s41598-018-30371-3
Chopin, A., Chopin, M.-C., Moillo-Batt, A., and Langella, P. (1984). Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11, 260–263. doi: 10.1016/0147-619x(84)90033-7
Crutz-Le Coq, A.-M., Cesselin, B., Commissaire, J., and Anba, J. (2002). Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148, 985–1001. doi: 10.1099/00221287-148-4-985
Curti, E., Smerdon, S. J., and Davis, E. O. (2007). Characterization of the helicase activity and substrate specificity of Mycobacterium tuberculosis UvrD. J. Bacteriol. 189, 1542–1555. doi: 10.1128/JB.01421-06
Davey, M. J., Jeruzalmi, D., Kuriyan, J., and O’Donnell, M. (2002). Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell. Biol. 3, 826–835. doi: 10.1038/nrm949
Deveau, H., Labrie, S. J., Chopin, M.-C., and Moineau, S. (2006). Biodiversity and classification of lactococcal phages. Appl. Environ. Microbiol. 72, 4338–4346. doi: 10.1128/AEM.02517-05
Dong, F., Gogol, E. P., and von Hippel, P. H. (1995). The phage T4-coded DNA replication helicase (gp41) forms a hexamer upon activation by nucleoside triphosphate. J. Biol. Chem. 270, 7462–7473. doi: 10.1074/jbc.270.13.7462
Egelman, E. H., Yu, X., Wild, R., Hingorani, M. M., and Patel, S. S. (1995). Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Nat. Acad. Sci. U.S.A. 92, 3869–3873. doi: 10.1073/pnas.92.9.3869
Frickey, T., and Lupas, A. N. (2004). CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704. doi: 10.1093/bioinformatics/bth444
Garneau, J. E., and Moineau, S. (2011). Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell. Fact. 10:S20. doi: 10.1186/1475-2859-10-S1-S20
George, T., Wen, Q., Griffiths, R., Ganesh, A., Meuth, M., and Sanders, C. M. (2009). Human Pif1 helicase unwinds synthetic DNA structures resembling stalled DNA replication forks. NAR 37, 6491–6502. doi: 10.1093/nar/gkp671
Gibson, T. J. (1984). Studies on the Eppstein-Barr Virus Genome. Ph.D thesis, England: Cambridge University.
Giraldo-Suárez, R., Fernández-Tresguerres, E., Díaz-Orejas, R., Malki, A., and Kohiyama, M. (1993). The heat-shock DnaK protein is required for plasmid R1 replication and it is dispensable for plasmid ColE1 replication. NAR 21, 5495–5499. doi: 10.1093/nar/21.23.5495
Guo, H., Li, M., Wu, H., Wang, W., Yu, F., and He, J. (2019). Crystal structures of phage NrS-1 N300-dNTPs-Mg2+ complex provide molecular mechanisms for substrate specificity. Biochem. Biophys. Res. Commun. 515, 551–557. doi: 10.1016/j.bbrc.2019.05.162
Halgasova, N., Mesarosova, I., and Bukovska, G. (2012). Identification of a bifunctional primase–polymerase domain of corynephage BFK20 replication protein gp43. Vir. Res. 163, 454–460. doi: 10.1016/j.virusres.2011.11.005
He, Z. G., and Richardson, C. C. (2004). Effect of single-stranded DNA-binding proteins on the helicase and primase activities of the bacteriophage T7 gene 4 protein. J. Biol. Chem. 279, 22190–22197. doi: 10.1074/jbc.M401100200
Hickman, A. B., and Dyda, F. (2005). Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 15, 77–85. doi: 10.1016/j.sbi.2004.12.001
Huttner, D., and Hickson, I. D. (2013). “Helicases,” in The Brenner’s Encyclopedia of Genetics, eds S. Maloy and K. Hughes (Cambridge, US: Academic Press), 406–408. doi: 10.1016/B978-0-12-374984-0.00687-2
Iyer, L. M., Leipe, D. D., Koonin, E. V., and Aravind, L. (2004). Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31. doi: 10.1016/j.jsb.2003.10.010
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. NAR 30, 3059–3066. doi: 10.1093/nar/gkf436
Konieczny, I. (2003). Strategies for helicase recruitment and loading in bacteria. EMBO Rep. 4, 37–41. doi: 10.1038/sj.embor.embor703
Kraemer, J. A., Sanderlin, A. G., and Laub, M. T. (2019). The stringent response inhibits DNA replication initiation in E. coli by modulating supercoiling of oriC. mBio 10, e1330–e1319. doi: 10.1128/mBio.01330-19
Kulczyk, A. W., and Richardson, C. C. (2016). “The replication system of bacteriophage T7,” in The Enzymes, Vol. 39, eds K. S. Kaguni and M. T. Oliveira (Cambridge, US: Academic Press), 89–136. doi: 10.1016/bs.enz.2016.02.001
Labrie, S. J., Josephsen, J., Neve, H., Vogensen, F. K., and Moineau, S. (2008). Morphology, genome sequence, and structural proteome of type phage P335 from Lactococcus lactis. Appl. Environ. Microbiol. 74, 4636–4644. doi: 10.1128/AEM.00118-08
Li, W., Jaroszewski, L., and Godzik, A. (2001). Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 17, 282–283. doi: 10.1093/bioinformatics/17.3.282
Lohman, T. M., and Bjornson, K. P. (1996). Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65, 169–214. doi: 10.1146/annurev.bi.65.070196.001125
Longo, L. M., Jabłońska, J., Vyas, P., Kanade, M., Kolodny, R., Ben-Tal, N., et al. (2020). On the emergence of P-Loop NTPase and Rossmann enzymes from a Beta-Alpha-Beta ancestral fragment. Elife 9:e64415. doi: 10.7554/eLife.64415
Marcelli, B., de Jong, A., Janzen, T., Serrano, M., Kok, J., and Kuipers, O. P. (2020). Complete genome sequences of 28 lactococcal bacteriophages isolated from failed dairy fermentation processes. Microbiol. Resour. Announc. 9, e01535–19. doi: 10.1128/MRA.01535-19
Marie, L., Rapisarda, C., Morales, V., Bergé, M., Perry, T., Soulet, A. L., et al. (2017). Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension. Nat. Commun. 8:15638. doi: 10.1038/ncomms15638
Marintcheva, B., Hamdan, S. M., Lee, S. J., and Richardson, C. C. (2006). Essential residues in the C terminus of the bacteriophage T7 gene 2.5 single-stranded DNA-binding protein. J. Biol. Chem. 281, 25831–25840. doi: 10.1074/jbc.M604601200
McGrath, S., Seegers, J. F., Fitzgerald, G. F., and van Sinderen, D. (1999). Molecular characterization of a phage-encoded resistance system in Lactococcus lactis. Appl. Environ. Microbiol. 65, 1891–1899. doi: 10.1128/AEM.65.5.1891-1899.1999
Meijer, W. J., Horcajadas, J. A., and Salas, M. (2001). Phi29 family of phages. Microbiol. Mol. Biol. Rev. 65, 261–287. doi: 10.1128/MMBR.65.2.261-287.2001
Mensa-Wilmot, K., Seaby, R., Alfano, C., Wold, M. C., Gomes, B., and McMacken, R. (1989). Reconstitution of a nine-protein system that initiates bacteriophage lambda DNA replication. J. Biol. Chem. 264, 2853–2861. doi: 10.1016/s0021-9258(19)81691-5
Miller, J. M., and Enemark, E. J. (2016). Fundamental characteristics of AAA+ protein family structure and function. Archaea 14:9294307. doi: 10.1155/2016/9294307
Muhammed, M. K., Kot, W., Neve, H., Mahony, J., Castro-Mejía, J. L., Krych, L., et al. (2017). Metagenomic analysis of dairy bacteriophages: extraction method and pilot study on whey samples derived from using undefined and defined mesophilic starter cultures. Appl. Environ. Microbiol. 83, e00888–17. doi: 10.1128/AEM.00888-17
Niedenzu, T., Röleke, D., Bains, G., Scherzinger, E., and Saenger, W. (2001). Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol. 306, 479–487. doi: 10.1006/jmbi.2000.4398
Oliveira, J., Mahony, J., Hanemaaijer, L., Kouwen, T. R. H. M., and van Sinderen, D. (2018). Biodiversity of bacteriophages infecting Lactococcus lactis starter cultures. J. Dairy Sci. 101, 96–105. doi: 10.3168/jds.2017-13403
Ostergaard, S., Brøndsted, L., and Vogensen, F. K. (2001). Identification of a replication protein and repeats essential for DNA replication of the temperate lactococcal bacteriophage TP901-1. Appl. Environ. Microbiol. 67, 774–781. doi: 10.1128/AEM.67.2.774-781.2001
Patel, S. S., and Hingorani, M. M. (1993). Oligomeric structure of bacteriophage T7 DNA primase/helicase proteins. J. Biol. Chem. 268, 10668–11075. doi: 10.1016/s0021-9258(18)82249-9
Patel, S. S., and Picha, K. M. (2000). Structure and function of hexameric helicases. Annu. Rev. Biochem. 69, 651–697. doi: 10.1146/annurev.biochem.69.1.651
Richardson, C. C. (1983). Bacteriophage T7: minimal requirements for the replication of a duplex DNA molecule. Cell 33, 315–317. doi: 10.1016/0092-8674(83)90411-7
Sawaya, M. R., Guo, S., Tabor, S., Richardson, C. C., and Ellenberger, T. (1999). Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell 99, 167–177. doi: 10.1016/s0092-8674(00)81648-7
Scherzinger, E., Haring, V., Lurz, R., and Otto, S. (1991). Plasmid RSF1010 DNA replication in vitro promoted by purified RSF1010 RepA, RepB and RepC proteins. NAR 19, 1203–1211. doi: 10.1093/nar/19.6.1203
Scherzinger, E., Ziegelin, G., Bárcena, M., Carazo, J. M., Lurz, R., and Lanka, E. (1997). The RepA protein of plasmid RSF1010 is a replicative DNA helicase. J. Biol. Chem. 272, 30228–30236. doi: 10.1074/jbc.272.48.30228
Seco, E. M., Zinder, J. C., Manhart, C. M., Lo Piano, A., McHenry, C. S., and Ayora, S. (2013). Bacteriophage SPP1 DNA replication strategies promote viral and disable host replication in vitro. NAR 41, 1711–1721. doi: 10.1093/nar/gks1290
Short, J. M., Liu, Y., Chen, S., Soni, N., Madhusudhan, M. S., Shivji, M. K., et al. (2016). High-resolution structure of the presynaptic RAD51 filament on single-stranded DNA by electron cryo-microscopy. NAR 44, 9017–9030. doi: 10.1093/nar/gkw783
Singleton, M. R., Dillingham, M. S., and Wigley, D. B. (2007). Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50. doi: 10.1146/annurev.biochem.76.052305.115300
Trakselis, M. A. (2016). Structural mechanisms of hexameric helicase loading, assembly, and unwinding. F1000Res 5, F1000FacultyRev–111. doi: 10.12688/f1000research.7509.1
Tuteja, N., and Tuteja, R. (2006). Helicases as molecular motors: An insight. Physica A. 372, 70–83. doi: 10.1016/j.physa.2006.05.014
Valjavec-Gratian, M., Henderson, T. A., and Hill, T. M. (2005). Tus-mediated arrest of DNA replication in Escherichia coli is modulated by DNA supercoiling. Mol. Microbiol. 58, 758–773. doi: 10.1111/j.1365-2958.2005.04860.x
Weigel, C., and Seitz, H. (2006). Bacteriophage replication modules. FEMS Microbiol. Rev. 30, 321–381. doi: 10.1111/j.1574-6976.2006.00015.x
West, S. (1996). DNA helicases: new breeds of translocating motors and molecular pumps. Cell 86, 177–180. doi: 10.1016/s0092-8674(00)80088-4
Wickner, S., Hoskins, J., and McKenney, K. (1991). Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature 350, 165–167. doi: 10.1038/350165a0
Ye, J., Osborne, A. R., Groll, M., and Rapoport, T. A. (2004). RecA-like motor ATPases – lessons from structures. Biochim. Biophys. Acta Bioenergetics. 1659, 1–18. doi: 10.1016/j.bbabio.2004.06.003
Zhang, H., Lee, S. J., Zhu, B., Tran, N. Q., Tabor, S., and Richardson, C. C. (2011). Helicase-DNA polymerase interaction is critical to initiate leading-strand DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 108, 9372–9377. doi: 10.1073/pnas.1106678108
Zhao, Z., De-Donatis, G. M., Schwartz, C., Fang, H., Li, J., and Guo, P. (2016). An arginine finger regulates the sequential action of asymmetrical hexameric ATPase in the double-stranded dna translocation motor. Mol. Cell. Biol. 36, 2514–2523. doi: 10.1128/MCB.00142-16
Ziegelin, G., Scherzinger, E., Lurz, R., and Lanka, E. (1993). Phage P4 alpha protein is multifunctional with origin recognition, helicase and primase activities. EMBO J. 12, 3703–3708. doi: 10.1002/j.1460-2075.1993.tb06045.x
Zimmermann, L., Stephens, A., Nam, S. Z., Rau, D., Kübler, J., Lozajic, M., et al. (2018). A completely reimplemented mpi bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243. doi: 10.1016/j.jmb.2017.12.007
Zúñiga, M., Franke-Fayard, B., Venema, G., Kok, J., and Nauta, A. (2002). Characterization of the putative replisome organizer of the lactococcal bacteriophage r1t. J. Virol. 76, 10234–10244. doi: 10.1128/jvi.76.20.10234-10244.2002
Keywords: Lactococcus lactis, sk1-like bacteriophage, P-loop NTPase, DNA unwinding, DNA replication
Citation: Chmielewska-Jeznach M, Steczkiewicz K, Kobyłecki K, Bardowski JK and Szczepankowska AK (2022) An Adenosine Triphosphate- Dependent 5′-3′ DNA Helicase From sk1-Like Lactococcus lactis F13 Phage. Front. Microbiol. 13:840219. doi: 10.3389/fmicb.2022.840219
Received: 20 December 2021; Accepted: 31 January 2022;
Published: 15 March 2022.
Edited by:
Heejoon Myung, Hankuk University of Foreign Studies, South KoreaReviewed by:
Yuliang Wu, University of Saskatchewan, CanadaSergey Korolev, Saint Louis University, United States
Copyright © 2022 Chmielewska-Jeznach, Steczkiewicz, Kobyłecki, Bardowski and Szczepankowska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agnieszka K. Szczepankowska, agaszczep@ibb.waw.pl
†Present address: Magdalena Chmielewska-Jeznach, Department of Medical Biology, Faculty of Health Sciences, Medical University of Warsaw, Warsaw, Poland
 Magdalena Chmielewska-Jeznach
Magdalena Chmielewska-Jeznach Kamil Steczkiewicz
Kamil Steczkiewicz Kamil Kobyłecki
Kamil Kobyłecki Agnieszka K. Szczepankowska
Agnieszka K. Szczepankowska