- 1Laboratory of Antimicrobial Resistance, Institute of Environmental and Agricultural Biology (X-BIO), Tyumen State University, Tyumen, Russia
- 2A.V. Zhirmunsky National Scientific Center of Marine Biology, Vladivostok, Russia
- 3Gause Institute of New Antibiotics, Moscow, Russia
- 4Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry Russian Academy of Sciences, Moscow, Russia
Gausemycin A is the first member of the novel lipoglycopeptides family produced by Streptomyces roseoflavus INA-Ac-5812. Gausemycin A has a pronounced bactericidal activity against methicillin-resistant Staphylococcus aureus. However, the ability of S. aureus to be resistant to gausemycin A has not been investigated yet. Using serial passaging, we have obtained the resistant variant S. aureus 5812R, which is 80 times more resistant compared to the parent strain. Susceptibility testing of S. aureus 5812R revealed the acquisition of cross-resistance to daptomycin, cefazolin, tetracycline, and gentamicin, while the resistance to vancomycin, nisin, and ramoplanin was absent. Whole genome sequencing revealed single nucleotide polymorphism (SNP) and deletions in S. aureus 5812R, among which are genes encoding efflux pump (sepA), the two-component Kdp system (kdpE), and the component of isoprenoid biosynthesis pathway (hepT). Phenotypically, S. aureus 5812R resembles a small-colony variant, as it is slow-growing, forms small colonies, and is deficient in pigments. Profiling of fatty acids (FA) composition constituting the cytoplasmic membrane of S. aureus 5812R revealed the prevalence of anteiso-branched FA, while straight FA was slightly less present. The evidence also showed that the gausemycin A-resistant strain has increased expression of the cls2 gene of the cardiolipin synthase. The performed checkerboard assay pointed out that the combination of gausemycin A and ciprofloxacin showed a synergistic effect against S. aureus 5812R.
Introduction
Antimicrobial resistance (AMR) poses a serious global threat to human, animal, and environmental health. The emergence and rapid spread of pathogenic microorganisms with multidrug resistance (MDR) to antibiotics, observed in recent decades, is especially dangerous (Bhullar et al., 2012; Perry et al., 2016; Aslam et al., 2018). The list of the main Gram-positive pathogens includes methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant S. aureus (VRSA), since they play a leading role in the etiology of a wide range of community-acquired and hospital-acquired human infections (DeLeo and Chambers, 2009; Watkins et al., 2012; Rapacka-Zdonczyk et al., 2019). Most of the new antibiotics discovered over the past 30 years have shown activity mainly against Gram-positive bacteria (Lewis, 2020), in this regard, antibiotic resistance among Gram-positive bacteria is an alarming phenomenon.
Antimicrobial peptides (AMPs) of various origins are considered a powerful weapon against a resistant form of pathogens (Sharma et al., 2014; Singh et al., 2014; Lum et al., 2015; Raman et al., 2015; Lyu et al., 2016; Gwynne and Gallagher, 2018). Non-ribosomal peptide antibiotics (NRPs) attract considerable interest from the standpoint of their therapeutic potential (Li, 2018). Glycopeptide vancomycin is a conventional antibiotic with a long clinical history originating from the 1950s, and the spread of vancomycin-resistant bacterial strains occurred in 1980s (Wilhelm and Estes, 1999). Lipopeptide daptomycin was discovered in 1985 and approved by the US Food and Drug Administration (FDA) in 2003 (Mishra et al., 2012). Although daptomycin has become available in medical practice relatively recently, daptomycin-resistant isolates have already widely appeared (Bayer et al., 2013). Ramoplanin is a glycolipodepsipeptide, which contains a carbohydrate molecule and a fatty acid, both attached to a peptide core. Ramoplanin was discovered in the 1980s (Cavalleri et al., 1984), and the last available clinical trial status is phase III for VRE treatment (de la Cruz et al., 2017). Ramoplanin-resistant S. aureus strain was obtained in vitro in 2010 (Schmidt et al., 2010).
Recently, new lipoglycopeptides have been revealed among metabolites of actinomycete Streptomyces roseoflavus INA-Ac-5812. These lipoglycopeptides were firstly described in 2016 (Lapchinskaya et al., 2016), and consequently named gausemycins A and B (Tyurin et al., 2021). Gausemycin A is a macrocyclic peptide containing 14 amino acids, where tyrosine is glycosylated with arabinose, which makes gausemycin A unique among natural peptide antibiotics (Tyurin et al., 2021). The closest structurally related compounds are depsipeptides (daptomycin, taromycins, and cadazides) and cyclopeptides (amphomycin, rumycins, and malacidins) (Tyurin et al., 2021). Gausemycin A demonstrates rapid membrane disturbing activity against planktonic S. aureus. Gausemycin A was able to inhibit biofilm-embedded staphylococcal cells, while daptomycin was inactive (Vasilchenko et al., 2020).
Thus, the above-mentioned properties of gausemycin A make it an alternative to existing antibiotics for the treatment of infectious diseases caused by S. aureus. However, before a new substance will move to preclinical investigations, it is necessary to evaluate its ability to generate antibiotic-resistant bacterial variants (Parachin and Franco, 2014).
In this regard, this work aims to evaluate the ability of model strain S. aureus FDA209P to develop resistance to new lipoglycopeptide gausemycin A, and recognize the genomic and phenotypic changes occurring in the S. aureus population under the treatment.
Materials and methods
Bacterial culture and chemicals
Staphylococcus aureus FDA209P (gausemycin A-susceptible) was used in this study. Daptomycin, vancomycin, ramoplanin, nisin, cefazolin, tetracycline, gentamicin, ampicillin, and ciprofloxacin were purchased from Sigma-Aldrich (US).
Gausemycin A was obtained as described (Vasilchenko et al., 2020). Briefly, the culture medium after S. roseoflavus INA-Ac-5812 fermentation was applied to Amberlite XAD2 (Serva, UK) low-pressure column, and an unbound liquid was collected and further extracted by n-butanol in a ratio of 1:1 (v/v). The obtained butanolic extract was fully evaporated and resolved in 70% water-ethanol. Gausemycin A isolation was made using semi-preparative high-performance liquid chromatography on XBridge BEH C18 column (10 x 250 mm) (Waters, Ireland). The target compound was detected at a wavelength of 364 nm, which is optimum for 4-L-chloro-kenyrenine amino acid residue. The purity of gausemycin A was confirmed by re-chromatography with an analytical reversed-phase HPLC column (XBridge BEH C18 column, 4.6 x 250 mm, Waters, Ireland) and absorbance was monitored at 214 nm.
Determination of minimum inhibitory concentrations
Minimal inhibitory concentration (MIC) was determined as previously described in Fuchs et al. (2000), following the Clinical and Laboratory Standards Institute (CLSI) guidelines for broth microdilution MIC assay. The bacteria were incubated in a 96-well microtiter plate (Eppendorf, Germany) containing 90 μL of inoculum prepared in growth media at 106 CFU/ml with 10 μL of twofold dilutions of the antibiotics. The results were evaluated after 24 h of cultivation at 370C. The bacterial growth was assessed by scanning the absorbance data at 600 nm obtained by spectrophotometer (Multiscan GO, Thermo Fisher Scientific, US).
Serial passage experiments
Gausemycin A-resistant variant was obtained by serial passaging as described previously (Pollard et al., 2012). Isolated colonies of S. aureus FDA209P were inoculated into Mueller-Hinton II broth (CaCl2 50 mg/L) containing twofold dilutions of gausemycin A. The cultures were incubated at 37 °C with aeration for 24 h. After incubation, bacterial cells growing at the highest concentration of gausemycin A (below the MIC) were harvested and used for the next passage. The process was repeated for 20 passages. The dynamic of the developing resistance to gausemycin A was evaluated in comparison with daptomycin and ramoplanin. The experiments were performed in two independent series on separate days with three technical replicates each.
Resistance stability testing
The overnight culture of S. aureus 5812R was sub-cultured in gausemycin A-free Muller-Hinton broth and incubated overnight at 37°C with shaking. The obtained culture was evaluated on susceptibility to gausemycin A in the MIC assay as described previously. Passaging in the drug-free medium was repeated 17 times. The experiments were performed in two independent series on separate days with three technical replicates each.
Assessment of phenotypical changes in S. aureus 5812R-resistant strain
Quantification of staphylococcal pigment
S. aureus strains were grown for 24 h at 37°C on agarose Mueller Hinton broth with the addition of sheep's blood (Tubby et al., 2013). Cells were washed from plates and harvested by centrifugation (4,500 g, 15 min); then washed twice with phosphate-buffered saline (PBS) (Sigma-Aldrich, US).
Staphyloxanthin and carotenoids were extracted and quantified as previously described with slight modification (Zhang et al., 2018). Cultures of S. aureus were equilibrated for optical density (OD600nm 0.1) and then subjected to 50% ethanol. Washed cells were re-suspended in 2 mL 50% ethanol, heated in ultrasonic bath at 55°C for at least 5 min while being stirred gently, until all visible pigment of bacteria was extracted and dissolved into ethanol. After that, the ethanol extract was cooled and centrifuged. Pigment content was then quantified by the absorbance profile of carotenoids as determined spectrophotometrically at an optical density of 450 nm (OD450). The experiments were performed in three independent series on separate days with three technical replicates each. The means plus standard deviations of the means of results from three independent series with three technical replicates are shown.
Atomic force microscopy
S. aureus strains were grown for 24 h at 37°C on agarose Mueller Hinton broth with the addition of sheep's blood (Tubby et al., 2013). Cells were washed from plates and harvested by centrifugation (4,500 g, 15 min); then washed twice with double distilled water.
Atomic force microscopy (AFM) was performed using Integra NT-MDT (NT-MDT, Russia) in the tapping mode. The microscopy investigation was carried out using the NSG01 cantilever (Tipsnano, Estonia) with a spring constant of ~5.1 N/m. Preliminarily, wide scanning of about 70 × 70 μm was conducted to produce a reference map of the sample surface in order to help localize cells. The scan size was then set with a sampling of 512 by 512 points and a scan rate of 0.5 Hz.
Growth rates and biochemical properties of studied strains
S. aureus 5812R and S. aureus FDA209P strains were incubated in a 96-well microtiter plate (Eppendorf, Germany) containing 10 μL of inoculum (107 CFU/ml) and 90 μl of drug-free Muller-Hinton media. The dynamics of bacterial growth were assessed by scanning and plotting the absorbance data at 600 nm obtained by spectrophotometer (Multiscan GO, Thermo Scientific, US). The experiments were performed using two independent series with three technical replicates each.
The biochemical properties were assayed using the ID32 Staph strip (BioMerieux, France), which consists of a set of wells containing dried biochemical media for colorimetric tests.
Zeta potential measurement
The electrophoretic mobility (EPM) of bacterial cells was measured with a zeta potential analyzer (Litesizer 500, Anton Paar, Austria) before being converted to zeta potentials using the Helmholtz–Smoluchowski theory as described in Shireen et al. (2013). Prior to sample analysis, the electrodes were polarized using the test solution in the absence of cells under identical conditions to establish consistent sample conductance readings. Between each measurement, electrodes were rinsed with copious amounts of ethanol and double distilled water, followed by the test bacterial suspension. The assay was performed twice on separate days.
Permeabilization of the plasma membrane and microscopy
Membrane integrity was assessed using the fluorescent probes SYTO 9 and propidium iodide (PI) (LIVE/DEAD BacLight Bacterial Viability Kit, Molecular Probes, USA). The test strains were grown to the mid-log phase. The obtained cultures were centrifuged at 10,000 rpm for 10 min, the supernatant was discarded and the pellet was dispersed in 10 mM HEPES buffer supplemented with 1.25 mM Ca 2+ to an optical density of 0.2 (OD 600) that corresponded to 108 CFU/ml. Bacterial cells in 50 μl were transferred to a 96-well microtiter plate (Eppendorf, US). SYTO 9 fluorescence kinetics was measured using Fluoroscan Ascent FL plate reader (Thermo Scientific, US) at 485 nm excitation and 535 nm of emission wavelengths as described earlier (Vasilchenko et al., 2020). The assay was performed twice in three technical replicates.
Microscopy was performed using a Zeiss Axio Imager A2 fluorescent microscope (Carl Zeiss, Germany) equipped with filter sets useful for simultaneous viewing of SYTO 9 and PI stains.
Membrane lipid extraction, fractioning, and methylation
Fatty acid (FA) composition was determined using biomass of S. aureus FDA209P and S. aureus 5812R. Bacterial cultures were grown to the stationary phase in Muller-Hinton medium with CaCl2 at 37°C with shaking. Bacterial cells were harvested by centrifugation, washed twice with PBS, and lyophilized.
Lipid extraction was performed according to the Folch method (Folch and Lees, 1957). Chloroform: methanol (2:1, v/v) was added to each sample according to the original Folch procedure allowing for a CHCl3:MeOH:H2O ratio of 8:4:3 (v/v/v). Briefly, ice-cold methanol and chloroform were added to the sample. The suspension was vortexed occasionally to bring about physical mixing and the sample was incubated on ice for 30 min. After the addition of water, which was used to separate the aqueous and organic layers, the suspension was incubated on ice for additional 10 min. Samples were centrifuged at 2,000 rpm for 5 min at 4°C. The lower phase (organic) layer was transferred to a new tube. The aqueous layer was re-extracted with 1 mL of 2:1 (v/v) chloroform/methanol. The chloroform layers were combined for analysis. The aqueous layer was centrifuged at 2,000 rpm for 5 min at 4 °C and collected for metabolite analysis. Extracts were then dried under nitrogen.
Fatty acid methyl esters (FAME) were obtained by the treatment of the total lipids with 2% H2SO4 in MeOH in a screw-capped vial (2 h, 80°C) under Argon and purified by thin-layer chromatography development in benzene (Carreau and Dubacq, 1978). The gas chromatography (GC) analysis of FAME was carried out on Shimadzu GC-2010 chromatograph (Kyoto, Japan) with a flame ionization detector on an Equity-5 (Supelco, Bellefonte, PA, US) capillary column (30 m/0.25 mm ID), at 160 °C with a 2 °C/min ramp to 240°C that was held for 20 min. Injector and detector temperatures were 250 °C. FAME were identified by gas chromatography-mass spectrometry (GC–MS) using Shimadzu GCMS-QP5050A instrument (Kyoto, Japan) (electron impact at 70 eV) with an SPB-5 (Supelco, Bellefonte, PA, US) capillary column (30 m /0.25 mm ID). The GC–MS analysis of FAME was performed at 160°C with a 2°C/min ramp to 240°C that was held for 20 min. Injector and detector temperatures were 250°C. Spectra were compared with the NIST library and FA mass spectra archive (The AOCS Lipid Library 2015). The assay was performed three times on separate days.
Assessment of genetic changes in the S. aureus 5812R strain
DNA extraction, genome sequencing, and assembly
Genomic DNA was isolated from overnight broth culture using Monarch HMW DNA Extraction Kit (New England Biolabs, USA). DNA concentration was estimated using Qubit 4.0 fluorimeter (Thermo Scientific, Germany). DNA quality was evaluated using NanoPhotomete N120 spectrophotometer (Implen, Germany). Genomic libraries were constructed using the NEBNext Ultra II FS library preparation kit (New England Biolabs, USA) according to the manufacturer's instruction for the MiSeq (Illumina, US) sequencing. Sequencing was carried out in the Center of Shared Scientific Equipment “Microorganisms Persistence” of the Institute of Cellular and Intracellular Symbiosis of the Ural Branch of the Russian Academy of Sciences using a MiSeq reagent kit V3 with 2×300-bp reads.
Nanopore sequencing libraries were generated using the Native Barcoding Kit SQK-NBD112.24 (Oxford Nanopore Technologies, UK) protocol and sequenced on a SpotON flow cell vR9 (catalog number FLO-MIN106) for 48 h.
The quality of the raw Illumina reads was analyzed by FastQC software (Andrews, 2010). Trimming of the raw reads was done by Trimmomatic v. 0.39 software (Bolger et al., 2014). Reads were filtered by minimum quality (Q = 20) and length (removing 10 nucleotides from both ends). The obtained nanopore long reads were trimmed for minimal length (5,000 bp) and quality (q = 9) with Nanofilt (Coster et al., 2018). Genome assembly was performed using Unicycler v.0.4.9b (Wick et al., 2017) in normal mode. The draft assembly was then polished using error-corrected Illumina reads with Pilon 1.23 (Walker et al., 2014).
Annotation and subsystem analysis
The annotation of genome was carried out using the default Prokka pipeline (https://github.com/tseemann/prokka). The predicted CDSs were annotated based on COG (Cluster of Orthologous Groups of proteins) and KEGG (Kyoto Encyclopedia of Gene and Genomes) (Kanehisa and Goto, 2000) databases, using eggNOG 5.0 (Huerta-Cepas et al., 2019) and KASS (Moriya et al., 2007), respectively. A graphical circular map of the genome was performed with the CGview server (Grant and Stothard, 2008).
SNP identification
Identification of SNP was performed using the Snippy software (https://github.com/tseemann/snippy). The fastq clean reads of resistant strain S. aureus 5812R were aligned against complete genome of wild-type strain S. aureus FDA209P.
RNA extraction and real-time quantitative PCR analysis
Bacterial strains were grown in Mueller Hinton Broth in the absence of gausemycin A. S. aureus FDA209P and S. aureus 5812R cells were collected at mid-exponential, late-exponential, and stationary growth phases and diluted to an optical density corresponding to 106 CFU/ml.
Total RNA was isolated using the RNeasy Mini kit (Qiagen, US) according to the manufacturer's instructions. Total RNA was further treated with DNase I (New England Biolabs, US) followed by the RNeasy MinElute Cleanup kit (Qiagen, US) according to the manufacturer's instructions. RNA was quantified using Qubit 4.0 (Thermo Fisher Scientific, US) and the quality of the extracted RNA was assessed by TapeStation 4150 (Agilent Technologies, US). Samples with preserved 16S and 23S peaks and RIN values >8 were selected for gene expression analyses. The complementary DNA was obtained using the iScript reversed transcription supermix for RT-qPCR reagent (Bio-Rad, US) in accordance with the manufacturer's protocol. Then quantitative PCR was carried out using SsoAdvanced Universal SYBR Green Supermix reagent (Bio-Rad, US). Each reaction mixes with a volume of 20 μL was prepared with 300 nM each primer (final concentration) and 20 ng of template RNA.
LightCycler96 Real-Time PCR detection system (Roche, Switzerland) was used with the following thermal cycling conditions: denaturation at 950C for 1 min, followed by 40 cycles of denaturation at 950C for 10 s and annealing/extension at 600C for 15 s. After the last amplification cycle, a melting curve analysis was carried out by heating from 65 to 95 0C in increments of 0.50C /s. Negative controls (without template or reverse transcriptase enzyme) were included in each run.
The design of oligonucleotide primer sequences for the gene responsible for the synthesis of cardiolipin (cls) was carried out using the online service “Integrated DNA technologies” (https://www.idtdna.com/Primerquest/Home/Index) using gene sequences present in the GenBank database. Fold changes in the gene expression levels were normalized in relation to the levels of gyrB mRNA (Supplementary Table S1).
The relative changes in gene expression were quantified using the Pfaffl method (Pfaffl, 2001):
where E target is the amplification efficiency of the target (gene of interest); E reference is the amplification efficiency of reference (gyrB); Ct is the point at which the fluorescence rises above the background fluorescence; ΔCt target is the Ct deviation of the control minus the sample of the target gene transcript; and ΔCt reference is the Ct deviation of the control minus the sample of the reference gene transcript.
Checkerboard technique
The effect of gausemycin A in combination with other antibiotics was evaluated using the checkerboard technique with modifications (Nwabor et al., 2021). Briefly, 80 μL of diluted bacterial suspension (106 CFU/mL) was added to wells containing 10 μL of sub-inhibitory concentrations of gausemycin A and 10 μL of sub-inhibitory concentrations of conventional antibiotics. The plates were incubated for 24 h at 37°C. The effects of the antimicrobial combination were defined according to the fractional inhibitory concentration index (FICI). The ΣFICs were calculated as follows:
ΣFIC = FIC A + FIC B,
where FIC A is the MIC of drug A in the combination/MIC of drug A alone; and FIC B is the MIC of drug B in the combination/MIC of drug B alone. The result was interpreted as FIC ≤ 0.5 = Synergism, FIC > 0.5 to 1 = additivity, FIC >1 to 2 = indifference, and FIC >2 antagonism.
Statistical processing
The obtained results were statistically manipulated using Origin 2021 (OriginLab Corporation, Northampton, MA, USA) software. The pair-sample Student's t-test was used to estimate the significance of the differences between the pairs.
Results
The dynamics of the resistance formation
The first goal of this work is to evaluate the dynamics of the resistance development of S. aureus FDA209P to gausemycin A in comparison with ramoplanin and daptomycin, which are similar in structure and mechanism of action, respectively.
The resistant population of S. aureus FDA209P was obtained by multiple serial passages with sub-inhibitory concentrations of gausemycin A. The formation of the resistant variant of S. aureus FDA209P was established within 20 passages (Table 1).
From the 12th passage with gausemycin A, the bacteria became significantly less sensitive. The obtained clone of S. aureus FDA209P at the 13th passage was resistant to the antibiotic concentration in the medium equal to 50 μg/mL.
The adapted population was found to display up to 80-fold increment in their minimum inhibitory concentration (MIC) relative to the wild-type strain. S. aureus FDA209P developed resistance to ramoplanin within 10 passages. Right from the 5th passage of cultivation with ramoplanin, the bacteria became significantly less sensitive to this antibiotic. Eight passages were enough to reach MIC value for 100 μg/mL. At the same time, the increase in resistance of S. aureus FDA209P to daptomycin during the series of passages was less pronounced. Daptomycin‘s minimum inhibitory concentration (MIC) increased from 1.25 μg/mL to 5.0 μg/mL due to 20 passages (Table 1).
Cross-resistant of S. aureus 5812R
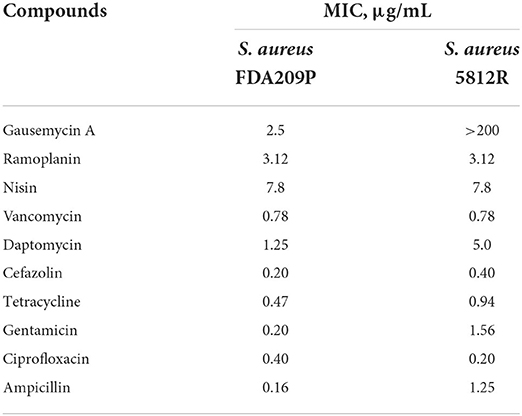
The cross-resistance of the obtained S. aureus 5812R strain was evaluated against the main class of antibiotics targeting Gram-positive bacterial barrier structures (cell wall and/or cytoplasmic membrane). Antibiotics with other targets of action were also used, including those affecting the 30S subunit of the ribosome and inhibiting bacterial DNA topoisomerases and gyrase.
It was found that the most significant difference was acquired in susceptibility to daptomycin, which was 4-fold more potent against S. aureus FDA209P compared to S. aureus 5812R. Surprisingly, the two compared strains did not differ in their susceptibility to vancomycin, ramoplanin, and nisin (Table 2). MIC values for non-peptide antibiotics changed differently. The most pronounced shift occurred with gentamycin, S. aureus 5812R demonstrated phenotype which is 7.8-fold resistant, compared to its parent strain (Table 2). MIC values for cefazolin, tetracycline, and ampicillin increased by twice for S. aureus 5812R, while MIC value for ciprofloxacin, on the contrary, decreased twice.
Phenotype of the resistant clone
The resistance of S. aureus 5812R was stable, since multiple (17) passaging throughout the gausemycin A-free media did not change the MIC value for gausemycin A.
The phenotype of S. aureus 5812R grown on plate resembles a small-colony variant, since it forms colonies a few times smaller than the parent strain (Figures 1A,B), and is less pigmented (Figure 1C) (Proctor et al., 2006; Sulaiman et al., 2021). The AFM revealed no significant difference in cell diameter: 1.001±0.24 μm (S. aureus FDA209P) versus 1.0056±0.19 μm (S. aureus 5812R) (p-0.9498, Figures 1A,B). The growth rate of S. aureus 5812R in liquid medium was compared with the parent strain, but the final cells density was lower (Figure 1D). S. aureus 5812R did not differ from its parent strain in their biochemical features (Supplementary Table S2).
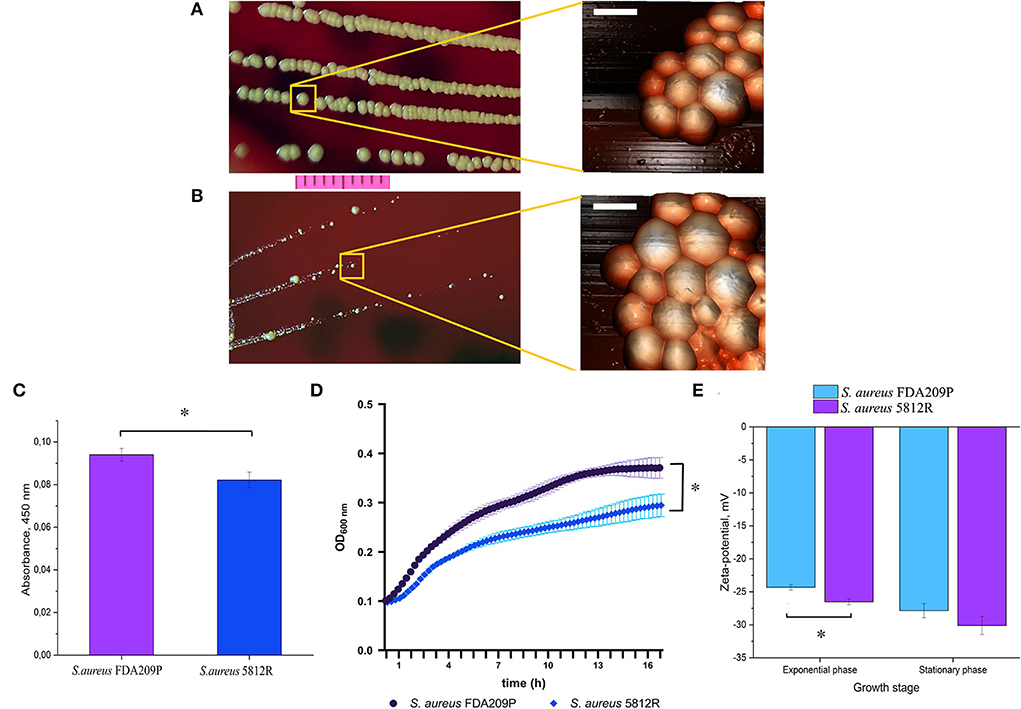
Figure 1. The phenotypical characteristics of the gausemycin A-resistant S. aureus 5812R strain. Morphology of colonies and bacterial cells of S. aureus FDA209P (A) and S. aureus 5812R (B), which were formed for 24 h. Optical microscopy (bar-−1 cm) and atomic force microscopy (bar – 1 μm) were used for revealing the morphology of bacterial colonies and cells, respectively. (C) The amount of pigment which was extracted from colonies and expressed in absorbance units. The means plus standard deviations of the means of results from three independent experiments with three technical replicates are shown (* p < 0.05). (D) The growth kinetics of S. aureus FDA209P and S. aureus 5812R on Muller-Hinton medium without gausemycin A. (E) The charges of bacterial cells at various growth stages, which were estimated in zeta-values. The means plus standard deviations of the means of results from two independent experiments with three technical replicates are shown (*p < 0.05).
Zeta potential
Zeta potential measurements revealed that the growing S. aureus FDA209P population exhibited a slightly more positive charge (−24.32 ± 0.40 mV) than the strain S. aureus 5812R (−26.52 ± 0.44 mV). The same relation was obtained for bacterial cells taken from the stationary phase: −27.85 ± 1.08 mV (S. aureus FDA209P), and −30.12 ± 1.36 mV (S. aureus 5812R) (Figure 1E).
Membrane integrity assay
Our previous study showed that the mechanism of action of gausemycin A (5812-A/C) is through disrupting the structural integrity of bacterial membranes (Vasilchenko et al., 2020). The method of detecting membrane-disturbing action is simple and it is based on the detection of quenching of SYTO9 green fluorescence, when red-fluorescent dye propidium iodide influx through disordered bacterial membrane. If the resistance of staphylococci is associated with a change in the membrane structures of cells, then there will be no quenching of SYTO9 fluorescence. The study showed that adding gausemycin A into the reaction medium with S. aureus FDA209P quenched SYTO9 fluorescence in a dose-dependent manner (Figure 2A), and complete quenching was achieved at 50 μg/mL. On the contrary, quenching of SYTO9 fluorescence in the cells of S. aureus 5812R was not achieved at all taken concentrations (Figure 2A).
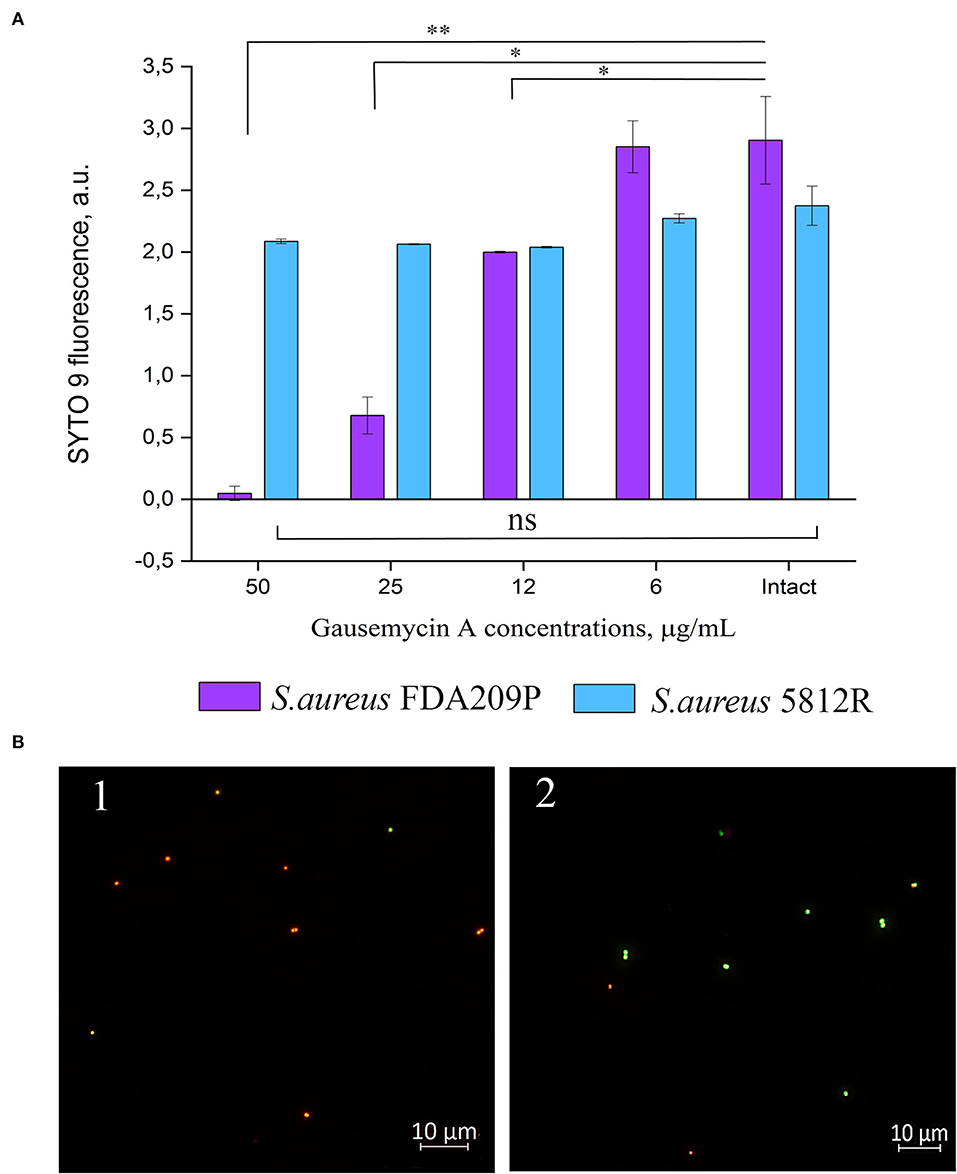
Figure 2. The evaluation of membrane disturbance of S. aureus FDA209P and S. aureus 5812R using Live/Dead fluorescent dyes. The data show the intensity of SYTO 9 fluorescence at the end-point of measurement (10 min) (A). Epifluorescence microscopy images of S. aureus FDA209P (1) and S. aureus 5812R (2) after treatment with gausemycin A (B). The means plus standard deviations of the means of results from two independent experiments with three technical replicates are shown (*p < 0.05, **p < 0.01, ns—values are not significantly different).
The performed microscopical visualization has revealed that S. aureus FDA209P (Figure 2B.1) consisted of numerous cells with permeabilized membranes (glowing red), while cells of S. aureus 5812R were predominantly unaffected (glowing green) (Figure 2B.2). Collectively, the obtained data have shown that resistance to gausemycin A is related to membrane modifications.
Fatty acid profiles of S. aureus FDA209P and S. aureus 5812R
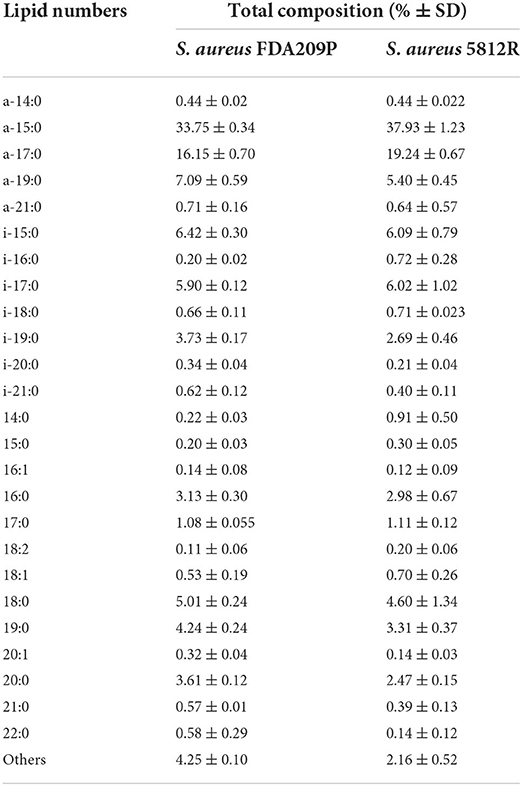
In total, 26 fatty acids were identified in the extracts of S. aureus cells (Table 3). The membrane of S. aureus FDA209P is composed by straight chain fatty acids (SCFAs) (17.56 ± 2.03 %) and branched-chain fatty acids (BCFAs) (76.0 ± 0.18 %), including anteiso-BCFAs (58.14 ± 0.37%), and iso-BCFAs (17.86 ± 0.20%) (Figure 3).
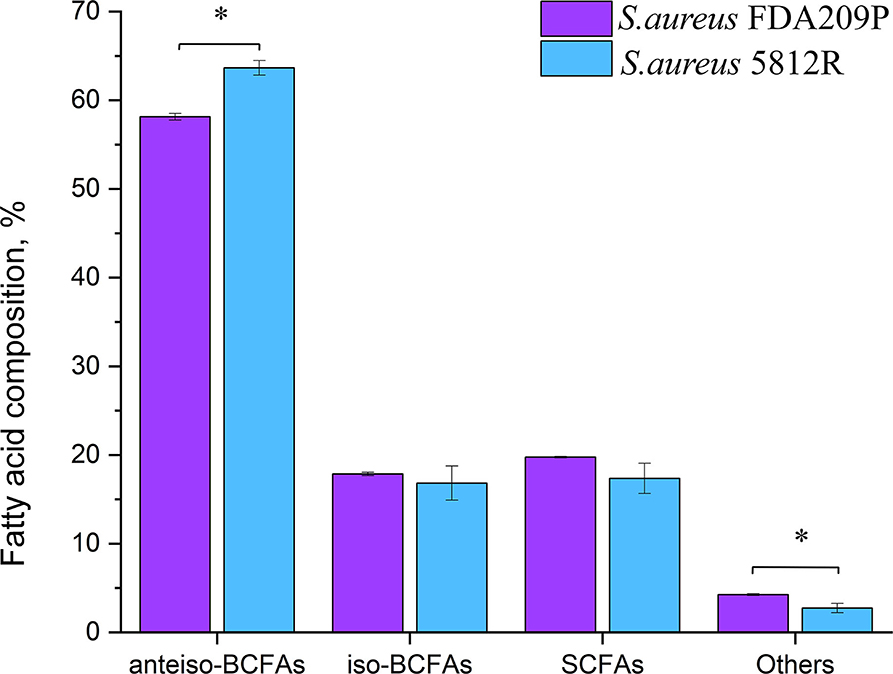
Figure 3. The fatty acid composition of cytoplasmic membrane S. aureus FDA209P and S. aureus 5812R. The measurements were performed using gas chromatography–mass spectrometry. Y-axis is a relative proportion (%) of each group of fatty acids in relation to the total amount of identified fatty acids. The means plus standard deviations of the means of results from three independent experiments are shown (*p < 0.05).
The membrane of S. aureus 5812R consists of insignificantly less amount of SCFAs (17.36 ± 1.07 %, p-0.13239). Comparatively, the decrease of SCFAs in cells of S. aureus 5812R was mostly related to C18:0, C19:0, C20:0, and C22:0 fatty acids, which were 0.5–2.0 times less present in comparison with S. aureus FDA209P (Table 3). On the contrary, the branched-chain fatty acids (BCFAs) were in prevalence (80.48 ± 1.51 %, p−0.03186), among them there were significantly more anteiso-BCFAs (63.64 ± 0.82 %, p−0.0048), whereas the proportion of iso-BCFAs was insignificantly less present (16.83±1.93 %, p−0.46085).
Our finding suggests that the change in the proportions of BCFAs in S. aureus 5812R is possibly related to the resistance phenomenon.
Genetic basis of the antibiotic resistance
The DNA sequencing of S. aureus FDA209P and S. aureus 5812R was performed using both Nanopore sequencing and Next generation sequencing technologies. Performed sequencing of the S. aureus FDA209P genome generated 18,875 reads (465,232,550 bases, 167 times coverage) with MinION Mk1B, and 1,849,663 reads (969,265,770 bases, 349 times coverage) with MiSeq. At the same time, for the gausemycin A-resistant S. aureus 5812R strain, 15,814 reads (394,758,410 bases, 142 times coverage) and 1,998,645 reads (1,047,191,454 bases, 377 times coverage) were obtained by MinION and MiSeq, respectively (Supplementary Table S3). The assembled genomes were released in GenBank NCBI for further study and annotation (S. aureus FDA209P JANQDW000000000; S. aureus 5812R JANPYH000000000).
About 1512 (59.4%) annotated protein-coding genes were classified according to KEGG categories. The most represented functional identified categories were genetic information processing (359), carbohydrates (178), signaling and cellular processing (169), environmental information processing (129), metabolism of cofactors and vitamins (87), amino acids metabolism (84), nucleotide metabolism (64), and energy metabolism (45). Among 2,547 protein-coding genes, seven genes are associated with the resistance to cationic antimicrobial peptides (CAMPs) (Supplementary Table S4).
Comparing the nucleotide sequences of both strains, we have found one single nucleotide polymorphism (SNP) and deletions of three genes in S. aureus 5812R genome (Table 4, Figure 4A). SNP was non-synonymous and it was found in the vraT gene (previously called yvqF) (C → G at the nucleotide 370), which encodes the membrane protein with unknown biological function. The hepT gene, encoding heptaprenyl diphosphate synthase component II has the single nucleotide deletion at position 940. The sepA gene, which encodes the multidrug-resistant efflux pump protein SepA, has five nucleotide deletions between 347-351. This mutation has caused a frameshift in sepA and—presumably—premature termination. Also, the sequencing revealed deletions of five nucleotides between 585 and 589 positions in the kdpE gene, which is a transcriptional activator in the two-component system KdpD/KdpE.
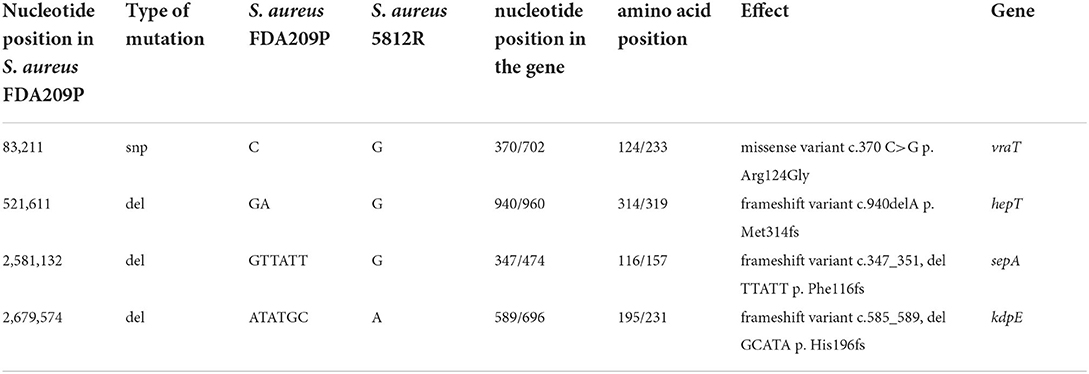
Table 4. Genetical differences between the wild-type S. aureus FDA209P and its mutant variant S. aureus 5812R.
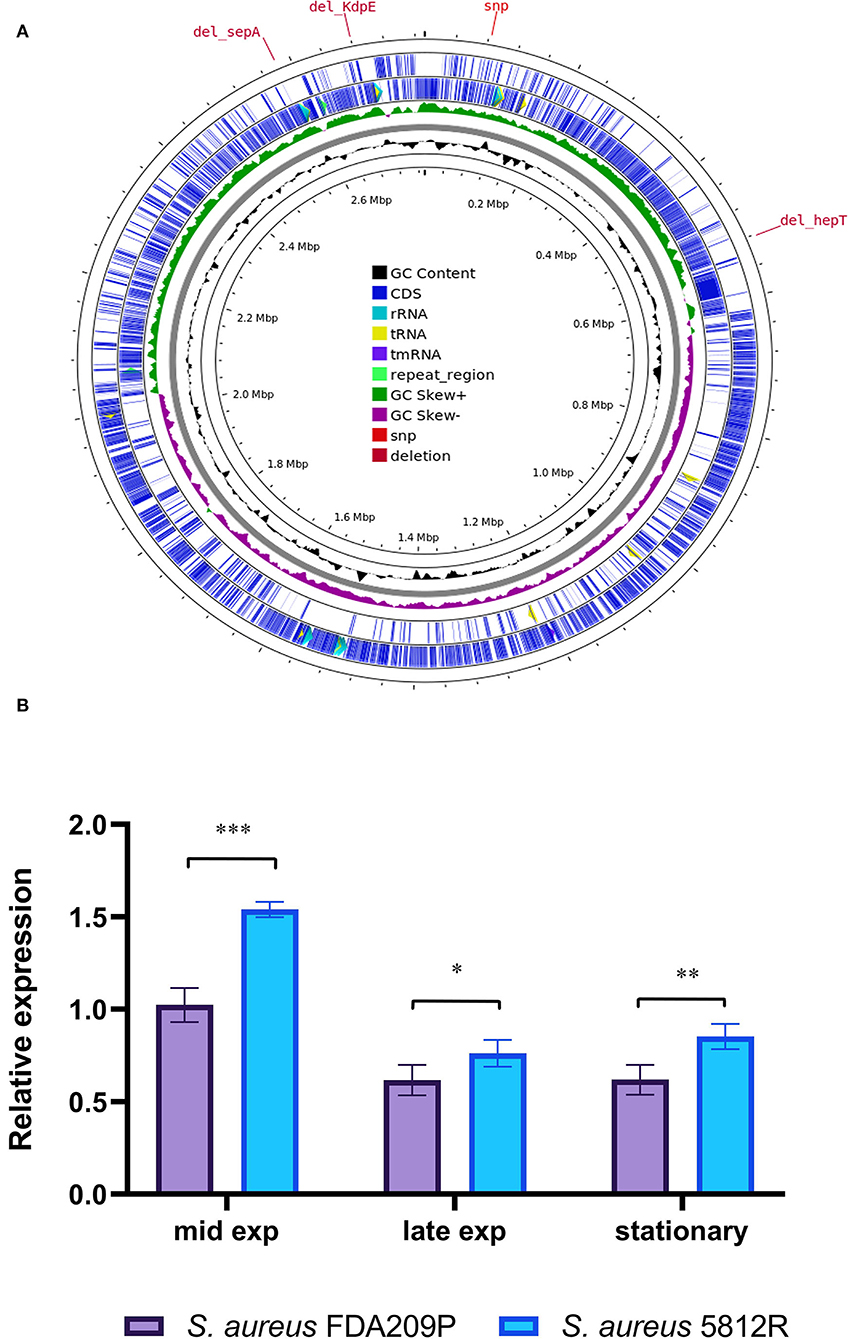
Figure 4. Graphical circular map of the S. aureus 5812R chromosome (A). The map was generated using CGView Server (http://cgview.ca/). The expression analysis of gene responsible for the synthesis of cardiolipin (cls) (B). Transcript levels of the analyzed genes were determined by RT-qPCR in relation to the gyrB expression. Mid Exp—the mid-exponential growth phase; Late Exp—the late exponential growth phase; Stationary—the stationary growth phase. The means plus standard deviations of the means of results from three technical replicates are shown (*p < 0.05, **p < 0.01, ***p < 0.001).
The comparison of cardiolipin synthase (Cls2) gene expression with S. aureus FDA209P and S. aureus 5812R
Cardiolipin plays a critical role in bacterial physiology, especially for cytokinesis and antibiotic resistance (Tiwari et al., 2020). The synthesis of cardiolipin in bacteria is catalyzed by the cardiolipin synthases Cls, which provide the condensation of two phosphatidylglycerol molecules to yield cardiolipin and glycerol. We analyzed the expression of the cls2 (cardiolipin synthase) gene in both strains in a comparison manner.
It was found that the most active expression of the cls2 gene occurred in the mid-log phase of bacterial growth. Continued growth was associated with a double decrease in the expression level. The difference between the compared strains was a higher level of cls2 expression in the resistant strain 5812R. In the mid-log phase, the relative expression of the cls2 gene was higher in S. aureus 5812R cells (1.54 ± 0.04), compared to S. aureus FDA209P (1.02 ± 0.06, p-0.00115). The following bacterial growth was accompanied by the reduction in the cls2 gene relative expression. In the late-exponential phase, the relative expression decreased to 0.61 ± 0.03 (S. aureus FDA209P) and 0.76 ± 0.05 (S. aureus 5812R) (p-0.03876); in the stationary phase, the relative expression was 0.62 ± 0.05 for S. aureus FDA209P, and 0.85 ± 0.04 for S. aureus 5812R (p−0.00985) (Figure 4B).
Thus, we showed that gausemycin A resistance is accompanied by the increased relative expression of the cls2 gene in the S. aureus 5812R cells.
The antibacterial effect of gausemycin A in combination with conventional antibiotics
The antibacterial activity of four combinations of antibiotics against S. aureus 5812R strain was tested using the checkerboard method. The effects of combinations of antimicrobials were determined by the index of fractional inhibitory concentration (FICI). The results of in vitro antibacterial activity of gausemycin A in combination with other antibiotics against S. aureus 5812R are presented in Table 5. Three antibiotic combinations showed a synergistic activity resulting in the reduction in MICs. The combination of Cip + Gau showed a pronounced synergistic antibacterial effect against S. aureus 5812R (FICI 0.49).
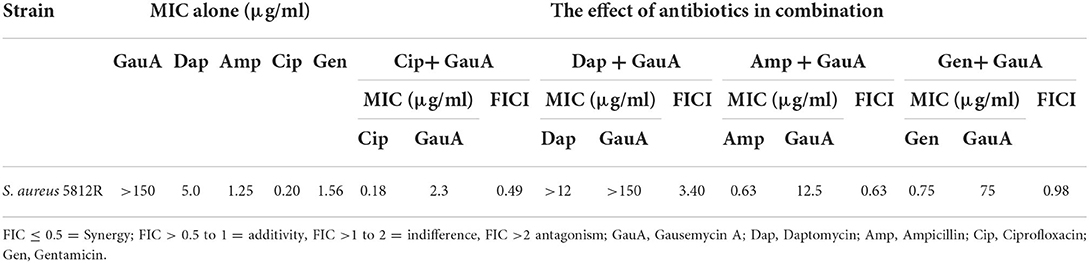
Table 5. The in vitro antibacterial activity of gausemycin A in the combination with other antibiotics against S. aureus 5812R.
The combinations of Amp + Gau (FICI 0.63) and Gen + Gau (FICI 0.98) showed an additive effect. The combination of Dap + Gau demonstrated an antagonistic effect (FICI 3.40).
Discussion
Natural lipoglycopeptides are rare and are grouped in a small number of structural classes including gausemycins (Freitas et al., 2022). The present work is the only second evidence of bacterial resistance to lipoglycopeptides after Schmidt's study of ramoplanin-resistant S. aureus (Schmidt et al., 2010). Ramoplanin and daptomycin are peptide antibiotics, which are similar in structure and/or mechanism of action, and also are available as commercial drugs. The obtained results indicate that the development of the resistance to gausemycin A was much faster than to daptomycin, but much slower than to ramoplanin.
Since the structure of gausemycin A molecule is unique, it would be interesting to determine whether the S. aureus 5812R strain would be cross-resistant to the closest analogs and other antibiotics. It was found that the resistance to gausemycin A does not provide resistance to nisin, vancomycin, and ramoplanin. These peptide antibiotics are united together by the same target within bacterial cells. For example, nisin has a membrane-destroying effect upon binding to a receptor, which is lipid II (Paiva et al., 2011). Vancomycin is a cationic glycopeptide that kills bacteria by binding to the C-terminal D-Ala–D-Ala residues of the peptidoglycan precursor lipid II, and prevents the use of the precursor for cell wall synthesis (Hu et al., 2016). The pyrophosphate pocket of lipid II is also the target for ramoplanin (Cheng et al., 2014).
Our primary objective was to find out what kind of changes could be responsible for staphylococcal resistance to gausemycin A. First of all, it was necessary to evaluate the changes at the genome level. The comparison of S. aureus FDA209P and S. aureus 5812R genomes revealed deletions. Among the mutations, hepT and kdpE are the genes which could have the most contribution in gausemycin A resistance.
The kdpE gene is a transcriptional activator that is a part of the two-component system KdpD/KdpE (Kdp system) (Freeman et al., 2013). The Kdp system has a function in S. aureus as a K+ uptake system that directly regulates virulence and stress response. KdpE can bind directly to the DNA promoters of a range of virulence genes. It was found that deletion of kdpD/kdpE altered the level of transcription of over 100 genes (Zhao et al., 2010). Existing studies link kdpD/kdpE with resistance to antimicrobial stresses, since gene knockouts increase bacterial susceptibility to polymyxins and chitosan, compared to the wild strains (Alegado et al., 2011; Mellegård et al., 2011). Our results suggest further studies to uncover the role of the Kdp system in bacterial resistance to peptide antibiotics.
The resistance of S. aureus 5812R to gausemycin A, as well as to daptomycin and other antibiotics could be explained by the transition of cells into the special phenotype, known as a small-colony variant (SCV), that is a variant subpopulation of S. aureus characterized by the deficiency in electron transport, and the deficiency in thymidine biosynthesis (Proctor et al., 1998; Watkins et al., 2012; Johns et al., 2015). The reduced electrochemical gradient in SCV cells promotes resistance to gentamycin and other antibiotics (Proctor et al., 1998). The prerequisite for this was found in the analysis of genetic differences between the S. aureus FDA209P and the S. aureus 5812R strains. Sequencing revealed single nucleotide deletion in the gene hepT which encodes protein associated with the isoprenoid biosynthesis pathway (Liu et al., 2016). Isoprenoids are involved in the production of carotenoids, quinones, hemes, and other compounds, which are necessary for the development of normal colony morphotypes (Liu et al., 2016). Deletions in hepT lead to blockage in menaquinone biosynthesis (Wakeman et al., 2012). Thus, this nucleotide deletion could be associated with the transition of the S. aureus population to the SCV morphotype. The genome analysis also gives a possible explanation why S. aureus 5812R is more susceptible to ciprofloxacin than the parent strain. Nucleotide deletions in the sepA gene could be associated with the susceptibility of S. aureus 5812R to ciprofloxacin, since SepA is an efflux pump that allows bacteria to be resistant to this antibiotic (Hassanzadeh et al., 2017).
Our secondary objective was to find the phenotypic changes that are associated with staphylococcal resistance to gausemycin A. We evaluated the structural changes that occurred with the cytoplasmic membrane, since it is the main target for gausemycin A (Vasilchenko et al., 2020). The bacterial membrane equilibrium between rigidity and fluidity is driven by multiple factors that impact membrane order (water, metal ions, proteins, carotenoids, staphyloxanthin, etc.), among these factors the composition of phospholipid fatty acids is the main factor. (Boudjemaa et al., 2018).
In the case of S. aureus, bacterial membranes mainly consist of straight-chain (SCFAs) and branched-chain (BCFAs) saturated fatty acids. SCFAs pack together to produce a bilayer with low permeability properties, while branched-chain iso or/and anteiso methyl species promote a more fluid membrane structure (Zhang and Rock, 2008). The development of resistance of S. aureus 5812R to gausemycin A has been accompanied by an increase in the relative proportion of anteiso-BCFAs in cell membranes and by a decrease in the level of iso-BCFAs. At the same time, the percentage of saturated fatty acids in the cell membrane of the resistant strain decreased slightly compared to the parent strain. This result is inconsistent with previous studies showing that S. aureus responded to higher concentrations of daptomycin and vancomycin by lowering the biosynthesis of iso and anteiso branched-chain fatty acids while increasing the levels of saturated fatty acids (Mirani and Jamil, 2013; Boudjemaa et al., 2018).
The main S. aureus membrane lipids consist negatively charged phospholipids: phosphatidylglycerol (PG) and cardiolipin (CL) (Jung et al., 2008). Cardiolipin synthases (Cls) are the critical enzymes for the synthesis of CL in bacteria. It is known that mutation in csl2 gene increases daptomycin susceptibility in S. aureus (Peleg et al., 2012), while the enhanced levels of cardiolipin are associated with daptomycin resistance in Enterococci (Palmer et al., 2011).
We found out that the S. aureus 5812R strain showed an increased relative expression of the cls2 gene. It seems that cardiolipin plays a more important role in the resistance of staphylococci to gausemycin A, than daptomycin which could be explained by the differences in their physicochemical properties. Gausemycin A molecule has a more negative net charge compared to daptomycin. Zeta potential measurements revealed the S. aureus FDA209P population consisted of cells that are slightly more positively charged compared to the S. aureus 5812R population. Although the difference is not high, it may contribute to the resistance to gausemycin A.
Thereby, antimicrobial peptides are increasingly recognized as a promising class of compounds that are used in combination with conventional antibiotics. In this regard, we have tried to overcome gausemycin A resistance by choosing that combination, where a pair of antibiotics affects another cellular target, for example, DNA replication or protein synthesis. Besides, we tested the combination of gausemycin A and daptomycin, and, surprisingly, revealed the antagonism between them for an unknown reason. Similar work showed that the combinations of daptomycin with other peptide antibiotics resulted in indifferent interactions, while the combination of daptomycin + ciprofloxacin had a synergistic effect against S. aureus (Mataraci and Dosler, 2012). Numerous studies demonstrated the effectiveness of the combination of daptomycin with antibiotics, which target nucleic acid replication (Rand and Houck, 2004; Kelesidis et al., 2011; Kamble et al., 2022). In our study, the combination of ciprofloxacin and gausemycin A has been found to have synergistic activity against gausemycin A-resistant S. aureus. Thus, it has been revealed that such a combination could be successful to suppress the gausemycin A-resistant S. aureus.
Conclusion
In this study, we showed the ability of S. aureus FDA209P to form gausemycin A-resistant phenotype. However, the revealed phenomenon does not mean that this peptide antibiotic has no prospects as a therapeutic agent. Ramoplanin showed an even more pronounced ability to form a resistant S. aureus phenotype in this comparative experiment. In addition, S. aureus 5812R did not form significant cross-resistance to antibiotics that other cellular targets have, which allows considering gausemycin A in combination therapy using conventional antibiotics like ciprofloxacin.
The obtained results also deal with the mechanism of action and the bacterial response of gausemycin A. We found that S. aureus 5812R was cross-resistant to daptomycin. This finding confirms the similarity between gausemycin A and daptomycin in their modes of action. At the same time, the identified features of genetic and phenotypic changes associated with bacterial resistance suggest that the response to gausemycin A is specific due to the uniqueness of the structure of gausemycin A.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary materials, further inquiries can be directed to the corresponding author.
Author contributions
DP, ER, SK, and AV conducted the experiments. DP, SK, AV, VS, and OL performed analyses. DP, ER, and AV wrote the manuscript and prepared the figures. AV conceived, designed the experiments, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Russian Science Foundation (project number 21-74-00123).
Acknowledgments
We are grateful to Nadezhda N. Zhuravleva (Center for Academic Writing, Tyumen State University) for the valuable comments which improved the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.963979/full#supplementary-material
References
Alegado, R. A., Chin, C.-.Y, Monack, D. M., and Tan, M-, W. (2011). The two-component sensor kinase KdpD is required for salmonella typhimurium colonization of caenorhabditis elegans and survival in macrophages. Cell. Microbiol. 13, 1618–1637. doi: 10.1111/j.1462-5822.2011.01645.x
Andrews, S. (2010). FastQC a Quality Control Tool for High Throughput Sequence Data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed July 2022).
Aslam, B., Wang, W., Arshad, M. I., Khurshid, M., Rasool, M. H., Nisar, M. A., et al. (2018). Antibiotic resistance : a rundown of a global crisis. Infect. Drug Resist. 2018, 1645–1658. doi: 10.2147/IDR.S173867
Bayer, A. S., Schneider, T., and Sahl, H. G. (2013). Mechanisms of daptomycin resistance in Staphylococcus aureus: Role of the cell membrane and cell wall. Ann. N. Y. Acad. Sci. 1277, 139–158. doi: 10.1111/j.1749-6632.2012.06819.x
Bhullar, K., Waglechner, N., Pawlowski, A., Koteva, K., Banks, E. D., Johnston, M. D., et al. (2012). Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE. 7, 1–11. doi: 10.1371/journal.pone.0034953
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boudjemaa, R., Cabriel, C., Dubois-Brissonnet, F., Bourg, N., Dupuis, G., Gruss, A., et al. (2018). Impact of Bacterial Membrane Fatty Acid Composition on the Failure of Daptomycin To Kill Staphylococcus aureus. Antimicrob. Agents Chemother. 62, e00023–e00018. doi: 10.1128/AAC.00023-18
Carreau, J. P., and Dubacq, J. P. (1978). Note Adaptation of a macro-scale method to the micro-scale transesterification of biological lipid extracts. J. Chromatogr. 151, 384–390. doi: 10.1016/S0021-9673(00)88356-9
Cavalleri, B., Hermes, P., Giancarlo, V., Enrico, S., and Franco, P. (1984). A-16686, a new antibiotic from Actinoplanes. I. Fermentation, isolation and preliminary physico-chemical characteristics. J. Antibiot. 37, 309–317. doi: 10.7164/antibiotics.37.309
Cheng, M., Huang, J. X., Ramu, S., Butler, M. S., and Cooper, M. A. (2014). Ramoplanin at bactericidal concentrations induces bacterial membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 58, 6819–6827. doi: 10.1128/AAC.00061-14
Coster, W. D. e, D'Hert, S., Schultz, D. T., Cruts, M., and Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 34, 2666–2669. doi: 10.1093/bioinformatics/bty149
de la Cruz, M., González, I., Parish, C. A., Onishi, R., Tormo, J. R., Martín, J., et al. (2017). Production of Ramoplanin and Ramoplanin Analogs by Actinomycetes. Front. Microbiol. 8, 343. doi: 10.3389/fmicb.2017.00343
DeLeo, F. R., and Chambers, H. F. (2009). Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J. Clin. Invest. 119, 2464–2474. doi: 10.1172/JCI38226
Folch, J., and Lees, M. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509. doi: 10.1016/S0021-9258(18)64849-5
Freeman, Z. N., Dorus, S., and Waterfield, N. R. (2013). The KdpD/KdpE two-component system: integrating K? homeostasis and virulence. PLoS Pathog. 9, e1003201. doi: 10.1371/journal.ppat.1003201
Freitas, S., Castelo-Branco, R., Wenzel-Storjohann, A., Vasconcelos, V. M., Tasdemir, D., and Leão, P. N. (2022). Structure and biosynthesis of desmamides A-C, Lipoglycopeptides from the endophytic cyanobacterium desmonostoc muscorum. LEGE. 85, 1704–1714. doi: 10.1021/acs.jnatprod.2c00162
Fuchs, P. C., Barry, A. L., and Brown, S. D. (2000). Daptomycin susceptibility tests: interpretive criteria, quality control, and effect of calcium on in vitro tests. Diagn. Microbiol. Infect. Dis. 38, 51–58. doi: 10.1016/S0732-8893(00)00164-4
Grant, J. R., and Stothard, P. (2008). The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36. W181–W184. doi: 10.1093/nar/gkn179
Gwynne, P. J., and Gallagher, M. P. (2018). Light as a broad-spectrum antimicrobial. Front. Microbiol. 9, 1–9. doi: 10.3389/fmicb.2018.00119
Hassanzadeh, S., Mashhadi, R., Yousefi, M., Askari, E., Saniei, M., Pourmand, M. R., et al. (2017). Frequency of efflux pump genes mediating ciprofloxacin and antiseptic resistance in methicillin-resistant Staphylococcus aureus isolates. Microb. Pathog. 111, 71–74. doi: 10.1016/j.micpath.2017.08.026
Hu, Q., Peng, H., and Rao, X. (2016). Molecular events for promotion of vancomycin resistance in vancomycin intermediate Staphylococcus aureus. Front. Microbiol. (2016) 7. doi: 10.3389/fmicb.2016.01601
Huerta-Cepas, J., Szklarczyk, D., Heller, D., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, 309–314. doi: 10.1093/nar/gky1085
Johns, B. E., Purdy, K. J., Tucker, N. P., and Maddocks, S. E. (2015). Phenotypic and genotypic characteristics of small colony variants and their role in chronic infection. Microbiol Insights. 8, 15–23. doi: 10.4137/MBI.S25800
Jung, D., Powers, J. P., Straus, S. K., and Hancock, R. E. W. (2008). Lipid-specific binding of the calcium-dependent antibiotic daptomycin leads to changes in lipid polymorphism of model membranes. Chem. Phys. Lipids. 154, 120–8. doi: 10.1016/j.chemphyslip.2008.04.004
Kamble, E., Sanghvi, P., and Pardesi, K. (2022). Synergistic effect of antibiotic combinations on Staphylococcus aureus biofilms and their persister cell populations. Biofilm. 4, 100068. doi: 10.1016/j.bioflm.2022.100068
Kanehisa, M., and Goto, S. K. (2000). Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kelesidis, T., Humphries, R., Ward, K., Lewinski, M. A., and Yang, O. O. (2011). Combination therapy with daptomycin, linezolid, and rifampin as treatment option for MRSA meningitis and bacteremia. Diagn. Microbiol. Infect. Dis. 71, 286–290. doi: 10.1016/j.diagmicrobio.2011.07.001
Lapchinskaya, O. A., Katrukha, G. S., Gladkikh, E. G., Kulyaeva, V. V., Coodan, P. V., Topolyan, A. P., et al. (2016). Investigation of the complex antibiotic INA-5812. Russ. J. Bioorganic Chem. 42, 664–671. doi: 10.1134/S1068162016060078
Lewis, K. (2020). The science of antibiotic discovery. Cell. 181, 29–45. doi: 10.1016/j.cell.2020.02.056
Li, Y. X. (2018). Resistance to nonribosomal peptide antibiotics mediated by d-stereospecific peptidases. Nat. Chem. Biol. 14, 381–387 doi: 10.1038/s41589-018-0009-4
Liu, Y. L., Wei, H., Liu, W., Ko, T. P., Guo, R. T., Oldfield, E., et al. (2016). Structure, Function, and Inhibition of Staphylococcus aureus Heptaprenyl Diphosphate Synthase. ChemMedChem. 11, 1915–1923. doi: 10.1002/cmdc.201600311
Lum, K. Y., Tay, S. T., Le, C. F., Lee, V. S., Sabri, N. H., Velayuthan, R. D., et al. (2015). Activity of novel synthetic peptides against Candida albicans. Sci. Rep. 5. doi: 10.1038/srep09657
Lyu, Y., Yang, Y., Lyu, X., Dong, N., and Shan, A. (2016). Antimicrobial activity, improved cell selectivity and mode of action of short PMAP-36-derived peptides against bacteria and Candida. Sci. Rep. 6, 1–12. doi: 10.1038/srep27258
Mataraci, E., and Dosler, S. (2012). In vitro activities of antibiotics and antimicrobial cationic peptides alone and in combination against methicillin-resistant Staphylococcus aureus biofilms. Antimicrob. Agents Chemother. 56, 6366–6371. doi: 10.1128/AAC.01180-12
Mellegård, H., Lindbäck, T., Christensen, B. E., Kuipers, O. P., and Granum, P. E. (2011). Transcriptional responses of Bacillus cereus towards challenges with the polysaccharide chitosan. PLoS ONE. 6, e24304. doi: 10.1371/journal.pone.0024304
Mirani, Z. A., and Jamil, N. (2013). Effect of vancomycin on the cytoplasmic membrane fatty acid profile of vancomycin-resistant and -susceptible isolates of Staphylococcus aureus. J. Infect. Chemother. 19, 24–33. doi: 10.1007/s10156-012-0447-y
Mishra, N. N., Bayer, A. S., Tran, T. T., Shamoo, Y., Mileykovskaya, E., Dowhan, W., et al. (2012). Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS ONE. 7, 1–10. doi: 10.1371/journal.pone.0043958
Moriya, Y., Itoh, M., Okuda, S., et al. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. doi: 10.1093/nar/gkm321
Nwabor, O. F., Terbtothakun, P., Voravuthikunchai, S. P., Chusri, S., and Notarbartolo, M. (2021). Evaluation of the synergistic antibacterial effects of fosfomycin in combination with selected antibiotics against carbapenem-resistant Acinetobacter baumannii. Pharmaceuticals. 14, 185. doi: 10.3390/ph14030185
Paiva, A. D., Breukink, E., and Mantovani, H. C. (2011). Role of lipid II and membrane thickness in the mechanism of action of the lantibiotic bovicin HC5. Antimicrob. Agents Chemother. 55, 5284–5293. doi: 10.1128/AAC.00638-11
Palmer, K. L., Daniel, A., Hardy, C., Silverman, J., and Gilmore, M. S. (2011). Genetic basis for daptomycin resistance in enterococci. Antimicrob. Agents Chemother. 55, 3345–3356. doi: 10.1128/AAC.00207-11
Parachin, N. S., and Franco, O. L. (2014). New edge of antibiotic development: antimicrobial peptides and corresponding resistance. Front. Microbiol. 5, 147. doi: 10.3389/fmicb.2014.00147
Peleg, A. Y., Miyakis, S., Ward, D. V., Earl, A. M., Rubio, A., Cameron, D. R., et al. (2012). Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS ONE. 7. doi: 10.1371/journal.pone.0028316
Perry, J., Waglechner, N., and Wright, G. (2016). The prehistory of antibiotic resistance. Cold Spring Harb. Perspect. Med. 6, a025197. doi: 10.1101/cshperspect.a025197
Pfaffl, M. W. A. (2001). new mathematical model for relative quantification in real-time RT – PCR. Nucleic Acids Res. 29, 16–21. doi: 10.1093/nar/29.9.e45
Pollard, J. E., Snarr, J., Chaudhary, V., Jennings, J. D., Shaw, H., Christiansen, B., et al. (2012). In vitro evaluation of the potential for resistance development to ceragenin CSA-13. J. Antimicrob. Chemother. 67, 2665–2672. doi: 10.1093/jac/dks276
Proctor, R. A., Kahl, B., von Eiff, C., Vaudaux, P. E., Lew, D. P., Peters, G., et al. (1998). Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin. Infect. Dis. 27, S68–74. doi: 10.1086/514906
Proctor, R. A., von Eiff, C., Kahl, B. C., Becker, K., McNamara, P., Herrmann, M., et al. (2006). Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305. doi: 10.1038/nrmicro1384
Raman, N., Lee, M. R., Lynn, D. M., and Palecek, S. P. (2015). Antifungal activity of 14-helical β-peptides against planktonic cells and biofilms of candida species. Pharmaceuticals. 8, 483–503. doi: 10.3390/ph8030483
Rand, K. H., and Houck, H. (2004). Daptomycin synergy with rifampicin and ampicillin against vancomycin-resistant enterococci. J. Antimicrob. Chemother. 53, 530–532. doi: 10.1093/jac/dkh104
Rapacka-Zdonczyk, A., Wozniak, A., Pieranski, M., Woziwodzka, A., Bielawski, K. P., Grinholc, M., et al. (2019). Development of Staphylococcus aureus tolerance to antimicrobial photodynamic inactivation and antimicrobial blue light upon sub-lethal treatment. Sci. Rep. 9, 9423. doi: 10.1038/s41598-019-45962-x
Schmidt, J. W., Greenough, A., Burns, M., Luteran, A. E., and Mccafferty, D. G. (2010). Generation of ramoplanin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 310, 104–11 doi: 10.1111/j.1574-6968.2010.02051.x
Sharma, D., Mandal, S. M., and Manhas, R. K. (2014). Purification and characterization of a novel lipopeptide from Streptomyces amritsarensis sp. nov. active against methicillin-resistant Staphylococcus aureus. AMB Express. 4, 1–9. doi: 10.1186/s13568-014-0050-y
Shireen, T., Singh, M., Das, T., and Mukhopadhyay, K. (2013). Differential adaptive responses of Staphylococcus aureus to in vitro selection with different antimicrobial peptides. Antimicrob. Agents Chemother. 57, 5134–5137. doi: 10.1128/AAC.00780-13
Singh, P. K., Sharma, S., Kumari, A., and Korpole, S. A. (2014). Non-pediocin low molecular weight antimicrobial peptide produced by Pediococcus pentosaceus strain IE-3 shows increased activity under reducing environment. BMC Microbiol. 14, 1–9. doi: 10.1186/s12866-014-0226-2
Sulaiman, J. E., Long, L., Wu, L., Qian, P. Y., and Lam, H. (2021). Comparative proteomic investigation of multiple methicillin-resistant Staphylococcus aureus strains generated through adaptive laboratory evolution. iScience. 6, 102950. doi: 10.1016/j.isci.2021.102950
Tiwari, K. B., Gatto, C., and Wilkinson, B. J. (2020). Plasticity of coagulase-negative staphylococcal membrane fatty acid composition and implications for responses to antimicrobial agents. Antibiotics. 9, 3–12. doi: 10.3390/antibiotics9050214
Tubby, S., Wilson, M., Wright, J. A., et al. (2013). Staphylococcus aureus small colony variants are susceptible to light activated antimicrobial agents. BMC Microbiol. 13, 201. doi: 10.1186/1471-2180-13-201
Tyurin, A. P., Alferova, V. A., Paramonov, A. S., Shuvalov, M. V., Kudryakova, G. K., Rogozhin, E. A., et al. (2021). Gausemycins A,B: cyclic lipoglycopeptides from Streptomyces sp. Angew Chemie - Int Ed. 60, 18694–18703. doi: 10.1002/anie.202104528
Vasilchenko, A. S., Julian, W. T., Lapchinskaya, O. A., Katrukha, G. S., Sadykova, V. S., Rogozhin, E. A. A., et al. (2020). Novel peptide antibiotic produced by Streptomyces roseoflavus Strain INA-Ac-5812 with directed activity against gram-positive bacteria. Front. Microbiol. 11, 1–13. doi: 10.3389/fmicb.2020.556063
Wakeman, C. A., Hammer, N. D., Stauff, D. L., Attia, A. S., Anzaldi, L. L., Dikalov, S. I., et al. (2012). Menaquinone biosynthesis potentiates haem toxicity in Staphylococcus aureus. Mol. Microbiol. 86, 1376–1392. doi: 10.1111/mmi.12063
Walker, B. J., Abeel, T., Shea, T., Priest, M., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 9, e112963. doi: 10.1371/journal.pone.0112963
Watkins, R. R., David, M. Z., and Salata, R. A. (2012). Richard Watkins rwatkins CR. Current concepts on the virulence mechanisms of meticillin-resistant Staphylococcus aureus. J. Med. Microbiol. 61, 1179–1193 doi: 10.1099/jmm.0.043513-0
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Wilhelm, M. P., and Estes, L. (1999). Symposium on antimicrobial agents—Part XII. vancomycin. Mayo Clin. Proc. 74, 928–935. doi: 10.4065/74.9.928
Zhang, J., Suo, Y., Zhang, D., Jin, F., and Zhao, H. (2018). and Shi C. Genetic and virulent difference between pigmented and non-pigmented Staphylococcus aureus. Front. Microbiol. 9, 598. doi: 10.3389/fmicb.2018.00598
Zhang, Y. M., and Rock, C. O. (2008). Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6, 222–233. doi: 10.1038/nrmicro1839
Keywords: gausemycins, lipoglycopeptides, Streptomyces roseoflavus INA-Ac-5812, antibiotic resistance, Staphylococcus aureus
Citation: Poshvina DV, Dilbaryan DS, Kasyanov SP, Sadykova VS, Lapchinskaya OA, Rogozhin EA and Vasilchenko AS (2022) Staphylococcus aureus is able to generate resistance to novel lipoglycopeptide antibiotic gausemycin A. Front. Microbiol. 13:963979. doi: 10.3389/fmicb.2022.963979
Received: 08 June 2022; Accepted: 14 September 2022;
Published: 29 September 2022.
Edited by:
Mona I. Shaaban, Mansoura Universiy, EgyptReviewed by:
Jordy Evan Sulaiman, University of Wisconsin-Madison, United StatesJhih-Hang Jiang, Monash University, Australia
Aroonlug Lulitanond, Khon Kaen University, Thailand
Copyright © 2022 Poshvina, Dilbaryan, Kasyanov, Sadykova, Lapchinskaya, Rogozhin and Vasilchenko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexey S. Vasilchenko, avasilchenko@gmail.com
 Darya V. Poshvina
Darya V. Poshvina Diana S. Dilbaryan1
Diana S. Dilbaryan1 Vera S. Sadykova
Vera S. Sadykova Eugene A. Rogozhin
Eugene A. Rogozhin Alexey S. Vasilchenko
Alexey S. Vasilchenko