- 1School of Nursing, Johns Hopkins University, Baltimore, MD, United States
- 2National Institute of Nursing Research, National Institutes of Health, Bethesda, MD, United States
- 3School of Nursing, University of Texas Health Science Center San Antonio, San Antonio, TX, United States
- 4Henry M. Jackson Foundation for the Advancement of Military Medicine, Bethesda, MD, United States
- 5Department of Neurosurgery, Medical College of Wisconsin, Milwaukee, WI, United States
- 6Center for Neuroscience and Regenerative Medicine, Uniformed Services University of the Health Sciences, Bethesda, MD, United States
- 7Predictiv Care, Mountain View, CA, United States
- 8Michigan Concussion Center, University of Michigan, Ann Arbor, MI, United States
- 9Department of Psychiatry, Indiana University School of Medicine, Indianapolis, IN, United States
- 10Departments of Pediatrics and Neurosurgery, University of California, Los Angeles, Los Angeles, CA, United States
- 11UCLA Steve Tisch BrainSPORT Program, University of California, Los Angeles, Los Angeles, CA, United States
- 12Department of Epidemiology and Biostatistics School of Public Health-Bloomington, Indiana University, Bloomington, IN, United States
- 13John A. Feagin Sports Medicine Fellowship, Keller Army Hospital, West Point, NY, United States
- 14United States Air Force Academy, Colorado Springs, CO, United States
- 15Matthew Gfeller Center, Department of Exercise and Sport Science, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 16Department of Orthopedics, Division of Sports Medicine, School of Medicine and Public Health, University of Wisconsin-Madison, Madison, WI, United States
- 17Department of Biomedical Engineering and Mechanics, Virginia Tech, Blacksburg, VA, United States
- 18Department of Neurology, Johns Hopkins University, Baltimore, MD, United States
Objective: To investigate the plasma proteomic profiling in identifying biomarkers related to return to sport (RTS) following a sport-related concussion (SRC).
Methods: This multicenter, prospective, case-control study was part of a larger cohort study conducted by the NCAA-DoD Concussion Assessment, Research, and Education (CARE) Consortium, athletes (n = 140) with blood collected within 48 h of injury and reported day to asymptomatic were included in this study, divided into two groups: (1) recovery <14-days (n = 99) and (2) recovery ≥14-days (n = 41). We applied a highly multiplexed proteomic technique that uses DNA aptamers assay to target 1,305 proteins in plasma samples from concussed athletes with <14-days and ≥14-days.
Results: We identified 87 plasma proteins significantly dysregulated (32 upregulated and 55 downregulated) in concussed athletes with recovery ≥14-days relative to recovery <14-days groups. The significantly dysregulated proteins were uploaded to Ingenuity Pathway Analysis (IPA) software for analysis. Pathway analysis showed that significantly dysregulated proteins were associated with STAT3 pathway, regulation of the epithelial mesenchymal transition by growth factors pathway, and acute phase response signaling.
Conclusion: Our data showed the feasibility of large-scale plasma proteomic profiling in concussed athletes with a <14-days and ≥ 14-days recovery. These findings provide a possible understanding of the pathophysiological mechanism in neurobiological recovery. Further study is required to determine whether these proteins can aid clinicians in RTS decisions.
Introduction
Each year, an estimated 8 million collegiate athletes participate in organized sports in the United States (1). The increasing sports-related concussion (SRC) rate has become a major public health concern due to the subtle nature of the injury, which makes diagnosis and return to sport (RTS) decision-making challenging (2). Premature RTS may adversely affect the neurobiological recovery process and increase risk for subsequent SRC, which has been linked to short or long-term neurological consequences (3, 4). Repetitive SRC has been associated with memory and cognitive deficits and risks of chronic traumatic encephalopathy (5, 6). The lack of prognostic biomarkers of recovery poses a significant challenge concerning assessing an athlete's readiness for RTS. Biomarkers are needed to guide clinical decision-making, thereby mitigating risk for exposure to head impacts before resolution of prior SRC. Concerning individuals at risk for prolonged recovery and poor outcomes, prognostic biomarkers as well as objective measures of neurobiological recovery are critically needed to facilitate early risk-stratification and ensure effective clinical management of SRC.
Peripheral blood-based biomarkers can play a vital role in identifying the intrinsic factors influencing SRC symptoms and recovery time. Serum glial fibrillary acidic protein (GFAP) and ubiquitin c-terminal hydrolase L1 (UCH-L1) have been shown to be elevated after a concussion, with higher levels observed in athletes with more severe acute injury characteristics (e.g., loss of consciousness, posttraumatic amnesia) (7). Further, both GFAP and tau have been associated with prolonged recovery (i.e., ≥14-days) after SRC (8). In addition to brain-specific biomarkers, altered inflammatory cytokine levels at acute and subacute timepoints can serve as prognostic markers of the neurobiological recovery. A recent study showed that higher levels of serum cytokines interleukin (IL) 1 receptor antagonist, and IL-6 within 6 h of injury were associated with greater symptom duration in concussed athletes (9). At the subacute timepoint, plasma monocyte chemoattractant protein-1 and 4 proteins were associated with recovery in SRC (10). Although these studies provide evidence that protein changes relate to SRC occurrence and recovery time, they are limited by a hypothesis-based approach that limits the identification of unique biomarkers changes that might provide a more in-depth characterization of relationships.
To date, biomarker studies related to RTS decisions have relied upon a hypothesis driven-approach, screening for markers with known involvement with traumatic brain injury (TBI) pathophysiology. Therefore, we implemented a proteomic approach to discover the candidate biomarkers for neurobiological recovery at 14 days or more after SRC occurrence. In addition, our study was to examine the protein networks and potential pathways involving neurobiological recovery. A 14-day cutoff was based on NCAA guidelines for management of SRC (11). We used the SOMAscan assay from SomaLogic to measure more than a thousand proteins in a scalable manner in a single specimen, which greatly accelerates biomarker discovery (12).
Materials and Methods
Participants and Clinical Data
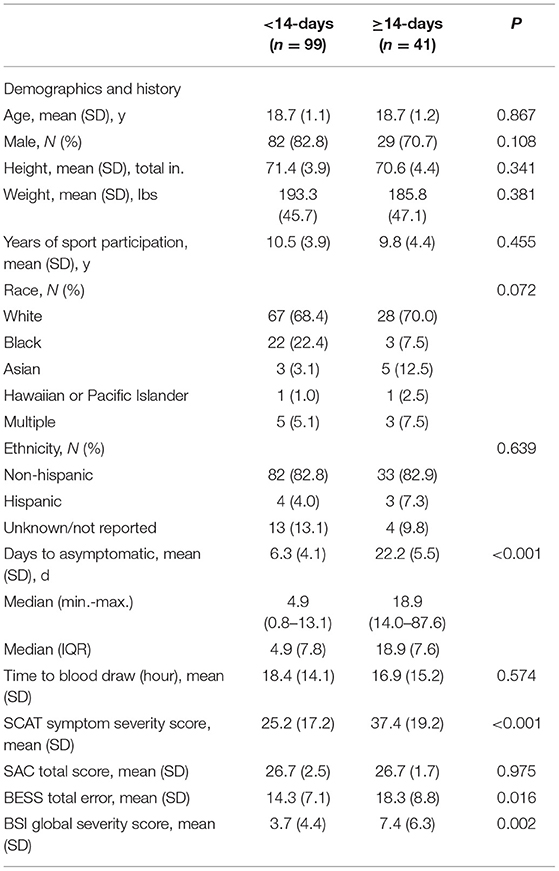
A total of 140 concussed athletes include in this study were part of a larger cohort enrolled in the NCAA–Department of Defense Concussion Assessment, Research and Education (CARE) Consortium. The CARE Consortium was approved by the Medical College of Wisconsin Institutional Review Board and the Human Research Protection Office at the US Army Medical Research and Material Command and written informed consent was obtained for all participants. Concussion was defined according to the consensus definition from the US Department of Defense evidence based-guidelines (13). After an athlete was diagnosed with concussion, a single blood plasma sample was collected within 48 h, and a clinical battery consisting of the Sport Concussion Assessment Tool-Third Edition (SCAT-3), the Standardized Assessment of Concussion (SAC), the Balance Error Scoring System (BESS), and the Brief Symptom Inventory 18 (BSI-18). Concussed athletes were divided into two groups on the basis of the number of days following SRC that asymptomatic status was reported: (1) recovery <14-days, n = 99 and (2) recovery ≥14-days, n = 44. The group were matched by age, sex, height, weight, race, ethnicity, and years of participation.
Blood Sampling Processing
A single timepoint of non-fasting blood samples were collected by venipuncture from athletes within 48 h following injury. Blood was collected in 10 ml plastic dipotassium ethylenediaminetetraacetic acid (EDTA) tubes, immediately placed on ice, centrifuged (15 min, 1,500 g, room temperature), and frozen (−27°C) in aliquots within 60 min of sample collection. Samples were shipped on dry ice to CARE Consortium biorepository at Indiana University School of Medicine and were stored at −80°C until analysis.
SOMAscan Assay
Proteomic analysis was performed in plasma samples using the SOMAscan assay (SomaLogic Inc.), an aptamer-based assay that allows for the simultaneous measurement and quantification of a large number of proteins (1,305 proteins based on the version used here). The assay uses chemically modified nucleotides to transform a protein signal into a nucleotide signal quantified using relative fluorescence on microarrays (14, 15). Sample identifications were randomized in the SOMAscan plate prior to loading to minimize the potential for artifacts due to specific loci in a microarray and following the recommended sample preparation protocols. Samples were adequately stored in −80°C freezer before use, thawed, and then loaded into the barcoded vials within the plate using pipette techniques and 55 μl aliquots, keeping all samples and the plate on ice (4°C) throughout the process. The plate was analyzed by SomaLogic, Inc. and the standardization results were produced as relative fluorescent units (RFUs). Standardized results consisted of the following steps: hybridization control normalization, median signal normalization, and calibration normalization. Sample data were first normalized to remove hybridization variation within a run. Median signal normalization was performed across calibrator samples to remove other assay biases within the run. Overall scaling was then performed on a per-plate basis to remove overall intensity differences between runs. Calibration was then performed to remove assay differences between runs.
Statistical Analysis
Data were analyzed using SPSS statistical software (Armonk, NY: IBM Corp.) and GraphPad prism 8.4 was used to generate graphs (La Jolla, CA: GraphPad Software). Descriptive statistics were used to describe the demographic and clinical characteristics of the participants. Chi-square (χ2) test and Mann–Whitney U-test were performed to determine the group differences. The log-fold changes were calculated by using an interactive Shiny web tool framework developed by the National Institutes of Health's Center for Human Immunology, Autoimmunity, and Inflammation (CHI) facility (16). Adjusted p-values were calculated by using Benjamini–Hochberg's false discovery rate (FDR). Dysregulated proteins were uploaded into the Ingenuity Pathway Analysis (IPA) software (Qiagen IPA) to explore the mechanistic networks most significantly associated with the study outcome. The significance level was set at 0.05 in all tests.
Results
The demographic and clinical characteristics for concussed athletes with recovery <14-days and ≥14-days are shown in Table 1. The recovery group ≥14-days and <14-days group were between 17 and 23 years of age. The groups did not significantly differ on age, sex, race, and ethnicity. The median number of days to attainment of asymptomatic status for the <14-days group was 4.9 days and 18.9 days for ≥14-days group. Concussed athletes with recovery ≥14-days had significantly higher SCAT symptom severity scores, BESS total scores, and BSI-18 global severity scores compared with the recovery <14-days group.
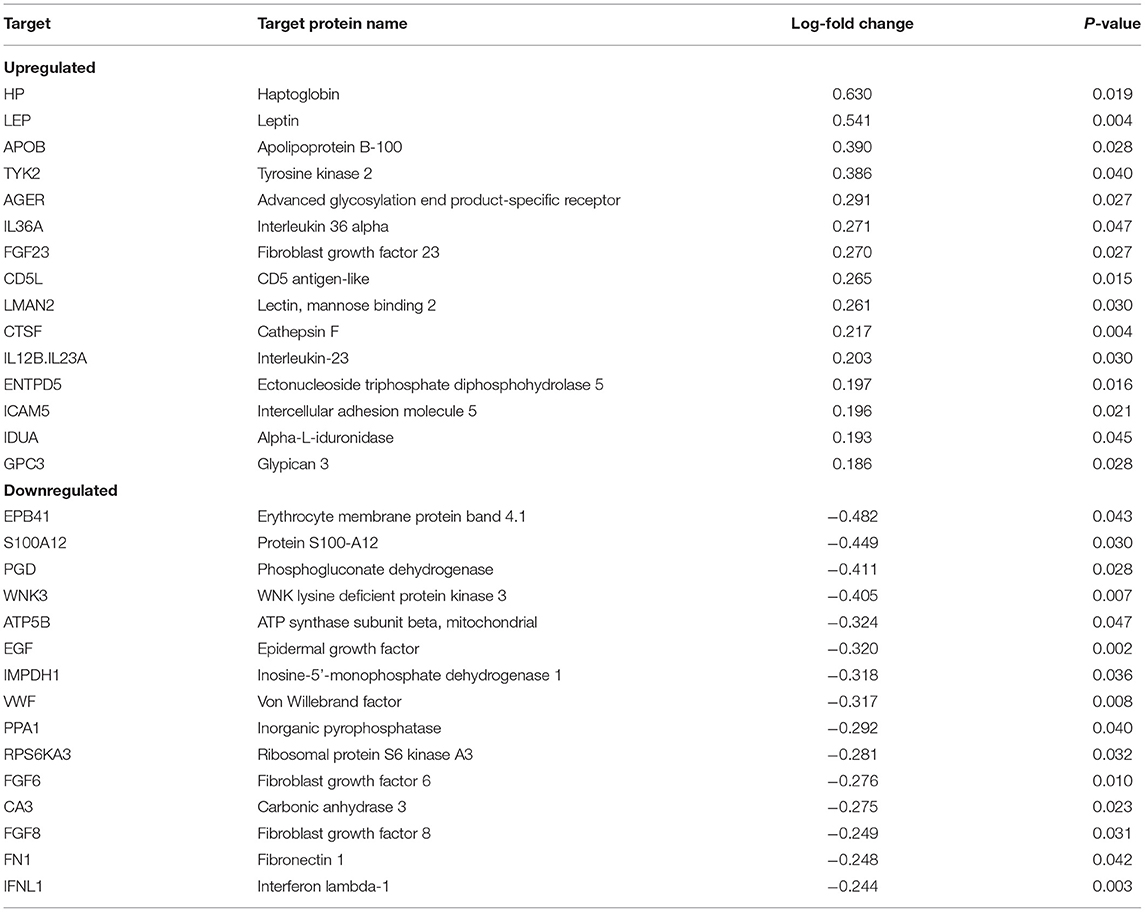
We found 87 proteins that were significantly dysregulated in recovery ≥14-days relative to recovery <14-days. The top proteins upregulated in recovery ≥14-days were haptoglobin (HP), leptin (LEP), apolipoprotein B-100 (APOB), tyrosine kinase 2 (TYK2), advanced glycosylation end product-specific receptor (AGER), and IL36A. Top proteins that were downregulated in recovery ≥14-days were erythrocyte membrane protein band 4.1 (EPB41), protein S100-A12 (S100A12), WNK lysine deficient protein kinase 3 (WNK3), ATP synthase subunit beta, mitochondrial (ATP5B), and epidermal growth factor (EGF) (Table 2). All proteins that were significantly different in concussed athletes with recovery <14-days relative to recovery ≥14-days are listed in the Supplementary Table 1.
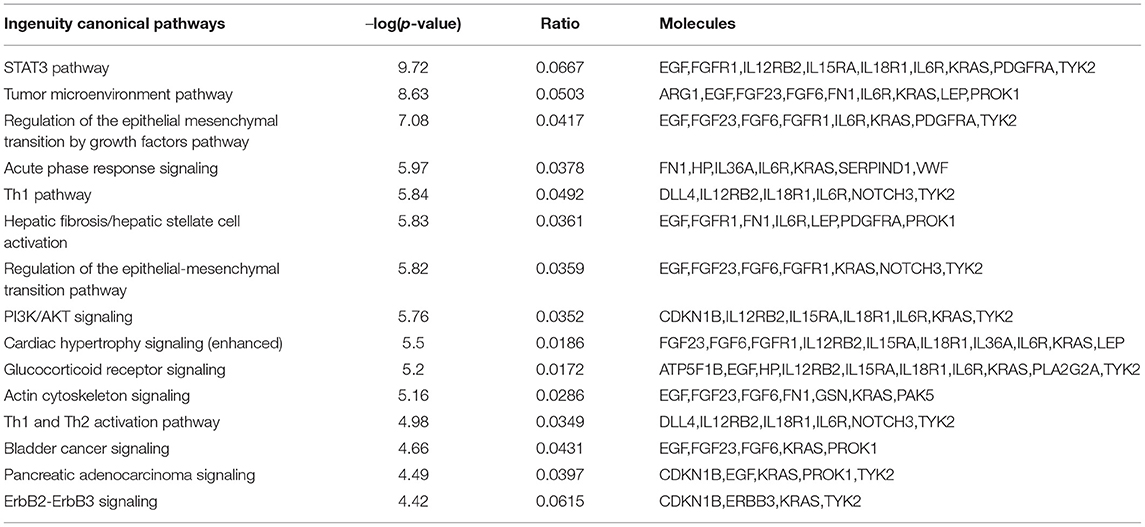
The 30 proteins that were most significantly dysregulated were uploaded into IPA software for core expression analysis. The pathway analysis revealed the molecular mechanisms underlying the prolonged recovery in SRC. The top enrichment of the canonical pathway and the associated proteins in each pathway are presented in Table 3. Top canonical pathways associated with recovery were Signal Transducer and Activator of Transcription 3 (STAT3) Pathway, Tumor Microenvironment Pathway, Regulation of the Epithelial Mesenchymal Transition by Growth Factors Pathway, and Acute Phase Response Signaling. Top IPA mechanistic network analysis revealed the protein-protein interaction in the network associated with Hepatic System Development and Function, Cellular Movement, and Organismal Injury and Abnormalities (Figure 1).
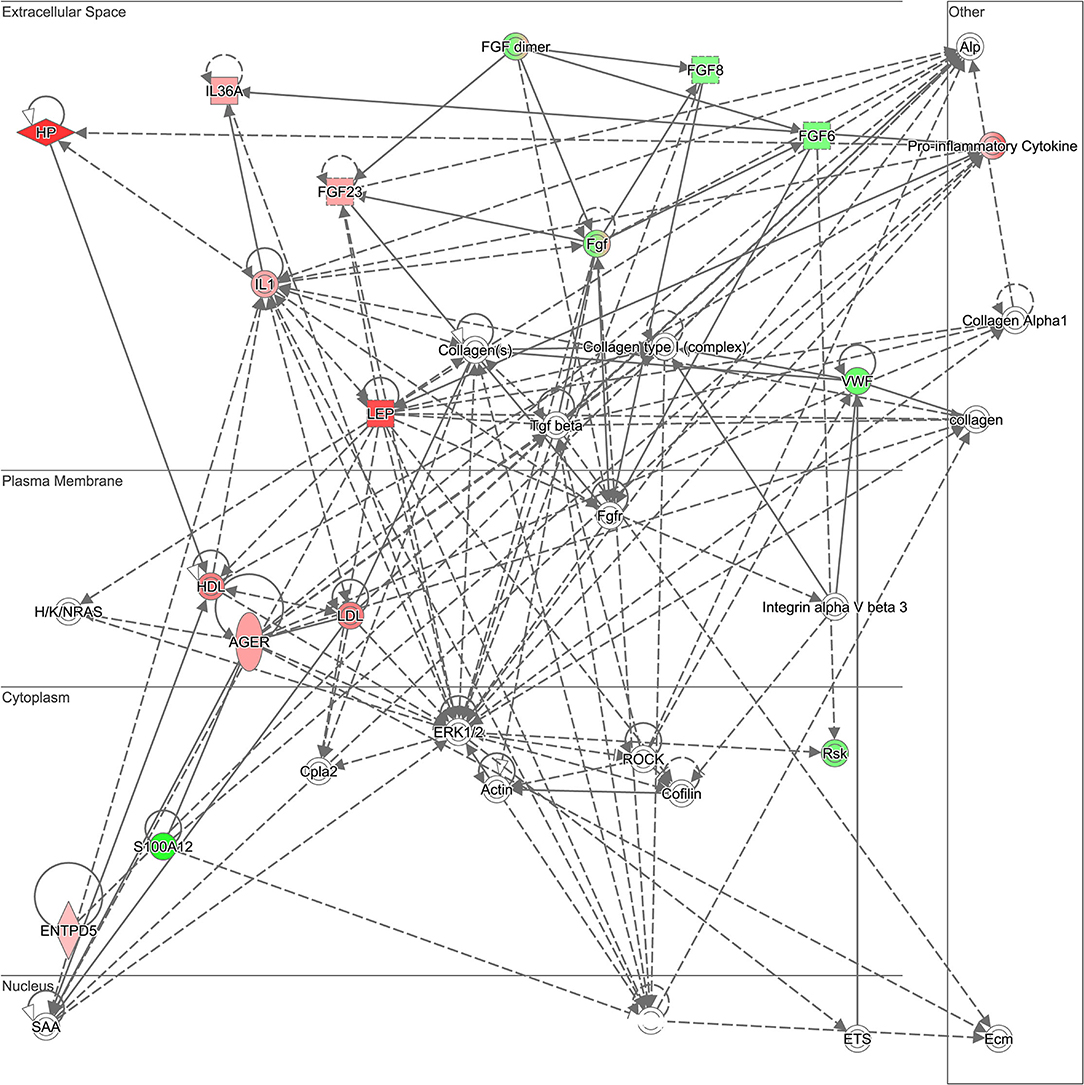
Figure 1. Ingenuity Pathway Analysis (IPA) mechanistic network. Top Ingenuity Pathway Analysis (IPA) mechanistic network analysis revealed the protein-protein interaction in the network associated with Hepatic System Development and Function, Cellular Movement, and Organismal Injury and Abnormality. Green indicates that the protein is downregulated and red indicates that the protein is upregulated, with increased color saturation representing more extreme measurement in the dataset. Proteins in gray indicate the direct relationship with differential expression proteins in the network. Solid lines indicate direct interaction and dashed lines indicated indirect interactions.
Discussion
The RTS decision is important for the safety of collegiate athletes to avoid the short- and long-term impact of repetitive concussions. Here, using a proteomic approach, we were able to identify several dysregulated plasma protein biomarkers in concussed athletes with recovery ≥14-days relative to concussed athletes with recovery <14-days. When evaluated within 48 h of injury, the SCAT-3, BESS, and BSI-18 scores were significantly higher in ≥14-days compared with <14-days. These findings provide a better understanding of pathophysiological mechanisms of sports injuries for exploring potential prognostic and recovery markers related to SRC.
Beyond the clinical implications of the findings, they also advance understanding of the pathophysiological mechanism in neurobiological recovery process following concussion. Pathway analysis showed that the most significant proteins were associated with STAT3 pathway. Signal transducer and activator of transcription (STAT3) is a member of the STAT family and deregulated STAT3 pathway may contribute to the proinflammatory response in SRC (17, 18). The proteins represented in this pathway are listed in Table 3. In addition, the proteins involved in the mechanistic network include LEP, HP, VWF, AGER, S100A4, fibroblast growth factor 6 (FGF6), and FGF23. In particular, LEP is a 16 kDa protein secreted predominantly by white adipose tissue which regulates energy homeostasis, neuroendocrine function, metabolism, and inflammatory response (19). In the context of brain injury, LEP and its receptor protein may be expressed from many brain regions, which may be involved in regulation of the central nervous system (CNS) (20). However, another plausible explanation is that LEP is released from adipocytes which may cross the blood brain barrier (BBB) to regulate many functions in the CNS such as synaptic plasticity and spatial learning and memory (21). The elevation of LEP levels in the prolonged recovery group is consistent with the findings of previous studies related to neurodegeneration. An observational study has identified an association between LEP level with cognitive decline in healthy older age population (22). Furthermore, cerebrospinal fluid LEP was higher in AD population compared with healthy controls (23). Conversely, not all studies support these findings, in a 15 years longitudinal study showed that higher levels of LEP proteins are associated with reduced incidence of dementia and Alzheimer's disease (AD) (24). Also, higher serum LEP levels at baseline are protective against cognitive decline after 5 years of follow-up in the healthy older population (25). As such, LEP protein may act as a neuroprotective role in the CNS system. In support of this, in vitro study suggested that LEP reduced tau phosphorylation levels (26), which is a hallmark of neurodegenerative diseases including AD and chronic traumatic encephalopathy through the aggregation of neurofibrillary tangles (27–29). A preclinical study of mice showed that LEP has a positive effect in neuronal protection against ischemic brain injury through extracellular signal-related kinase (ERK) 1/2 signaling pathway (30). LEP may have a therapeutic effect following spinal cord injury by enhancing the expression of neuroprotective and anti-inflammatory genes (31). Further study is needed to investigate the effect of LEP protein as a prognostic marker for concussion.
Disruption of the BBB following SRC may lead to increased release of proteins into peripheral circulation and recruitment of immune cells or proteins to the injury site (32). We identified several vascular injury markers such as vascular endothelial growth factor C (VEGFC), VWF, Platelet derived growth factor receptor alpha (PDGFRA), and FN1 proteins that may be involved in BBB damaged following concussion. In particular, VEGF plays an important role in angiogenesis and neuroprotection (33, 34), and is mainly expressed from endothelial cells to preserves intact BBB. Increased peripheral circulating levels of VEGF have been linked to alterations in BBB permeability following a TBI and infiltration of immune cells (35, 36). Our lab has recently demonstrated that VEGF protein was dysregulated in concussed athletes and associated with neuroimage findings in mild TBI patients (37). VWF is an acute phase protein response to vascular injury and has been reported in several diseases such as acute coronary syndrome (38), cancer (39), and TBI (40). Elevated plasma VWF was correlated with unfavorable outcomes in severe TBI patients (41). However, we found inconsistent results of plasma VWF proteins to previous studies. We reported lower VWF protein levels in the recovery ≥14-days group compared with recovery <14-days group. The difference in these findings may be attributed to different demographic population of the study, injury type (i.e., sports injury) and the type of protein assay used to quantify the data. Increased plasma FN1 proteins predominantly reflect vascular injury after a concussion, which has an important role in the repair of tissue damage and protection against neuron cell death by focal ischemia (42). A preclinical study has shown that TBI model with FN deficiency group had the worst motor and cognitive ability (43). Our data indicate that dysregulation of these protein expressions in concussed athletes with recovery ≥14-days may affect neuronal damage and delay neurobiological recovery after concussion.
Moreover, AGER is a multiligand receptor and a member of the immunoglobin superfamily which plays a major role in inflammation response in binding of its ligands, including amyloid beta, damage-associated molecule patterns, S100 proteins, and high-mobility group box-1 protein (44, 45). Elevated AGER concentration in our data may reflect the severity of injury which caused the prolonged recovery. Another plausible explanation is that imbalanced production of pro-inflammatory and neuroprotective protein that may cause secondary injury and lead to a worsened pathophysiological response following sport concussion. In support of this, our data have shown that S100A4 protein was significantly downregulated in concussed athletes with recovery ≥14-days. The effect of S100A4 protein expression following TBI in protecting against neuron cell death has been shown in the preclinical model (46). Downregulation of S100A4 protein expression may prolong the neurological recovery process. Also, we reported a higher expression of FGF23 in recovery ≥14-days group. FGF23 is a circulating hormone that regulates phosphate homeostasis and vitamin D metabolism. Elevated serum FGF23 concentration is a potential prognostic marker of chronic kidney disease (47), and is associated with dementia and AD (48). Moreover, elevated plasma FGF23 concentration has been implicated as a potential risk factor for stroke and intracerebral hemorrhage (49). Upregulation of FGF23 may serve as a prognostic marker of prolonged recovery.
This study has several limitations. First, our results may not be applicable to other populations (e.g., non-athletes, younger or older athletes). Our study participants were well matched between the group, although they had a relatively small number of athletes who sustained SRC with the prolonged recovery group. Due to the expense of the SOMAscan analysis, blood from a single time point was analyzed. Future studies should conduct this prognostic analysis as part of a longitudinal study with a larger cohort. Injury mechanisms may vary widely across the injury cohort and could plausibly impact the brain neurophysiology recovery as a result. Finally, these proteins were not significant after multiple corrections and the results should be interpreted cautiously.
Although this study has several limitations, the large-scale proteomic analysis strengthens our results. Our data represent the first exploratory biomarkers derived from a large-scale proteomic analysis to determine the neurobiological recovery after concussion. In addition, our findings indicate the feasibility and utility of proteomic profiling in identifying the blood-based biomarkers for neurobiological recovery following concussion. Our data provide a better understanding of physiological mechanisms associated with concussion recovery. Further research is required to determine the eventual clinical utility of these candidate proteins.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Medical College of Wisconsin Institutional Review Board and the Human Research Protection Office at the US Army Medical Research. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
RV, MM, CG, KC, PP, SB, JH, KG, TMc, and JG: conceptualization. RV, JG, CL, and H-SK: methodology. RV: software and visualization. RV, JG, CL, H-SK, SY, and CD: formal analysis. RV, CL, JG, MM, SB, and CG: investigation. JG, MM, CG, and PP: resources. RV and CD: data curation. RV and JG: writing—original draft preparation. RV, SM, CD, TMe, CL, SY, SB, TMc, CG, H-SK, DH, JH, KC, GM, JJ, KG, JM, AB, SD, SR, LN, PP, MM, and JG: writing—review and editing. JG: supervision. JG, SB, TMc, CG, DH, KG, JM, LN, and MM: project administration. JG, MM, SB, TMc, JM, and PP: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Grand Alliance Concussion Assessment, Research, and Education (CARE) Consortium and funded in parts by the National Collegiate Athletic Association (NCAA) and the Department of Defense (DOD). The U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick MD 21702-5014 is the awarding and administering acquisition office. This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Combat Casualty Care Research Program, endorsed by the Department of Defense, under Award No. W81XWH1420151. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the DOD.
Conflict of Interest
SY was employed by Predictiv Care, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would also like to thank the research and medical staff at each of the CARE participation sites.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2022.901238/full#supplementary-material
References
1. Pierpoint LA, Collins C. Epidemiology of sport-related concussion. Clin Sports Med. (2021) 40:1–18. doi: 10.1016/j.csm.2020.08.013
2. Langlois JA, Rutland-Brown W, Wald MM. The epidemiology and impact of traumatic brain injury: a brief overview. J Head Trauma Rehabil. (2006) 21:375–8. doi: 10.1097/00001199-200609000-00001
3. Manley G, Gardner AJ, Schneider KJ, Guskiewicz KM, Bailes J, Cantu RC, et al. A systematic review of potential long-term effects of sport-related concussion. Br J Sports Med. (2017) 51:969–77. doi: 10.1136/bjsports-2017-097791
4. Guskiewicz KM, Marshall SW, Bailes J, Mccrea M, Harding HP, Matthews A, et al. Recurrent concussion and risk of depression in retired professional football players. Med Sci Sports Exerc. (2007) 39:903–9. doi: 10.1249/mss.0b013e3180383da5
5. McKee AC, Cantu RC, Nowinski CJ, Hedley-Whyte ET, Gavett BE, Budson AE, et al. Chronic traumatic encephalopathy in athletes: progressive tauopathy after repetitive head injury. J Neuropathol Exp Neurol. (2009) 68:709–35. doi: 10.1097/NEN.0b013e3181a9d503
6. Veksler R, Vazana U, Serlin Y, Prager O, Ofer J, Shemen N, et al. Slow blood-to-brain transport underlies enduring barrier dysfunction in American football players. Brain. (2020) 143:1826–42. doi: 10.1093/brain/awaa140
7. McCrea M, Broglio SP, McAllister TW, Gill J, Giza CC, Huber DL, et al. Association of blood biomarkers with acute sport-related concussion in collegiate athletes: findings from the NCAA and Department of Defense CARE Consortium. JAMA Netw Open. (2020) 3:e1919771. doi: 10.1001/jamanetworkopen.2019.19771
8. Pattinson CL, Meier TB, Guedes VA, Lai C, Devoto C, Haight T, et al. Plasma biomarker concentrations associated with return to sport following sport-related concussion in collegiate athletes-a concussion assessment, research, and education (care) consortium study. JAMA Netw Open. (2020) 3:e2013191. doi: 10.1001/jamanetworkopen.2020.13191
9. Meier TB, Huber DL, Bohorquez-Montoya L, Nitta ME, Savitz J, Teague TK, et al. A prospective study of acute blood-based biomarkers for sport-related concussion. Ann Neurol. (2020) 87:907–20. doi: 10.1002/ana.25725
10. Di Battista AP, Churchill N, Rhind SG, Richards D, Hutchison MG. Evidence of a distinct peripheral inflammatory profile in sport-related concussion. J Neuroinflammation. (2019) 16:17. doi: 10.1186/s12974-019-1402-y
11. NCAA Sport Science Institute,. Diagnosis Management of Sport-Related Concussion Best Practices. (2016). Available online at: https://www.ncaa.org/sites/default/files/SSI_ConcussionBestPractices_20170616.pdf
12. Mosley JD, Benson MD, Smith JG, Melander O, Ngo D, Shaffer CM, et al. Probing the virtual proteome to identify novel disease biomarkers. Circulation. (2018) 138:2469–81. doi: 10.1161/CIRCULATIONAHA.118.036063
13. Carney N, Ghajar J, Jagoda A, Bedrick S, Davis-O'Reilly C, Du Coudray H, et al. Concussion guidelines step 1: systematic review of prevalent indicators. Neurosurgery. (2014) 75:S3–S15. doi: 10.1227/NEU.0000000000000433
14. Mehan MR, Ostroff R, Wilcox SK, Steele F, Schneider D, Jarvis TC, et al. Highly multiplexed proteomic platform for biomarker discovery, diagnostics, and therapeutics. In: Lambris J, Holers V, Ricklin D, editors. Complement Therapeutics. Advances in Experimental Medicine and Biology. New York, NY: Springer (2013). p. 283–300. doi: 10.1007/978-1-4614-4118-2_20
15. Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. (2010) 5:e15004. doi: 10.1371/journal.pone.0015004
16. Cheung F, Fantoni G, Conner M, Sellers BA, Kotliarov Y, Candia J, et al. Web tool for navigating and plotting somalogic ADAT files. J Open Res Softw. (2017) 5:20. doi: 10.5334/jors.166
17. Oliva AA, Kang Y, Sanchez-Molano J, Furones C, Atkins CM. STAT3 signaling after traumatic brain injury. J Neurochem. (2012) 120:710–720. doi: 10.1111/j.1471-4159.2011.07610.x
18. Ding Y, Qian J, Li H, Shen H, Li X, Kong Y, et al. Effects of SC99 on cerebral ischemia-perfusion injury in rats: selective modulation of microglia polarization to M2 phenotype via inhibiting JAK2-STAT3 pathway. Neurosci Res. (2019) 142:58–68. doi: 10.1016/j.neures.2018.05.002
19. Kelesidis T, Kelesidis I, Chou S, Mantzoros CS. Narrative review: The role of leptin in human physiology: emerging clinical applications. Ann Intern Med. (2010) 152:93–100. doi: 10.7326/0003-4819-152-2-201001190-00008
20. Shioda S, Funahashi H, Nakajo S, Yada T, Maruta O, Nakai Y. Immunohistochemical localization of leptin receptor in the rat brain. Neurosci Lett. (1998) 243:41–4. doi: 10.1016/S0304-3940(98)00082-2
21. Harvey J. Leptin and Alzheimer's disease. In: Martin CR, Preedy VR, editors. San Diego, CA: Academic Press (2015). p. 457–67. doi: 10.1016/B978-0-12-407824-6.00041-0
22. Gunstad J, Spitznagel MB, Keary TA, Glickman E, Alexander T, Karrer J, et al. Serum leptin levels are associated with cognitive function in older adults. Brain Res. (2008) 1230:233–6. doi: 10.1016/j.brainres.2008.07.045
23. Bonda DJ, Stone JG, Torres SL, Siedlak SL, Perry G, Kryscio R, et al. Dysregulation of leptin signaling in Alzheimer disease: evidence for neuronal leptin resistance. J Neurochem. (2014) 128:162–72. doi: 10.1111/jnc.12380
24. Lieb W, Beiser AS, Vasan RS, Tan ZS, Au R, Harris TB, et al. Association of plasma leptin levels with incident Alzheimer disease and MRI measures of brain aging. JAMA. (2009) 302:2565–72. doi: 10.1001/jama.2009.1836
25. Holden KF, Lindquist K, Tylavsky FA, Rosano C, Harris TB, Yaffe K. Serum leptin level and cognition in the elderly: findings from the Health ABC Study. Neurobiol Aging. (2009) 30:1483–9. doi: 10.1016/j.neurobiolaging.2007.11.024
26. Greco SJ, Sarkar S, Johnston JM, Zhu X, Su B, Casadesus G, et al. Leptin reduces Alzheimer's disease-related tau phosphorylation in neuronal cells. Biochem Biophys Res Commun. (2008) 376:536–41. doi: 10.1016/j.bbrc.2008.09.026
27. Janelidze S, Mattsson N, Palmqvist S, Smith R, Beach TG, Serrano GE, et al. Plasma P-tau181 in Alzheimer's disease: relationship to other biomarkers, differential diagnosis, neuropathology and longitudinal progression to Alzheimer's dementia. Nat Med. (2020) 26:379–86. doi: 10.1038/s41591-020-0755-1
28. Katsumoto A, Takeuchi H, Tanaka F. Tau pathology in chronic traumatic encephalopathy and alzheimer's disease: similarities and differences. Front Neurol. (2019) 10:980. doi: 10.3389/fneur.2019.00980
29. Rajmohan R, Reddy PH. Amyloid-beta and phosphorylated tau accumulations cause abnormalities at synapses of Alzheimer's disease neurons. J Alzheimers Dis. (2017) 57:975–99. doi: 10.3233/JAD-160612
30. Zhang F, Wang S, Signore AP, Chen J. Neuroprotective effects of leptin against ischemic injury induced by oxygen-glucose deprivation and transient cerebral ischemia. Stroke. (2007) 38:2329–36. doi: 10.1161/STROKEAHA.107.482786
31. Fernández-Martos CM, González P, Rodriguez FJ. Acute leptin treatment enhances functional recovery after spinal cord injury. PLoS ONE. (2012) 7:e35594. doi: 10.1371/journal.pone.0035594
32. Wu Y, Wu H, Guo X, Pluimer B, Zhao Z. Blood–brain barrier dysfunction in mild traumatic brain injury: evidence from preclinical murine models. Front Physiol. (2020) 11:1030. doi: 10.3389/fphys.2020.01030
33. Zacchigna S, Lambrechts D, Carmeliet P. Neurovascular signalling defects in neurodegeneration. Nat Rev Neurosci. (2008) 9:169–81. doi: 10.1038/nrn2336
34. G. Argandona E, Bengoetxea H, Ortuzar N, Bulnes S, Rico-Barrio I, Vicente Lafuente J. Vascular endothelial growth factor: adaptive changes in the neuroglialvascular unit. Curr Neurovasc Res. (2012) 9:72–81. doi: 10.2174/156720212799297119
35. Sulhan S, Lyon KA, Shapiro LA, Huang JH. Neuroinflammation and blood–brain barrier disruption following traumatic brain injury: pathophysiology and potential therapeutic targets. J Neurosci Res. (2020) 98:19–28. doi: 10.1002/jnr.24331
36. Nasr IW, Chun Y, Kannan S. Neuroimmune responses in the developing brain following traumatic brain injury. Exp Neurol. (2019) 320:112957. doi: 10.1016/j.expneurol.2019.112957
37. Edwards KA, Pattinson CL, Guedes VA, Peyer J, Moore C, Davis T, et al. Inflammatory cytokines associate with neuroimaging after acute mild traumatic brain injury. Front Neurol. (2020) 11:348. doi: 10.3389/fneur.2020.00348
38. Spiel AO, Gilbert JC, Jilma B. Von Willebrand factor in cardiovascular disease: focus on acute coronary syndromes. Circulation. (2008) 117:1449–59. doi: 10.1161/CIRCULATIONAHA.107.722827
39. Wang WS, Lin JK, Lin TC, Chiou TJ, Liu JH, Yen CC, et al. Plasma von Willebrand factor level as a prognostic indicator of patients with metastatic colorectal carcinoma. World J Gastroenterol. (2005) 11:2166–70. doi: 10.3748/wjg.v11.i14.2166
40. Sandsmark DK, Bogoslovsky T, Qu BX, Haber M, Cota MR, Davis C, et al. Changes in plasma von willebrand factor and cellular fibronectin in MRI-defined traumatic microvascular injury. Front Neurol. (2019) 10:246. doi: 10.3389/fneur.2019.00246
41. De Oliveira CO, Reimer AG, Da Rocha AB, Grivicich I, Schneider RF, Roisenberg I, et al. Plasma von Willebrand factor levels correlate with clinical outcome of severe traumatic brain injury. J Neurotrauma. (2007) 24:1331–8. doi: 10.1089/neu.2006.0159
42. Sakai T, Johnson KJ, Murozono M, Sakai K, Magnuson MA, Wieloch T, et al. Plasma fibronectin supports neuronal survival and reduces brain injury following transient focal cerebral ischemia but is not essential for skin-wound healing and hemostasis. Nat Med. (2001) 7:324–30. doi: 10.1038/85471
43. Tate CC, García AJ, LaPlaca MC. Plasma fibronectin is neuroprotective following traumatic brain injury. Exp Neurol. (2007) 207:13–22. doi: 10.1016/j.expneurol.2007.05.008
44. Neeper M, Schmidt AM, Brett J, Shi Du Yan, Wang F, Pan YCE, et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. (1992) 267:14998–5004. doi: 10.1016/s0021-9258(18)42138-2
45. Yan SF, Ramasamy R, Schmidt AM. Mechanisms of Disease: Advanced glycation end-products and their receptor in inflammation and diabetes complications. Nat Clin Pract Endocrinol Metab. (2008) 4:285–93. doi: 10.1038/ncpendmet0786
46. Dmytriyeva O, Pankratova S, Owczarek S, Sonn K, Soroka V, Ridley CM, et al. v Nat Commun. (2012) 3:1197. doi: 10.1038/ncomms2202
47. Gutiérrez OM, Mannstadt M, Isakova T, Rauh-Hain JA, Tamez H, Shah A, et al. Fibroblast growth factor 23 and mortality among patients undergoing hemodialysis. N Engl J Med. (2008) 359:584–92. doi: 10.1056/nejmoa0706130
48. McGrath ER, Himali JJ, Levy D, Conner SC, Pase MP, Abraham CR, et al. Circulating fibroblast growth factor 23 levels and incident dementia: the Framingham heart study. PLoS ONE. (2019) 14:e0213321. doi: 10.1371/journal.pone.0213321
Keywords: sport injuries, concussion, return to sport (RTS), biomarker, proteomic
Citation: Vorn R, Mithani S, Devoto C, Meier TB, Lai C, Yun S, Broglio SP, McAllister TW, Giza CC, Kim H-S, Huber D, Harezlak J, Cameron KL, McGinty G, Jackson J, Guskiewicz KM, Mihalik JP, Brooks A, Duma S, Rowson S, Nelson LD, Pasquina P, McCrea MA and Gill JM (2022) Proteomic Profiling of Plasma Biomarkers Associated With Return to Sport Following Concussion: Findings From the NCAA and Department of Defense CARE Consortium. Front. Neurol. 13:901238. doi: 10.3389/fneur.2022.901238
Received: 21 March 2022; Accepted: 08 June 2022;
Published: 19 July 2022.
Edited by:
Ryusuke Takechi, Curtin University, AustraliaReviewed by:
Gary B. Wilkerson, University of Tennessee at Chattanooga, United StatesAndrew P. Lavender, Federation University Australia, Australia
Copyright © 2022 Vorn, Mithani, Devoto, Meier, Lai, Yun, Broglio, McAllister, Giza, Kim, Huber, Harezlak, Cameron, McGinty, Jackson, Guskiewicz, Mihalik, Brooks, Duma, Rowson, Nelson, Pasquina, McCrea and Gill. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jessica M. Gill, jessicagill@jhu.edu
†These authors share last authorship
 Rany Vorn
Rany Vorn Sara Mithani
Sara Mithani Christina Devoto
Christina Devoto Timothy B. Meier
Timothy B. Meier Chen Lai2,6
Chen Lai2,6 Sijung Yun
Sijung Yun Steven P. Broglio
Steven P. Broglio Christopher C. Giza
Christopher C. Giza Hyung-Suk Kim
Hyung-Suk Kim Daniel Huber
Daniel Huber Jaroslaw Harezlak
Jaroslaw Harezlak Kenneth L. Cameron
Kenneth L. Cameron Gerald McGinty
Gerald McGinty Kevin M. Guskiewicz
Kevin M. Guskiewicz Jason P. Mihalik
Jason P. Mihalik Lindsay D. Nelson
Lindsay D. Nelson Michael A. McCrea
Michael A. McCrea Jessica M. Gill
Jessica M. Gill