- 1Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
- 2College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China
The development of central nervous system (CNS) and enteric nervous system (ENS) is under precise and strict control in vertebrates. Whether and how the Polycomb repressive complex 1 (PRC1) is involved in it remain unclear. To investigate the role of PRC1 in the nervous system development, using CRISPR/Cas9 technology, we have generated mutant zebrafish lines for the rnf2 gene which encodes Ring1b, the enzymatic component of the PRC1 complex. We show that rnf2 loss of function leads to abnormal migration and differentiation of neural crest and neural precursor cells. rnf2 mutant embryos exhibit aganglionosis, in which the hindgut is devoid of neurons. In particular, the formation of 5-HT serotonin neurons and myelinating glial cells is defective. Furthermore, ectopic expression of ENS marker genes is observed in forebrain of rnf2 mutant embryos. These findings suggest that the rnf2 gene plays an important role in the migration and differentiation of neural precursor cells, and its absence leads to abnormal development of ENS and CNS in zebrafish.
Introduction
During neural development, the migration and differentiation of progenitor cells must be precisely regulated to ensure that various neuronal cell types are generated in a tightly controlled manner. Enteric nervous system (ENS) is completely derived from neural crest cells (NCs), a transitional cell group with multiple differentiation potential (Nagy and Goldstein, 2017). The NCs can also differentiate into craniofacial tissues, pigment cells, neurons, and glial cells from sensory ganglion, and sympathetic ganglion (Rogers et al., 2012). In zebrafish, the ENS is derived from part of the vagal crest cells (Elworthy et al., 2005; Kenneth and Michael, 2005; Olden et al., 2008; Nagy and Goldstein, 2017; Julia, 2018). Neural crest stem cells (NCSCs) form between the non-neural ectoderm and the edge of the neural plate under the induction of signaling molecules (Avantaggiato et al., 1994). After the specification, they leave the neural crest and migrate, and a portion of them finally colonize the intestine. Once they reach the gut, these cells can be named as enteric NCSCs, also called the ENS progenitor cells, and are capable of differentiating into the neurons of the ENS (Jain et al., 2004). The ENS contains more than 1 million neurons, which can be divided into at least 18 functional subtypes, including four main neuron types: motor neurons, intrinsic primary afferent neurons, enteric neurons, and interneurons (Furness, 2000; Simon, 2001; Schemann, 2005).
Previous studies have shown that the migration and differentiation of NCSCs are under control of signal pathways and transcription factors, disruption of which may lead to abnormal development of ENS. For instance, in mice and birds, Sox10, Foxd3, Phox2b, Pax3, and other transcription factors have been shown to play key roles in the development of ENS (Jhiang et al., 1996; Hansford and Mulligan, 2000; Shepherd et al., 2004; Kwon et al., 2011). Patients with PHOX2B mutation show deficiency of ganglion cells (Benailly et al., 2010). In mice, Ret and Gdnf can regulate the survival, proliferation, and differentiation of ENS precursor cells (Shepherd et al., 2004; Amiel and Lyonnet, 2008). Either Ret or Gdnf gene knockout results in enteric aponeurosis ganglion cell and renal dysplasia (Wilson et al., 2002). In zebrafish, ablation of either ret or gdnf gene leads to the defective formation of enteric ganglion cells (Shimotake et al., 2001).
The central nervous system (CNS) of zebrafish originates from the ectodermal epithelium on the dorsal side of the embryo, called the neural plate (Wilson et al., 2002). During the development of zebrafish CNS, the primitive neural tube gradually develops into a mature system with more function-specific cell types (Kimmel et al., 1995; Wilson et al., 2002). The cells in the anterior region of the neural tube proliferate and differentiate into the primordia of the fore-, mid-, and hindbrain, while the posterior neural tube differentiates into the spinal cord (Wilson et al., 2002; Schier and Talbot, 2005; Durston and Zhu, 2015).
The Polycomb group (PcG) is composed of a variety of transcription inhibitors, mainly through epigenetic regulation of its target genes at the chromatin level (Bulyzhenkov et al., 1974; Croce and Helin, 2013). The PcG proteins primarily form two principal complexes, the Polycomb repressive complex 1 (PRC1), and the Polycomb-repressive complex 2 (PRC2). PRC2 is responsible for the trimethylation of Lys27 on histone H3 (H3K27me3) via the enzymatic subunit EZH1 or EZH2, while PRC1 catalyzes the ubiquitination of Lys119 on histone H2A (H2AK119ub) through the E3 ligase RING1. The biological functions of PRCs are under intensive investigation and are believed to be involved in stem cells maintenance, cell differentiation, cell cycle, aging, × chromosome inactivation, and tumorigenesis (Gil and O’Loghlen, 2014; Dupret et al., 2016; Almeida et al., 2017; Schuettengruber et al., 2017; Zhao et al., 2017; Chan et al., 2018; King et al., 2018). Two homologous subtype genes Ring1a and Ring1b have been found in mammalian genome (Schoorlemmer et al., 1997; Vidal, 2009). Knockout of Ring1b leads to the cessation of gastrulation and death of mouse embryos (Voncken et al., 2003; Napoles et al., 2004). Ring1a and Ring1b in mouse embryonic stem cells (ESCs) maintain its undifferentiated state by inhibiting differentiation genes (Endoh et al., 2008; van der Stoop et al., 2008). Patients with RING1 dysfunction show neurogenic psychosis, developmental abnormalities, and cognitive impairment (Pierce et al., 2018), suggesting that it has an important function in neural development. Zebrafish has only one rnf2 gene, which is high homology to human RING1B (Le Faou et al., 2011). rnf2 mutant zebrafish embryos show pleiotropic phenotypes, including lack of pectoral fin, craniofacial cartilage defects, edema, and stringy heart with abnormal sarcomere assembly (Velden et al., 2012, 2013; Chrispijn et al., 2019; Peng et al., 2021). However, the role of rnf2 in neural development remains unclear.
In this work, we hypothesize that the rnf2 gene of PRC1 plays key role in the development of central and ENS s. By generating rnf2 mutant zebrafish, we show that rnf2 loss of function affects the migration and differentiation of neural precursor cells during the development of both ENS and CNS. Our results provide important insights into the roles of rnf2 in embryonic development and diseases.
Materials and methods
Zebrafish strain and husbandry
The AB strain zebrafish and their embryos were maintained and raised in recirculation system at 28.5°C under a 14-h light, 10-h dark photoperiod. Developmental stages of zebrafish embryos were determined as previously described (Kimmel et al., 1995).
Generation of the rnf2 mutant line
The zebrafish rnf2 mutants were generated using the CRISPR/Cas9 system (Hwang et al., 2013; Jao et al., 2013; Peng et al., 2021). The guide RNA (gRNA) was designed targeting the exon 3 of the rnf2 gene (Peng et al., 2021). Embryos at the 1-cell stage were co-injected with 200 ng/μL Cas9 mRNA and 80 ng/μL gRNA. The genomic DNA of 30 embryos at 24 hpf was extracted and subjected to PCR amplification. The DNA fragment containing the rnf2 target site was amplified by PCR using the primers 5′-TTGAGGTAGTTGCTCCCAAAG-3′ and 5′-GGCATTCCTTGGTGGTCATA-3′, and the genotype was determined by DNA sequencing.
Whole-mount in situ hybridization
Embryos at different developmental stages were sampled and fixed in 4% paraformaldehyde (PFA) at 4°C overnight and then transferred in 100% MeOH. Whole-mount in situ hybridization was carried out according to a standard protocol (Thisse and Thisse, 2008). The DIG-labeled anti-sense probes were generated using a DIG RNA Labeling Kit (SP6/T7) (Roche). INT/BCIP (Roche) were used as alkaline phosphatase substrates. The primers of the probes were showed in the table of Supplementary Table S1. For embryos at or after 48 hpf, the homozygotes were separated from their siblings according to their heart edema and pectoral fin phenotype. For embryos before 48 hpf, as it was difficult to separate homozygous mutants from the heterozygous and wild-type siblings, WISH was performed for all progeny of rnf2± parents. After the WISH, each embryo was photographed and genotyped separately. The photographs were taken under a stereomicroscope (Leica Z16 APO) with a digital camera (Leica DFC450). The number and phenotype of embryos in each group were recorded, and then the offspring produced by rnf2± self-cross were genotyped.
Generation of phox2b: Enhanced green fluorescent protein transgenic line
We cloned a phox2b-promoter into pT2AL200R150G according to the Tol2 methods (Asakawa and Kawakami, 2009). The promoter contains approximately 4.8 kb of sequence upstream of transcription start site in the phox2b gene.1 Next, we co-injected linearized plasmid DNA and transposase mRNAs into 1-cell stage zebrafish embryos, and raised the injected embryos to adulthood. The injected embryos were crossed with wild type fish, and day 1 F1 embryos were screened for GFP under a dissecting microscope MZ 16FA (Leica). Embryos with GFP expression were raised up to adulthood. F1 adults were crossed to obtain F2 embryos with stable expression of GFP.
Whole-mount immunostaining
For immunohistochemistry processing, embryos were fixed overnight at 4°C in 4% paraformaldehyde (PFA). Next day, embryos were washed three times in 1 × PBS and then transferred into tubes with 100% MeOH. Fluorescent immunostaining for 96 hpf embryos was performed using the polyclonal zebrafish Rnf2 antiserum (1:300). The DyLight 488 goat anti-rabbit IgG (1:350, Abbkine, United States) was used as secondary antibody. The neurons were detected by using rabbit anti-HuC/D (1:200) as primary antibody and DyLight 555 goat anti-mouse IgG (1:350, Abbkine, United States) as secondary antibody. For whole-mount immunostaining, embryos were digested in PBS containing 10 g/ml proteinase K, 0.1% Tween20 and blocked in PBS containing 10% normal goat serum, 0.5% DMSO and 0.3% Triton X-100 (Wang et al., 2013). Images were taken with a confocal laser scanning microscope (Leica SP8 DLS).
Image, quantification, and statistical analysis
Images of fluorescent signals were taken with a confocal laser scanning microscope (Leica SP8 DLS). The signal density of the images was analyzed by Image J software (National Institutes of Health). The values are presented as mean ± SEM. The p-values were calculated by Origin 9.0 with two-tailed Student’s test, * represent p ≤ 0.05.
Results
rnf2 is expressed in enteric nervous system and central nervous system of zebrafish embryos
The expression pattern of rnf2 during early embryogenesis has been described (Figure 1A; Velden et al., 2012). In addition to the brain, we found that rnf2 was abundantly expressed in gut regions at 72 hpf (Figure 1B), suggesting that it may be involved in the development of ENS.
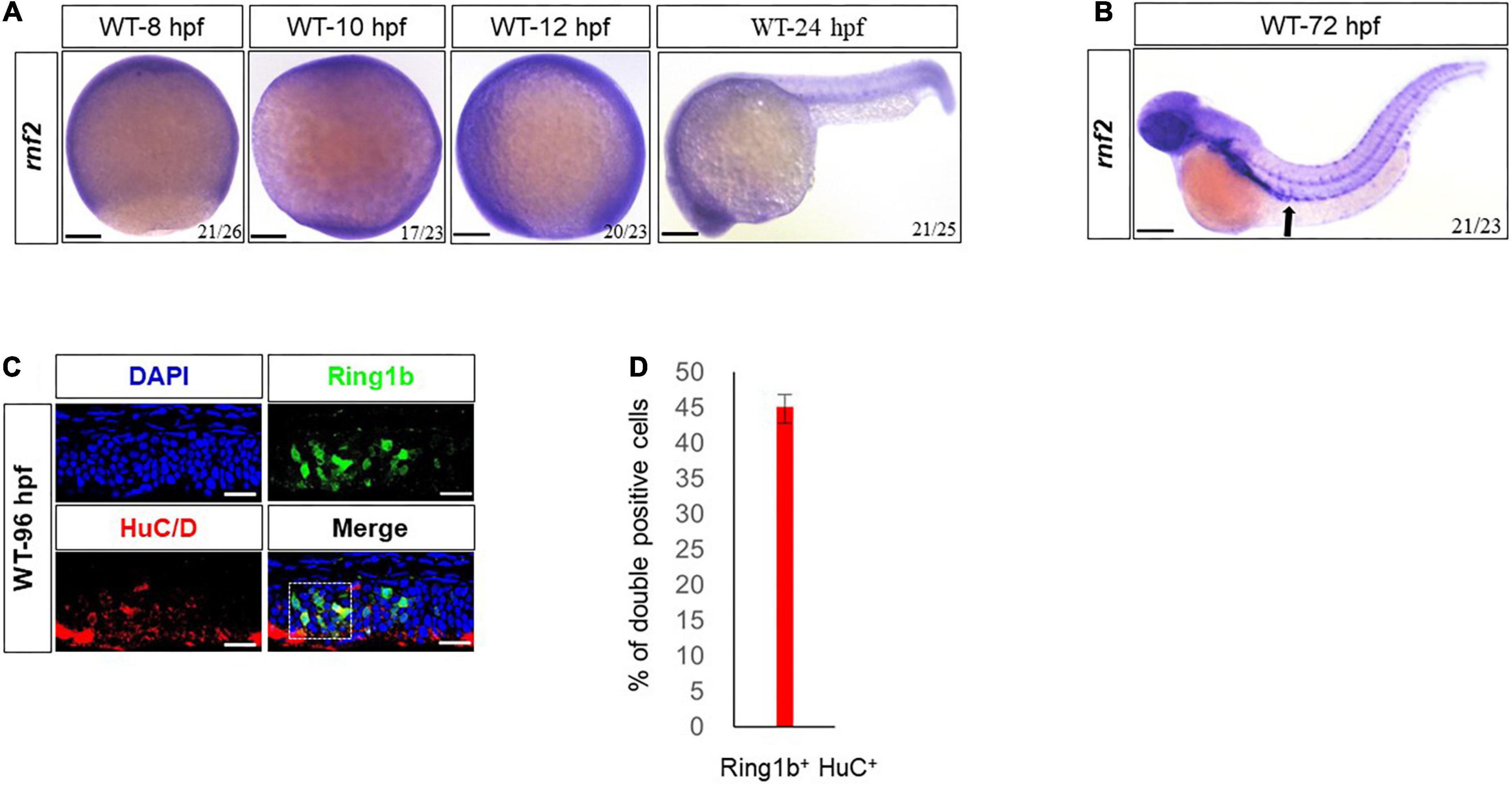
Figure 1. Expression patterns of rnf2 in zebrafish embryos. (A) Whole mount in situ hybridization (WISH) of rnf2 at the indicated time points. (B) The expression of rnf2 at 72 hpf WT embryos. Black arrows showing the expression of rnf2 in the gut of zebrafish embryos. (C) Immunohistochemistry images of Rnf2 and HuC/D in zebrafish gut (40 × oil, embryos direction: anterior is to the left). The overlapping of Rnf2 and HuC/D expression is marked by the white box. (D) Quantitation of percentage of Rnf2/HuC double positive cells (n = 29). The experiments were repeated at least three times. Scale bar: 0.1 mm.
To confirm this, double fluorescence in situ hybridization for Rnf2 and the pan neural marker HuC/D was performed in 96 hpf embryos. The results showed that Rnf2 was expressed in the gut and its surrounding environment, and was colocalized with a portion of HuC/D+ neuronal cells (Figures 1C,D). These data suggested that rnf2 was involved in the development of ENS and CNS in zebrafish embryos.
rnf2 is required for the migration of enteric neural precursor cells
To investigate the role of the rnf2 gene, we generated rnf2 mutant zebrafish using the CRISPR/Cas9 technology (Peng et al., 2021). The 5 bp deletion mutant line was used for the most of the experiments. No obvious phenotypes between WT (Wild type) and mutant embryos were observed at 24 hpf (Peng et al., 2021). After 72 hpf, rnf2 mutants displayed pleiotropic phenotypes, including craniofacial defects, cardiac edema, and lack of pectoral fins (Velden et al., 2012, 2013; Chrispijn et al., 2019; Peng et al., 2021). No Ring1b protein was detected in rnf2–/– embryos, and as expected, H2AK119ub level was markedly decreased. The rnf2–/– embryos usually die within a week (Peng et al., 2021).
First, we investigated the role of rnf2 in ENS development. During zebrafish ENS development, enteric neural precursor cells (ENPCs) migrate as two chains from the postotic vagal regions to the caudal end along the two sides of the gut (Olden et al., 2008). Phox2b, which is mainly expressed in neural precursor cells (Garcia-Barceló et al., 2003; Fitze et al., 2010; Kwon et al., 2011), can be used to mark the enteric neural precursors (Elworthy et al., 2005). In order to investigate whether rnf2 is required for the migration of neural precursor cells, whole mount in situ hybridization (WISH) was performed for phox2b. At 24 hpf, phox2b mRNAs were detected in the hindbrain and vagal regions, and the expression was comparable between WT and mutant embryos (Figure 2A). In 48 hpf WT, phox2b was expressed in the vagal region and the anterior part of the gut (Figure 2B). In 48 hpf rnf2 mutant, however, phox2b was mainly distributed in the vagal region, and barely detected in the gut, suggesting that the migration of Phox2b-positive ENPCs was defective (Figure 2B). Of note, phox2b was barely expressed in forebrain and eye regions in WT embryos, while it was ectopically expressed in mutant embryos.
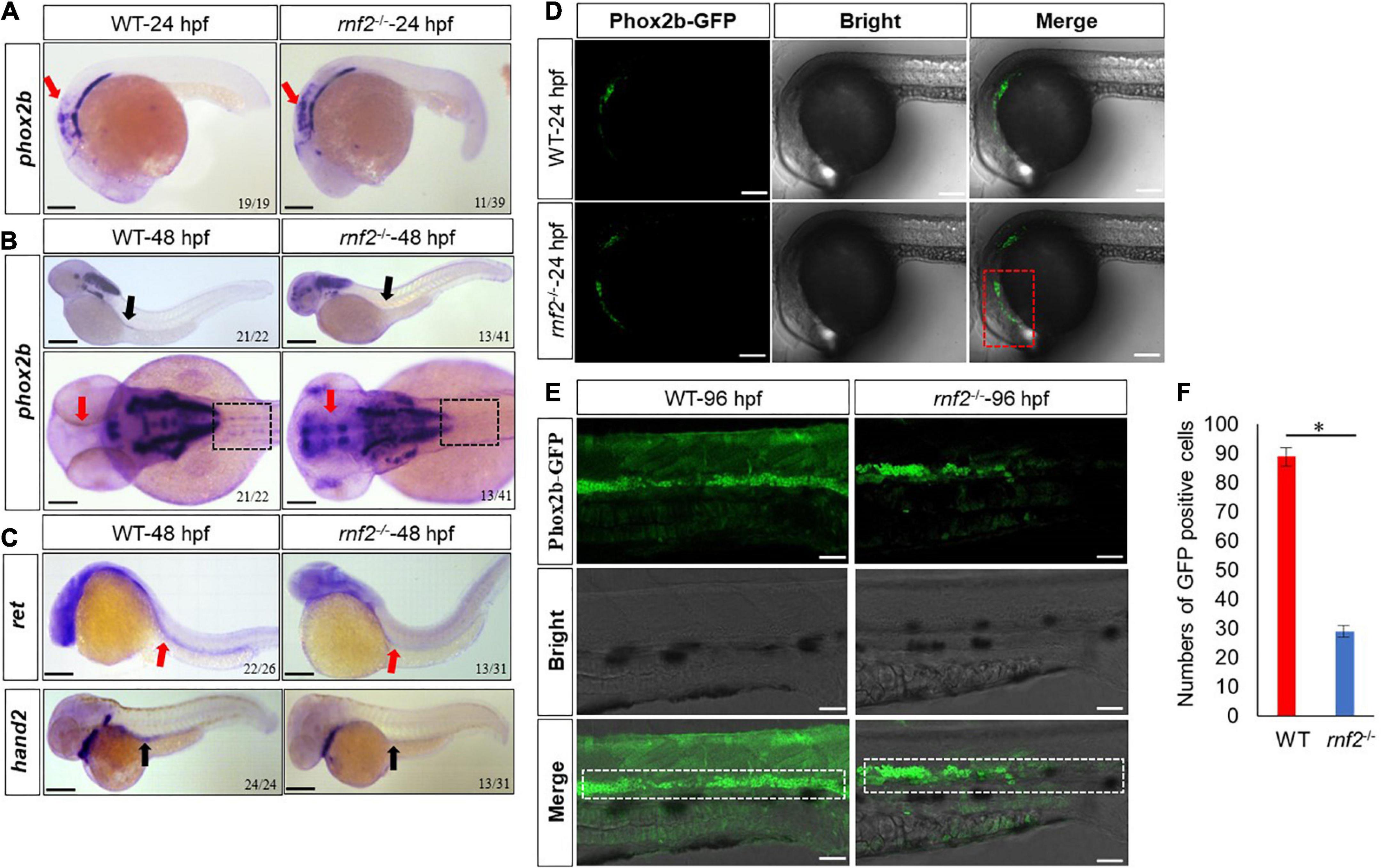
Figure 2. The effect of rnf2 deficiency on the migration of enteric neural precursor cells in zebrafish embryos. (A) The expression of phox2b in rnf2–/– and WT embryos at 24 hpf. Red arrows showing the reduced expression of phox2b in intestinal bulb of mutant embryos. Scale bar = 0.1 mm. (B) The expression of phox2b in rnf2–/– and WT embryos at 48 hpf. Black arrows showing the reduced expression of phox2b in the gut of mutant embryos, red arrows showing the ectopic expression of phox2b in the brain regions. The dashed black box showing the compromised migration of ENPCs. Upper: lateral view; bottom: dorsal view. (C) The expression of ret and hand2 in rnf2–/– and WT embryos at 48 hpf. Black arrows showing the reduced expression of hand2 in the gut of mutant embryos; red arrows showing the reduced expression of ret. The dashed black box showing the compromised migration of ENPCs. Upper: lateral view; bottom: lateral view. (D) The expression of Phox2b-GFP in the brain regions of rnf2–/– and WT embryos at 24 hpf (40 × oil, lateral view). The numbers of samples were 28 and 24 in 24 hpf WT and rnf2–/– embryos, respectively. The red dashed box showing the expansion of GFP signals. (E) The expression of Phox2b-GFP in gut regions of rnf2–/– and WT embryos at 96 hpf (40 × oil, lateral view and head is to the left). The white dashed box showing the different distribution of GFP+ signals in the gut of WT and rnf2–/– embryos at 96 hpf. The numbers of samples were 30 and 27 in 96 hpf WT and rnf2–/– embryos, respectively. (F) Quantitation of GFP positive cells in (E). The star indicates significant differences at p ≤ 0.05. The experiments were repeated at least three times. Scale bar: 0.1 mm.
Next, we performed WISH for more enteric neural precursor markers, such as ret and hand2 (Figure 2C). The results showed that the expression of ret and hand2 was decreased significantly in the gut regions of 48 hpf mutant embryos, compared to the controls.
To further investigate this, we examined enteric neural crest migration by using the Tg [phox2b: enhanced green fluorescent protein (EGFP)] transgenic line. In 24 hpf WT transgenic embryos, the Phox2b-GFP+ cells were mainly confined to the position of intestinal bulb (Figure 2D). In 24 hpf mutant transgenic embryos, Phox2b-GFP+ cells were more broadly distributed. Then we examined embryos at 96 hpf, when the ENPCs had finished the migration along the gut (Olden et al., 2008). In WT embryos, the Phox2b-GFP+ cells were distributed throughout the whole gut. In mutant embryos, however, the Phox2b-GFP+ cells were detected only in the foregut and midgut but not in hindgut (Figure 2E). Consistently, the number of Phox2b-GFP positive cells was much smaller in hindgut of mutant embryos compared to controls (Figure 2F). These results indicated that loss of rnf2 leads to abnormal migration of enteric neural precursors.
rnf2 loss of function decreases enteric neurons
During the migration toward the gut, the ENPCs will gradually differentiate into different type of neurons (Oppenheim, 1991; Chelyshev et al., 2000). In zebrafish, the first differentiating enteric neurons are detected along the anterior gut at ∼55 hpf (Velden et al., 2012; Chrispijn et al., 2019), and by 74 hpf, HuC/D+ enteric neurons appear at the most caudal end of the gut, near the anus (Velden et al., 2012; Bergeron et al., 2013). Based on the above results, we speculated that the enteric neuron formation may be affected in the absence of rnf2. To test this, we performed whole mount immunohistochemistry for WT and rnf2–/– embryos at 96 hpf, using antibodies against the pan-neuronal marker HuC/D. In WT embryos, HuC/D+ neurons were observed along the entire length of the gut, all the way to the anus (Figure 3). In rnf2–/– embryos, however, HuC/D+ neurons were found only within the foregut and midgut regions (Figures 3A,B), indicating that loss of rnf2 resulted in aganglionosis (Bergeron et al., 2013). Even in the midgut, the number of HuC/D+ neurons were smaller in rnf2–/– embryos than in the controls (Figure 3C). These data suggested that rnf2 is required for efficient colonization of ENPCs to the entire gut, and its loss causes aganglionosis.
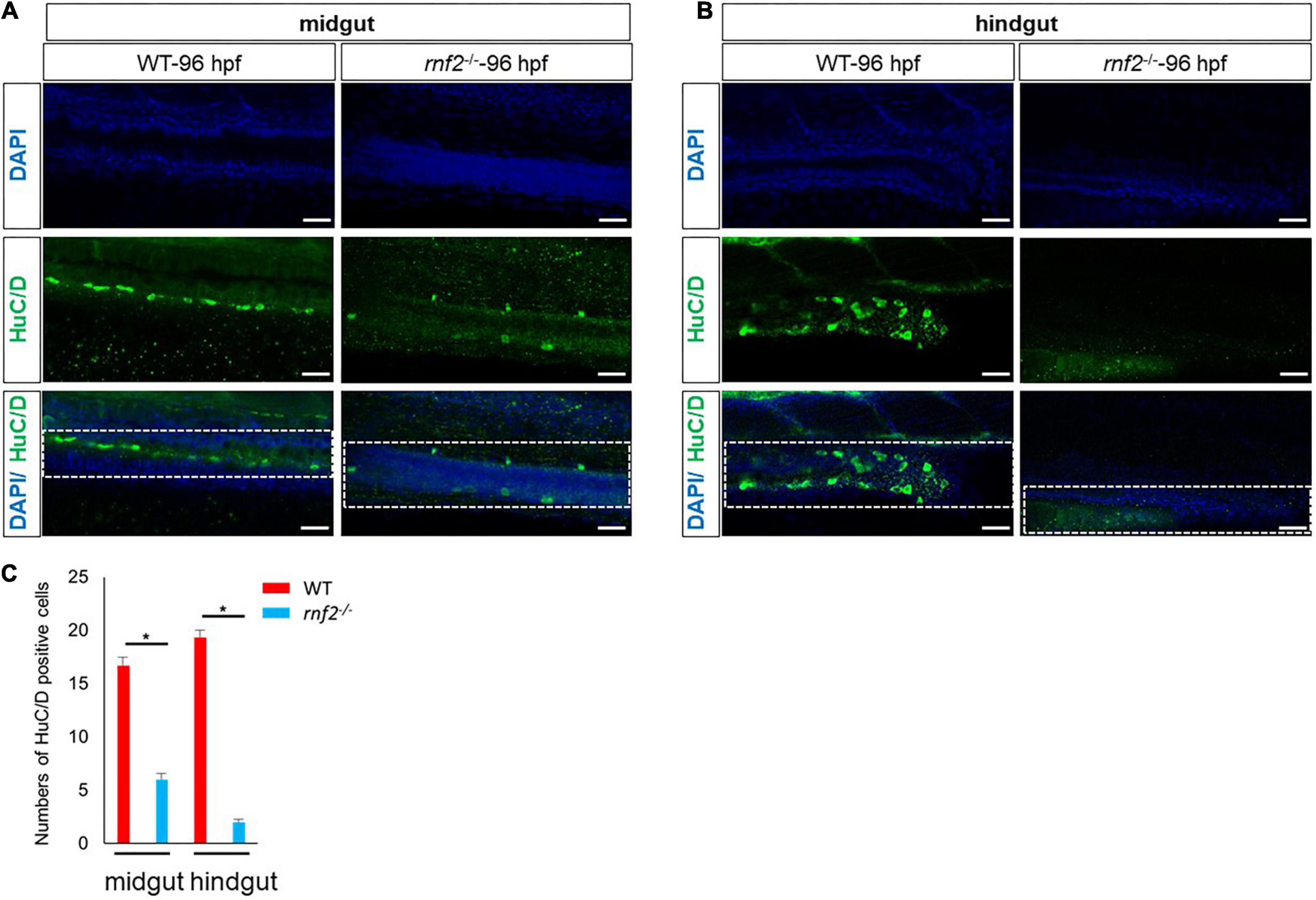
Figure 3. The deficiency of rnf2 decreased the enteric neurons in zebrafish embryos. (A) Confocal images showing HuC/D positive enteric neurons in midgut of rnf2–/– and WT embryos at 96 hpf (40 × oil, lateral view and head is to the left). The white dashed box showing the different distribution of HuC/D+ signals in the midgut of WT and rnf2–/– embryos at 96 hpf. (B) Confocal images showing HuC/D positive enteric neurons in hindgut of rnf2–/– and WT embryos at 96 hpf (40 × oil, lateral view and head is to the left). The white dashed box showing the different distribution of HuC/D+ signals in the hindgut of WT and rnf2–/– embryos at 96 hpf. Scale bar: 0.1 mm. (C) Quantitation of HuC positive cells in (A,B). The numbers of samples were 26 and 21 in 96 hpf WT and rnf2–/– embryos, respectively. The star indicates significant differences at p ≤ 0.05. The experiments were repeated at least three times. Scale bar: 0.1 mm.
Loss of rnf2 affects the specification of neural crest cells
Next, we asked whether loss of rnf2 affects the initiation and specification of NCs, by examining the expression of foxd3, nestin, sox9b, and sox1b in 10 and 12 hpf embryos (Li et al., 2002; Mahler and Driever, 2007; Hochgreb-Hägele and Bronner, 2013; Andrzejczuk et al., 2018). The results showed that at 10 hpf and 12 hpf, the expression of foxd3, nestin, sox9b, and sox1b was changed to different extent (Figure 4). In 10 hpf WT embryos, the foxd3-labeled NCs migrated linearly from the animal pole to the plant pole, and formed three distinct migration pathways. In rnf2–/– embryos, there was a global similar expression of foxd3, but the migration chains of NCs were shorter, compared to controls (Figure 4A). At 12 hpf, the expression of nestin, sox9b was decreased significantly (Figures 4C,D), while sox1b showed an ectopic expression (Figure 4B). These observations suggested that the migration and distribution of NCs were disturbed in the absence of Ring1b.
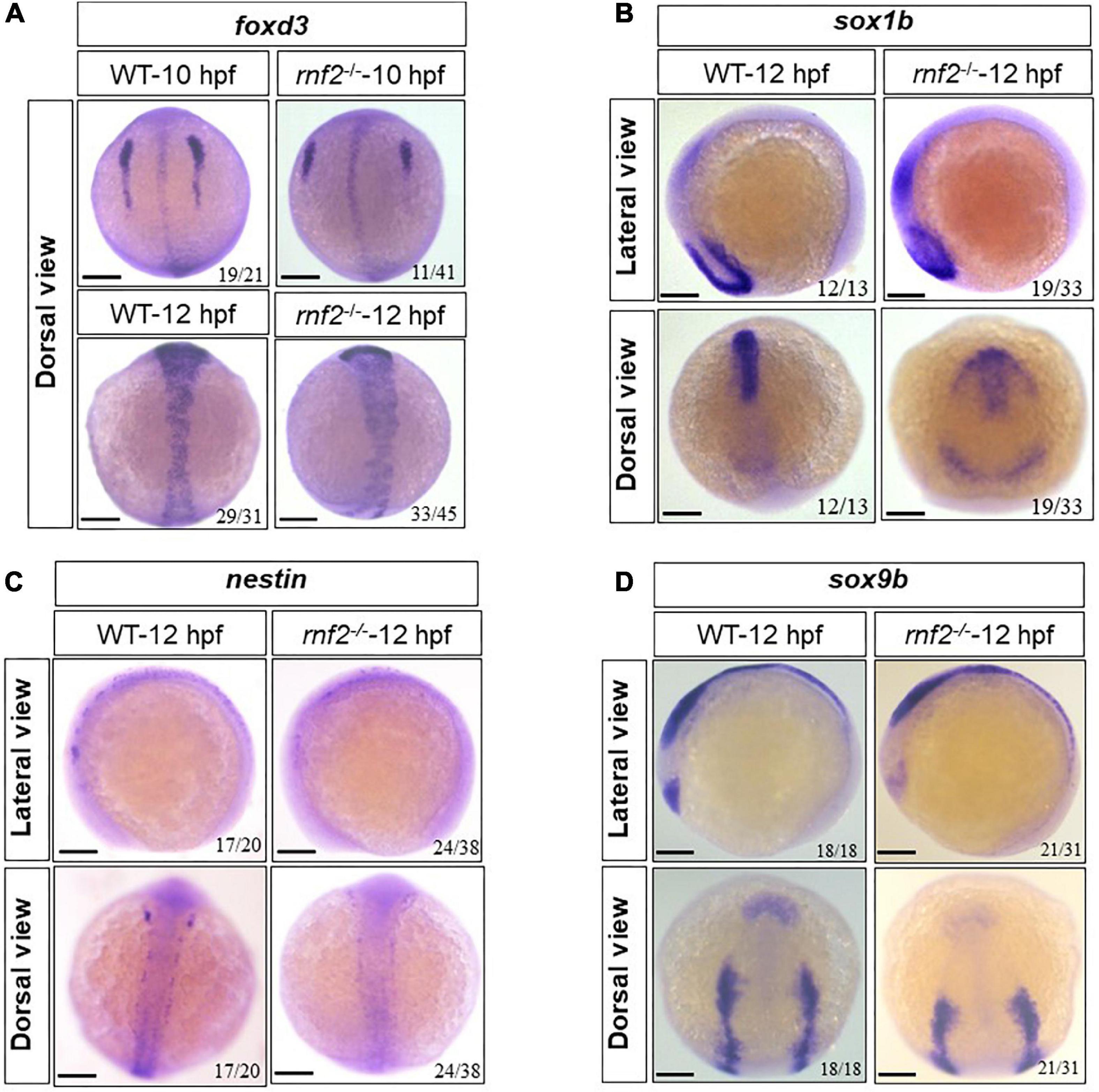
Figure 4. The effect of loss of rnf2 on the specification of neural crest cells. (A) The expression of foxd3 in rnf2–/– and WT embryos at 10 and 12 hpf. Dorsal view. (B) The expression of sox1b in rnf2–/– and WT embryos at 12 hpf. Upper: lateral view, bottom: dorsal view. (C) The expression of nestin in rnf2–/– and WT embryos at 12 hpf. Upper: lateral view, bottom: dorsal view. (D) The expression of sox9b in rnf2–/– and WT embryos at 12 hpf. Upper: lateral view, bottom: dorsal view. The experiments were repeated at least three times. Scale bar = 0.1 mm.
Then we investigated the genes related to the early differentiation of NCs, including prdm1a, cxcr4a, and vgll2a (Olesnicky et al., 2010; Johnson et al., 2011; Chen et al., 2018). The expression of prdm1a, cxcr4a, and vgll2a was decreased significantly with varying degrees (Figure 5). In 12 hpf embryos, cxcr4a expression levels in neural crest progenitors were reduced in rnf2–/– embryos (Figure 5A). vgll2a was strongly expressed along the two lines of the neural crest in WT embryos. In rnf2–/– embryos, however, the expression of vgll2a was barely detected (Figure 5B). In 12 hpf WT embryos, prdm1a formed two expression chains around the vagal region, and exhibited 4 parallel chains around the neural crest and neural plate (Figure 5C). In 12 hpf rnf2–/– embryos, the anterior expression of prdm1a in neural plate was hardly detected, and its posterior expression domains were closer to the midline, compared to the controls (Figure 5C). All these results indicated that rnf2 knockout leads to abnormal migration and differentiation of NCs.
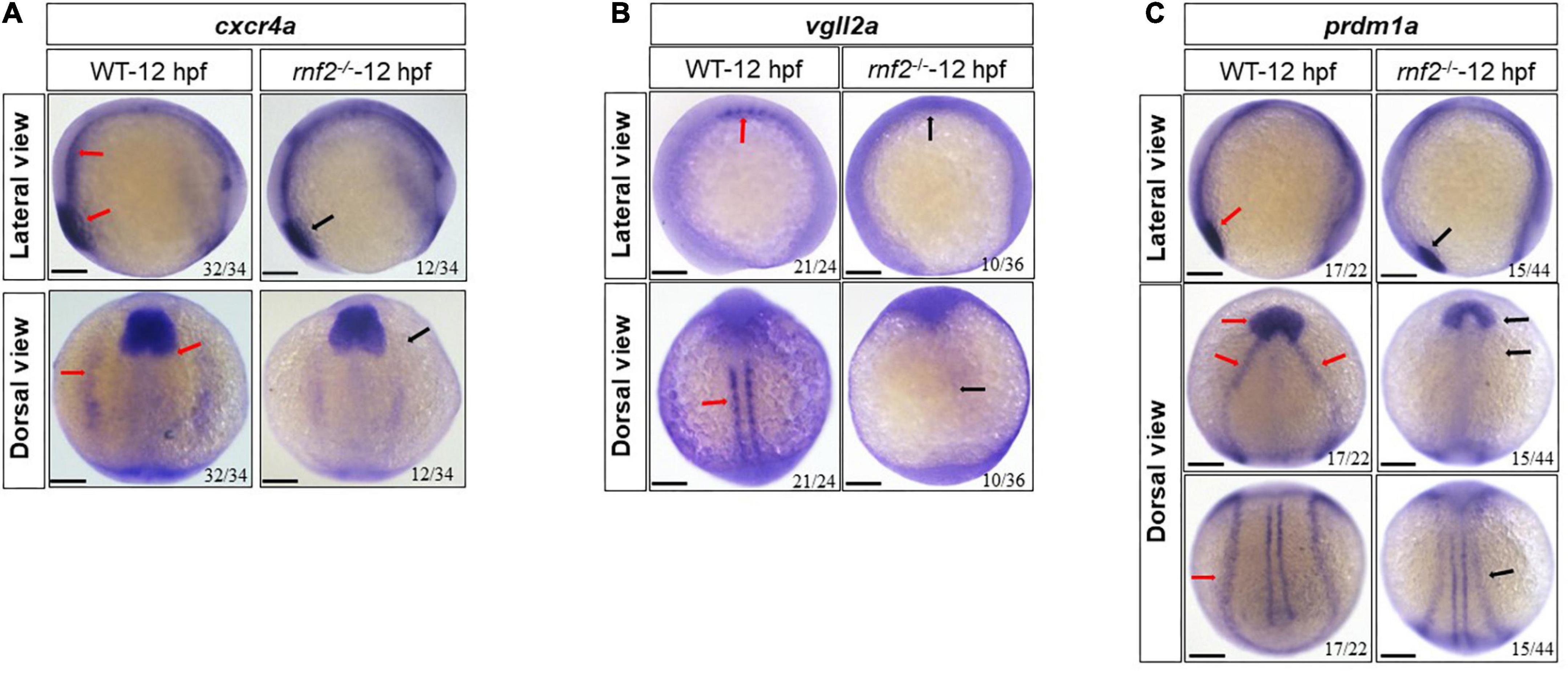
Figure 5. The effect of rnf2 deficiency on differentiation of neural crest in zebrafish embryos. (A) The expression of cxcr4a in rnf2–/– and WT embryos at 12 hpf. Upper: lateral view, bottom: dorsal view. (B) The expression of vgll2a in rnf2–/– and WT embryos at 12 hpf. Upper: lateral view, bottom: dorsal view. (C) The expression of prdm1a in rnf2–/– and WT embryos at 12 hpf. Upper: lateral view; middle: dorsal anterior view; bottom: dorsal posterior view. Red arrows showing the expression of cxcr4a, vgll2a, and prdm1a in 12 hpf WT embryos, black arrows showing the reduced expression of cxcr4a, vgll2a, and prdm1a in 12 hpf rnf2–/– mutant embryos. The experiments were repeated at least three times. Scale bar = 0.1 mm.
rnf2 loss of function affects central nervous system development
We next investigated whether rnf2 loss of function disrupted normal CNS development in zebrafish embryos. To this end, we performed in situ hybridization against neurod1, egr2b, tfap2a, and phox2a in 48 hpf control and rnf2 mutant embryos (Figure 6). neurod1, egr2b, tfap2a, and phox2a have been shown to be related to CNS development (Tahayato et al., 2003; Uehara et al., 2007; Zhang et al., 2017; Kousa et al., 2019; Song et al., 2020; Elliott et al., 2021). The results showed that the expression of tfap2a was slightly reduced in rnf2–/– embryos compared to controls, especially in the middle and hindbrain regions (Figure 6A). phox2a was down-regulated in the eyes in the rnf2–/– embryos compared to the controls (Figure 6B). Tfap2a and Phox2a are associated with the differentiation of adrenergic neurons and noradrenergic neurons, respectively (Kousa et al., 2019; Song et al., 2020). In WT embryos, neurod1 was abundantly expressed in the forebrain, the midbrain, the hindbrain, and the midbrain-hindbrain boundary (MHB). In rnf2–/– embryos, however, the expression domains of neurod1 were lost in midbrain and MHB (Figure 6C). Consistently, in rnf2–/– embryos, there was a reduced expression of egr2b, a marker of MHB (Figure 6D). These results suggested that rnf2 is involved in the differentiation of neurons in the CNS.
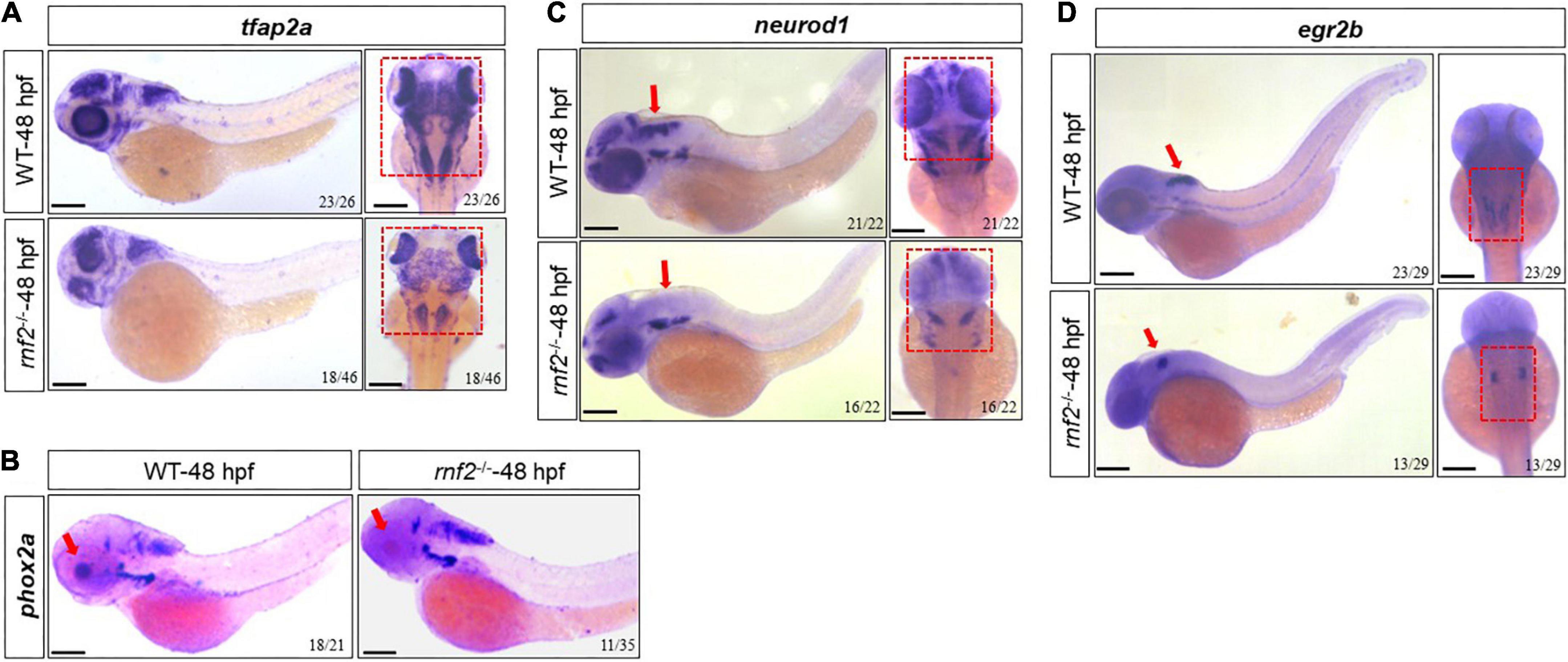
Figure 6. The effect of rnf2 deficiency on central nervous system development in zebrafish embryos. (A) The expression of tfap2a in rnf2–/– and WT embryos at 48 hpf. Left: lateral view; right: dorsal view of the brain. The red dashed boxes showing the different expansion of tfap2a in the brain regions of rnf2–/– embryos at 48 hpf. (B) The expression of phox2a in rnf2–/– and WT embryos at 48 hpf. The red boxes and arrows showing the loss of phox2a expression in the brain regions of mutant embryos. Lateral view. (C) The expression of neurod1 in rnf2–/– and WT embryos at 48 hpf. The red boxes and arrows showing the loss of neurod1 expression in MHB and hindbrain of mutant embryos. Left: lateral view; right: dorsal view of the brain. (D) The expression of egr2b in rnf2–/– and WT embryos at 48 hpf. The red arrows showing the reduced egr2b expression in MHB of mutant embryos. Left: lateral view; right: dorsal view of the brain. The experiments were repeated at least three times. Scale bar = 0.1 mm.
Next, we examined the expression of genes related to CNS progenitor development, including pax2a, egr2b, cyp26c1, and ngn1 (Korzh et al., 1998; Tahayato et al., 2003; Uehara et al., 2007; Ota et al., 2016; Zhang et al., 2017). As shown in Figure 7, globally, rnf2 knockout resulted in decreased expression of these marker genes. In WT embryos, pax2a displayed a V-type expression pattern. In rnf2–/– embryos, the two expression domains along the midline were disconnected (Figure 7A). The expression of egr2b was decreased in 12 hpf rnf2–/– embryos, especially in the hindbrain (Figure 7B). Similar results were observed for cyp26c1 (Figure 7C). The expression of ngn1 was markedly reduced in rnf2–/– embryos compared to controls (Figure 7D).

Figure 7. The effect of ring1b deficiency on the early development of brain in zebrafish embryos. (A) The expression of pax2a in rnf2–/– and WT embryos at 12 hpf. Upper: lateral view, bottom: dorsal view. The red and white arrows, respectively, showing the different expression of pax2a in WT and rnf2–/– embryos at 12 hpf. (B) The expression of egr2b in rnf2–/– and WT embryos at 12 hpf. Upper: lateral view, bottom: dorsal view. The red arrows showing the different expression of egr2b in WT and rnf2–/– embryos at 12 hpf. (C) The expression of cyp26c1 in rnf2–/– and WT embryos at 12 hpf. Upper: lateral view, bottom: dorsal view. The red arrows showing the different expression of cyp26c1 in WT and rnf2–/– embryos at 12 hpf. (D) The expression of ngn1 in rnf2–/– and WT embryos at 12 hpf. The experiments were repeated at least three times. Scale bar = 0.1 mm.
To investigate whether rnf2 loss of function affects further differentiation of function-specific neurons, we analyzed 96 hpf WT and rnf2–/– embryos by performing whole-mount immunofluorescence staining with antibodies against HuC/D and 5-HT (Figure 8A). 5-HT marks the serotonin neurons. In both WT and mutant embryos, 5-HT+ neurons were overlapped with a portion of HuC/D neurons. The expression of HuC/D was markedly reduced in the brain of rnf2 mutants compared to the controls, particularly in the midbrain and hindbrain. In the brain of control embryos, 5-HT+ serotonin neurons were bilaterally localized and centralized distributed. By contrast, in rnf2–/– embryos, 5-HT+ neurons were mis-localized and displayed a dispersed manner (Figure 8A). Furthermore, the percentage of 5-HT/HuC double positive cells was slightly smaller in rnf2 mutant embryos than in controls (Figure 8B). These results indicated that 5-HT serotonin neuronal differentiation may still occur, but its migration was abnormal.
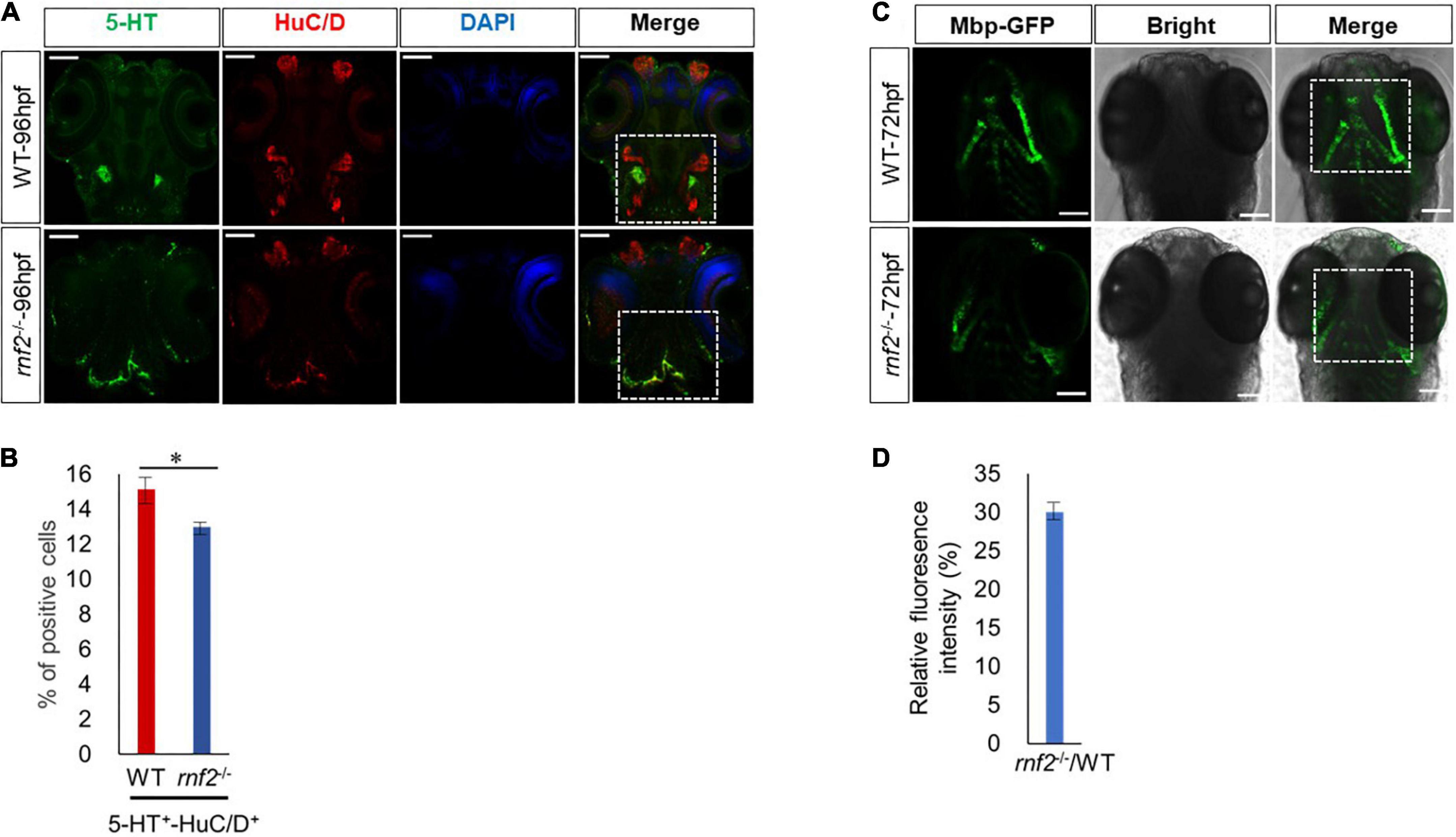
Figure 8. Loss of Rnf2 leads to neuronal differentiation defects in zebrafish embryos. (A) Confocal images showing the expression of HuC/D and 5-HT in zebrafish brain at 96 hpf (40 × oil). Dorsal view. The white dashed boxes showing the overlapping of HuC/D+ and 5-HT+ signals in the brain of WT and rnf2–/– embryos at 96 hpf. (B) Quantitation of percentage of 5-HT/HuC double positive cells in (A). The numbers of samples were 20 and 18 in 96 hpf WT and rnf2–/– embryos, respectively. The star indicates significant differences at p ≤ 0.05. (C) Confocal images showing the expression of Mbp-GFP in zebrafish brain at 72 hpf (40 × oil). Dorsal view. The white dashed boxes showing the distribution of Mbp+ signals in the brain of WT and rnf2–/– embryos at 72 hpf. (D) Quantitation of relative fluorescence intensity in (C). The numbers of samples were 28 and 20 in 72 hpf WT and rnf2–/– embryos, respectively. The experiments were repeated three times. Scale bar = 0.1 mm.
Finally, we asked whether loss of rnf2 leads to defective formation of myelinating glial cells. To this end, we utilized a transgenic zebrafish line Tg (mbp: EGFP), in which the myelinating glial cells are labeled by GFP (von Jonquieres et al., 2013). In 72 hpf WT embryos, Mbp-GFP signals appeared in hindbrain and bilateral symmetry (Figure 8C). In 72 hpf rnf2–/– embryos, however, Mbp-GFP signals in these regions were reduced and mis-localized (Figures 8C,D). These results showed that rnf2 loss of function may affect the migration and differentiation of myelinating glial cells.
Discussion
Polycomb group proteins (PcGs) are important epigenetic repressors that play important roles in the regulation of ESCs and hematopoietic stem cells (HSCs), cell fate determination, cell lineage restriction and organogenesis (Chittock et al., 2017). The role of PRC1 in the nervous system development is not well studied.
In this work, we show that rnf2 loss of function is sufficient to perturb enteric neural crest migration and differentiation along the developing gut in zebrafish embryos. As a result, fewer enteric neurons were born in the ENS, leading to colonic aganglionosis. This clearly shows that the rnf2 gene plays an important role in ENS development and highlights it as a novel candidate gene in Hirschsprung’s disease. We found that rnf2 loss of function decreases the expression of phox2b, ret, hand2, and crestin. Previous reports have shown that in Phox2b knockout mice and zebrafish, ENS precursor cells can reach the foregut, but cannot continue to migrate or differentiate, and the expression of enteric neural precursor cell markers is abnormal (Fitze et al., 2010; Kwon et al., 2011). Phox2b is crucial to the formation of all autonomic ganglia, including enteric ganglia, and is suggested to promote the proliferation and survival of ENPCs. Consistently, its depletion leads to complete enteric aganglionosis (Alexandre et al., 1999). Interestingly, phox2b was ectopically expressed in the brain regions of rnf2 mutant embryos. In zebrafish and mice, knockout of the ret gene leads to abnormal development of ENS (Carrasquillo et al., 2002; Carniti et al., 2006). The expression of early neural crest markers such as foxd3, nestin, sox9b, and sox1b, was altered in the absence of rnf2. The results of in situ hybridization showed that rnf2 mutation disrupts the migration of ENPCs to the gut; the immunofluorescence assay showed that rnf2 mutation results in the decrease of enteric neurons. These results suggest that rnf2 loss of function leads to deficits in the specification of NCs, and affects the differentiation of subsequent enteric neurons by regulating the migration of ENPCs.
Mutations in PRC1 members such as PHC1 and PCGF2 lead to human neurodevelopmental disorders (Awad et al., 2013; Fitzgerald et al., 2015). Patients with RING1 mutation have neurological psychosis, developmental abnormalities and cognitive impairment (Pierce et al., 2018). A recent study has shown that RING1 p.R95Q, which alters a conserved arginine residue in the catalytic RING domain, results in syndromic neurodevelopmental disabilities of a 13-year-old girl (Pierce et al., 2018). These observations suggest that RING1 also plays a critical role in CNS development beyond ENS. In order to study the function of rnf2 in zebrafish CNS development, we compared the neuronal differentiation in control and mutant embryos. The results of immunofluorescence further showed that 5-HT+ neurons were mis-localized and displayed a dispersed manner. In 72 hpf rnf2–/– embryos, Mbp-GFP signals in these regions are reduced and mis-localized. This result shows that knockout of rnf2 gene also disrupts the development of myelinated glial cells (von Jonquieres et al., 2013). We also showed that formation of adrenergic and noradrenergic neurons was abnormal in rnf2–/– embryos. This observation indicated that rnf2 is required for proper differentiation of neurons in the CNS (Kousa et al., 2019; Song et al., 2020). Taken together, we concluded that rnf2 is important for the proper migration and differentiation of neural precursor cells, and its absence leads to abnormal development of ENS and CNS in zebrafish.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
YS: conceptualization and funding acquisition. GF: formal analysis, investigation, resources, and visualization. GF and YS: manuscript. Both authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Strategic Priority Research Program of Chinese Academy of Sciences (grant no. XDB31000000) and the National Natural Science Foundation of China (grant nos. 31671526 and 32000384).
Acknowledgments
We thank the members of the Sun Laboratory. We are grateful for Fang Zhou for confocal imaging, who is from the analysis and testing center of Institute of Hydrobiology, Chinese Academy of Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2022.960149/full#supplementary-material
Footnotes
- ^ https://asia.ensembl.org/Danio_rerio/Gene/Sequence?db=core;g=ENSDARG00000091029;r=14:17064126-17072736
References
Alexandre, P., Xavier, M., Harold, C., Christo, G., and Jean-FranÇois, B. (1999). The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature 399, 366–370. doi: 10.1038/20700
Almeida, M., Pintacuda, G., Masui, O., Koseki, Y., Gdula, M., Cerase, A., et al. (2017). PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 356, 1081–1084. doi: 10.1126/science.aal2512
Amiel, J., and Lyonnet, S. (2008). Hirschsprung disease, associated syndromes, and genetics: A review. J. Med. Genet. 38, 729–39. doi: 10.1136/jmg.38.11.729
Andrzejczuk, L. A., Banerjee, S., England, S. J., Voufo, C., Kamara, K., and Lewis, K. E. (2018). Tal1, Gata2a, and Gata3 Have Distinct Functions in the Development of V2b and Cerebrospinal Fluid-Contacting KA Spinal Neurons. Front. Neurosci. 12:170. doi: 10.3389/fnins.2018.00170
Asakawa, K., and Kawakami, K. (2009). The Tol2-mediated Gal4-UAS method for gene and enhancer trapping in zebrafish. Methods 49, 275–281. doi: 10.1016/j.ymeth.2009.01.004
Avantaggiato, V., Dathan, N. A., Grieco, M., Fabien, N., Lazzaro, D., Fusco, A., et al. (1994). Developmental expression of the RET protooncogene. Cell Growth Dif. 5, 305–311.
Awad, S., Al-Dosari, M. S., Al-Yacoub, N., Colak, D., Salih, M. A., Alkuraya, F. S., et al. (2013). Mutation in PHC1 implicates chromatin remodeling in primary microcephaly pathogenesis. Hum. Mol. Genet. 22, 2200–2213. doi: 10.1093/hmg/ddt072
Benailly, H. K., Lapierre, J. M., Laudier, B., Amiel, J., Attié, T., Blois, M. C. D., et al. (2010). PMX2B, a new candidate gene for Hirschsprung’s disease. Clin. Genet. 64, 204–209. doi: 10.1034/j.1399-0004.2003.00105.x
Bergeron, K. F., Silversides, D. W., and Pilon, N. (2013). The developmental genetics of Hirschsprung’s disease. Clin. Genet. 83, 15–22. doi: 10.1111/cge.12032
Bulyzhenkov, V. E., Ginter, E. K., and Ivanov, V. I. (1974). Interaction of the homoeotic mutations aristapedia and polycomb during ontogenesis of Drosophila melanogaster. Ontogenez 5, 634–641.
Carniti, C., Belluco, S., Riccardi, E., Cranston, A. N., Mondellini, P., Ponder, B. A. J., et al. (2006). The Ret (C620R) mutation affects renal and enteric development in a mouse model of Hirschsprung’s disease. Am. J. Pathol. 168, 1262–1275. doi: 10.2353/ajpath.2006.050607
Carrasquillo, M. M., McCallion, A. S., Puffenberger, E. G., Kashuk, C. S., Nouri, N., and Chakravarti, A. (2002). Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat. Genet. 32, 237–244. doi: 10.1038/ng998
Chan, H. L., Beckedorff, F., Zhang, Y., Garcia-Huidobro, J., Jiang, H., Colaprico, A., et al. (2018). Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat. Commun. 9:3377. doi: 10.1038/s41467-018-05728-x
Chelyshev, Y. A., Cherepnev, G. V., and Saĭtkulov, K. I. (2000). Apoptosis in the nervous system. Nature 32, 118–129.
Chen, J. Y., Tan, X., Wang, Z. H., Liu, Y. Z., Zhou, J. F., Rong, X. Z., et al. (2018). The ribosome biogenesis protein Esf1 is essential for pharyngeal cartilage formation in zebrafish. FEBS J. 285, 3464–3484. doi: 10.1111/febs.14622
Chittock, E. C., Latwiel, S., Miller, T. C. R., and Müller, C. W. (2017). Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans. 45, 193–205. doi: 10.1042/BST20160173
Chrispijn, N. D., Elurbe, D. M., Mickoleit, M., Aben, M., de Bakker, D. E. M., Andralojc, K. M., et al. (2019). Loss of the Polycomb group protein Rnf2 results in derepression of tbx-transcription factors and defects in embryonic and cardiac development. Sci. Rep. 9:4327. doi: 10.1038/s41598-019-40867-1
Croce, L. D., and Helin, K. (2013). Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155. doi: 10.1038/nsmb.2669
Dupret, B., Völkel, P., Le Bourhis, X., and Angrand, P. O. (2016). The Polycomb Group Protein Pcgf1 Is Dispensable in Zebrafish but Involved in Early Growth and Aging. PLoS One 11:e0158700. doi: 10.1371/journal.pone.0158700
Durston, A. J., and Zhu, K. (2015). A time space translation hypothesis for vertebrate axial patterning. Semin. Cell Dev. Biol. 42, 86–93. doi: 10.1016/j.semcdb.2015.06.001
Elliott, K. L., Pavlinkova, G., Chizhikov, V. V., Yamoah, E. N., and Fritzsch, B. (2021). Neurog1, Neurod1, and Atoh1 are essential for spiral ganglia, cochlear nuclei, and cochlear hair cell development. Faculty Rev. 10:47. doi: 10.12703/r/10-47
Elworthy, S., Pinto, J. P., Pettifer, A., Cancela, M. L., and Kelsh, R. N. (2005). Phox2b function in the enteric nervous system is conserved in zebrafish and is sox10-dependent. Mechanisms Dev. 122, 659–669. doi: 10.1016/j.mod.2004.12.008
Endoh, M., Endo, T. A., Endoh, T., Fujimura, Y., Ohara, O., Toyoda, T., et al. (2008). Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135, 1513–1524. doi: 10.1242/dev.014340
Fitze, G., König, I. R., Paditz, E., Serra, A., Schläfke, M., Roesner, D., et al. (2010). Compound effect of PHOX2B and RET gene variants in congenital central hypoventilation syndrome combined with Hirschsprung disease. Am. J. Med. Genet. Part A 146A, 1486–1489. doi: 10.1002/ajmg.a.32300
Fitzgerald, T. W., Gerety, S. S., Jones, W. D., Kogelenberg, M. V., King, D. A., McRae, J., et al. (2015). Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228. doi: 10.1038/nature14135
Furness, J. B. (2000). Types of neurons in the enteric nervous system. J. Auto. Nervous Syst. 81, 87–96. doi: 10.1016/S0165-1838(00)00127-2
Garcia-Barceló, M., Sham, M. H., Lui, V. C. H., Chen, B. L. S., Ott, J., and Tam, P. K. H. (2003). Association study of PHOX2B as a candidate gene for Hirschsprung’s disease. Gut 52, 563–567. doi: 10.1136/gut.52.4.563
Gil, J., and O’Loghlen, A. (2014). PRC1 complex diversity: Where is it taking us? Trends Cell Biol. 24, 632–641. doi: 10.1016/j.tcb.2014.06.005
Hansford, J. R., and Mulligan, L. M. (2000). Multiple endocrine neoplasia type 2 and RET: From neoplasia to neurogenesis. J. Med. Genet. 37, 817–827. doi: 10.1136/jmg.37.11.817
Hochgreb-Hägele, T., and Bronner, M. E. (2013). A novel FoxD3 gene trap line reveals neural crest precursor movement and a role for FoxD3 in their specification. Dev. Biol. 374, 1–11. doi: 10.1016/j.ydbio.2012.11.035
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229. doi: 10.1038/nbt.2501
Jain, S., Naughton, C. K., Yang, M., Strickland, A., Vij, K., Encinas, M., et al. (2004). Mice expressing a dominant-negative Ret mutation phenocopy human Hirschsprung disease and delineate a direct role of Ret in spermatogenesis. Development 131, 5503–5513. doi: 10.1242/dev.01421
Jao, L.-E., Wente, S. R., and Chen, W. (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U.S.A. 110, 13904–13909. doi: 10.1073/pnas.1308335110
Jhiang, S. M., Sagartz, J. E., Tong, Q., Parker-Thornburg, J., Capen, C. C., Cho, J. Y., et al. (1996). Targeted expression of the ret/PTC1 oncogene induces papillary thyroid carcinomas. Endocrinology 137, 375–378. doi: 10.1210/endo.137.1.8536638
Johnson, C. W., Hernandez-Lagunas, L., Feng, W., Melvin, V. S., Williams, T., and Artinger, K. B. (2011). Vgll2a is required for neural crest cell survival during zebrafish craniofacial development. Dev. biol. 357, 269–281. doi: 10.1016/j.ydbio.2011.06.034
Julia, G. (2018). Gut feelings: Studying enteric nervous system development, function, and disease in the zebrafish model system. Dev. Dynamics 247, 268–278. doi: 10.1002/dvdy.24597
Kenneth, N. W., and Michael, P. (2005). Intestinal growth and differentiation in zebrafish. Mechanisms Dev. 122, 157–173. doi: 10.1016/j.mod.2004.10.009
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dynamics 203, 253–310. doi: 10.1002/aja.1002030302
King, H. W., Fursova, N. A., Blackledge, N. P., and Klose, R. J. (2018). Polycomb repressive complex 1 shapes the nucleosome landscape but not accessibility at target genes. Genome Res. 28, 1494–1507. doi: 10.1101/gr.237180.118
Korzh, V., Sleptsova, I., Liao, J., He, J., and Gong, Z. (1998). Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev. Dynamics 213, 92–104.
Kousa, Y. A., Zhu, H., Fakhouri, W. D., Lei, Y., Kinoshita, A., Roushangar, R. R., et al. (2019). The TFAP2A-IRF6-GRHL3 genetic pathway is conserved in neurulation. Hum. Mol. Genet. 28, 1726–1737. doi: 10.1093/hmg/ddz010
Kwon, M., Lee, G. H., Lee, M. K., Kim, J. Y., Yoo, H. S., Ki, C. S., et al. (2011). PHOX2B mutations in patients with Ondine-Hirschsprung disease and a review of the literature. Eur. J. Pediatrics 170, 1267–1271. doi: 10.1007/s00431-011-1434-5
Le Faou, P., Völkel, P., and Angrand, P. O. (2011). The zebrafish genes encoding the Polycomb repressive complex (PRC) 1. Gene 475, 10–21. doi: 10.1016/j.gene.2010.12.012
Li, M., Zhao, C., Wang, Y., Zhao, Z., and Meng, A. (2002). Zebrafish sox9b is an early neural crest marker. Dev. Genes Evol. 212, 203–206. doi: 10.1007/s00427-002-0235-2
Mahler, J., and Driever, W. (2007). Expression of the zebrafish intermediate neurofilament Nestin in the developing nervous system and in neural proliferation zones at postembryonic stages. BMC Dev. Biol. 7:89. doi: 10.1186/1471-213X-7-89
Nagy, N., and Goldstein, A. M. (2017). Enteric nervous system development: A crest cell’s journey from neural tube to colon. Semin. Cell Dev. Biol. 66, 94–106. doi: 10.1016/j.semcdb.2017.01.006
Napoles, M. D., Mermoud, J. E., Wakao, R., Tang, Y. A., Endoh, M., Appanah, R., et al. (2004). Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7, 663–676. doi: 10.1016/j.devcel.2004.10.005
Olden, T., Akhtar, T., Beckman, S. A., and Wallace, K. N. (2008). Differentiation of the zebrafish enteric nervous system and intestinal smooth muscle. Genesis 46, 484–498. doi: 10.1002/dvg.20429
Olesnicky, E., Hernandez-Lagunas, L., and Artinger, K. B. (2010). prdm1a Regulates sox10 and islet1 in the development of neural crest and Rohon-Beard sensory neurons. Genesis 48, 656–666. doi: 10.1002/dvg.20673
Oppenheim, R. W. (1991). Cell death during development of the nervous system. Ann. Rev.Neurosci. 14, 453–501. doi: 10.1146/annurev.ne.14.030191.002321
Ota, S., Taimatsu, K., Yanagi, K., Namiki, T., Ohga, R., Higashijima, S. I., et al. (2016). Functional visualization and disruption of targeted genes using CRISPR/Cas9-mediated eGFP reporter integration in zebrafish. Sci. Rep. 6:34991. doi: 10.1038/srep34991
Peng, X., Feng, G., Zhang, Y., and Sun, Y. (2021). PRC1 Stabilizes Cardiac Contraction by Regulating Cardiac Sarcomere Assembly and Cardiac Conduction System Construction. Int. J. Mol. Sci. 22:11368. doi: 10.3390/ijms222111368
Pierce, S. B., Stewart, M. D., Gulsuner, S., Walsh, T., Dhall, A., McClellan, J. M., et al. (2018). De novo mutation in RING1 with epigenetic effects on neurodevelopment. Proc. Natl. Acad. Sci. U.S.A. 117, 1558–1563. doi: 10.1073/pnas.1721290115
Rogers, C. D., Jayasena, C. S., Nie, S., and Bronner, M. E. (2012). Neural crest specification: Tissues, signals, and transcription factors. Wiley interdisciplinary reviews. Dev. Biol. 1, 52–68. doi: 10.1002/wdev.8
Schemann, M. (2005). Control of gastrointestinal motility by the “gut brain”–the enteric nervous system. J. Pediatric Gastroenterol. Nutr. 41:S4–S6. doi: 10.1097/01.scs.0000180285.51365.55
Schier, A. F., and Talbot, W. S. (2005). Molecular genetics of axis formation in zebrafish. Ann. Rev. Genet. 39, 561–613. doi: 10.1146/annurev.genet.37.110801.143752
Schoorlemmer, J., Marcos-Gutiérrez, C., Were, F., Martínez, R., García, E., Satijn, D. P., et al. (1997). Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 16, 5930–5942. doi: 10.1093/emboj/16.19.5930
Schuettengruber, B., Bourbon, H. M., Croce, L. D., and Cavalli, G. (2017). Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 171, 34–57. doi: 10.1016/j.cell.2017.08.002
Shepherd, I. T., Pietsch, J., Elworthy, S., Kelsh, R. N., and Raible, D. W. (2004). Roles for GFRalpha1 receptors in zebrafish enteric nervous system development. Development 131, 241–249. doi: 10.1242/dev.00912
Shimotake, T., Go, S., Inoue, K., Tomiyama, H., and Iwai, N. (2001). A homozygous missense mutation in the tyrosine kinase domain of the RET proto-oncogene in an infant with total intestinal aganglionosis. Am. J. Gastroenterol. 96, 1286–1291. doi: 10.1111/j.1572-0241.2001.03714.x
Simon, J. H. B. (2001). Classes of enteric nerve cells in the guinea-pig small intestine. Anat. Rec. 262, 58–70. doi: 10.1002/1097-0185(20010101)262:1<58::AID-AR1011>3.0.CO;2-V
Song, N. N., Ma, P., Zhang, Q., Zhang, L., Wang, H., Zhang, L., et al. (2020). Rnf220/Zc4h2-mediated monoubiquitylation of Phox2 is required for noradrenergic neuron development. Development 147:dev185199. doi: 10.1242/dev.185199
Tahayato, A., Dollé, P., and Petkovich, M. (2003). Cyp26C1 encodes a novel retinoic acid-metabolizing enzyme expressed in the hindbrain, inner ear, first branchial arch and tooth buds during murine development. Gene Exp. Patterns 3, 449–454. doi: 10.1016/s1567-133x(03)00066-8
Thisse, C., and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protocols 3, 59–69. doi: 10.1038/nprot.2007.514
Uehara, M., Yashiro, K., Mamiya, S., Nishino, J., Chambon, P., Dolle, P., et al. (2007). CYP26A1 and CYP26C1 cooperatively regulate anterior-posterior patterning of the developing brain and the production of migratory cranial neural crest cells in the mouse. Dev. Biol. 302, 399–411. doi: 10.1016/j.ydbio.2006.09.045
van der Stoop, P., Boutsma, E. A., Hulsman, D., Noback, S., Heimerikx, M., Kerkhoven, R. M., et al. (2008). Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS One 3:e2235. doi: 10.1371/journal.pone.0002235
Velden, Y. U. V. D., Wang, L., Cano, L. Q., and Haramis, A.-P. G. (2013). The Polycomb group protein Ring1b/Rnf2 is specifically required for craniofacial development. Plos One 8:e73997. doi: 10.1371/journal.pone.0073997
Velden, Y. U. V. D., Wang, L., Lohuizen, M. V., and Haramis, A.-P. G. (2012). The Polycomb group protein Ring1b is essential for pectoral fin development. Development 139, 2210–2220. doi: 10.1242/dev.077156
Vidal, M. (2009). Role of Polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Int. J. Dev. Biol. 53, 355–370. doi: 10.1387/ijdb.082690mv
von Jonquieres, G., Mersmann, N., Klugmann, C. B., Harasta, A. E., Lutz, B., Teahan, O., et al. (2013). Glial promoter selectivity following AAV-delivery to the immature brain. PLoS One 8:e65646. doi: 10.1371/journal.pone.0065646
Voncken, J. W., Roelen, B. A. J., Roefs, M., Vries, S. D., Verhoeven, E., Marino, S., et al. (2003). Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. U.S.A. 100, 2468–2473. doi: 10.1073/pnas.0434312100
Wang, X., Yu, Q., Wu, Q., Bu, Y., Chang, N. N., Yan, S., et al. (2013). Genetic interaction between pku300 and fbn2b controls endocardial cell proliferation and valve development in zebrafish. J. Cell Sci. 126, 1381–1391. doi: 10.1242/jcs.116996
Wilson, S. W., Brand, M., and Eisen, J. S. (2002). Patterning the zebrafish central nervous system. Results Problems Cell Dif. 40, 181–215. doi: 10.1007/978-3-540-46041-1_10
Zhang, S., Xu, J., Kuang, X., Li, S., Li, X., Chen, D., et al. (2017). Biological impacts of glyphosate on morphology, embryo biomechanics and larval behavior in zebrafish (Danio rerio). Chemosphere 181, 270–280. doi: 10.1016/j.chemosphere.2017.04.094
Keywords: PRC1, rnf2, ENS, CNS, neural crest, neural precursor cells
Citation: Feng G and Sun Y (2022) The Polycomb group gene rnf2 is essential for central and enteric neural system development in zebrafish. Front. Neurosci. 16:960149. doi: 10.3389/fnins.2022.960149
Received: 02 June 2022; Accepted: 03 August 2022;
Published: 01 September 2022.
Edited by:
Debra Silver, Duke University, United StatesReviewed by:
Yan Zhou, Wuhan University, ChinaJun Wang, Hubei University of Technology, China
Hong Chen, Tongji Hospital, China
Copyright © 2022 Feng and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Feng, 1562400205@qq.com; Yuhua Sun, sunyh@ihb.ac.cn
 Gang Feng
Gang Feng Yuhua Sun
Yuhua Sun