DHA Suppresses Hepatic Lipid Accumulation via Cyclin D1 in Zebrafish
- 1China-Norway Joint Lab on Fish Gastrointestinal Microbiota, Institute of Feed Research, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Norway-China Joint Lab on Fish Gastrointestinal Microbiota, Institute of Biology, Norwegian University of Science and Technology, Trondheim, Norway
- 3Key Laboratory for Feed Biotechnology of the Ministry of Agriculture and Rural Affairs, Institute of Feed Research, Chinese Academy of Agricultural Sciences, Beijing, China
With the widespread use of high-fat diets (HFDs) in aquaculture, fatty livers are frequently observed in many fish species. The aim of this study was to investigate if docosahexaenoic acid (DHA) could be used to reduce the fatty liver in zebrafish generated by a 16% soybean oil-HFD over 2 weeks of feeding. The DHA was added to iso-lipidic HFD at 0.5, 1.0, and 2.0% of diet. Supplementation of DHA reduced growth and feed efficiency in a dose dependent manner being lowest in the HFDHA2.0 group. Hepatic triglyceride (TG) in zebrafish fed 0.5% DHA-supplemented HFD (HFDHA0.5) was significantly lower than in the HFD control. Transcriptional analyses of hepatic genes showed that lipid synthesis was reduced, while fatty acid β-oxidation was increased in the HFDHA0.5 group. Furthermore, the expression of Cyclin D1 in liver of zebrafish fed HFDHA0.5 was significantly reduced compared to that in fish fed HFD. In zebrafish liver cells, Cyclin D1 knockdown and blocking of Cyclin D1-CDK4 signal led to inhibited lipid biosynthesis and elevated lipid β-oxidation. Besides, DHA-supplemented diet resulted in a rich of Proteobacteria and Actinobacteriota in gut microbiota, which promoted lipid β-oxidation but did not alter the expression of Cyclin D1 in germ-free zebrafish model. In conclusion, DHA not only inhibits hepatic lipid synthesis and promotes lipid β-oxidation via Cyclin D1 inhibition, but also facilitates lipid β-oxidation via gut microbiota. This study reveals the lipid-lowering effects of DHA and highlights the importance of fatty acid composition when formulating fish HFD.
Introduction
Feeding high-fat diets (HFDs) is common in many farmed fish species as it increases growth rate and feed conversion. However, HFDs are also known to generate fatty livers in many fish (1–6). Although it is unclear to what extent the fatty acid compositions of the oils affect the severity of fatty livers, it has been demonstrated that oils rich in saturated (SFA) or monounsaturated fatty acids (MUFAs) are more obesogenic than oils containing highly unsaturated n – 3 fatty acids (HUFAs) (7, 8). It appears that docosahexaenoic acid, 22:6n – 3 (DHA) in particular has a distinct anti-obesity effect as shown in both mice and zebrafish (7, 9). If this is correct, then the extensive replacement of fish oil in farmed fish diets by soybean oil, palm oil and coconut oil that are deficient in HUFAs (10–13) may predispose for a nutritional condition favoring the development of fatty livers.
In mammals, dietary interventions with DHA reduce the severity of non-alcoholic fatty liver disease (NAFLD) and acute ethanol-induced hepatic steatosis. This is linked to reduced lipid synthesis and increased fatty acid oxidation (14–18). The mechanism appears, in part, to be due to modulations of transcription factors including Pparα, Nrf2, Srebp-1c, and Nfκb (19, 20). DHA has therefore been suggested as a potential treatment for fatty liver disease (21). Current data on the potential anti-obesogenic effects of DHA in fish has focused on adipose tissues. The DHA requirement of warm water fish is about 0.5-2.6% of the diet (22). Dietary supplementation of 0.6% DHA reduces lipid accumulation via induction of adipocyte apoptosis (23). Using primary cultures of preadipocytes from cobia showed that DHA hinders preadipocyte differentiation and lipid accumulation from cobia (24). There is however a general lack of understanding of the underlying mechanisms on how DHA regulates hepatic lipid metabolism.
The main fate of dietary DHA in cells is to incorporate into membrane phospholipids (25–27). DHA can regulate basic membrane properties such as fluidity, permeability and protein activity (26), thus affecting cell growth and proliferation (28). The first gap (G1) phase is a period between nuclear division and DNA synthesis, during which various signals derived from metabolism, stress and environmental factors are integrated to determine the developmental program including cell division, differentiation or death (29). Cyclin D1, a critical target of proliferative signals in the G1 phase, plays an important role in the transition at end of G1 phase when cells move into the synthesis (S) phase just before cell division (30). In addition to its role in cell cycle progression, the high expression of Cyclin D1 in NAFLD and livers with microsteatosis suggests its possible importance in fatty liver diseases and lipid metabolism (31, 32). Moreover, the drug, metformin, used for treating NAFLD leads to inhibition of Cyclin D1 (33). A recent study in blunt snount bream (Megalobrama amblycephala) showed that dietary DHA induces liver cell cycle arrest at G1 phase (34). However, whether similar links exist between Cyclin D1 and lipid metabolism in fish is still uncertain.
In this study, we evaluated the effects of DHA on growth and hepatic lipid accumulation in zebrafish fed with three levels of DHA (0.5, 1.0, and 2.0%) in diets. We also measured the expression levels of genes related to lipid synthesis, fatty acid oxidation in the liver. The effects of DHA on the basic cell life processes were evaluated in zebrafish liver (ZFL) cells, which were further validated by analyzing the expression of Cyclins. The roles of Cyclin D1 were also validated in a gene-knockdown ZFL model. Finally, the effect of gut microbiota and possible consequences of microbial changes on fish physiology were further assessed in HFD and 0.5% DHA-supplemented HFD (HFDHA0.5) groups by transferring gut micobiota to a germ-free (GF) zebrafish model.
Materials and Methods
Fish Husbandry
Zebrafish of the Tübingen strain were maintained at the zebrafish facility of the Institute of Feed Research, Chinese Academy of Agricultural Sciences (Beijing, China). The size of each tank was 25.5 × 18.5 × 18.0 cm. During the 2-wk feeding period, the water in the rearing system was kept running, the rearing temperature was 25-28°C, the dissolved oxygen was >6.0 mg/L, the pH was 7.0-7.2, the nitrogen content was <0.50 mg/L and the nitrogen content (as NO2) was <0.02 mg/L. Zebrafish were maintained at a 14:10 L:D cycle.
Diets and Feeding Trial
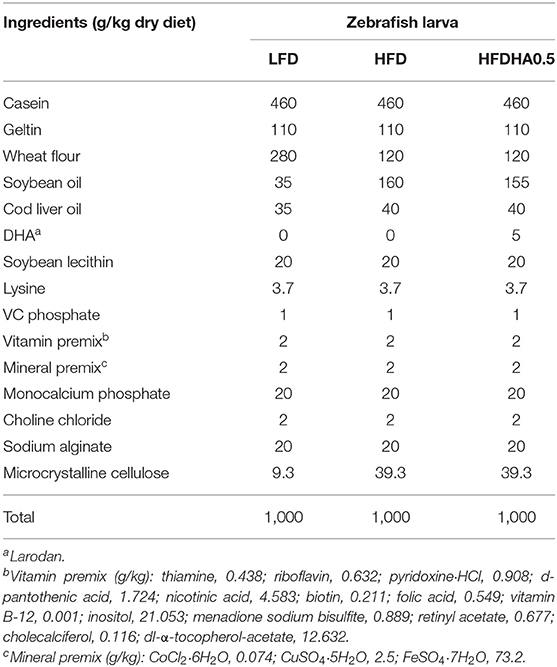
Feed formulation and fatty acid composition of diets are presented in Tables 1, 2. Casein, soybean oil and wheat flour were used as dietary protein, lipid and carbohydrate sources, respectively. The low-fat diet (LFD) was supplemented with soybean oil with 60 g/kg and the HFD was supplemented with soybean oil with 160 g/kg (Table 1). The DHA used in the feeding trial was at purity of 90% (Larodan, 10-2206-90-13). Increased DHA levels were compensated by decreasing equal levels of soybean oil. All dry ingredients were ground through a 60-mesh screen. The diets were prepared by mixing the dry ingredients with the oil and water manually. Then each diet was extruded in a manual extruder with a 2.5-mm aperture. The extruded pellets were freeze-dried and stored at −20°C in plastic bags in small quantities. Before feeding, the feed pellets were ground through a 30-mesh screen.
During the feeding trial, healthy, uniformly sized 1-month-old zebrafish (1.160 ± 0.005 g/20 fish) were divided into five groups at random and fed the LFD, HFD, or 0.5, 1.0, 2.0% DHA-supplemented HFDs (HFDHA0.5, HFDHA1.0, HFDHA2.0, respectively; Table 1). The feed amount was at 6% of body weight, twice a day at 9:00 and 16:00. Each group contained three tanks with 20 fish per tank.
Sample Collection and Analysis
All fish were anesthetized with tricaine methanesulfonate (MS222) before sampling. At the end of the 2-wk feeding trial, the survival rate in each group was recorded (100 × final survival individuals/initial individuals); the fish in each tank were weighed to calculate weight gain (100 × [final body weight – initial body weight]/initial body weight) and feed efficiency (100 × [final body weight – initial body weight]/feed intake). The liver was collected at 24 h after the last feeding for analysis of histology (H&E), triacylglycerol (TG) content, fatty acid compositions detection and gene expression. The gut contents were collected at the 4-6 h after the last feeding. The gut contents were collected under aseptic conditions. The digesta samples from the six fish were pooled as a replicate. The muscle was collected at 24 h after the last feeding for analysis of TG content and gene expression.
Histological Analysis
The livers of zebrafish were rinsed with sterilized PBS, fixed in 4% formalin solution, and embedded in paraffin. For histological analysis, the embedded livers were cut into 5-8 μm sections and placed on slides. After dewaxing, hematoxylin and eosin were used to stain nucleus and cytoplasm, respectively. Following dehydration by alcohol and being transparent by xylene. For oil red staining, the embedded livers were cut into 10 μm sections and placed on slides. Liver sections were immersed by 60% isopropanol for 2 min. Then liver sections were stained by oil red solution and hematoxylin. After staining, liver sections were mounted by resin. Images were obtained Zeiss microscope (AXIO Observer A1).
TG Detection
Liver TG was extracted by chloroform: methanol (2: 1) and TG of ZFL cells was extracted by n-hexane: isopropanol (3: 2). Fresh livers were homogenized in 1 mL PBS, then mixed with 5 mL chloroform: methanol (2: 1) vigorously. After standing for 10 min, TG-contained sublayer was extracted and heated with nitrogen at 70°C. ZFL cells were broken by sonication in 1 ml cell lysis, and then mixed with 5 ml n-hexane: isopropanol (3: 2) vigorously. After standing for 10 min, TG-contained uplayer was extracted and heated with nitrogen at 70°C. TG was dispersed by deionized water. The TG content was determined by a quantitative enzymatic method as described in a previous study (35) and was expressed as TG weight per gram protein (mg. g protein−1).
Fatty Acid Extraction and Analysis
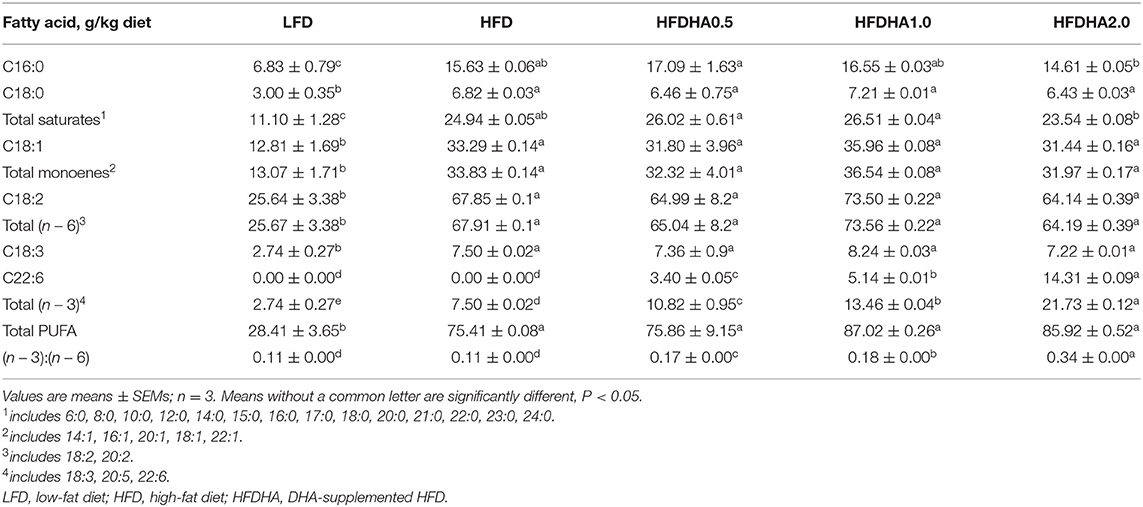
Total lipid was extracted from feed and liver freeze-dried powder in chloroform/methanol (2:1, v/v), methylated, and trans-esterified with boron trifluoride in methanol. FAMEs were separated and quantified by GC (Agilent 6890; Agilent, Savage, MD, USA) equipped with a flame ionization detector and a 60.0 m × 250 μm × 0.25 μm fused silica capillary column (DB-23). Helium was used as the carrier gas at a flow rate of 2 ml/min. The temperature was initially 50–220°C at 4°C/min, and then held at 220°C for 35 min. The injector and detector temperatures were set at 260 and 270°C, respectively. Individual methyl esters were identified by comparison to known standards (12, 36).
DHA and Oleic Acid Solution Preparation
The DHA (D2534, CAS#6217-54-5) and oleic acid (OA, CAS#143-19-1) used in cell culture was purchased from Sigma (USA). A solution of 10 mM DHA and OA were prepared by dilution in 5% fatty acid-free BSA (Sigma, CAS#9048-46-8) and storaged in −20°C. The prepared DHA and OA solutions were subjected to vortexing before use in cell culture.
Cell Culture and Treatments
The ZFL cell line was purchased from American Type Culture Collection (Manassas, VA, USA), and cultured according to established protocols (37). All media, L-15 (CAS#10045CVR), DMEM (LOT#14318009) and DMEM/F12 (LOT#35220009), were obtained from Corning Inc. (New York, NY, USA). Penicillin-Streptomycin solution (LOT#30002304) and bovine insulin (CAS#11070-73-8) were purchased from Sigma (St. Louis, MO, USA). Murine epidermal growth factor (LOT#0221179) was purchased from Peprotech (Rocky Hill, NJ, USA). Rainbow trout serum (LOT#05191027) was purchased from Caisson Labs (USA). For the detection of cell growth by Real-Time Cell Analyzer (RTCA) xCelligence System (ACEA Biosciences Ins, Santiage, USA), the treatment of DHA was lasting for 48 h. For TG quantification and qPCR analysis, the treatment periods were 24 h.
RTCA xCelligence System (ACEA Biosciences Ins, Santiage, USA) was used to detect real-time growth of ZFL cells treated with different doses of DHA (prepared in 1% BSA) for 48 hrs. ZFL cells were seeded at a density of 3 × 104 cells/well on an E-plate. The treatment started 24 h post-seeding (sub-confluence) by adding DHA to fresh medium at final concentrations of 10 and 50 μM. CI values were monitored in 15 min intervals to 48 h after treatments.
For cell cycle and cell apoptosis analysis, ZFL cells were seeded at a density of 1 × 105 cells/well on a six-well plate. The treatment started 24 h post-seeding by adding DHA to fresh medium at final concentrations of 10 and 50 μM. After 24-h treatment, cells were harvested for cell cycle and cell apoptosis analysis by Guava easyCyte Flow Cytometer (Merck Millipore, Stafford, VA, USA).
For the determination of intracellular TG level and gene expression, ZFL cells were were seeded at a density of 1 × 105 cells/well on a six-well plate and treated with 100 μM OA (OA100) and mixture of 50 μM OA and 50 μM DHA (ODHA) at 24 h post-seeding. After 24-h treatment, cells were harvested for the detection of intracellular TG level and gene expression.
LY2835219 (Selleck) prepared in water, an effective inhibitor of Cyclin D1-CDK4 pathway, was used to block Cyclin D1-CDK4 signal. ZFL cells were seeded at a density of 1 × 105 cells/well on a six-well plate. After 24 h, LY2835219 was added to cell medium at 2 nM. Cells were harvested at 24 h post-treatment. Then expression of genes related to lipid metabolism were detected by real-time fluorescence quantitative PCR (qPCR).
Cell Growth by Real-Time Cell Impedance Analysis
The RTCA xCelligence System (ACEA Biosciences Inc., San Diego, CA, USA) was used to detect real-time cell growth at different doses of DHA. The RTCA software supplied by the manufacturer was used to analyze cell growth and calculate the doubling time of the cells based on the cell index. ZFL cells were seeded at a density of 3 × 104 cells/well on an E-plate. DHA treatment started at 24 h post-seeding. Cell index values were monitored in 15-min interval during 48 h-treatment. The results were plotted as lines by the RTCA software.
Cell Apoptosis and Cell Cycle Analysis With Flow Cytometry
Cell apoptosis detection was performed with Annexin V-fluorescein isothiocyante (FITC) kits (Sigma, LOT#095M4121V). After exposure to the indicated concentrations of DHA for 24 hrs, cells were collected and incubated with Annexin V-FITC and propidium iodide (PI) in binding buffer in the dark for 10 min at room temperature. The analysis was conducted by the Guava easyCyte Flow Cytometer (Merck Millipore, Stafford, VA, USA). The fluorescence intensity was measured at an excitation wavelength of 488 nm using GRN (525 nm) and RED (690 nm) filters. Data analysis was performed using Flow Guava software.
For cell cycle assay, ZFL cells were fixed with 70% ethanol at 4°C for 24 h and then stained with PI in binding buffer at 37°C in dark for 30 min. Cell cycle was detected by measuring the DNA contents in cells using guava easyCyte flow cytometer. The proportions of cells in the SUBG1, G1, S, and G2/M phases were calculated by Flow guava software. Fluorescence was measured with an excitation wavelength of 488 nm through RED filters (690 nm). Data analysis was performed using Flow Guava software.
Bodipy Staining
ZFL cells were washed with PBS three times, fixed with 4% paraformaldehyde for 1 hr, and stained with bodipy 493/503 (1 μg / mL in PBS) (Invitrogen, CAS#D3992) in the dark for 15 min at room temperate. The bodipy-stained cells were re-stained with Hochest (1 μg/ml in PBS) (Invitrogen, CAS#H21486) for 5 min following with PBS rinse for three times. Image was acquired with Leica microscope (Leica DMIL-LED).
Gene Silencing With siRNA
Negative control (scrambled siRNA, NC), Cyclin D1 siRNA (Table 3) were synthesized by GenePharma Co. Ltd. (Shanghai, China). Cells were first seeded on 6-well plates (Corning) and incubated for 24 h to sub-confluence. Then the medium was removed and the cells were transfected with the scrambled siRNA and Cyclin D1 siRNA using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, LOT#2098845). The concentration of siRNA stock solution was 20 μM. One transfection dispersion for one well consisted of 25 pmol siRNA. After 24 h, efficiency of the siRNA was determined by qPCR.
Gut Microbiota Analysis
The 16s V3–V4 region was amplified using the primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-CCATTGTAGCACGTGTGTAGCC-3′). The 16S ribosomal RNA gene sequencing was performed by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China) using Illumina MiSeq PE300 platform (Illumina, San Diego, USA). Then the raw pair-end readings were subjected to a quality-control procedure using the UPARSE-operational taxonomic unit (OTU) algorithm (38). The qualified reads were clustered to generate OTUs at the 97% similarity level using the USEARCH sequence analysis tool (38). A representative sequence of each OTU was assigned to a taxonomic level in the Ribosomal Database Project (RDP) database using the RDP classifier (39).
Germ-Free Zebrafish Culture and Treatment
GF zebrafish were generated from normal parent zebrafish through disinfecting with antibiotics, PVP and sodium hypochlorite in sequence, and reared following established protocols (40). Zebrafish larvae hatched from their chorions at 3 days postfertilization (dpf). Each group had six bottles with 20 fish per bottle. At 5 dpf, the yolk was largely absorbed and the GF-zebrafish was subjected with feeding and transferring trials. In the feeding trial, GF zebrafish were fed with LFD, HFD and HFDHA0.5 (Table 4). All dry ingredients were ground through a 100-mesh screen. The diets were prepared by mixing the dry ingredients with the oil and water manually. Then each diet was extruded in a manual extruder with a 2.5-mm aperture. The extruded pellets were freeze-dried and stored at −20°C in plastic bags in small quantities. Before sterilization, the feed pellets were ground through a 60-mesh screen. The diets for GF zebrafish were sterilized by irradiation with 20 kGy gamma ray (Institute of Food Science and Technology, Chinese Academy of Agricultural Sciences, Beijing, China).
In the transferring trial, GF zebrafish were transferred with gut microbiota deprived from conventional zebrafish fed LFD, HFD and HFDHA0.5. At 12 dpf, GF zebrafish in the feeding trial were collected for for oil red staining and qPCR. At 8 dpf, zebrafish larvae transferred with LFD, HFD and HFDHA0.5-gut microbiota were collected for oil red staining and qPCR.
Total RNA Extraction, Reverse Transcription, and QPCR
Total RNA was isolated using Trizol reagent (LOT#03877/35120) (Cwbio, Beijing, China) and then reversed transcribed to cDNA by FastKing gDNA Dispelling RT SuperMix (LOT#W9813) (Tiangen, Beijing, China). The qPCR was performed using SYBR®Green Supermix (LOT#W9413) according to the manufacturer's instructions (Tiangen, Beijing, China). The results were stored, managed, and analyzed using LightCycler 480 software (Roche, Basel, Switzerland). The qPCR primers used are listed in Table 3.
Data Analysis
The statistical analyses were conducted using GraphPad Prism 5 softw4are (GraphPad Software Inc., San Diego, CA, USA). Results are expressed as the means ± standard errors of the means (SEMs). Comparisons between two groups were analyzed using the Student's t-test, and comparisons between multiple groups were analyzed using one-way ANOVA followed by a Duncan's test. The statistical significance was set at P < 0.05 and P < 0.01.
Results
The Effects of DHA on Survival, Body Weight Gain, Feed Efficiency
At the end of 2-wk feeding trial, survival was similar among groups (Figure 1A). The body weight gain in HFD-fed zebrafish was moderately higher than that of fish fed LFD (64.56 vs. 54.44%; Figure 1B). Zebrafish fed HFDHA0.5 had moderate lower body weight gain than that in zebrafish fed HFD (55.21 vs. 64.56%) (Figure 1B). However, zebrafish fed HFDHA1.0 and HFDHA2.0 had significantly lower body weight gain as compared to those fed HFD (53.42, 48.77 vs. 64.56%, respectively; P < 0.05; Figure 1B). The feed efficiency of zebrafish fed HFDHA0.5, HFDHA1.0, and HFDHA2.0 were 45.65, 44.16, and 40.52%, respectively (Figure 1C). Consistently, the feed efficiency in HFDHA0.5 group was non-significantly lower than that in HFD group (Figure 1C). The feed efficiency in HFDHA1.0 and HFDHA2.0 groups were significantly lower than that in HFD group (P < 0.05; Figure 1C). These results suggested that the supplementation of DHA to HFD can inhibit fish growth, especially at the additive amount higher than 0.5%.
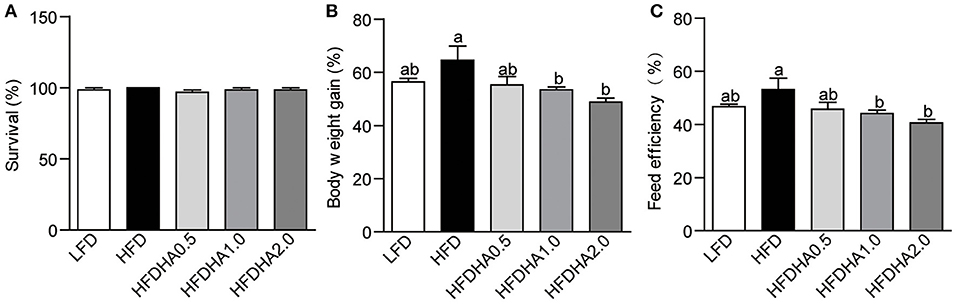
Figure 1. Effects of DHA supplementation on (A) Survival, (B) body weight, (C) feed efficiency. Values are means ± SEMs (n = 3 biological replicates). Means without a common letter are significantly different, P < 0.05.
The Effects of DHA on Hepatic Lipid Metabolism
Zebrafish fed HFD developed a large vesicular intracellular lipid droplets as revealed by H&E staining and oil red staining (Figures 2A,B). Zebrafish fed HFDHA0.5, HFDHA1.0, and HFDHA2.0 showed a tendency of alleviated hepatic steatosis as compared to those fed HFD (Figures 2A,B). However, only HFD supplemented with 0.5% DHA significantly led to a 40.8% reduction in hepatic TG level (P < 0.05; Figure 2C). Thus, only the 0.5% DHA supplementation was used for the further analysis in this study. This also appeared in a 4-wk feeding trial (Supplementary Figure 1).
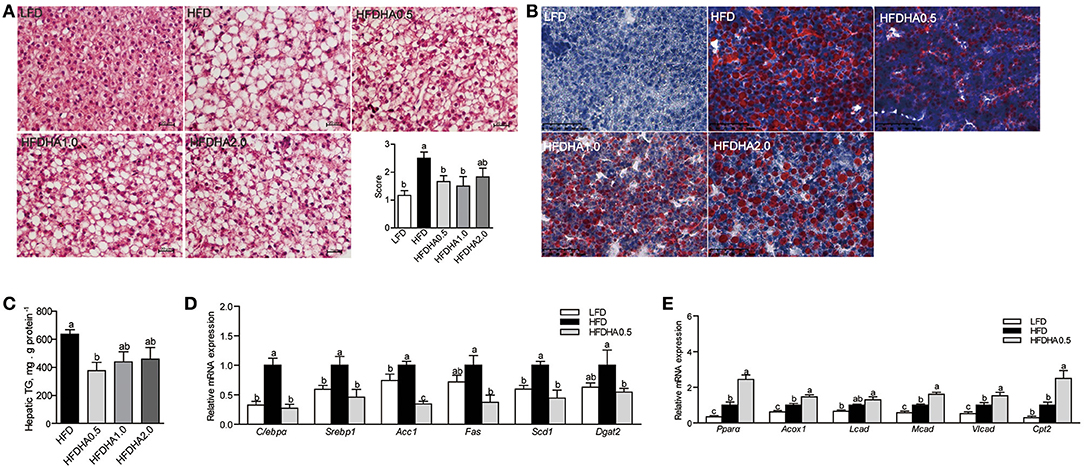
Figure 2. Effects of DHA supplementation on hepatic lipid metabolism. (A) H&E staining of liver sections of LFD, HFD, and HFDHAs-fed zebrafish for 2 wks. The scale bar is 20 μm. (B) Oil red staining of liver sections of LFD, HFD, and HFDHAs-fed zebrafish for 2 wks. The scale bar is 50 μm. (C) Hepatic TG content of zebrafish fed HFD and HFDHAs for 2 wks. Relative mRNA expression of genes related to (D) lipid synthesis and (E) β-oxidation. Values are means ± SEMs (n = 3-5 biological replicates). Means without a common letter are significantly different, P < 0.05.
Relative to LFD, feeding HFD significantly upregulated liver mRNA expression of CCAAT/enhancer binding protein α (C/ebpα), sterol regulatory element binding transcription factor 1 (Srebp1), acetyl-CoA carboxylase (Acc1), stearoyl-Coenzyme A desaturase 1 (Scd1) and diacylglycerol O-acyltransferase 2 (Dgat2) (P < 0.05; Figure 2D). When the fish were fed the HFDHA0.5 and compared to the HFD, the relative mRNA expressions of C/ebpα, Srebp1, Acc1, Fas, Scd1, and Dgat2 in liver were reduced and for most parts similar to that of LFD-fed fish (P < 0.05; Figure 2D). Feeding HFD led to a small increase in peroxisome proliferator activated receptor α (Pparα), acyl-CoA oxidase 1 (Acox1) and medium-chain acyl-CoA dehydrogenase (Mcad), but not long-chain acyl-CoA dehydrogenase (Lcad), carnitine palmitoyltransferase 2 (Cpt2) when compared to those fed LFD (Figure 2E), however, feeding HFDAH0.5 led to massive increases in the relative mRNA expression of all these genes (P < 0.05; Figure 2E). Although the total hepatic PUFA content in HFD and HFDHA0.5 groups showed no significant difference, the concentration of n – 3 PUFA in HFDHA0.5 group was 46.7% higher than that in HFD group (P < 0.01; Table 5). Moreover, zebrafish fed HFDHA0.5 had higher hepatic DHA level than fish fed HFD (P < 0.05; Table 5). These results suggested that the supplementation of 0.5% DHA can regulate hepatic lipid metabolism partly by inhibiting hepatic lipid synthesis and promoting lipid β-oxidation, and increase the accumulation of DHA in liver.
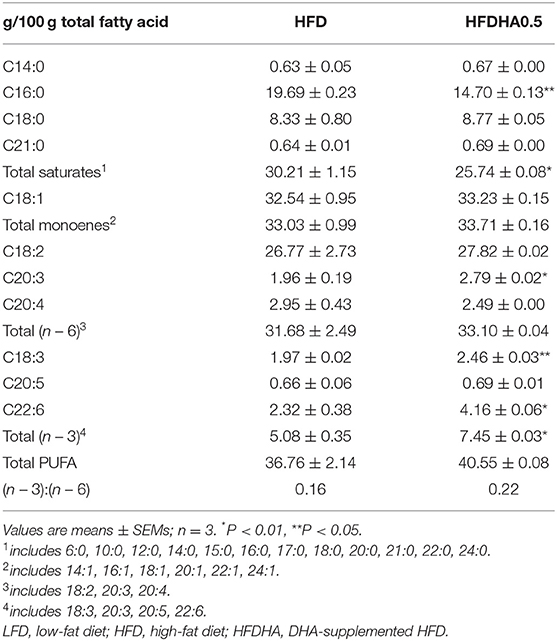
Table 5. Fatty acid composition in the liver of 1-month-old zebrafish fed diets containing DHA for 2-wk.
We also confirmed the effect of dietary DHA on muscle, the largest part of fish body. The muscular TG level and the mRNA expression of Scd1 and Dgat2 were non-significantly reduced while muscular Acox1 and Cpt2 were non-significantly increased in HFDHA0.5 group as compared to HFD group (Supplementary Figures 2A–C). Thus, 0.5% DHA supplementation had no significant effect on muscle lipid metabolism.
DHA Modulates Lipid Metabolism in ZFL Cell Model
The results showed that the intracellular lipid droplets, marked by the green fluorescencent probe BODIPY 493/503, were reduced in cells treated with ODHA (Figure 3A). Moreover, the TG content in ZFL cells treated with ODHA was 22.6% lower than that in OA100-treated cells (P < 0.05; Figure 3B). The relative mRNA expression of lipid synthesis-related genes (C/ebpα, Scd1 and Dgat2) in cells treated with ODHA were significantly lower than that in OA100-treated cells (0.53-, 0.33-, and 0.30-fold; P < 0.05; Figure 3C), whereas the relative mRNA expressions of the fatty acid β-oxidation-related genes Pparα, Acox1, and Cpt2 were significantly higher than that in OA100-treated cells (2.33-, 1.59-, and 1.65-fold; P < 0.05; Figure 3D). These results indicated that DHA can also regulate lipid metabolism by inhibiting lipid synthesis and promoting β-oxidation in vitro.
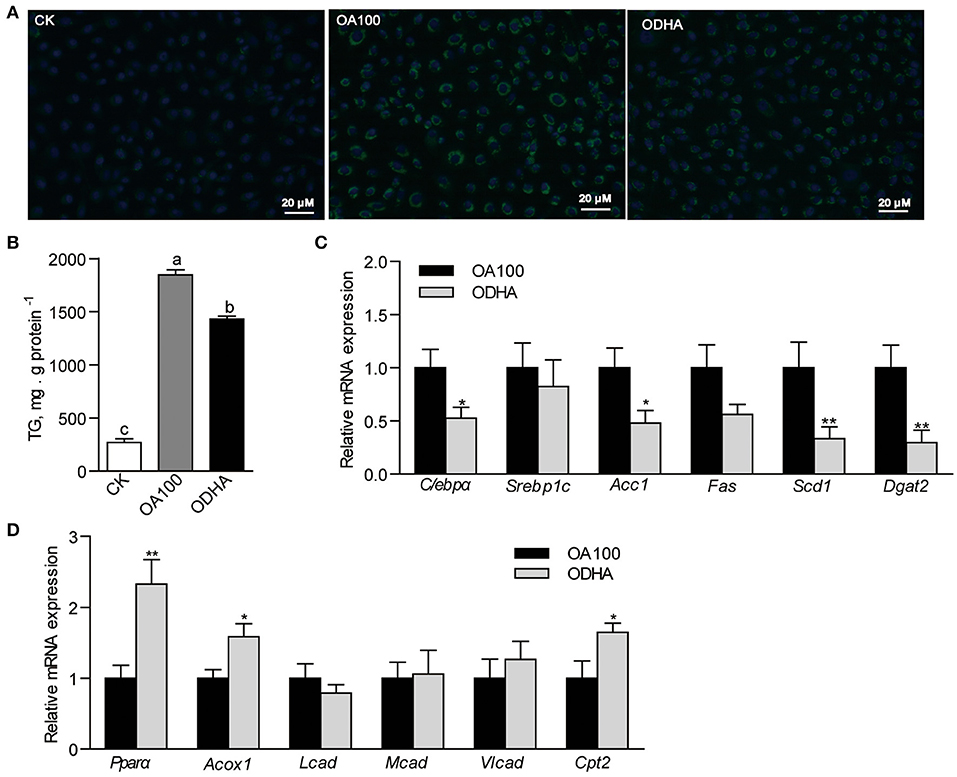
Figure 3. Modulation of DHA on lipid metabolism in ZFL cells. (A) Lipid droplets accumulation in ZFL cells with lipid treatment. The scale bar is 20 μm. (B) TG content in ZFL cells treated with CK, OA, and ODHA. (C) Relative mRNA expression of genes related to lipid synthesis and (D) β-oxidation in control and lipid treated ZFL cells. Values are means ± SEMs (n = 5-6 biological replicates). Means without a common letter are significantly different, P < 0.05. *P < 0.05; **P < 0.01. CK, controls; OA, 100 μM oleic acid; ODHA, mix of 50 μM OA and 50 μM DHA.
DHA Modulates Cyclin D1 Expression in Liver
Results showed that DHA inhibited cell growth in a concentration-dependent manner (Figure 4A). At 24 h, the growth of ZFL cells treated with 10 and 50 μM DHA was inhibited by 24.5 and 51.4%, respectively (Figure 4A). However, DHA did not induce cell apoptosis (Figure 4B). Flow cytometry analysis of the cell cycle indicated that 50 μM DHA arrested ZFL cells in the G1 phase at 24 h (Figure 4C). The relative mRNA expression of Cyclin D1 in 10 and 50 μM DHA-treated ZFL cells were significantly reduced as compared to control cells, respectively (P < 0.05; Figure 4D).
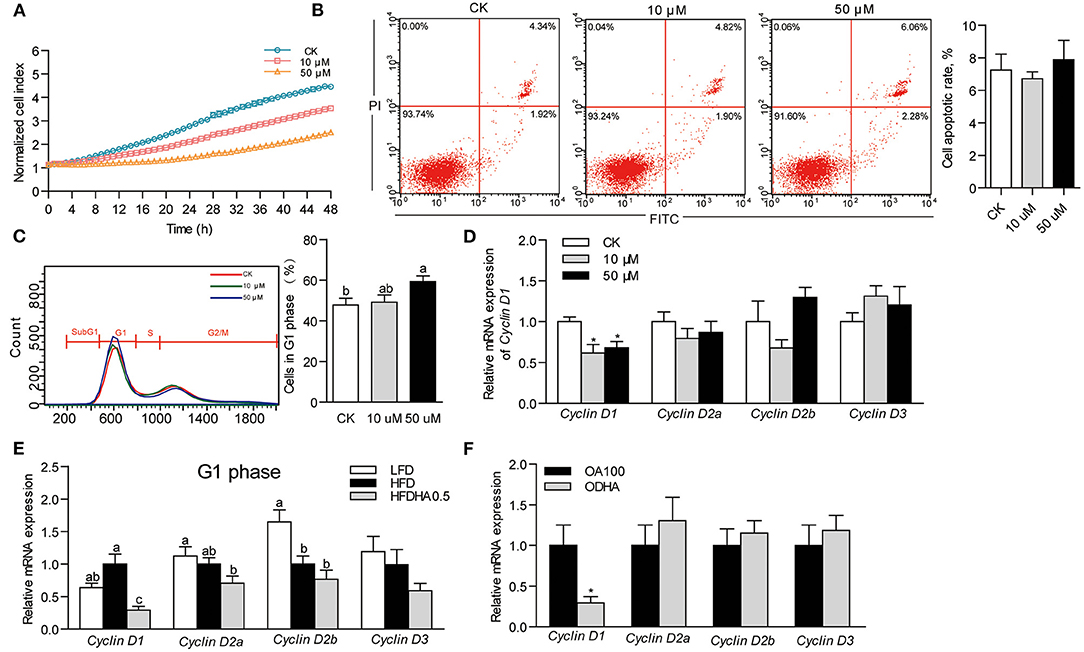
Figure 4. Effects of DHA on cell growth, cell apoptosis and cell cycle. (A) Growth of ZFL cells treated with DHA for 48 h. (B) Cell apoptosis and (C) cell cycle of ZFL treated with DHA at 24 h. (D) Relative mRNA expression of genes encoding D-type Cyclins in ZFL cells. (E) Relative mRNA expression of genes encoding D-type Cyclins in the liver of LFD, HFD, and HFDHA0.5-fed zebrafish. (F) Relative mRNA expression of genes encoding D-type Cyclins in ZFL cells treated with OA and ODHA. Values are means ± SEMs (n = 3-5 biological replicates). Means without a common letter are significantly different, P < 0.05. *P < 0.05.
Accordingly, we analyzed cell cycle by analyzing the expression of genes encoding cell cycle modulators of zebrafish liver. Changing from LFD to HFD had little effect on the mRNA expression of the G1 phase-related modulators Cyclin D1, Cyclin D2a and Cyclin D3 but reduced the expression of Cyclin D2b (Figure 4E). Replacing HFD to HFDHA0.5 led to a clear tendency of further reduction of all genes but was only significant for Cyclin D1 (0.29-fold; P < 0.05; Figure 4E). The relative mRNA expression of the G2 phase-related modulator, Cyclin B, was similar between zebrafish fed HFD and HFDHA0.5 (Supplementary Figure 3A). Besides, the relative mRNA expression of Cyclin D1 in ODHA-treated ZFL cells was significantly reduced as compared to that in OA100-treated cells (0.29- fold; P < 0.05; Figure 4F). These results indicated that DHA can induce cell cycle arrest at G1 phase both in liver tissue and in vitro cell model.
The Suppression of Cyclin D1 Modulates Lipid Metabolism in ZFL Cell Model
To determine the relationship between Cyclin D1 and lipid metabolism, we repressed the expression of Cyclin D1 by transfecting siRNA into ZFL cells. The growth of Cyclin D1-knockdown ZFL cells was slower than that in NC cells (Figure 5A). However, Cyclin D1 knockdown showed no significant influence on cell apoptosis (Figure 5B). The results showed that siRNA of Cyclin D1 reduced its mRNA expression by 70.7% as compared to NC (P < 0.05; Figure 5C). The expression of the lipid synthesis-related genes (C/ebpα, Scd1, and Dgat2) in Cyclin D1-knockdown ZFL cells were significantly lower than that in NC-treated ZFL cells (0.43-, 0.43- and 0.56-fold; P < 0.05; Figure 5D), while the relative mRNA expression of the fatty acid β-oxidation-related genes Pparα, Acox1, and Cpt2 were significantly higher (2.11-, 1.45-, and 1.59-fold; P < 0.05; Figure 5E). Furthermore, when we used the Cyclin D1-Cdk4 inhibitor, LY2835219, to induce cell arrest in G1 phase, we observed decreased expression of genes related to lipid synthesis (C/ebpα, Scd1, Acc1, and Dgat2) (Figure 5F) while the expression of genes related to lipid β-oxidation (Pparα, Acox1, and Cpt2) increased (Figure 5G). These results suggested that Cyclin D1 and Cyclin D1-Cdk4 signal were involved with, at least partly, the modulation of genes expression involved in lipid metabolism.
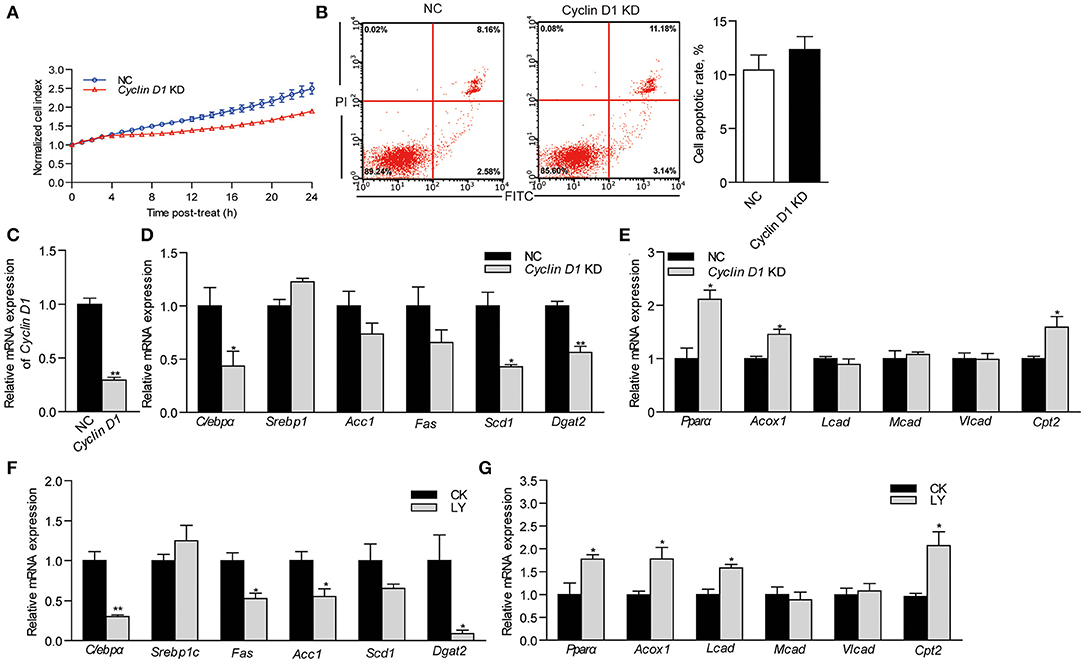
Figure 5. Effects of Cyclin D1 knockdown and Cyclin D1-Cdk4 blocking on lipid metabolism. (A) Growth of NC and Cyclin D1-knockdown ZFL cells. (B) Apoptosis of NC and Cyclin D1-knockdown ZFL cells. (C) Efficiency of siRNA targeting cyclin D1 in ZFL cells. (D) Relative mRNA expression of genes related lipid synthesis in ZFL cells with Cyclin D1 knockdown. (E) Relative mRNA expression of genes related β-oxidation in ZFL cells with Cyclin D1 knockdown. Relative mRNA expression of genes related (F) lipid synthesis and (G) lipid β-oxidation in ZFL cells treated with LY2835219. Values are means ± SEMs (n = 3 biological replicates). *P < 0.05; **P < 0.01.
Dietary DHA Directly Modulates Lipid Metabolism in Germ-Free Zebrafish Model
To investigate the effects of dietary DHA with no involvement of gut microbiota, HFDHA0.5 was fed to GF-zebrafish larvae. After feeding for 1-wk, HFDHA0.5-fed GF zebrafish had reduced hepatic steatosis compared to HFD-fed zebrafish (Figure 6A) and reduced relative mRNA expression of Cyclin D1 (P < 0.05; Figure 6B). Furthermore, the relative expression of the lipid synthesis-related genes tested were significantly lower in the HFDHA0.5 group compared to those fed HFD (P < 0.05; Figure 6C), while the expression of some lipid β-oxidation-related including Pparα, Acox1 and Cpt2 were upregulated (P < 0.05; Figure 6D). These results suggested that DHA can directly modulate lipid metabolism in zebrafish.
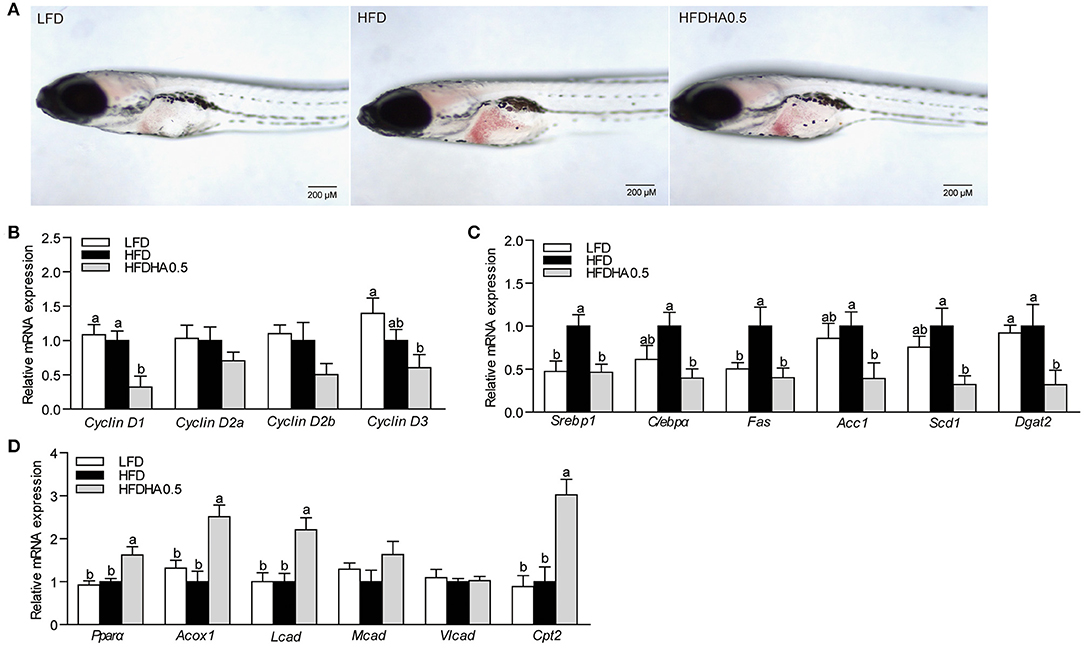
Figure 6. Effects of DHA supplementation on lipid metabolism in GF zebrafish. (A) Whole-mount oil red staining in GF zebrafish fed LFD, HFD and HFDHA0.5. The scale bar is 200 μm. (B) Relative mRNA expression of CyclinDs. Relative mRNA expression of (C) lipid synthesis-related, and (D) lipid β-oxidation-related genes. Values are means ± SEMs (n = 4 biological replicates). Means without a common letter are significantly different, P < 0.05.
Dietary DHA Supplementation Leads to Gut Microbial Community Changes in Zebrafish
At the end of the 2-wk feeding trial, we analyzed the effects of DHA supplementation on the gut microbial community. The results clearly showed that adding DHA to the HFD significantly altered the composition as shown through clustering analysis in Figure 7A. Proteobacteria (95.9%) was the dominant phylum in HFD-fed zebrafish (Figure 7B), while Proteobacteria (50.2%) and Actinobacteriota (47.3%) were the predominant phylums in HFDHA0.5-fed zebrafish (Figure 7B). Relative abundance analysis at the genus level showed significant enrichment of Mycobacterium in the HFDHA0.5 group, whereas the abundance of Pseudomonas was significantly reduced in the HFDHA0.5 group as compared to the HFD group (P < 0.05; Figure 7C; Table 6). The diversity indexes of the gut microbial community including Shannon-, Ace- and Chao-indexes were higher in the HFDHA0.5 (P < 0.05), while the Simpson-index was lower (P < 0.05) that in HFD group (Table 7).
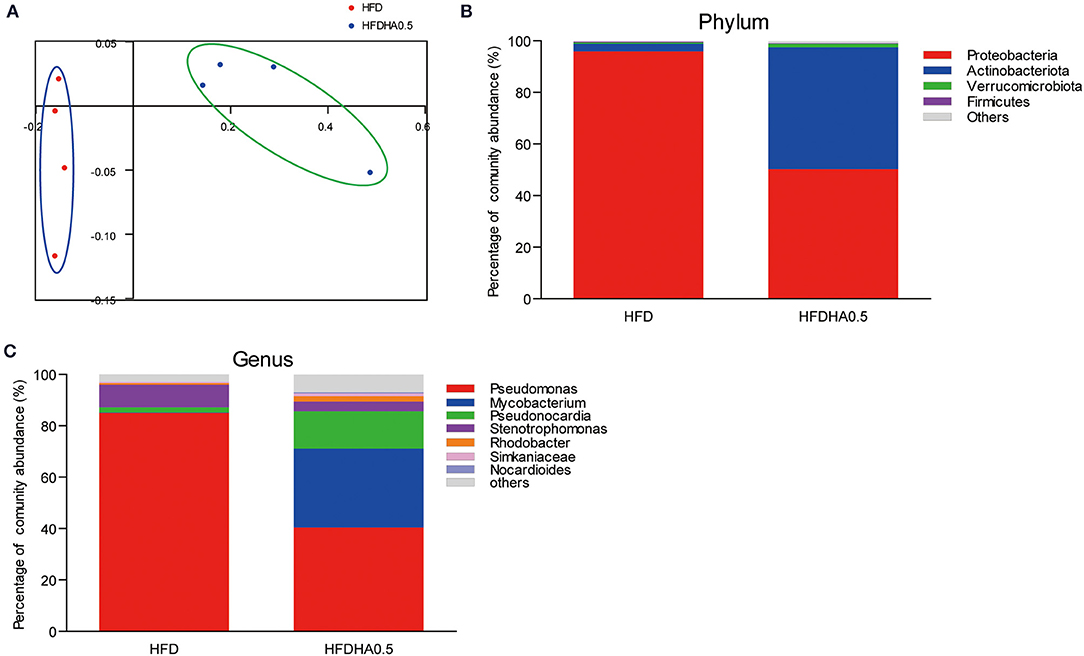
Figure 7. Effects of DHA supplementation on gut microbial community in zebrafish. (A) PCoA analysis of gut microbiota in HFD- and HFDHA0.5-fed zebrafish. (B) Relative abundance at the phylum level of gut microbial community from HFD- and HFDHA0.5-fed zebrafish (C) Relative abundance at the genus level of gut microbial community from HFD- and HFDHA0.5-fed zebrafish. Values are means ± SEMs (n = 4 biological replicates).
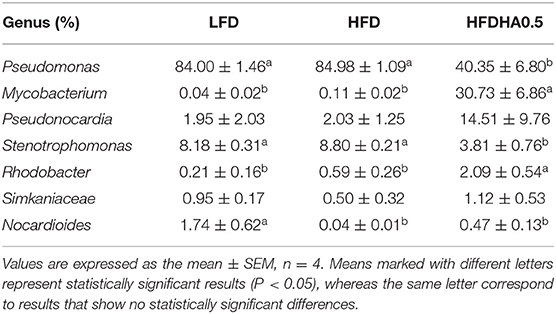
Table 6. The predominant gut bacterial genus in zebrafish fed the LFD, HFD, or HFDHA 0.5 for 2 weeks based on V3–V4 sequences.
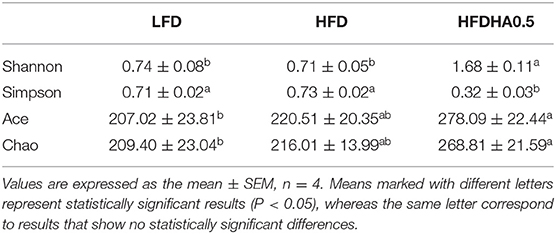
Table 7. Diversity index of gut bacteria of zebrafish fed with the LFD, HFD or HFDHA0.5 for 2 weeks.
DHA-Altered Gut Microbiota Contribute to Lipid Oxidation in Germ-Free Zebrafish
To investigate the microbiota-mediated effects, gut microbiota from the HFD and HFDHA0.5 groups were transferred to GF-zebrafish. HFDHA0.5 microbiota-colonized GF zebrafish showed lower hepatic steatosis than HFD microbiota-colonized zebrafish (Figure 8A). The colonization of HFDHA0.5-microbiota for 72 h showed no effect on the expression of Cyclin Ds (Cyclin D1, D2a, D2b, and D3) (Figure 8B) and genes related to lipid synthesis (Figure 8C), but led to higher expression of the lipid β-oxidation-related genes Pparα and Acox1(P < 0.05; Figure 8D) as compared to HFD-microbiota. These results showed that the gut microbial community induced by DHA-supplemented diet can modulate lipid metabolism by promoting lipid β-oxidation independent of Cyclin D1.
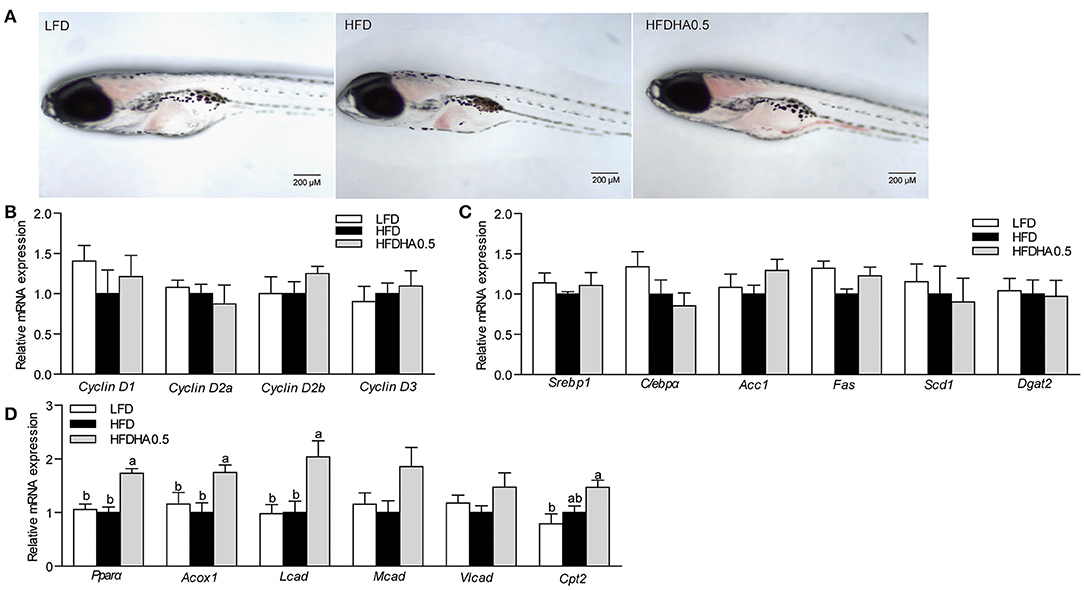
Figure 8. Effects of gut microbiota on lipid metabolism in GF zebrafish. (A) Whole-mount oil red staining in GF zebrafish in LFD, HFD, and HFDHA0.5. The scale bar is 200 μm. (B) Relative mRNA expression of CyclinDs. Relative mRNA expression of (C) lipid synthesis-related, and (D) lipid β-oxidation-related genes. Values are means ± SEMs (n = 4 biological replicates). Means without a common letter are significantly different, P < 0.05.
Discussion
In this study, the short-term feeding (2 wks) of HFD induced hepatic fat accumulation in zebrafish. In line with these studies in mammals, the current study clearly showed that DHA can reduce the hepatic lipid content in HFD-fed zebrafish. In mammals, DHA is known to have several positive effects of diseases related to fatty liver (14–16). DHA reduces lipid content by downregulating fatty acid synthesis and increase fatty acid β-oxidation in non-alcoholic fatty liver disease (NAFLD) models (19, 20). The mechanism appears in part to be regulated through reduction of the transcription factor SREBP1c, which leads to a downstream reduction of the activity of lipid synthesis genes like Fas, Scd1, Dgats, and an upregulation of Pparα which stimulates peroxisomal and mitochondrial lipid transport and β-oxidation (e.g., Acox and Cpt2) (15, 41, 42).
To determine the effects of DHA supplementation on hepatic lipid metabolism, we analyzed the expression of genes related to lipid synthesis and β-oxidation. In this study, the transcription factor for lipid synthesis, C/ebpα, was reduced along with genes in the TG synthesis pathway, while Pparα was upregulated along with β-oxidation genes. Besides, the effects of DHA were also confirmed in ZFL cells. The results showed that the modulation of DHA on hepatic lipid accumulation in zebrafish appears to be similar to that in mammals. Moreover, we treated ZFL cells with 100 μM OA and ODHA and found that the partially replacing OA with DHA led to reduced lipid accumulation with decreased lipid synthesis and decreased β-oxidation. However, it is well-known that DHA is relatively resistant to β-oxidation (43, 44) and frequently incorporates to membranes in the form of phospholipids (27, 45). This means DHA might not be the main substrate for β-oxidation. Thus, the increased capacity of β-oxidation induced by DHA might increase the turnover of other obesogenous fatty acids, such as OA. Taken together, these results confirmed a conservative lipid-lowering effect of DHA in zebrafish.
DHA is known to have effects on cell growth and proliferation (28). The increased DHA incorporation into the phospholipids of the nucleus membrane may produce a direct influence on modulation of genes, RNA synthesis and release, and the activation of DNA polymerase α, all that can affect cell proliferation (46). Thus, the effects of DHA on cellular processes including growth, apoptosis and proliferation in zebrafish liver cells (ZFL) were examined after treating ZFL cells with 0, 10, and 50 μM DHA up to 48 h. In the present study, we convincingly showed that DHA inhibited cell growth and led to cell cycle arrest by increasing the number of cells in the G1 phase and reducing mRNA expression of Cyclin D1. This agrees with previous reports in both fish and mammals where DHA induced cell cycle arrest via Cyclin-Cdk manners (34, 47, 48).
Moreover, Cyclin D1 has its own functions in modulating lipid metabolism via its regulation of nuclear receptors and transcriptional modulation (49–53). The direct role of dietary DHA was also tested in the GF zebrafish model where dietary DHA not only inhibited the expression of Cyclin D1, but also modulated lipid metabolism similar to that found in the conventional feeding trial. To determine the relationship between Cyclin D1 and lipid metabolism, we repressed the expression of Cyclin D1 by transfecting Cyclin D1 siRNA into ZFL cells. In Cyclin D1-knockdown ZFL cells, we observed that lipid synthesis was inhibited and lipid β-oxidation was promoted. Moreover, the inhibitor for Cyclin D1-Cdk4, LY2835219, exerted similar effects. These results showed that DHA might reduce hepatic lipid accumulation via blocking of the Cyclin D1-Cdk4 signal.
Curiously, this appeared to partly contradict to previous studies where Cyclin D1 knockdown was shown to promote hepatic lipogenesis in a well-differentiated AML12 cell line cultured at high density (52). But some indirect evidences suggested the positive role of Cyclin D1 in promoting fatty liver development. The inhibition of Cdk4, the downstream kinase of Cyclin D1, can prevent the development of hepatic steatosis by reducing C/ebpα phosphorylation and its target gene, Fasn, Acc, Scd, Gpat, Dgat1, and Dgat2 (54), which is in line with the effect of LY2835219 (inhibitor for Cyclin D1-Cdk4) in this study. Cell cycle arrest in G1 phase significantly attenuates Srebp-mediated lipid synthesis (55). Due to the scarcity of Cyclin D1 regulatory mode, further studies are needed to fully illustrate the sophisticated mechanism upon Cyclin D1-mediated lipid metabolism. It nevertheless appears quite clear that there is a link between the cell cycle machinery and the regulation of hepatic lipid metabolism.
In our previous studies, HFD could lead to alteration in gut microbial community and reduced diversity in zebrafish (35, 56), which also was observed in this study. In addition, DHA supplementation partly diminished the effects of HFD on the gut microbial community and increased diversity of species. This result was partly in agreement with studies in mice and zebrafish, where n–3 PUFA or fish oil supplementation reduced the growth of Pseudomonas (57, 58). The formation of biofilm might play an important role in Pseudomonas's adherence and growth on any surface, including gut mucosa (59). Although the effect of DHA on Pseudomonas biofilm has not been studied, it is clear that DHA can disturb biofilm formation in Streptococcus mutans and inhibit its growth (60). Thus, the reduction of Pseudomonas caused by DHA supplementation might be due to the anti-bacterial effect mediated by biofilm damage. However, what out of expectation was that DHA supplementation also increased the growth of Mycobacterium. Mycobacterium is the pathogen causing mycobacteriosis in zebrafish and the gastrointestinal tract is the primary route of infection (61). Although some Mycobacterium spp isolated from zebrafish do not cause significant mortalities, granulomatous lesions in visceral organs have been reported in zebrafish (62). No significant mortality was observed in the short-term feeding trial, but further studies are needed to reveal the presence or not of granulomatous lesions in visceral organs so that the effect of increased Mycobacterium could be defined.
Thus, to investigate the microbiota-mediated effects on lipid metabolism, gut microbiota from the HFD and HFDHA0.5 groups were respectively transferred to GF-zebrafish. In this study, DHA supplementation subsequently altered the effect of gut microbiota on lipid metabolism, as shown by boosting lipid oxidation independent of Cyclin D1, in zebrafish host. Although both nutrients and gut microbiota play important role in host metabolism, the composition and metabolism of the gut microbiota are mainly drived by diets (63). Thus, the effect gut microbiota in this study was the result of a secondary microbiota-host interaction drived by dietary DHA. The lipid-lowering effects mediated by Cyclin D1 was related to DHA entering hepatocytes, which was directly linked to the supplementation of DHA in diet. Thus the effect of DHA without involvement of gut microbiota is a primary effect. Moreover, the effect of DHA without involvement of gut microbiota modulated both lipid synthesis and β-oxidation, while the effect of gut microbiota modulated β-oxidation. This suggested a more comprehensive role of Cyclin D1 in modulating lipid metabolism. The effects of gut microbiota on host lipid metabolism might be mediated by its bacteria-derived components (64–66). Its dysbiosis can inhibit phosphorylation of AMPK thereby negatively influencing hepatic fatty oxidation and favoring lipogenesis resulting in excessive fat storage in the liver and obesity (67). However, the probiotic, Lactobacillus, and the microbial metabolite, butyrate, can activate AMPK and increase fatty acid oxidation (68). Further studies about the metabolites generated from DHA-altering gut microbial community might be beneficial to clarify the mechanism underlying the effects of gut microbita on lipid β-oxidation.
Conclusion
Taken together, we described a regulatory mechanism in which DHA alleviates hepatic lipid synthesis and promotes lipid oxidation via the down-regulation of Cyclin D1. On the other hand, the gut microbiota formed by DHA-supplemented diet enhanced lipid oxidation via a mode independent on Cyclin D1. These results suggested when we use HFD in fish feeding, fatty acid composition in diet also should be taking into consideration to make sure containing appropriate DHA.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA771550.
Ethics Statement
The animal study was reviewed and approved by Institute of Feed Research of Chinese Academy of Agricultural Sciences Animal Care Committee.
Author Contributions
ZZho designed the research. QD wrote the paper and performed experiments and acquired data. ZZha gave conceptual advice for the paper and assisted in gut microbiota analysis. ER and REO reviewed and helped to revise the manuscript. QH and QZ participated in zebrafish husbandry and sampling. CR, YY, and ZZha co-analyzed and discussed the results. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC 31925038 and 32061133004) and the National Key R&D Program of China (2018YFD0900400).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.797510/full#supplementary-material
References
1. Karalazos V, Bendiksen EA, Bell JG. Interactive effects of dietary protein/lipid level and oil source on growth, feed utilisation and nutrient and fatty acid digestibility of Atlantic salmon. Aquaculture. (2011) 311:193–200. doi: 10.1016/j.aquaculture.2010.11.022
2. Karalazos V, Bendiksen EA, Dick JR, Tocher DR, Bell JG. Influence of the dietary protein: lipid ratio and fish oil substitution on fatty acid composition and metabolism of Atlantic salmon (Salmo salar) reared at high water temperatures. Br J Nutr. (2011) 105:1012–25. doi: 10.1017/S0007114510004605
3. Shapawi R, Ebi I, Yong ASK, Ng WK. Optimizing the growth performance of brown-marbled grouper, Epinephelus fuscoguttatus (Forskal), by varying the proportion of dietary protein and lipid levels. Anim Feed Sci Technol. (2014) 191:98–105. doi: 10.1016/j.anifeedsci.2014.01.020
4. Ruyter B, Moya-Falcon C, Rosenlund G, Vegusdal A. Fat content and morphology of liver and intestine of Atlantic salmon (Salmo salar): Effects of temperature and dietary soybean oil. Aquaculture. (2006) 252:441–52. doi: 10.1016/j.aquaculture.2005.07.014
5. Lu KL, Xu WN, Li JY, Li XF, Huang GQ, Liu WB. Alterations of liver histology and blood biochemistry in blunt snout bream Megalobrama amblycephala fed high-fat diets. Fisher Sci. (2013) 79:661–71. doi: 10.1007/s12562-013-0635-4
6. Wang XX, Li YJ, Hou CL, Gao Y, Wang YZ. Physiological and molecular changes in large yellow croaker (Pseudosciaena crocea R.) with high-fat diet-induced fatty liver disease. Aquacult Res. (2015) 46:272–82. doi: 10.1111/are.12176
7. Meguro S, Hasumura T. Fish oil suppresses body fat accumulation in zebrafish. Zebrafish. (2018) 15:27–32. doi: 10.1089/zeb.2017.1475
8. Ide T. Interaction of fish oil and conjugated linoleic acid in affecting hepatic activity of lipogenic enzymes and gene expression in liver and adipose tissue. Diabetes. (2005) 54:412–23. doi: 10.2337/diabetes.54.2.412
9. Mori T, Kondo H, Hase T, Tokimitsu I, Murase T. Dietary fish oil upregulates intestinal lipid metabolism and reduces body weight gain in C57BL/6J mice. J Nutr. (2007) 137:2629–34. doi: 10.1093/jn/137.12.2629
10. Turchini GM, Francis DS, Senadheera SPSD, Thanuthong T, De Silva SS. Fish oil replacement with different vegetable oils in Murray cod: Evidence of an “omega-3 sparing effect” by other dietary fatty acids. Aquaculture. (2011) 315:250–9. doi: 10.1016/j.aquaculture.2011.02.016
11. Alhazzaa R, Nichols PD, Carter CG. Sustainable alternatives to dietary fish oil in tropical fish aquaculture. Rev Aquacult. (2019) 11:1195–218. doi: 10.1111/raq.12287
12. Bell JG, Henderson RJ, Tocher DR, McGhee F, Dick JR, Porter A, et al. Substituting fish oil with crude palm oil in the diet of Atlantic salmon (Salmo salar) affects muscle fatty acid composition and hepatic fatty acid metabolism J Nutr. (2002) 132:222–30. doi: 10.1093/jn/132.2.222
13. Bransden MP, Carter CG, Nichols PD. Replacement of fish oil with sunflower oil in feeds for Atlantic salmon (Salmo salar L.): effect on growth performance, tissue fatty acid composition and disease resistance. Comparat Biochem Physiol B Biochem Mol Biol. (2003) 135:611–25. doi: 10.1016/S1096-4959(03)00143-X
14. Hodson L, Bhatia L, Scorletti E, Smith DE, Jackson NC, Shojaee-Moradie F, et al. Docosahexaenoic acid enrichment in NAFLD is associated with improvements in hepatic metabolism and hepatic insulin sensitivity: a pilot study. Eur J Clin Nutr. (2017) 71:973–9. doi: 10.1038/ejcn.2017.9
15. Soni NK, Nookaew I, Sandberg AS, Gabrielsson BG. Eicosapentaenoic and docosahexaenoic acid-enriched high fat diet delays the development of fatty liver in mice. Lipids Health Dis. (2015) 14:74. doi: 10.1186/s12944-015-0072-8
16. Nobili V, Bedogni G, Alisi A, Pietrobattista A, Rise P, Galli C, et al. Docosahexaenoic acid supplementation decreases liver fat content in children with non-alcoholic fatty liver disease: double-blind randomised controlled clinical trial. Arch Dis Child. (2011) 96:350–3. doi: 10.1136/adc.2010.192401
17. Albracht-Schulte K, Gonzalez S, Jackson A, Wilson S, Ramalingam L, Kalupahana NS, et al. Eicosapentaenoic acid improves hepatic metabolism and reduces inflammation independent of obesity in high-fat-fed mice and in HepG2 cells. Nutrients. (2019) 11:599. doi: 10.3390/nu11030599
18. Huang LL, Wan JB, Wang B, He CW, Ma H, Li TW, et al. Suppression of acute ethanol-induced hepatic steatosis by docosahexaenoic acid is associated with downregulation of stearoyl-CoA desaturase 1 and inflammatory cytokines. Prostaglandins Leukot Essent Fatty Acids. (2013) 88:347–53. doi: 10.1016/j.plefa.2013.02.002
19. Deng X, Dong Q, Bridges D, Raghow R, Park EA, Elam MB. Docosahexaenoic acid inhibits proteolytic processing of sterol regulatory element-binding protein-1c (SREBP-1c) via activation of AMP-activated kinase. Biochim Biophys Acta. (2015) 1851:1521–9. doi: 10.1016/j.bbalip.2015.08.007
20. Hernandez-Rodas MC, Valenzuela R, Echeverria F, Rincon-Cervera MA, Espinosa A, Illesca P, Munoz P, et al. Supplementation with docosahexaenoic acid and extra virgin olive oil prevents liver steatosis induced by a high-fat diet in mice through PPAR-alpha and Nrf2 upregulation with concomitant SREBP-1c and NF-kB Downregulation. Mol Nutr Food Res. (2017) 61:1700479. doi: 10.1002/mnfr.201700479
21. Verduci E, Lassandro C, Radaelli G, Soldati L. Docosahexaenoic acid and non-alcoholic fatty liver disease in obese children: a novel approach?. J Transl Med. (2015) 13:109. doi: 10.1186/s12967-015-0471-7
22. Toshio T. Essential fatty acid requirements of aquatic animals with emphasis on fish larvae and fingerlings. Rev Fisher Sci. (1997) 5:1. doi: 10.1080/10641269709388592
23. Jin A, Lei CX, Tian JJ, Sun J, Ji H. Dietary docosahexaenoic acid decreased lipid accumulation via inducing adipocytes apoptosis of grass carp, Ctenopharygodon idella. Fish Physiol Biochem. (2018) 44:197–207. doi: 10.1007/s10695-017-0424-6
24. Cheng YC, Chen HY. Effects of different fatty acids on cell differentiation and lipid accumulation in preadipocytes of warm water fish cobia (Rachycentron canadum Linnaeus, 1766). Aquacult Res. (2015) 46:590–601. doi: 10.1111/are.12204
25. Emery JA, Norambuena F, Trushenski J, Turchini GM. Uncoupling EPA and DHA in fish nutrition: dietary demand is limited in Atlantic Salmon and effectively met by DHA alone. Lipids. (2016) 51:399–412. doi: 10.1007/s11745-016-4136-y
26. Stillwell W, Wassall SR. Docosahexaenoic acid: membrane properties of a unique fatty acid. Chem Phys Lipids. (2003) 126:1–27. doi: 10.1016/S0009-3084(03)00101-4
27. Zhang ZJ, Wilcox HG, Elam MB, Castellani LW, Heimberg M. Metabolism of n-3 polyunsaturated fatty acids by the isolated perfused rat liver. Lipids. (1991) 26:504–11. doi: 10.1007/BF02536594
28. Michel GPF, Karibian D, Bonnaveiro N, Starka J. Is there a correlation between membrane phospholipid-metabolism and cell-division. Ann Inst Pasteur Microbiol. (1985) 136A:111–8. doi: 10.1016/S0769-2609(85)80031-4
29. Massague J. G1 cell-cycle control and cancer. Nature. (2004) 432:298–306. doi: 10.1038/nature03094
30. Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. (1993) 7:812–21. doi: 10.1101/gad.7.5.812
31. Dorn C, Engelmann JC, Saugspier M, Koch A, Hartmann A, Muller M, et al. Increased expression of c-Jun in nonalcoholic fatty liver disease. Lab Invest. (2014) 94:394–408. doi: 10.1038/labinvest.2014.3
32. Nunez KG, Gonzalez-Rosario J, Thevenot PT, Cohen AJ. Cyclin D1 in the liver: role of noncanonical signaling in liver steatosis and hormone regulation. Ochsner J. (2017) 17:56–65. doi: 10.1043/1524-5012-17.1.56
33. Mazza A, Fruci B, Garinis GA, Giuliano S, Malaguarnera R, Belfiore A. The role of metformin in the management of NAFLD. Exp Diab Res. (2012) 2012:716404. doi: 10.1155/2012/716404
34. Wang CC, Liu WB, Cao XF, Huang YY, Wang X, Xiao K, et al. Excess DHA induces cell cycle arrest by activating the P53/cycling pathway in blunt snout bream (Megalobrama amblycephala). Front Marine Sci. (2020) 7:286. doi: 10.3389/fmars.2020.00286
35. Zhang Z, Ran C, Ding QW, Liu HL, Xie MX, Yang YL, et al. Ability of prebiotic polysaccharides to activate a HIF1 alpha-antimicrobial peptide axis determines liver injury risk in zebrafish. Commun Biol. (2019) 2:274. doi: 10.1038/s42003-019-0526-z
36. Ackman RG. Simplification of analyses of fatty acids in fish lipids and related lipid samples. Acta Med Scand. (1987) 222:99–103. doi: 10.1111/j.0954-6820.1987.tb10644.x
37. Ghosh C, Zhou YL, Collodi P. Derivation and characterization of a zebrafish liver-cell line. Cell Biol Toxicol. (1994) 10:167–76. doi: 10.1007/BF00757560
38. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996–8 doi: 10.1038/nmeth.2604
39. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. (2007) 73:5261–7. doi: 10.1128/AEM.00062-07
40. Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. (2006) 127:423–33. doi: 10.1016/j.cell.2006.08.043
41. Xu PF, Wang H, Kayoumu A, Wang MY, Huang W, Liu G. Diet rich in Docosahexaenoic acid/Eicosapentaenoic acid robustly ameliorates hepatic steatosis and insulin resistance in seipin deficient lipodystrophy mice. Nutr Metab. (2015) 12:58. doi: 10.1186/s12986-015-0054-x
42. Shang TT, Liu L, Zhou J, Zhang MZ, Hu QL, Fang M, et al. Protective effects of various ratios of DHA/EPA supplementation on high-fat diet-induced liver damage in mice. Lipids Health Dis. (2017) 16:65. doi: 10.1186/s12944-017-0461-2
43. Crockett EL, Sidell BD. Substrate selectivities differ for hepatic mitochondrial and peroxisomal beta-oxidation in an Antarctic fish, Notothenia gibberifrons. Biochem J. (1993) 289:427–33. doi: 10.1042/bj2890427
44. Madsen L, Rustan AC, Vaagenes H, Berge K, Dyroy E, Berge RK. Eicosapentaenoic and docosahexaenoic acid affect mitochondrial and peroxisomal fatty acid oxidation in relation to substrate preference. Lipids. (1999) 34:951–63. doi: 10.1007/s11745-999-0445-x
45. Leray C, Andriamampandry M, Gutbier G, Raclot T, Groscolas R. Incorporation of N-3 fatty-acids into phospholipids of rat-liver and white and brown adipose tissues - a time-course study during fish-oil feeding. J Nutr Biochem. (1995) 6:673–80. doi: 10.1016/0955-2863(95)00151-4
46. Chapkin RS, Davidson LD, Davidson LA. Phospholipid molecular species composition of mouse liver nuclei. Influence of dietary n-3 fatty acid ethyl esters. Biochem J. (1992) 287:237–40. doi: 10.1042/bj2870237
47. Chen ZY, Istfan NW. Docosahexaenoic acid, a major constituent of fish oil diets, prevents activation of cyclin-dependent kinases and S-phase entry by serum stimulation in HT-29 cells. Prostaglandins Leukot Essent Fatty Acids. (2001) 64:67–73. doi: 10.1054/plef.2000.0239
48. Albino AP, Juan G, Traganos F, Reinhart L, Connolly J, Rose DP, et al. Cell cycle arrest and apoptosis of melanoma cells by docosahexaenoic acid: association with decreased pRb phosphorylation. Cancer Res. (2000) 60:4139–45.
49. Lee YJ, Dominy JE, Choi YJ, Jurczak M, Tolliday N, Camporez JP, et al. Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature. (2014) 510:547. doi: 10.1038/nature13267
50. Bhalla K, Liu WJ, Thompson K, Anders L, Devarakonda S, Dewi R, et al. Cyclin D1 represses gluconeogenesis via inhibition of the transcriptional coactivator PGC1 alpha. Diabetes. (2014) 63:3266–78. doi: 10.2337/db13-1283
51. Fu MF, Rao M, Bouras T, Wang CG, Wu KM, Zhang XP, et al. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem. (2005) 280:16934–41. doi: 10.1074/jbc.M500403200
52. Hanse EA, Mashek DG, Becker JR, Solmonson AD, Mullany LK, Mashek MT, et al. Cyclin D1 inhibits hepatic lipogenesis via repression of carbohydrate response element binding protein and hepatocyte nuclear factor 4 alpha. Cell Cycle. (2012) 11:2681–90. doi: 10.4161/cc.21019
53. Newberry EP, Kennedy SM, Xie Y, Luo J, Stanley SE, Semenkovich CF, et al. Altered hepatic triglyceride content after partial hepatectomy without impaired liver regeneration in multiple murine genetic models. Hepatology. (2008) 48:1097–105. doi: 10.1002/hep.22473
54. Jin JL, Valanejad L, Nguyen TP, Lewis K, Wright M, Cast A, et al. Activation of CDK4 triggers development of non-alcoholic fatty liver disease. Cell Rep. (2016) 16:744–56. doi: 10.1016/j.celrep.2016.06.019
55. Sanchez-Alvarez M, Zhang QF, Finger F, Wakelam MJO, Bakal C. Cell cycle progression is an essential regulatory component of phospholipid metabolism and membrane homeostasis. Open Biol. (2015) 5:150093. doi: 10.1098/rsob.150093
56. Ding Q, Zhang Z, Ran C, He S, Yang Y, Du Z, et al. The hepatotoxicity of palmitic acid in zebrafish involves the intestinal microbiota. J Nutr. (2018) 148:1217–28. doi: 10.1093/jn/nxy084
57. Mil-Homens D, Ferreira-Dias S, Fialho AM. Fish oils against burkholderia and pseudomonas aeruginosa: in vitro efficacy and their therapeutic and prophylactic effects on infected galleria mellonella larvae. J Appl Microbiol. (2016) 120:1509–19. doi: 10.1111/jam.13145
58. Arias-jayo N, Abecia L, Lavin JL, Tueros I, Pardo MA. Host-microbiome interactions in response to a high-saturated fat diet and fish-oil supplementation in zebrafish adult. J Funct Foods. (2019) 60:103416. doi: 10.1016/j.jff.2019.103416
59. Sharma G, Rao S, Bansal A, Dang S, Gupta S, Gabrani R. Pseudomonas aeruginosa biofilm: potential therapeutic targets. Biologicals. (2014) 42:1–7. doi: 10.1016/j.biologicals.2013.11.001
60. Sun M, Zhou Z, Dong J, Zhang J, Xia Y, Shu R. Antibacterial and antibiofilm activities of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) against periodontopathic bacteria. Microb Pathog. (2016) 99:196–203. doi: 10.1016/j.micpath.2016.08.025
61. Harriff MJ, Bermudez LE, Kent ML. Experimental exposure of zebrafish, danio rerio (hamilton), to mycobacterium marinum and mycobacterium peregrinum reveals the gastrointestinal tract as the primary route of infection: a potential model for environmental mycobacterial infection. J Fish Dis. (2010) 30:587–600. doi: 10.1111/j.1365-2761.2007.00839.x
62. Watral V, Kent ML. Pathogenesis of Mycobacterium spp. in zebrafish (Danio rerio) from research facilities. Comp Biochem Physiol C Toxicol Pharmacol. (2007) 145:55–60. doi: 10.1016/j.cbpc.2006.06.004
63. Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res. (2013) 69:52–60. doi: 10.1016/j.phrs.2012.10.020
64. Heianza Y, Sun DJY, Li X, DiDonato JA, Bray GA, Sacks FM, et al. Gut microbiota metabolites, amino acid metabolites and improvements in insulin sensitivity and glucose metabolism: the POUNDS Lost trial. Gut. (2019) 68:263–70. doi: 10.1136/gutjnl-2018-316155
65. van de Wouw M, Boehme M, Lyte JM, Wiley N, Strain C, O'Sullivan O, et al. Short-chain fatty acids: microbial metabolites that alleviate stress-induced brain-gut axis alterations. J Physiol. (2018) 596:4923–44. doi: 10.1113/JP276431
66. Askarian F, Sperstad S, Merrifield DL, Ray AK, Ringo E. The effect of different feeding regimes on enzyme activities of gut microbiota in Atlantic cod (Gadus morhua L.). Aquacult Res. (2013) 44:841–6. doi: 10.1111/j.1365-2109.2011.03079.x
67. Gerard P. Gut microbiota and obesity. Cell Mol Life Sci. (2016) 73:147–62. doi: 10.1007/s00018-015-2061-5
Keywords: high-fat diet, DHA, lipid accumulation, Cyclin D1, gut microbiota
Citation: Ding Q, Hao Q, Zhang Q, Yang Y, Olsen RE, Ringø E, Ran C, Zhang Z and Zhou Z (2022) DHA Suppresses Hepatic Lipid Accumulation via Cyclin D1 in Zebrafish. Front. Nutr. 8:797510. doi: 10.3389/fnut.2021.797510
Received: 01 November 2021; Accepted: 20 December 2021;
Published: 25 January 2022.
Edited by:
Anne Marie Minihane, University of East Anglia, United KingdomCopyright © 2022 Ding, Hao, Zhang, Yang, Olsen, Ringø, Ran, Zhang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Zhang, zhangzhen@caas.cn; Zhigang Zhou, zhouzhigang03@caas.cn
 Qianwen Ding1,2
Qianwen Ding1,2  Einar Ringø
Einar Ringø Zhen Zhang
Zhen Zhang Zhigang Zhou
Zhigang Zhou