- 1Centre for Biomedical Engineering, Indian Institute of Technology Delhi, New Delhi, India
- 2Medical Oncology, Dr. B.R.Ambedkar Institute Rotary Cancer Hospital, All India Institute of Medical Sciences, New Delhi, Delhi, India
- 3Department of Radiodiagnosis, All India Institute of Medical Sciences, New Delhi, Delhi, India
- 4Radiodiagnosis, Dr. B.R.Ambedkar Institute Rotary Cancer Hospital, All India Institute of Medical Sciences, New Delhi, Delhi, India
- 5Department of Biomedical Engineering, All India Institute of Medical Sciences, New Delhi, Delhi, India
The presence of lung metastases in patients with primary malignancies is an important criterion for treatment management and prognostication. Computed tomography (CT) of the chest is the preferred method to detect lung metastasis. However, CT has limited efficacy in differentiating metastatic nodules from benign nodules (e.g., granulomas due to tuberculosis) especially at early stages (<5 mm). There is also a significant subjectivity associated in making this distinction, leading to frequent CT follow-ups and additional radiation exposure along with financial and emotional burden to the patients and family. Even 18F-fluoro-deoxyglucose positron emission technology-computed tomography (18F-FDG PET-CT) is not always confirmatory for this clinical problem. While pathological biopsy is the gold standard to demonstrate malignancy, invasive sampling of small lung nodules is often not clinically feasible. Currently, there is no non-invasive imaging technique that can reliably characterize lung metastases. The lung is one of the favored sites of metastasis in sarcomas. Hence, patients with sarcomas, especially from tuberculosis prevalent developing countries, can provide an ideal platform to develop a model to differentiate lung metastases from benign nodules. To overcome the lack of optimal specificity of CT scan in detecting pulmonary metastasis, a novel artificial intelligence (AI)-based protocol is proposed utilizing a combination of radiological and clinical biomarkers to identify lung nodules and characterize it as benign or metastasis. This protocol includes a retrospective cohort of nearly 2,000–2,250 sample nodules (from at least 450 patients) for training and testing and an ambispective cohort of nearly 500 nodules (from 100 patients; 50 patients each from the retrospective and prospective cohort) for validation. Ground-truth annotation of lung nodules will be performed using an in-house-built segmentation tool. Ground-truth labeling of lung nodules (metastatic/benign) will be performed based on histopathological results or baseline and/or follow-up radiological findings along with clinical outcome of the patient. Optimal methods for data handling and statistical analysis are included to develop a robust protocol for early detection and classification of pulmonary metastasis at baseline and at follow-up and identification of associated potential clinical and radiological markers.
1 Introduction
The presence of lung metastases in patients with solid tumors is considered as an important criterion to direct appropriate treatment management and to further prognosticate. Computed tomography (CT) is the standard of care to detect pulmonary metastases and for staging of cancers (especially for intermediate- or high-grade tumors). Indeterminate pulmonary nodules are a very common finding and are often encountered in such clinical scenarios. However, CT is not very appropriate in differentiating metastatic nodules from benign nodules. This issue is even more glaring in developing countries in Asia and Africa where tuberculosis is highly prevalent, and it can be challenging, if not impossible, to differentiate tubercular granuloma from metastatic nodules. The benign and the malignant nodules, especially those at early stages (<5 mm) show very close resemblance to each other and there is a significant subjectiveness involved in making this distinction, requiring frequent follow-up imaging. This leads to increased financial and emotional burden over the healthcare facility and patients as well as unnecessary radiation exposure to patients. Even with the advent of 18F-fluoro-deoxyglucose positron emission technology-computed tomography (18F-FDG PET-CT), while extrapulmonary metastases may be additionally detected (1), the lung nodule conundrum remains a clinical problem. This is because not all metastatic nodules show FDG uptake on PET-CT, and at the same time, active benign nodules such as granulomas might show FDG uptake (1). In addition, small nodules (<5 mm) cannot be reliably evaluated using 18F-FDG PET-CT. Hence, currently there is no non-invasive imaging technique that can reliably characterize the malignant potential of lung nodules.
Lung is one of the favored sites of metastasis in sarcomas and 62%–83% of patients present with lung metastases during their entire disease course, with lung also being the most common site of relapse (2–6). Among patients with sarcomas of extremity, approximately 20% of patients develop isolated pulmonary metastasis at some point in their disease progression (2, 3). In patients who have multiple lung nodules detected with CT, 73% are reported to be metastases (7, 8). Hence, patients with sarcomas, especially those from low- to medium-income countries (LMICs), can provide an ideal platform to develop a model to differentiate metastases from benign nodules in the lung. Various consensus clinical guidelines exist for defining pulmonary nodules as metastatic. The Children Oncology Group definition of evident metastatic disease differs according to sarcoma type and is an area of constant evolution (9), which is left to the discretion of the treating physician (9, 10). For the subcentimeter lesions, optimal management remains unclear (11). The availability of thin slice CT technology introduced further uncertainty by detecting nodules <5 mm in diameter, thereby increasing the frequency of positive tests (12). There is no accurate non-invasive modality to determine the malignant potential of such smaller pulmonary nodules, and invasive biopsy of these small nodules is often not feasible. Moreover, in daily practice, overlapping radiologic features of metastases are frequently encountered, which makes distinction from other non-malignant pulmonary diseases difficult. For example, osteosarcoma metastases may appear as benign calcified pulmonary nodules, though as many as 40% of osteosarcoma lesions are not calcified and unusual forms of metastasis are also observed (7, 13).
To overcome the lack of optimal specificity of CT scan in detecting metastasis, it is imperative to develop a non-invasive imaging-based computational model to differentiate pulmonary nodules between benign and malignant etiologies. A lot of work has been reported for the detection and classification of primary lung cancer using machine learning (ML) and deep learning (DL) (14–20); however, predicting the malignant potential of the lung nodules in clinical scenarios of possible metastases from solid tumors is still an unmet need. Therefore, in this protocol study, we are proposing to utilize ML/DL techniques to identify lung nodules and develop a prediction model to characterize them as benign nodules or malignant lung metastases. The objectives of the proposed protocol are as follows: (a) identification of clinical and radiological markers for differentiation of lung metastases from benign lung nodules; (b) training and testing of classification model for lung metastases using retrospective training cohort; and (c) clinical validation of the developed classification model using both retrospective and prospective validation cohort.
2 Methods and analysis
2.1 Literature review on application of machine learning- and deep learning-based techniques for lung nodule characterization
Numerous studies have been reported for classification of primary lung cancer and benign lung nodules using ML/DL techniques (14–20). Extraction of effective discriminative features for lung nodule characterization is challenging due to complex anatomical structures in thoracic CT images. Many reported methods have shown improved classification performance toward this direction, and strategically, these methods can be categorized into two groups: traditional ML-based methods using feature engineering and DL-based methods.
The former methods employ feature engineering to extract various handcrafted features, like size, shape, margin, intensity, and statistical textural features in ROIs; next, identify effective discriminating features applying feature selection strategies (21, 22) and finally develop a classification model using ML algorithms, viz., logistic regression (LR), support vector machine (SVM), K-nearest neighbors (KNN), random forest (RF), linear discriminant analysis (LDA), deep neural network (DNN), and Adaboost (14–17). Various shape, margin, intensity, and textural features using GLCM in 2D axial plane and 3D volume of lung nodule (23–28), textural features using phylogenetic diversity (25), and wavelets (29, 30) were used to classify malignant and benign lung nodules using SVM, LDA, DNN, and naïve Bayes classifiers. Textural features using local binary pattern (LBP) and discrete cosine transform were extracted and used to train SVM and KNN models for lung nodule characterization (31). Shape, intensity, statistical textural features using GLCM, Gabor filters, and LBP were used to train RF models to classify lung nodules as malignant and benign (32, 33), and SVM, KNN, and LR models were used to classify different nodule types like solid, semi-solid, and ground-glass, respectively. A comparative study evaluating various ML models showed that an ensemble classifier combining SVM and RF produced the best classification performance for malignant nodules compared to KNN, LDA, and AdaBoost using shape, size, and texture-based features (34). Similarly, morphological and statistical features were applied to an ensemble of three classifiers utilizing multilayer perceptron (MLP), KNN, and SVM to classify benign or malignant lung nodules (35).
While handcrafted features need expert domain knowledge for pulmonary CT images, DL does not require explicit features extraction; it reveals intrinsic structural properties in input data by applying brain-inspired computing and has showed notable improvement in medical image analysis (18–20). Various frameworks based on convolutional neural networks (CNNs) have been developed for lung nodule detection and an elaborated review can be found in Refs (36–39). Applying reinforcement learning, region proposal network (40), faster region-based CNN (41), various advanced CNN models (42–45), and ensemble learners using multiple CNN models (46) have been designed for lung nodule detection and false-positive reduction. CNN-based dense convolutional binary-tree networks (47) and spatial pyramid dilated network (48) were developed to derive useful features from image data to discriminate malignant pulmonary nodules from benign nodules. Optimal Ensemble framework combining multiple CNN models using ResNet, AlexNet, DenceNet, InceptionNet, and SqueezeNet (49–53), transfer learning-based system (54–57), and hybrid CNN-based system (58–62) have been reported for classifying malignant and benign lung nodules. A research group proposed multi-view collaborative deep CNN models for incorporating prior knowledge about the association of nodule’s malignancy and its heterogeneity (49) and further showed learning from ambiguous labels for more accurate lung nodule malignancy prediction (51). 3D Deep CNN and SVM with multiple kernel learning algorithms was proposed to fuse the DL features with clinical information for lung nodule diagnosis (61). 3D CNN and RF were used to combine CT imagery with biomarker annotation and volumetric radiomics features for lung nodule malignancy prediction (62). CNN with adaptive morphology and textural features (63), 3D segmentation attention network-based systems integrating asymmetric convolution with a gradient boosting machine (64), and 3D non-local network-based systems incorporating channel attention and adaptive network growth algorithm (65) were reported for lung nodule classification. Lung nodule classification was performed using features learned from two deep 3D customized mixed link network with gradient boosting machine (66). Studies have shown improved classification accuracy for malignant nodules using optimal deep feature selection from different CNN-based convolution layers and fusion of the deep features for the final classifier (67–69). A multi-scale cost-sensitive neural network was proposed to mitigate the issue of insufficient labeled data and class imbalance (70). A soft activation mapping-based method meta-learning scheme was reported for interpretable lung nodule classification (71) and a meta ordinal set was further generated by the same research group by developing meta ordinal weighting network to explore the ordinal relationship between the data for lung nodule classification (72). Recently, DL models based on transformers (73, 74) or combined with CNN and transformers (75, 76) have been successfully applied for lung nodule detection and classification. A self-supervised region-based 3D transformer model was developed to identify lung nodules among a set of candidate regions (73). A local focus scheme was incorporated into a deformable dilated transformer to develop a multi-granularity dilated transformer to focus on the more discriminative local features to classify lung nodules in CT scans (74). TransUnet was developed based on the transformer to encode feature representations of input CT scans and the Unet network to decode the hidden feature for outputting the final classification results (75). Res-Trans was developed using convolutional operations to extract local features and transformer blocks with self-attention to capture global features (76).
Few research gaps have been identified from the above literature survey for lung nodule characterization. Firstly, all the studies reported development of detection and classification models for primary lung cancer nodules, while characterizing metastatic lung nodules, which are also prevalent among patients with various primary cancers, have not or rarely been explored. Secondly, it has been observed that clinical features, which are also informative for lung nodule characterization (6, 77, 78), have not been utilized by most of the reported ML and DL methods. Thirdly, temporal changes in lung nodules during treatment, which may be captured by radiological and/or radiomics features from follow-up CT scans and might be useful for predicting malignancy at early stage, have not been considered by the earlier studies. Fourthly, most of the studies have used publicly available retrospective thoracic CT datasets for training and validation of the proposed models; however, prospective datasets may be better suited to test the generalizability of the model’s performance. These issues have been addressed in the current study protocol.
2.2 Study design
It is an ambispective cohort study with a retrospective training cohort and a prospective validation cohort. Patients with bone and soft tissue sarcomas who registered for treatment between January 2011 and December 2023 in the Medical Oncology Clinic of Dr. B.R.Ambedkar Institutional Rotary Cancer Hospital (IRCH), All India Institute of Medical Sciences (AIIMS) New Delhi, India will be included in this study. CT scans at baseline and follow-up with the presence of lung nodules and clinical data available in institutional databases will be used as training and testing datasets for the proposed classification model. For the validation cohort, patients with bone sarcomas and soft tissue sarcomas will be prospectively recruited from the Medical Oncology Clinic of Dr. B.R.A. IRCH, AIIMS New Delhi, India and these prospectively performed CT scans and prospectively collected clinical data will be used for the validation of the proposed classification model.
2.3 Sample size
For a two-class classification problem, considering a group ratio of 1:1, to achieve an area under the receiver operating characteristics curve of 0.8 with a 20% absolute error margin in a two-sided 95% confidence interval, a minimum of 34 samples will need to be investigated. However, for supervised learning models, studies have shown that increasing sample size beyond 1,000/class demonstrated no further significant improvements in the overall classification accuracy (79). For any patient, each annotated nodule in the lung will serve as a sample in the training/testing process during model development. In the proposed model, CT scans of a minimum of 500 patients will be used retrospectively, and considering an average of 5 nodules (range: 1–10) per CT scan, a minimum of ~2,500 samples of lung nodules will be annotated. For validation of the proposed model, CT datasets of a minimum of 50 patients will be prospectively collected. For the development of the proposed prediction model, retrospective CT datasets will be analyzed, and for validation of the proposed model, both retrospective and prospective CT datasets will be analyzed. A total of 2,000–2,250 sample nodules from at least 450 patients from the retrospective cohort will be used for training and testing the prediction model for characterizing lung metastases of sarcoma. Nearly 250 nodules from 50 patients from the retrospective cohort and 250 sample nodules from 50 prospective cohorts comprising a total of 500 nodules will be used exclusively for validating the proposed prediction model.
2.4 Data collection
Patients with bone and soft tissue sarcomas who registered for treatment during January 2011 to December 2023 in the outpatient department (OPD) of Dr. B.R.Ambedkar IRCH, AIIMS New Delhi, India will be considered for inclusion in this project.
Inclusion criteria for the retrospective cohort (registered during January 2011–December 2021) will be as follows: (a) patients with biopsy-proven bone or soft tissue sarcoma and (b) having lung nodule(s) in the chest CT scan at the time of presentation or in the course of treatment. Exclusion criteria for the retrospective cohort will be as follows: (a) patients who could not be adequately followed up and decision about lung nodule(s)’s malignancy could not be reached and (b) patients did not undergo chest CT scan in the course of treatment.
For the prospective cohort (registered during January 2022–December 2023), inclusion criteria will be as follows: (a) patients with biopsy-proven bone or soft tissue sarcoma and (b) having lung nodule(s) in the chest CT scan at the time of presentation. Exclusion criteria for the prospective cohort will be as follows: (a) patient did not undergo CT scan in the course of treatment, (b) patients whose CT scan could not be retrieved, (c) refusal for informed consent, and (d) patient for whom decision about the nature of the lung nodule(s) could not be reached within the follow-up time. For the prospective cohort, follow-up time will be at least 1 year or till dropout due to death or other reason, whichever is earlier.
For the development of the proposed classification model, a retrospective dataset of patients who registered during 2011–2018 will be used. For validation, a retrospective dataset of patients who registered during 2019–2021 and a prospective dataset of patients who will be recruited during 2022–2023 in the Medical Oncology Clinic of Dr. B.R.Ambedkar IRCH, AIIMS New Delhi, India will be used.
2.5 Ground-truth annotation and labeling of lung nodules
2.5.1 Ground-truth region of interest for lung nodule
Conventionally, the demarcation of ground-truth region of interest (ROI) for lung nodules is performed by an expert radiologist by freehand manual drawing of ROI on CT scan using annotation software after thoroughly scanning the whole CT slices. As manual annotation tasks are very tedious and time-consuming for radiologists and due to severe workload and increased number of scans, radiologists’ decision-making may suffer from human error and inter- and intra-observer subjectivity. To mitigate these challenges, a semiautomated segmentation of lung nodules requiring only a minimal human input has been developed in-house at the Center for Biomedical Engineering, IIT Delhi, India. Using the developed semi-automated segmentation tool, demarcation of ground-truth ROI for lung nodules has been initiated under the Department of Radiodiagnosis, AIIMS, Delhi, India.
The developed lung nodule segmentation algorithm is based on the region-growing method and morphological image processing algorithms. It requires only manual input for selecting a seed-point inside the nodule on a CT image and then performs automatic segmentation of the nodule across the slices of the CT image. The algorithm is capable of automatically segmenting out the lung nodules having different shapes, sizes, locations, and characteristics like solitary, juxta-pleural, ground-glass, and juxta-vascular nodules and has shown promising results in limited datasets of 50 patients for initial assessment. This segmentation tool will be used by the radiologists for ground-truth annotation of lung nodules. Examples of segmentation results for lung nodules using the developed algorithm are shown in Figure 1. This segmentation will be verified by two expert radiologists with 12 and 20 years of experience, respectively. Any disagreements will be resolved by mutual discussion to build a consensus.
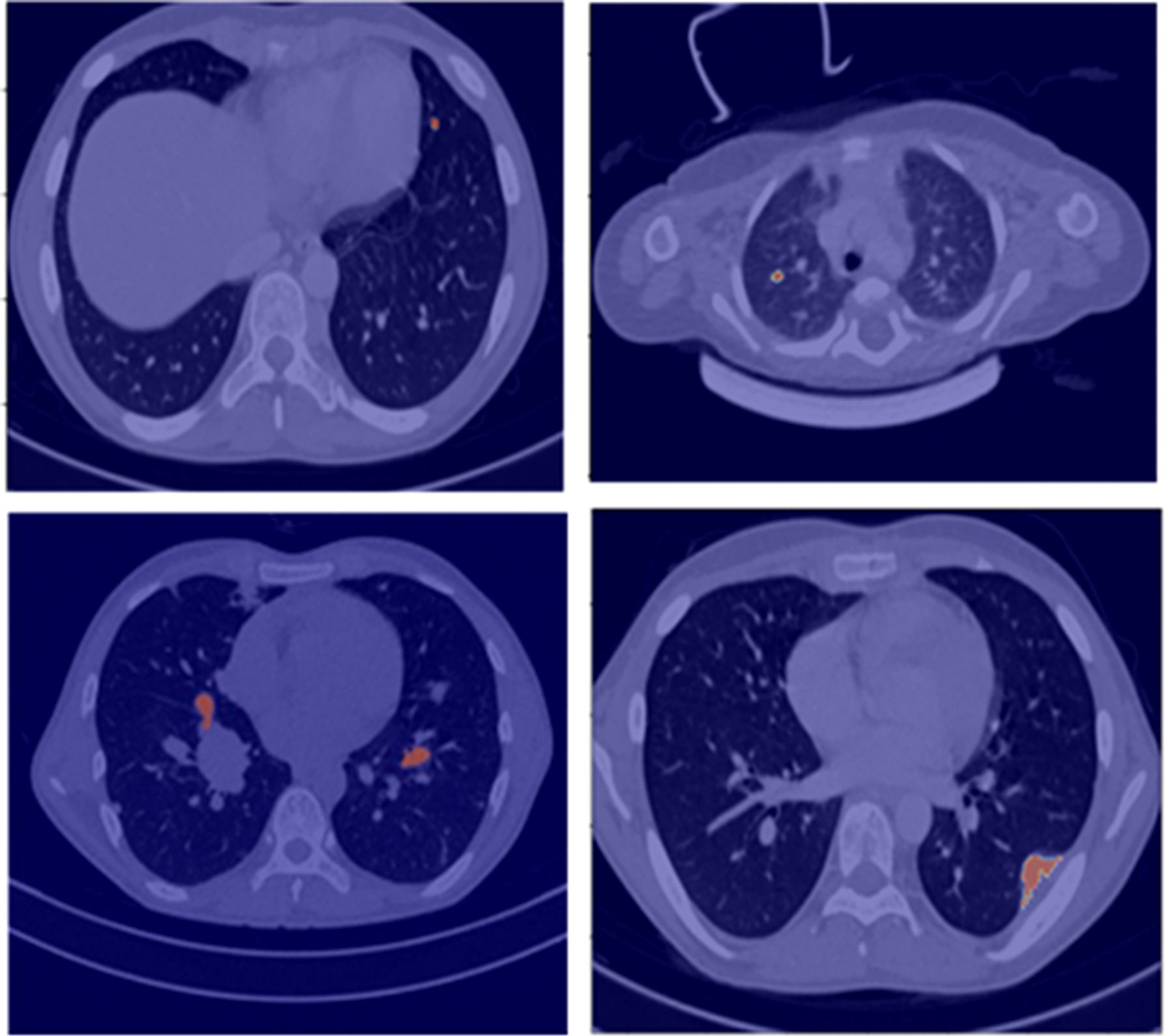
Figure 1 Segmentation results (red overlay) of lung nodules for a representative patient (25 years, male) with sarcoma.
2.5.2 Ground-truth labeling of lung nodule as metastatic or benign
Ground-truth labeling of lung nodules as metastatic or benign in CT scans will be performed based on a biopsy report if available or consensus decision depending on the clinical outcome of the patient and/or radiological changes in follow-up scans as summarized in Figure 2. At baseline imaging (prior to administration of chemotherapy), if consensus opinion at baseline suggests that the nodule can be clearly labeled as benign (no known malignancy, presence of calcification within the nodule in a patient with non-osteogenic sarcoma, history of tuberculosis, and presence of other features of tuberculosis), it will be labeled as benign. The label assigned at benign will be revised on the basis of follow-up imaging. After 6–12 months, if a lung nodule designated as benign at baseline remains stable, then it is assigned a definitive label of a benign lung nodule. On the other hand, if a lung nodule designated as malignant at baseline shows a size reduction after 6–12 months without undergoing chemotherapy, it is also assigned a definitive label of a benign lung nodule. If a malignant lung nodule at baseline shows a size reduction after chemotherapy or shows a size increase with or without chemotherapy, it is assigned a label of malignant lung nodule after 6–12 months. In contrast, if a malignant nodule at baseline remains stable during the next 6–12 months with or without chemotherapy, it is considered as a stable nodule and observed over the next 2–3 years for any change in size. If the size of the stable nodule continues to be stable over this time with or without chemotherapy, it is concluded as a benign nodule, likely granuloma. If the size of the stable nodule changes after 2–3 years whether being treated with chemotherapy or not, it is concluded as malignant.
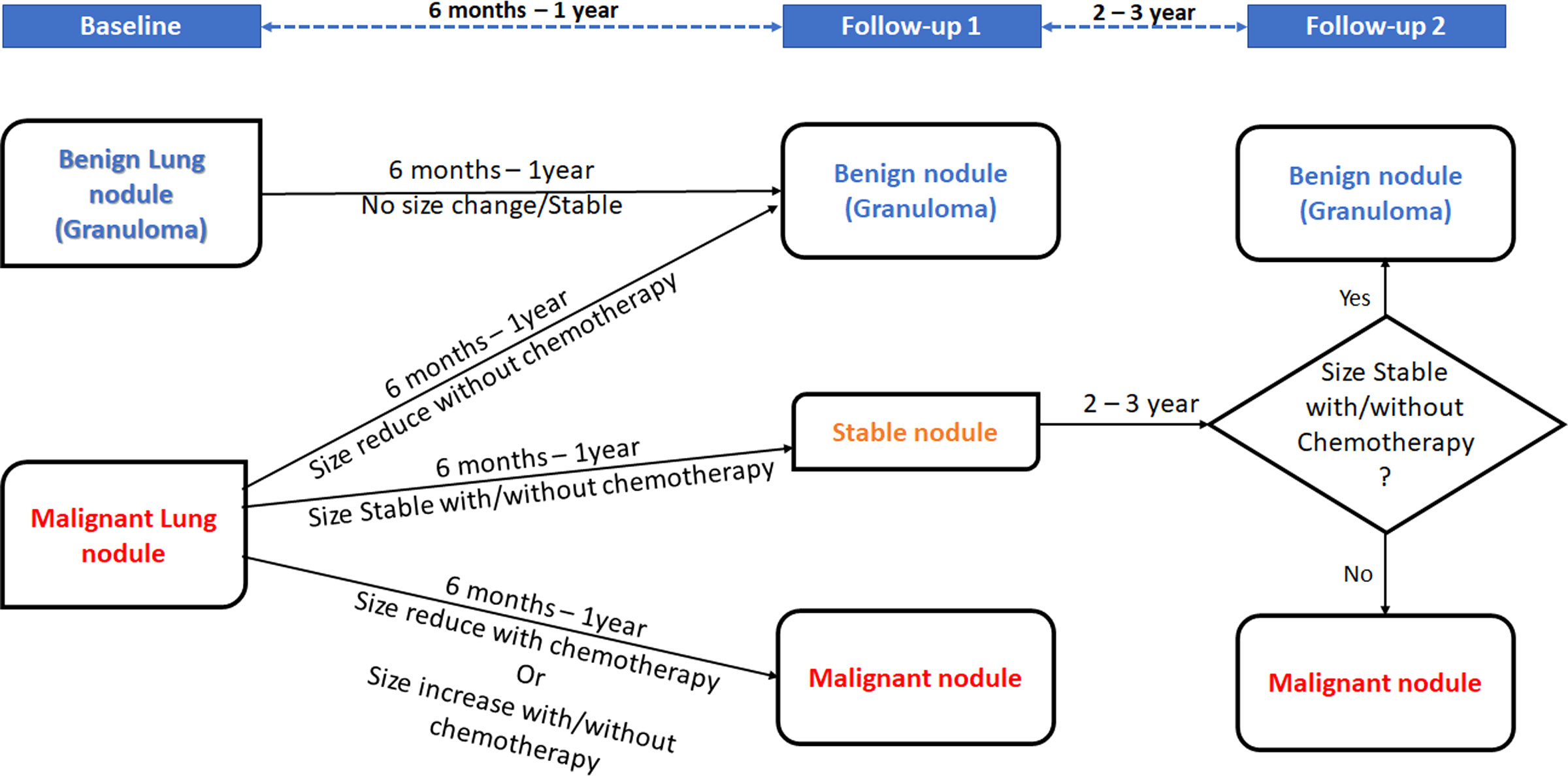
Figure 2 Decision rule for malignant lung nodules depending on patient outcome and/or radiological changes.
2.6 Workflow
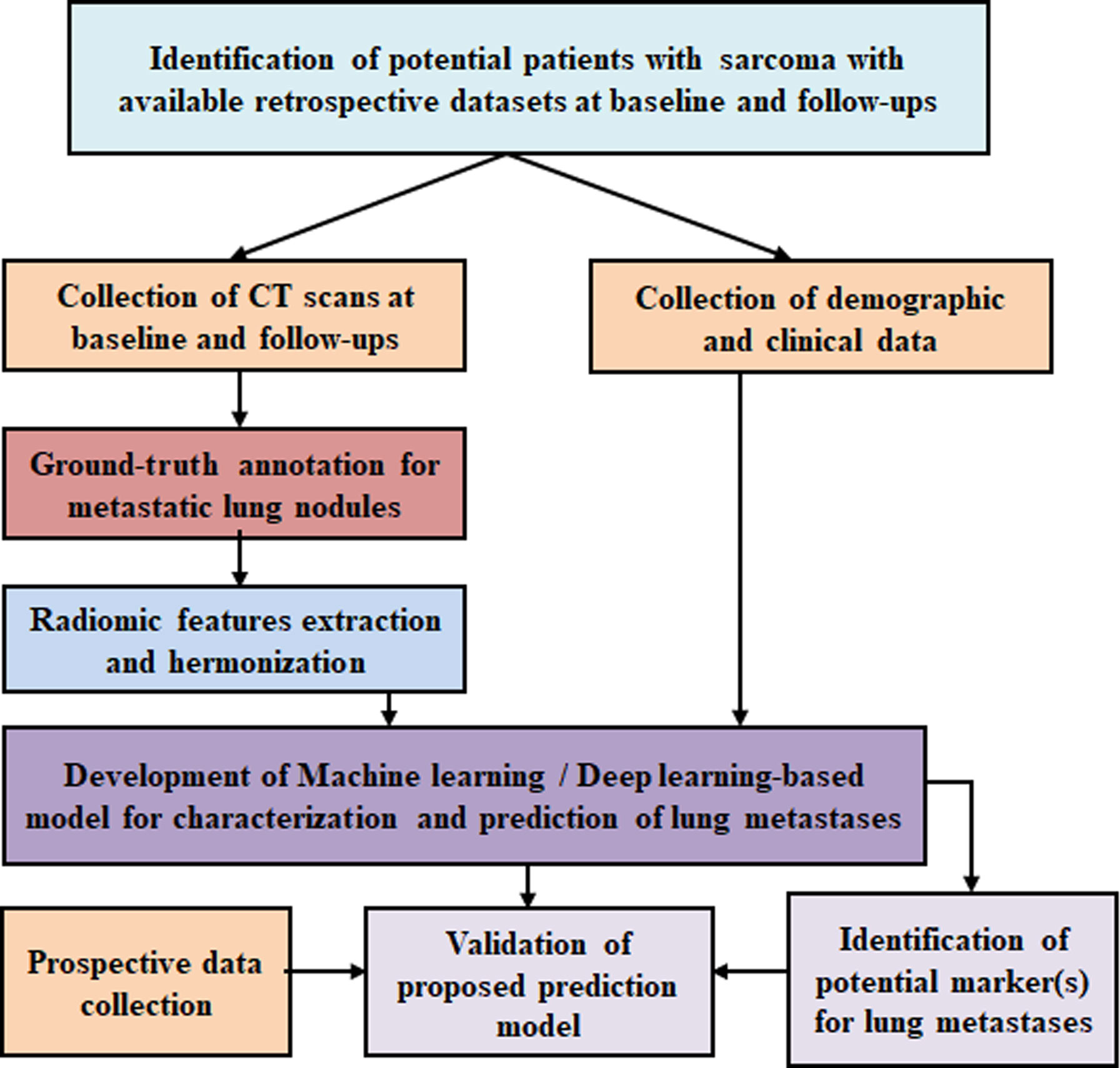
The proposed study is composed of the following steps and summarized in the flowchart in Figure 3:
a) Retrospective data collection
Screening of available retrospective CT scans and clinical data of soft tissue sarcoma and bone sarcoma patients with known clinical follow-up details registered at Dr. B.R.Ambedkar IRCH, AIIMS New Delhi, India, between January 2011 and December 2021 will be performed. We anticipate that datasets of at least 500 patients will be collected.
b) Prospective data collection
For validation of the proposed model, CT scans and clinical data of nearly 100 patients will be prospectively collected at Dr. B.R.Ambedkar IRCH, AIIMS New Delhi, India between January 2022 and December 2023.
c) Ground-truth preparation
Ground- truth annotation and collation of all individual CT scans at baseline and follow-ups will be performed using the in-house semi-automated lung nodule segmentation tool (Figure 1). Ground-truth labeling of lung nodules as metastatic or benign will be performed using histopathological results of biopsy or metastatectomy (if available) or will be based on multi-disciplinary opinion based on baseline and/or follow-up radiological findings along with the clinical outcome of the patient (Figure 2).
d) Radiological feature extraction
Radiological characteristics of lung nodules will be reviewed on CT scans at baseline and follow-up and relevant features like (i) nodule size, (ii) nodule position, (iii) change of size during follow-up, (iv) types of calcifications, (v) air bronchogram, (vi) surrounding ground-glass opacity, (vii) surrounding fibrosis, (viii) no feeding vessel sign, and (ix) bilaterality will be captured and analyzed.
Extraction of various radiomics features from the annotated ground-truth lung nodules will be performed to train the proposed classification model for identification of lung metastasis. The radiomics features consist of the following: (i) shape-based features, (ii) first-order textural features from histogram, (iii) second-order textural features from gray-level cooccurrence matrix (GLCM), and (iv) higher-order textural features from neighborhood gray-tone difference matrix (NGTDM) and run length matrix (RLM) will be evaluated. A detailed tentative list of radiomics features is listed in Table 1. Studies have shown that handcrafted features from tissues surrounding the lung nodule provide global representation of nodule CT images and can be informative for lung nodule characterization (80, 81). Therefore, the radiomics features in lung parenchyma surrounding the lung nodule will also be evaluated and analyzed. Temporal changes in semantic radiological and radiomics features in lung nodules will be evaluated from follow-up CT images (if available) during treatment and will be used along with baseline features. CT data are intrinsically dependent on the protocol acquisition parameters and pixel values in CT data are directly related to the physical characteristic of the tissues having different linear attenuation coefficients. Therefore, harmonization processes will be applied on extracted radiomics features to make them independent of scanner-specific parameter dependencies.
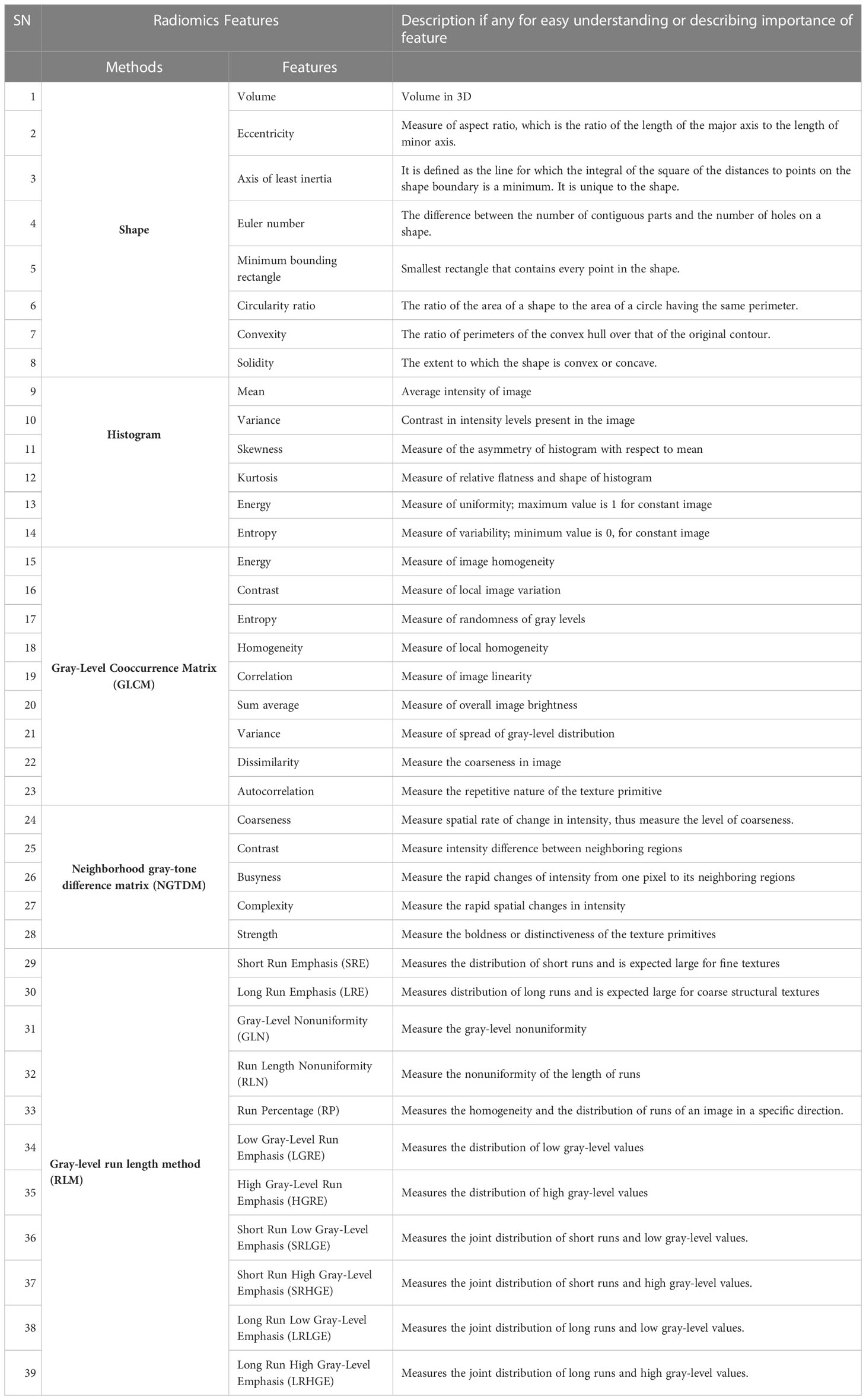
Table 1 List of radiomics features to be evaluated for lesion identification, characterization, diagnosis, monitoring prognosis or predicting outcome of patient.
e) Clinical features
The historical records of the eligible patient cohort will be reviewed, and all the relevant clinical details including demographic profile, histological variants, treatment details, and disease outcomes will be captured and analyzed. Data pertaining to the following clinical characteristics and laboratory parameters at baseline will be included: (i) age at the time of diagnosis, (ii) gender, (iii) histologic subtype of sarcoma (osteosarcoma, primitive neuroectodermal tumor, or soft tissue sarcoma), (iv) histological grade, (v) site of primary disease, (vi) the presence of metastases to other sites, (vii) symptom duration prior to presentation, (viii) performance status, (ix) hemoglobin, (x) total leucocyte count (TLC), (xi) platelet count, (xii) serum lactate dehydrogenase (LDH), (xiii) serum alkaline phosphatase (ALP), (xiv) serum C-reactive protein (CRP), and (xv) serum albumin. The baseline factors may aid in estimation of the disease burden and the likelihood of lung metastases. In addition, details pertaining to chemotherapy and local site therapy modalities received (surgery and/or radiotherapy) including the timeline of receipt, therapeutic responses, and event-free survival (EFS) and overall survival (OS) will be collected. The radiological changes in the lung nodules will be assessed in the context of the timeline of therapy received and clinical responses to therapy. The degree of clinician suspicion for lung metastases based on retrospective file review will be mentioned as “high”, “unclear”, or “low”. Clinical high suspicion of metastases is based on the progressive size of lung nodules in patients along with documented clinical deterioration or death or change of line of therapy. The degree of radiologist suspicion based on routine radiological findings will be mentioned separately by the radiologist. If there is a discrepancy between the two, a multidisciplinary discussion between the radiologist and the clinician will be carried out to reach a consensus. The ground truth as to whether the patient has lung metastases or not on imaging will be mentioned subsequently. For this study, OS will be defined as the time from treatment initiation till death from any cause. EFS will be defined as the time from treatment initiation till disease relapse/progression or death from any cause.
f) Feature selection
Dimensionality and redundancy reduction and identification of potential radiological and clinical feature(s) will be performed for training the proposed model. Feature selection methods like Fisher’s score, Lasso regularization, RF importance, and Recursive feature elimination will be applied on radiological and clinical feature sets separately for evaluating the importance of the features for the classification task and the final feature selection will be performed as the requirement of the training algorithm.
g) Machine learning/deep learning models for detection and classification of metastatic lung nodule
A number of studies have been reported for the classification of benign and malignant lung nodules using ML/DL techniques in primary lung cancer (14–20); however, classification of metastatic lung nodule using artificial intelligence (AI) has been rarely addressed. Various ML-based classification models for primary lung cancer have been reported, while SVM (23–25, 27, 29, 31, 34, 82) was applied mostly along with the other ML technologies like LR (82), KNN (31, 35, 82), RF (32, 33), LDA (27, 34), DNN (28, 30), and Adaboost (34). DL-based frameworks based on CNNs have been developed for lung nodule detection (36). Various advanced CNN models (41–45) and ensemble learners using multiple CNN models (46, 49–53), transfer learning-based systems (54–57), and hybrid CNN-based systems (58–62) have been reported for classifying malignant and benign lung nodules. CNN with adaptive morphology and textural features (63), deep feature selection from different convolution layers (67–69), and 3D segmentation attention network-based systems (64, 65) were reported for lung nodule classification. Recently, DL models based on transformers (73, 74) along with CNN (75, 76) have reported promising results for lung nodule detection and classification.
The plan in the current protocol is to train the proposed model using CT datasets of nearly 450 patients from a retrospective cohort. The aim is to detect and classify the metastatic lung nodules at baseline, i.e., even before commencement of treatment and at follow-up. A separate analysis will be carried out to train the classification model to predict the metastasis even for the smaller nodule (<5 mm) at its very early stage. For the classification model, ML-based algorithms like (i) multivariate LR, (ii) SVM, (iii) LDA, and (iv) RF and DL-based frameworks like (i) transformers with CNN, (ii) 3D CNN (CNN), (iii) recurrent CNN, and (iv) Spatial pyramid Pooling CNNs will be tested and validated. After features selection, relevant radiological features and relevant clinical features will be used separately and in combination to train the ML-based algorithms. For each ML algorithm, three separate training models will be prepared using (i) selected radiological features, (ii) selected clinical features, and (iii) selected radiological and clinical features in combination. For ML model training data normalization, data noise reduction and regularization techniques like ridge regularization and lasso regularization with k-fold cross-validation will be applied as applicable for the model to avoid overfitting and maintain generalizability of the model. The prediction accuracies of different training models for all ML-based algorithms will be compared to identify the best-performing prediction model. For DL-based models, convolutional layers will be used to extract the features from the labeled CT images, then the clinical features will be concatenated with the extracted features vectors from convolutional layers, which will be further feed to the fully connected layers to train the prediction model. Appropriate hyper-parameters like learning rate, dropout, batch size, loss function, momentum, and optimizer will be applied during DL model training. Data augmentation is an important step to overcome insufficiency of labeled data, prevent overfitting, and increase the training accuracy of a DL model. Many literatures reported that use of geometric transformations (55, 60, 65, 76, 83), kernel filtering, color and noise augmentation (50), and Generative Adversarial Networks (84, 85) for augmenting pulmonary CT data improved the accuracy for lung nodule characterization. Data augmentation may introduce inconsistency among data distribution between training and test data. To refine the trained model and maintain the model accuracy, the DL model will be trained on augmented data first, followed by data without any data augmentation. Prediction performance of applied DL-based techniques will be compared to determine the optimal prediction model balancing computational cost and accuracy. For ML-based model implementation, MATLAB® (MathWorks Inc., v2018, Philadelphia, USA) and Python version 3.9 (Python Software Foundation, https://www.python.org/) will be used, and for DL-based model development, programming environments PyTorch (https://pytorch.org/), TensorFlow (https://www.tensorflow.org/), and Keras (https://keras.io/) will be used.
h) Identification of potential marker(s) for lung metastases
The proposed model will be able to identify the potential clinical and/or radiological marker(s) for classifying the metastatic lung nodules from benign ones. The proposed prediction model for detecting lung metastases will be developed and tested for clinical and radiological marker(s) separately and in combination, and the importance of both in combination will be determined. This will help to improve the model performance, reducing the misdiagnosis and false-positive results particularly for the early-stage smaller metastatic lung nodules.
i) Validation
The validation of the proposed ML/DL-based prediction model will be performed using retrospective as well as prospective clinical and CT datasets of a total of 100 patients, 50 patients each from the retrospective and prospective cohort.
2.7 Statistical analysis
a) Student’s t-test will be used for continuous variables and the Chi-square test will be used for categorical variables. A p-value of <0.05 will be taken as significant.
b) Dimensionality reduction and identification of unique features to train the classification model for lung metastasis detection.
c) ML methods to identify independent radiological and clinical feature(s) as markers of lung metastases.
d) ROC curve analysis will be used to find which feature or combination of features would best classify the lung metastases from the other existing benign lung nodules.
e) True-negative and true-positive rate, positive predictive value or precision, recall, F score, and average false-positive rate per patient will be used to evaluate the performance of the proposed models.
Data analysis and development of an ML-based classification model for lung metastases detection will be performed at the laboratory facility of the Centre for Biomedical Engineering, IIT Delhi, India.
3 Discussion
The identification of metastatic disease at presentation is vital to the clinician since the expected prognosis and treatment outcomes are different in metastatic disease from that in localized disease. Furthermore, it allows the clinician to decide the intent of treatment with greater clarity. Thus, determining the nature of lung nodules in a clinical scenario of possible pulmonary metastases is of great importance.
Out of all the medical imaging techniques, CT is considered to be one of the most effective means of detecting lung cancer early (86, 87). Clinicians need to diagnose malignant nodules accurately by reading the patient’s lung CT image; however, reading a large number of CT images is not only time-consuming, and there is also a high probability of misdiagnosis. There has been a lot of research work done regarding the differentiation between benign and malignant solid lung nodules. Lung nodules can be evaluated according to diameter, area, or volume. Results from the literature agreed that volume measurement is a method with a better performance in nodule sizing, as well as in assessing the nodule’s growth (88). Mehta et al. (89) added volumetric nodule measurement to an existing prediction model for malignancy estimation of nodules, showing an improvement in the number of nodule classification. There are a number of other factors like tumor image intensity, shape, and texture that help in determining probability of malignancy in lung nodules (90–97). When evaluating individuals with lung nodules, the probability of malignancy is estimated on the basis of patient-related clinical factors like primary tumor grade, tumor size, and histology type (6, 78) and nodule characteristics, including size (77). As regards morphological characteristics of nodules, besides small size, diffuse, central, laminated, or popcorn calcifications, fat tissue density and perifissural location have been recognized as indicative of benign lesions. It has been found that pleural tags and contour may be identified as independent predictors of pulmonary metastases (98) or higher mean attenuation and larger diameter are significant predictors for pulmonary metastases, while higher mean attenuation is a significant predictor for small non-calcified pulmonary metastases (99). Studies have reported that radiological semantic features like lobulation, spiculation, subtlety, calcification, and texture were relevant along with automatically calculated image-based radiomics features for malignancy classification of pulmonary nodules, which is found to be in line with current clinical practice (83, 100). Inspired by the aforementioned work, in this study, both semantic radiological and clinical features of lung nodules will be combined with radiomics features to develop the proposed classification ML/DL predictive model for metastatic lung nodule detection. Harmonization of radiomics features is an important aspect to obtain standardization and reproducibility of the developed model. In this study, extracted radiomics features will be harmonized by the methods following the recent studies (101–104) as applicable.
ML/DL has achieved a series of satisfactory results in the field of medical imaging, and it also has made great progress in the detection and classification of lung nodules (14–20) and the prediction of nodule growth (105, 106) using ML/DL techniques in primary lung cancer. However, there has been a dearth of work reported on detection of lung metastases. There is no accurate non-invasive modality available to determine the malignant potential of a smaller (<5 mm) pulmonary nodule. Furthermore, functional imaging like 18F-FDG PET/CT scans are also not sensitive for lesions <1 cm. Because this ML/DL-based system will be trained using thousands of lung scans, it will be optimized to detect tiny malignant areas that specialists might overlook in the daily busy clinical routine. On successful completion of the project, the proposed model might be capable of assisting the radiologist to make a diagnostic decision combining the radiological screening with clinical data. The proposed classification model may shift the current clinical practice paradigms by utilizing novel non-invasive radiomics features of CT in combination with ML/DL techniques for characterizing indeterminate lung nodules in the patients with sarcomas. A working model for accurate detection of lung metastases may be effectively applicable for patients with different kinds of primary malignancy as well, especially those with predilection for lung metastases. Early detection of metastatic disease may help in planning personalized treatment protocol and may improve the OS rate.
The strength of the proposed study design is in analyzing an ambispective cohort with separate derivation and validation cohort to develop the proposed prediction and classification model. Moreover, combining radiological features and clinical features with various radiomics textural features may reduce the false-positive rate and help to produce a robust model. Furthermore, information about temporal changes in lung nodules during anti-cancer treatment will also be included into the feature map that will aid the prediction of the malignant potential of lung nodules. In addition, a number of ML and DL algorithms or a hybrid model combining both techniques will be implemented and tested to deliver an optimal performance.
There are few limitations in the proposed study. For demarcation of lung nodules in CT scans, an in-house-built segmentation tool has been used that has been tested on a limited number of 50 patients; however, the accuracy of the tool will be further evaluated in due course as larger data will be tested on the project. This project is multi-institutional and will be facilitated and carried out under AIIMS New Delhi and IIT Delhi in India. Retrospective and prospective datasets of patients with sarcoma will be collected from the institutional database at AIIMS New Delhi. In the future, a multi-institutional dataset may be considered for further improvement of the developed model. A comparison between limited ML and DL classification models will be performed for evaluating best performance; however, there might be a large number of ML/DL-based algorithms that need to be considered.
4 Ethics and dissemination
The ethical approval for the use of retrospective data and collection of prospective data of the patients with sarcomas has been obtained (IEC-234/09.04.2021, RP-03/2021) from the Institute Ethics Committee, AIIMS New Delhi, India. The details of data acquisition, processing, and sharing along with risks and benefits for participating in the study will be explained to all the patients before recruitment to the proposed project. Detailed patient information sheets written in English and Hindi (regional) languages have been reviewed and approved by the Institutional Ethics Committee.
The proposed study challenges and seeks to shift the current clinical practice paradigms by utilizing novel non-invasive radiomics features of CT combined with physiological symptoms and clinical biomarkers. As a result, misdiagnosis and false-positive rate might be reduced along with reduction in the total number of follow-up CT scans, which will subsequently reduce the time, cost, anxiety, and radiation exposure of the patients. Upon validation, the proposed model will be deployed in the local hospital settings and will be applied in the routine treatment protocol.
Ethics statement
The studies involving human participants were reviewed and approved by the Institute Ethics Committee, All India Institute of Medical Sciences New Delhi, India. Written informed consent is obtained for the prospective cohort from the participants in case of adult subjects (18 years and older), from the legal guardian in case of those below 18 years, and additionally assent from children who are 8 years and above. The Institute Ethics Committee waives off consent for the use of the participants' data for the retrospective cohort.
Author contributions
EBK, SG, KR, DK, SB, and AM conceptualized the study. Data curation and investigation were performed by EBK, SG, AS, SS, DDS, MS, and DK. Resources were provided by KR, DK, SB, and AM. Original manuscript was written by EBK. Reviewing and editing the manuscript were performed by EBK, SG, AS, SS, KR, DK, SB, and AM. All authors contributed to the article and read and approved the submitted version.
Funding
This research received external funding from Indian Council of Medical Research (ICMR), New Delhi, India (AI-Adhoc/06/2022-AI Cell) and intramural project funding under Multi-Institutional Faculty Interdisciplinary Research Project scheme in collaboration with All India Institute of Medical Sciences, Delhi, India (AI-56) and Indian institute of Technology Delhi, India (MI02654).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Singh T, Sharma A, Sharma A, Bakhshi S, Patel C, Pandey A, et al. Utility of 18F-FDG-PET/CT in management and prognostication of treatment naïve late-stage soft tissue sarcomas. Nucl Med Commun (2021) 42(7):818–25. doi: 10.1097/MNM.0000000000001401
2. Gadd MA, Casper ES, Woodruff JM. Development and treatment of pulmonary metastases in adult patients with extremity soft tissue sarcoma. Ann Surg (1993) 218(6):705–12. doi: 10.1097/00000658-199312000-00002
3. Billingsley KG, Burt ME, Jara E, Ginsberg RJ, Woodruff JM, Leung DHY, et al. Pulmonary metastases from soft tissue sarcoma analysis of patterns of disease and postmetastasis survival. Ann Surg (1999) 229(5):602–12. doi: 10.1097/00000658-199905000-00002
4. Biswas B, Rastogi S, Khan S, Shukla N, Deo S, Agarwala S, et al. Hypoalbuminaemia is an independent predictor of poor outcome in metastatic Ewing’s sarcoma family of tumours: a single institutional experience of 150 cases treated with uniform chemotherapy protocol. Clin Oncol (2014) 26(11):722–9. doi: 10.1016/j.clon.2014.05.006
5. Iqbal N, Shukla N, Deo S, Agarwala S, Sharma D, Sharma M, et al. Prognostic factors affecting survival in metastatic soft tissue sarcoma: an analysis of 110 patients. Clin Transl Oncol (2016) 18(3):310–6. doi: 10.1007/s12094-015-1369-9
6. Nataraj V, Rastogi S, Khan SA, Sharma MC, Agarwala S, Vishnubhatla S, et al. Prognosticating metastatic osteosarcoma treated with uniform chemotherapy protocol without high dose methotrexate and delayed metastasectomy: a single center experience of 102 patients. Clin Transl Oncol (2016) 18(9):937–44. doi: 10.1007/s12094-015-1467-8
7. Ciccarese F, Bazzocchi A, Ciminari R, Righi A, Rocca M, Rimondi E, et al. The many faces of pulmonary metastases of osteosarcoma: Retrospective study on 283 lesions submitted to surgery. Eur J Radiol [Internet] (2015) 84(12):2679–85. doi: 10.1016/j.ejrad.2015.09.022
8. Gross BH, Glazer GM, Bookstein FL. Multiple pulmonary nodules detected by computed tomography: diagnostic implications. J Comput Assist Tomogr (1985) 9(5):880–5. doi: 10.1097/00004728-198509000-00007
9. Meyers P. Trial of ADR, CDDP and MTX with and without ifos, with and without MTP-PE for treatment of osteogenic sarcoma(POG-9351). (1996), COG 7921.
10. Weiss A. COGARST1321: Pazopanib neoadjuvant trial in non-rhabdomyosarcoma soft tissue sarcomas (PAZNTIS): A phase II/III randomized trial of preoperative chemoradiation or preoperative radiation plus or minus pazopanib (NSC# 737754, IN. (2014). clinicaltrials.gov NCT No: NCT02180867.
11. Picci P, Vanel D, Briccoli A, Talle K, Haakenaasen U, Malaguti C, et al. Computed tomography of pulmonary metastases from osteosarcoma : The less poor technique. A study of 51 patients with histological correlation. Ann Oncol (2001) 12(11):1601–4. doi: 10.1023/a:1013103511633
12. Ginsberg MS, Panicek DM. Subcentimeter pulmonary nodules detected in patients with sarcoma. Sarcoma (2000) 4(1–2):63–6. doi: 10.1155/S1357714X00000116
13. Rastogi R, Garg R, Thulkar S, Bakhshi S, Gupta A. Unusual thoracic CT manifestations of osteosarcoma: review of 16 cases. Padiatr Radiol (2008) 38(5):551–8. doi: 10.1007/s00247-007-0735-3
14. Jassim MM, Jaber MM. Systematic review for lung cancer detection and lung nodule classification : Taxonomy, challenges, and recommendation future works. J Intell Syst (2022) 31:944–64. doi: 10.1515/jisys-2022-0062
15. Gu D, Liu G, Xue Z. On the performance of lung nodule detection, segmentation and classification. Comput Med Imaging Graph (2021) 89(January):101886. doi: 10.1016/j.compmedimag.2021.101886
16. De Margerie-mellon C, Chassagnon G. Artificial intelligence: A critical review of applications for lung nodule and lung cancer. Diagn Interv Imaging [Internet] (2023) 104(1):11–7. doi: 10.1016/j.diii.2022.11.007
17. Khawaja A, Bartholmai BJ, Rajagopalan S, Karwoski RA, Varghese C, Maldonado F, et al. Do we need to see to believe ?— radiomics for lung nodule classification and lung cancer risk stratification. J Thorac Dis (2020) 12(6):3303–16. doi: 10.21037/jtd.2020.03.105
18. Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med [Internet] (2019) 25:954–61. doi: 10.1038/s41591-019-0447-x
19. Hua K-L, Hsu C-H, Hidayati SC, Cheng W-H, Chen Y-J. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoloTargets Ther (2022) 8:2015–22. doi: 10.2147/OTT.S80733
20. Song Q, Zhao L, Luo X, Dou X. Using deep learning for classification of lung nodules on computed tomography images. J Healthc Eng (2017) 2017:8314740. doi: 10.1155/2017/8314740
21. Wu P, Xia K, Yu H. Correlation coefficient based supervised locally linear embedding for pulmonary nodule recognition. Comput Methods Programs BioMed (2016) 136:97–106. doi: 10.1016/j.cmpb.2016.08.009
22. Ciompi F, Jacobs C, Scholten ET, Wille MMW, De Jong PA, Prokop M, et al. Bag-of-frequencies : A descriptor of pulmonary nodules in computed tomography images. IEEE Trans Med Imaging (2015) 34(4):962–73. doi: 10.1109/TMI.2014.2371821
23. Kumar A, Sudipta D, Anirvan M, Mandeep D, Khandelwal N. A combination of shape and texture features for classification of pulmonary nodules in lung CT images. J Digit Imaging (2016) 29(4):466–75. doi: 10.1007/s10278-015-9857-6
24. Akram S, Javed MY, Hussain A, Riaz F. Intensity-based statistical features for classification of lungs CT scan nodules using artificial intelligence techniques. J Exp Theor Artif Intell ISSN (2015) 27(6):737–51. doi: 10.1080/0952813X.2015.1020526
25. De Carvalho Filho AO, Silva AC, de Paiva AC, Nunes RA, Gattass M. Computer-aided diagnosis of lung nodules in computed tomography by using phylogenetic diversity , genetic algorithm , and SVM. J Digit Imaging (2017) 30:812–22. doi: 10.1007/s10278-017-9973-6
26. Armato SG, Altman MB, Wilkie J, Sone S, Li F, Doi K, et al. Automated lung nodule classification following automated nodule detection on CT : A serial approach. Med P (2011) 30(6):1188–97. doi: 10.1118/1.1573210
27. Gong J, Liu J, Sun X, Zheng B, Niea S. Computer-aided diagnosis of lung cancer: the effect of training datasets on classification accuracy of lung nodules. Phys Med Biol (2018) 63(3):035036. doi: 10.1088/1361-6560/aaa610
28. Anand SKV. Segmentation coupled textural feature classification for lung tumor prediction, In: International conference on communication control and computing technologies, Nagercoil, India (2010). pp. 518–24. doi: 10.1109/ICCCCT.2010.5670607
29. Orozco HM, Osiris O, Villegas V, Guadalupe V, Sánchez C. Automated system for lung nodules classification based on wavelet feature descriptor and support vector machine. Biomed Eng Online (2015) 14(9):1–20. doi: 10.1186/s12938-015-0003-y
30. Lakshmanaprabu SK, Nandan S, Shankar K, Arunkumar N. Optimal deep learning model for classification of lung cancer on CT images. Futur Gener Comput Syst (2018) 92:374–82. doi: 10.1016/j.future.2018.10.009
31. Rehman A, Kashif M, Abunadi I, Ayesha N. Lung cancer detection and classification from chest CT scans using machine learning techniques, In: 2021 1st International Conference on Artificial Intelligence and Data Analytics (CAIDA), Riyadh, Saudi Arabia (2021). pp. 101–10. doi: 10.1109/CAIDA51941.2021.9425269
32. Li X, Li B, Tian L, Zhang L. Automatic benign and Malignant classification of pulmonary nodules in thoracic computed tomography based on RF algorithm. intitute Eng Technol (2018) 12(7):1072–291. doi: 10.1049/iet-ipr.2016.1014
33. Wu W, Hu H, Gong J, Li X, Huang G, Nie S. Malignant-benign classification of pulmonary nodules based on random forest aided by clustering analysis. Phys Med Biol (2019) 64(3):035017. doi: 10.1088/1361-6560/aafab0
34. Kaya A, Can AB. A weighted rule based method for predicting Malignancy of pulmonary nodules by nodule characteristics. J Biomed Inform (2015) 56:69–79. doi: 10.1016/j.jbi.2015.05.011
35. Farahani FV, Ahmadi A, Hossein M, Zarandi F. Hybrid intelligent approach for diagnosis of the lung nodule from CT images using spatial kernelized fuzzy c-means and ensemble learning learning. Math Comput Simul (2018) 149:48–68. doi: 10.1016/j.matcom.2018.02.001
36. Monkam P, Qi S, Ma HE, Yao Y, Qian WEI. Detection and classification of pulmonary nodules using convolutional neural networks : A survey. IEEE Access (2019) 7:78075–91. doi: 10.1109/ACCESS.2019.2920980
37. Gu Y, Chi J, Liu J, Yang L, Zhang B, Yu D, et al. A survey of computer-aided diagnosis of lung nodules from CT scans using deep learning. Comput Biol Med (2021) 137:104806. doi: 10.1016/j.compbiomed.2021.104806
38. Li R, Xiao C, Huang Y, Hassan H, Huang B. Deep learning applications in computed tomography images for pulmonary nodule detection and diagnosis : A review. Diagnostics (Basel) (2022) 12(2):298. doi: 10.3390/diagnostics12020298
39. Riquelme D, Akhloufi MA. Deep learning for lung cancer nodules detection and classification in CT scans. AI (2020) Ml):28–67. doi: 10.3390/ai1010003
40. Qin Y, Zheng H, Zhu Y, Yang J, Rd D, District M, et al. Simultaneous accurate detection of pulmonary nodules and false positive reduction using 3D CNNs, In: 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada (2018). pp. 1005–9. doi: 10.1109/ICASSP.2018.8462546
41. Tang H, Kim DR, Xie X. Automated pulmonary nodule detection using 3D deep convolutional neural networks, In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). (2017). pp. 3–6. doi: 10.48550/arXiv.1903.09876
42. Winkels M, Cohen TS. 3D G-CNNs for pulmonary nodule detection, In: 1st Conference on Medical Imaging with Deep Learning (MIDL 2018). (2018). pp. 1–11. doi: 10.48550/arXiv.1804.04656
43. Da Silva GLF, Valente TLA, Corres SA, De Paiva AC, Gattass M. Convolutional neural network-based PSO for lung nodule false positive reduction on CT images. Comput Methods Programs Biomed (2018) 162:109–18. doi: 10.1016/j.cmpb.2018.05.006
44. Monkam P, Qi S, Xu M, Han F, Zhao X, Qian W. CNN models discriminating between pulmonary micro − nodules and non − nodules from CT images. BioMed Eng Online (2018) 17(1):1–16. doi: 10.1186/s12938-018-0529-x
45. Jia AJ, Liu BJ, Gu CY. Computer-aided diagnosis of pulmonary nodules on CT scan images, In: 2018 10th International Conference on Modelling, Identification and Control (ICMIC), Guiyang, China (2018). pp. 2–4. doi: 10.1109/ICMIC.2018.8529939
46. Hamidian S, Sahiner B, Petrick N, Pezeshk A, Health R, Food US, et al. 3D convolutional neural network for automatic detection of lung nodules in chest CT. Proc SPIE Int Soc Opt Eng (2017) 10134:1013409. doi: 10.1117/12.2255795
47. Liu Y, Hao P, Zhang P, Xu X, Wu J, Chen WEI. Dense convolutional binary-tree networks for lung nodule classification. IEEE Access (2018) 6:49080–8. doi: 10.1109/ACCESS.2018.2865544
48. Zhang G, Luo Y, Zhu D, Xu Y, Sun Y, Lu J. Spatial pyramid dilated network for pulmonary nodule Malignancy classification, In: 2018 24th Int Conf Pattern Recognit, Beijing, China (2018). pp. 3911–6. doi: 10.1109/ICPR.2018.8546106
49. Xie Y, Xia Y, Zhang J, Song Y. Knowledge-based collaborative deep learning for benign-Malignant lung nodule classification on chest CT. IEEE Trans Med Imaging (2019) 38(4):991–1004. doi: 10.1109/TMI.2018.2876510
50. Ahmed I, Chehri A, Jeon G, Piccialli F. Automated pulmonary nodule classification and detection using deep learning architectures. IEEE/ACM Trans Comput Biol Bioinforma (2022) PP(X):1–12. doi: 10.1109/TCBB.2022.3192139
51. Liao Z, Xie Y, Hu S, Xia Y. Learning from ambiguous labels for lung nodule Malignancy prediction. IEEE Trans Med Imaging (2022) 41(7):1874–84. doi: 10.1109/TMI.2022.3149344
52. Liu H, Cao H, Song E. Multi-model ensemble learning architecture based on 3D CNN for lung nodule Malignancy suspiciousness classification. J Digit Imaging (2020) 1:1242–56. doi: 10.1007/s10278-020-00372-8
53. Zhang G, Yang Z, Gong L, Jiang S, Wang L, Zhang H. Classification of lung nodules based on CT images using squeeze − and − excitation network and aggregated residual transformations. Radiol Med [Internet] (2020) 125(4):374–83. doi: 10.1007/s11547-019-01130-9
54. Nóbrega RVMd, Peixoto SA, Silva SPPda, Filho PPR. Lung nodule classification via deep transfer learning in CT lung images, In: 2018 IEEE 31st International Symposium on Computer-Based Medical Systems Lung, Karlstad, Sweden (2018). pp. 244–9. doi: 10.1109/CBMS.2018.00050
55. Wu R, Liang C, Li Y, Shi X, Zhang J, Huang H. Self-supervised transfer learning framework driven by visual attention for benign – Malignant lung nodule classification on chest CT. Expert Syst Appl (2023) 215(November 2022):119339. doi: 10.1016/j.eswa.2022.119339
56. William I, Liawatimena S, Wawan T. Lung nodule detection and classification from Thorax CT-scan using RetinaNet with transfer learning. J King Saud Univ - Comput Inf Sci (2022) 34(3):567–77. doi: 10.1016/j.jksuci.2020.03.013
57. Huang H, Wu R, Li Y, Peng C. Self-supervised transfer learning based on domain adaptation for benign-Malignant lung nodule classification on thoracic CT. IEEE J BioMed Heal Informatics (2022) 26(8):3860–71. doi: 10.1109/JBHI.2022.3171851
58. Zhao X, Liu L, Qi S, Teng Y, Li J, Qian W. Agile convolutional neural network for pulmonary nodule classification using CT images. Int J Comput Assist Radiol Surg (2018) 13(4):585–95. doi: 10.1007/s11548-017-1696-0
59. Nishio M, Sugiyama O, Yakami M, Ueno S, Kubo T. Computer-aided diagnosis of lung nodule classification between benign nodule, primary lung cancer, and metastatic lung cancer at different image size using deep convolutional neural network with transfer learning. PLoS One (2018) 13(7):1–12. doi: 10.1371/journal.pone.0200721
60. Dodia S, Basava A, Anand MP. A novel receptive field-regularized V-net and nodule classification network for lung nodule detection. Int J Imaging Syst Technol (2021) 32(1):88–101. doi: 10.1002/ima.22636
61. Tong C, Liang B, Su Q, Yu M, Hu J, Bashir AK, et al. Pulmonary nodule classification based on heterogeneous features learning. IEEE J Sel AREAS Commun (2021) 39(2):574–81. doi: 10.1109/JSAC.2020.3020657
62. Mehta K, Jain A, Mangalagiri J, Menon S, Nguyen P. Lung nodule classification using biomarkers , volumetric radiomics , and 3D CNNs. J Digit Imaging (2021) 34(3):647–66. doi: 10.1007/s10278-020-00417-y
63. Halder A, Chatterjee S, Dey D. Biomedical Signal Processing and Control Adaptive morphology aided 2-pathway convolutional neural network for lung nodule classification. BioMed Signal Process Control (2022) 72(PB):103347. doi: 10.1016/j.bspc.2021.103347
64. Guo Z, Yang J, Zhao L, Yuan J, Yu H. 3D SAACNet with GBM for the classification of benign and Malignant. Comput Biol Med (2023) 153(January):106532. doi: 10.1016/j.compbiomed.2022.106532
65. Al-shabi M, Shak K, Tan M. ProCAN : Progressive growing channel attentive non-local network for lung nodule classification. Pattern Recognit (2022) 122:108309. doi: 10.1016/j.patcog.2021.108309
66. Nasrullah N, Sang J, Alam MS, Mateen M, Cai B, Hu H. Automated lung nodule detection and classification using deep learning combined with multiple strategies. Sensors (2019) 19(3722):1–19. doi: 10.3390/s19173722
67. Ali I, Muzammil M, Haq IUL, Khaliq AA, Abdullah S. Deep feature selection and decision level fusion for lungs nodule classification. IEEE Access (2021) 9:18962–73. doi: 10.1109/ACCESS.2021.3054735
68. Çoruh Ag, Yenigün B, Uzun Ç, Kahya Y, Büyükceran E, Orhan K, et al. A comparison of the fusion model of deep learning neural networks with human observation for lung nodule detection and classification. Br J Radiol (2021) 94:20210222. doi: 10.1259/bjr.20210222
69. Bruntha PM, Ahmed LJ, Pandian SIA, Abraham SS. Investigation of deep features in lung nodule classification. 2022 6th Int Conf Devices Circuits Syst (2022) :67–70. doi: 10.1109/ICDCS54290.2022.9780716
70. Xu X, Wang C, Guo J, Gan Y, Wang J, Bai H, et al. MSCS-DeepLN : Evaluating lung nodule Malignancy using multi-scale cost-sensitive neural networks. Med Image Anal (2020) 65:101772. doi: 10.1016/j.media.2020.101772
71. Lei Y, Tian Y, Shan H, Zhang J, Wang G, Kalra MK. Shape and margin-aware lung nodule classification in low-dose CT images via soft activation mapping. Med Image Anal (2020) 60:101628. doi: 10.1016/j.media.2019.101628
72. Lei Y, Shan H, Zhang J. Meta ordinal weighting net for improving lung nodule classification. IEEE Xplor (2021) 14(9):1210–4. doi: 10.1109/ICASSP39728.2021.9413622
73. Niu C, Wang G. Unsupervised contrastive learning based transformer for lung nodule detection Unsupervised contrastive learning based transformer for lung nodule detection. Phys Med Biol (2022) 67:204001. doi: 10.1088/1361-6560/ac92ba
74. Wu K, Peng B. Multi-granularity dilated transformer for lung nodule classification via local focus scheme. Appl Sci (2023) 13(337):1–12. doi: 10.3390/app13010377
75. Wang H, Zhu H, Ding L. Accurate classification of lung nodules on CT images using the TransUnet. Front Public Heal (2022) 10(1060798):1–9. doi: 10.3389/fpubh.2022.1060798
76. Liu D, Liu F, Tie Y, Qi L, Wang F. Res-trans networks for lung nodule classification. Int J Comput Assist Radiol Surg (2022) 17(6):1059–68. doi: 10.1007/s11548-022-02576-5
77. Gould MK, Donington J, Lynch WR, Mazzone PJ, Midthun DE, Naidich DP, et al. Evaluation of individuals with pulmonary nodules : when is it lung cancer? Diagnosis and management of lung cancer, 3rd ed : American college of chest physicians. Chest (2013) 143(5 Suppl):e93S–120S. doi: 10.1378/chest.12-2351
78. Xie L, Huang W, Wang H, Zheng C, Jiang J. Risk factors for lung metastasis at presentation with Malignant primary osseous neoplasms : a population-based study. J Orthop Surg Res (2020) 15(1):32. doi: 10.1186/s13018-020-1571-5
79. Balki I, Amirabadi A, Levman J, Martel AL, Emersic Z, Meden B, et al. Sample-size determination methodologies for machine learning in medical imaging research : A systematic review. Can Assoc Radiol J (2019) 70(4):344–53. doi: 10.1016/j.carj.2019.06.002
80. Calheiros JLL, de Amorim LBV, de Lima LL, de Lima Filho AF, Ferreira Júnior JR, de Oliveira MC. The effects of perinodular features on solid lung nodule classification. J Digit Imaging (2021) 34(4):798–810. doi: 10.1007/s10278-021-00453-2
81. Dilger SKN, Mott SL, Smith BJ, Newell JD, Hoffman EA, Sieren JC. Improved pulmonary nodule classification utilizing quantitative lung parenchyma features quantitative lung parenchyma features. J Med Imaging (2015) 2(4):041004. doi: 10.1117/1.JMI.2.4.041004
82. Kawathekar ID, Anu Areeckal S. Performance analysis of texture characterization techniques for lung nodule classification. J Physics:Conference Series (2022) 2161:012045. doi: 10.1088/1742-6596/2161/1/012045
83. Marques S, Schiavo F, Ferreira CA, Pedrosa J, Cunha A. A multi-task CNN approach for lung nodule Malignancy classification and characterization. Expert Syst Appl (2021) 184(September 2020):115469. doi: 10.1016/j.eswa.2021.115469
84. Apostolopoulos ID, Papathanasiou ND, Panayiotakis GS. Classification of lung nodule Malignancy in computed tomography imaging utilising generative adversarial networks and semi-supervised transfer learning. Biocybern BioMed Eng (2021) 41(4):1243–57. doi: 10.1016/j.bbe.2021.08.006
85. Ghosal SS, Sarkar I, Elhallaoui I. Lung nodule classification using Convolutional Autoencoder and Clustering Augmented Learning Method (CALM). HSDM@WSDM (2020).
86. Welter S, Grabellus F, Bauer S, Schuler M, Eberhardt W. Growth patterns of lung metastases from sarcoma : prognostic and surgical implications from histology. Interact Cardiovasc Thorac Surg (2012) 15(July):612–7. doi: 10.1093/icvts/ivs269
87. Digesu CS, Wiesel O, Vaporciyan AA, Yolonda L. Management of sarcoma metastases to the lung. Surg Oncol Clin N Am (2017) 25(4):721–33. doi: 10.1016/j.soc.2016.05.005
88. Manickavasagam R, Selvan S. Automatic detection and classification of lung nodules in CT image using optimized neuro fuzzy classifier with cuckoo search algorithm. J Med Syst (2019) 43(3):77. doi: 10.1007/s10916-019-1177-9
89. Jennings SG, Winer-muram HT, Tarver RD, Farber MO. Lung tumor growth : assessment with CT — Comparison of diameter and cross-sectional area with volume measurements. Radiology (2004) 231(3):866–71. doi: 10.1148/radiol.2313030715
90. MacMahon H, Naidich DP, Goo JM, Lee KS, Leung ANC, Mayo JR, et al. Guidelines for Management of incidental Pulmonary nodules Detected on cT images: From the Fleischner Society 2017. Radiology (2017) 284(1):228–43. doi: 10.1148/radiol.2017161659
91. Bankier AA, MacMahon H, Goo JM, Rubin GD, Schaefer-Prokop CM, Naidich DP. Recommendations for Measuring Pulmonary nodules at cT: A Statement from the Fleischner Society. Radiology (2017) 285(2):162894. doi: 10.1148/radiol.2017162894
92. Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Haibe-kains B, Grossmann P, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat C (2014) 5(1):4006. doi: 10.1038/ncomms5644
93. Cook GJR, Yip C, Siddique M, Goh V, Chicklore S, Roy A, et al. Are pretreatment 18F-FDG PET tumor textural features in non–small cell lung cancer associated with response and survival after chemoradiotherapy? J neuclear Med (2013) 54(1):19–26. doi: 10.2967/jnumed.112.107375
94. Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res (2003) 3(1):1157–82. doi: 10.1162/153244303322753616
95. Larici AR, Farchione A, Franchi P, Ciliberto M, Cicchetti G, Calandriello L, et al. Lung nodules : size still matters. Eur Respir Rev [Internet] (2017) 26(146):170025. doi: 10.1183/16000617.0025-2017
96. Shen J, Du H, Wang Y, Du L, Yang D, Wang L, et al. A novel nomogram model combining CT texture features and urine energy metabolism to differentiate single benign from Malignant pulmonary nodule. Front Oncol (2022) 12:1035307. doi: 10.3389/fonc.2022.1035307
97. He C, Liu J, Li Y, Lin L, Qing H. Quantitative parameters of enhanced dual-energy computed tomography for differentiating lung cancers from benign lesions in solid pulmonary nodules. Front Oncol (2022) 12:11027985. doi: 10.3389/fonc.2022.1027985
98. Guo R, Yan S, Wang F, Su H, Xie Q. A novel diagnostic model for differentiation of lung metastasis from primary lung cancer in patients with colorectal cancer. Front Oncol (2022) 12:1017618. doi: 10.3389/fonc.2022.1017618
99. Cho YJ, Kim WS, Choi YH, Ha JY, Lee S, Park SJ, et al. Computerized texture analysis of pulmonary nodules in pediatric patients with osteosarcoma : Differentiation of pulmonary metastases from non-metastatic nodules. PloS One (2019) 14(2):e0211969. doi: 10.1371/journal.pone.0211969
100. Hancock MC, Magnan JF. Lung nodule Malignancy classification using only radiologist-quantified image features as inputs to statistical learning algorithms : probing the Lung Image Database Consortium dataset with two statistical learning methods. J Med Imaging (Bellingham) (2017) 3(4):044504. doi: 10.1117/1.JMI.3.4.044504
101. Haga A, Takahashi W, Aoki S, Nawa K, Yamashita H, Abe O, et al. Standardization of imaging features for radiomics analysis. J Med Investig (2019) 66(1.2):35–7. doi: 10.2152/jmi.66.35
102. Fortin J, Parker D, Tunç B, Watanabe T, Elliott MA, Ruparel K, et al. Harmonization of multi-site diffusion tensor imaging data. Neuroimage (2018) 161:149–70. doi: 10.1016/j.neuroimage.2017.08.047
103. Pasini G, Bini F, Russo G, Comelli A, Marinozzi F, Stefano A. matRadiomics : A novel and complete radiomics framework, from image visualization to predictive model. J Imaging (2022) 8(8):221. doi: 10.3390/jimaging8080221
104. Bertolini M, Trojani V, Botti A, Cucurachi N, Galaverni M, Cozzi S, et al. Novel harmonization method for multi-centric radiomic studies in non-small cell lung cancer. Curr Oncol (2022) 29(8):5179–94. doi: 10.3390/curroncol29080410
105. Yang R, Hui D, Li X, Wang K, Li C, Li Z. Prediction of single pulmonary nodule growth by CT radiomics and clinical features — a one- year follow-up study. Front Oncol (2022) 12:1034817. doi: 10.3389/fonc.2022.1034817
Keywords: sarcoma, lung metastases, machine learning, deep learning, artificial intelligence, malignancy, radiomics, early diagnosis
Citation: Baidya Kayal E, Ganguly S, Sasi A, Sharma S, DS D, Saini M, Rangarajan K, Kandasamy D, Bakhshi S and Mehndiratta A (2023) A proposed methodology for detecting the malignant potential of pulmonary nodules in sarcoma using computed tomographic imaging and artificial intelligence-based models. Front. Oncol. 13:1212526. doi: 10.3389/fonc.2023.1212526
Received: 26 April 2023; Accepted: 31 July 2023;
Published: 21 August 2023.
Edited by:
Hongda Liu, Nanjing Medical University, ChinaReviewed by:
Yuting Ke, Massachusetts Institute of Technology, United StatesCui Youbin, Jilin University, China
Copyright © 2023 Baidya Kayal, Ganguly, Sasi, Sharma, DS, Saini, Rangarajan, Kandasamy, Bakhshi and Mehndiratta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit Mehndiratta, amit.mehndiratta@keble.oxon.org
 Esha Baidya Kayal1
Esha Baidya Kayal1 Shuvadeep Ganguly
Shuvadeep Ganguly Archana Sasi
Archana Sasi Sameer Bakhshi
Sameer Bakhshi Amit Mehndiratta
Amit Mehndiratta