- Institute of Biochemistry and Biophysics Polish Academy of Sciences, Warsaw, Poland
Multiple reports demonstrate associations between ethylene and sulfur metabolisms, however the details of these links have not yet been fully characterized; the links might be at the metabolic and the regulatory levels. First, sulfur-containing metabolite, methionine, is a precursor of ethylene and is a rate limiting metabolite for ethylene synthesis; the methionine cycle contributes to both sulfur and ethylene metabolism. On the other hand, ethylene is involved in the complex response networks to various stresses and it is known that S deficiency leads to photosynthesis and C metabolism disturbances that might be responsible for oxidative stress. In several plant species, ethylene increases during sulfur starvation and might serve signaling purposes to initiate the process of metabolism reprogramming during adjustment to sulfur deficit. An elevated level of ethylene might result from increased activity of enzymes involved in its synthesis. It has been demonstrated that the alleviation of cadmium stress in plants by application of S seems to be mediated by ethylene formation. On the other hand, the ethylene-insensitive Nicotiana attenuata plants are impaired in sulfur uptake, reduction and metabolism, and they invest their already limited S into methionine needed for synthesis of ethylene constitutively emitted in large amounts to the atmosphere. Regulatory links of EIN3 and SLIM1 (both from the same family of transcriptional factors) involved in the regulation of ethylene and sulfur pathway, respectively, is also quite probable as well as the reciprocal modulation of both pathways on the enzyme activity levels.
Introduction
Sulfur (S) is an important macronutrient for all organisms. Plants can metabolize inorganic sulfur that is taken up from the soil in the oxidized form (sulfate) and then it is reduced and incorporated into a broad range of primary and secondary metabolites. Some of them serve as precursors of other important (but not S-containing) cellular compounds. A schematic overview of the S assimilation pathway, including most of the related metabolites, is shown in Figure 1. The crosstalk between sulfur assimilation and ethylene signaling in plants attracts more attention because of the growing number of data concerning the influence of S nutrition on ethylene signaling and production, as well as the impact of ethylene on the expression of S genes, activity of S enzymes and level of S metabolites (Iqbal et al., 2013). Here, we briefly summarize the most important facts and observations related to the links between ethylene and S nutrition and propose a working model of the complex signaling and regulatory interplay between these two factors.
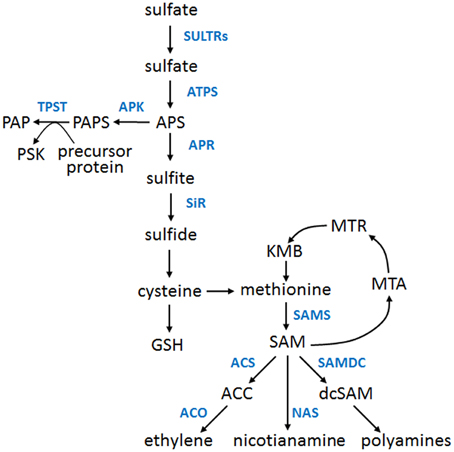
Figure 1. An overview of the sulfur assimilation pathway and major sulfur metabolites. Only the selected metabolites (black fonts) and selected enzymes (blue fonts) of the pathway are presented. ACC, 1-aminocyclopropane-1-carboxylate; ACO, ACC oxidase; ACS, ACC synthase; APK, APS kinase; APR, APS reductase; APS, adenosine 5′-phosphosulfate; ATPS, ATP sulfurylase; dcSAM, decarboxylated SAM; GSH, reduced glutathione; KMB, α-keto-γ-methylthiobutyric acid; MTA, S-methyl-5′-thioadenosine; MTR, S-methyl-5-thio-D-ribose; NAS, nicotianamine synthase; PAP, 3′-phosphoadenosine 5′-phosphate; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; PSK, phytosulfokine; SAM, S-adenosylmethionine; SAMDC, SAM decarboxylase; SAMS, SAM synthase; SiR, sulfite reductase; SULTR, sulfate transporter; TPST, tyrosylprotein sulfotransferase.
Sulfur Metabolites as Precursors in Ethylene Synthesis
Methionine (Met), a sulfur-containing amino acid is a substrate for S-adenosylmethionine synthase (SAMS) responsible for the synthesis of S-adenosylmethionine (SAM or AdoMet), an important metabolite in animals and plants (Fontecave et al., 2004; Roje, 2006). SAM serves as a donor of methyl, amino, ribosyl, and aminoalkyl groups. It is also a source of controlled 5′-deoxyadenosine radicals. In plants, SAM is a precursor of polyamines (PA), nicotianamine (NA) used to produce phytosiderophores, and ethylene. Production of ethylene is a two-step reaction with 1-aminocyclopropane-1-carboxylate (ACC), as a product of the first reaction, catalyzed by ACC synthase, and the substrate for the second reaction catalyzed by ACO (ACC oxidase; Figure 1). Met and SAM used for PA, NA and ethylene biosynthesis are recycled in the Met salvage cycle (known also as a Yang cycle). Noteworthy, soluble Met is apparently a rate-limiting metabolite of ethylene biosynthesis (Katz et al., 2006; Bürstenbinder et al., 2007), however for further details on the additional salvage cycles, regulatory circuits and complex relationships between the metabolites and enzymes, please see the reviews (Amir, 2010; Sauter et al., 2013). A new player, a plasma membrane receptor-like kinase, FERONIA, involved in the regulation of SAMS in Arabidopsis thaliana, has been recently reported (Mao et al., 2015).
Additional complexity is added by the fact that ACC seems to have more functions than just being the precursor of ethylene. It is a subject of short- and long-distance dedicated transport, can be conjugated to form three different derivatives. It also seems to be a signaling molecule by itself (Van De Poel and Van Der Straeten, 2014).
Sulfur Nutrition Affects Ethylene Synthesis During Various Stresses
Sulfur nutrition has been reported to modulate the stress response by increasing ethylene production in several stresses. The most intensively studied is cadmium (Cd)-induced stress. The results of experiments with mustard and wheat indicated that the reduced sensitivity of plants to ethylene due to Cd exposure is elevated with additional S supply. S application increased photosynthesis and dry mass, and resulted in the alleviation of oxidative stress by increasing the levels of antioxidant compounds, such as reduced glutathione (GSH; Masood et al., 2012; Asgher et al., 2014; Khan et al., 2015).
Drought stress has been shown to down-regulate S metabolism and ethylene enzymes in medicago roots and nodules (Larrainzar et al., 2014). Also in cassava grown during the dry season, the association of ethylene level with sulfur metabolism and GSH level in root cortex tissues was observed (Saithong et al., 2015). The regulatory aspect of various primary and secondary S metabolites in relation to drought response, including the role of 3′-phosphoadenosine 5′-phosphate (PAP) produced from 3′-phosphoadenosine 5′-phosphosulfate (PAPS) in retrograde signaling, were recently reviewed (Chan et al., 2013). Besides, the authors underline that various osmoprotectants (for example PA), accumulating during drought stress, require restoring the sulfur moiety in SAM through the Yang cycle.
Moreover, it has been shown that the effects of salt stress (inhibition of photosynthesis) in mustard can be reversed by excess S, and this reversal involved ethylene because the inhibition of ethylene biosynthesis counteracted the effects of S excess on salt stress alleviation (Nazar et al., 2014). The authors suggest that under salt stress, S was used for GSH synthesis instead of ethylene formation, while excess S resulted in increased ethylene, stimulating more efficient utilization of intracellular CO2 for photosynthesis (Nazar et al., 2014).
Several clusters of genes upregulated by iron (Fe) deficiency in Arabidopsis were reported; one of them contains genes with a function predominantly linked to S assimilation and genes induced by S deficiency (Ivanov et al., 2012). However, the regulatory links between Fe and S metabolism are still unclear. There are contradicting reports on the influence of Fe nutrition on the expression of genes encoding sulfate transporters. On one hand, genes encoding two high affinity sulfate transporters were induced during Fe starvation in tomatos (Paolacci et al., 2014), while, on the other hand, Fe starvation reduced the expression of SULTR1;1, encoding the high affinity sulfate transporter in the Arabidopsis roots (Forieri et al., 2013). Moreover, S deprivation limited Fe-deficiency responses in tomatos (Zuchi et al., 2009), however additional S nutrition ameliorated the damages in photosynthetic apparatus caused by Fe deficiency in oilseed rape (Muneer et al., 2014). The existence of co-regulation of S and Fe metabolism was recently discussed in terms of a possible role of several metabolic processes, including the involvement of [Fe–S] clusters in creating the important feedback signal leading to adjustment of the metabolism, for example, Fe and S uptake (Forieri et al., 2013). The role of ethylene in such co-regulation is unclear.
Transcriptomic analysis of grape berries treated with SO2 revealed the reprogramming of transcriptome after treatment. Transcripts involved in auxin, ethylene and jasmonate signaling were strongly upregulated, including transcripts encoding auxin responsive proteins, ACC synthase, ACC oxidase, ethylene responsive proteins and lipoxygenase (Giraud et al., 2012). In addition to the S supply, S limitation also results in induction of the ethylene pathway. For example, a short-term S limitation (2 days) resulted in increased expression of some ethylene-related genes (Lewandowska et al., 2010) and elevated ethylene level (Moniuszko et al., 2013) in tobacco and a long-term S limitation (35 days) resulted in an increased amount of ACS in oilseed rape plants (D'hooghe et al., 2013). Interestingly, no increase of ethylene synthesis was observed when tomato plants were starved for S and Fe simultaneously (Zuchi et al., 2009).
Vice Versa: Ethylene and Ethylene Signaling Affects Sulfur Metabolism
Accumulation of APR activity as a result of the treatment of Arabidopsis with 0.2 mM ACC has been shown (Koprivova et al., 2008). Additionally, ethylene has been shown to increase ATP-sulfurylase activity and S accumulation in mustard (Iqbal et al., 2012). However, these few reports cannot be extrapolated into a universal hypothesis that ethylene stimulates S metabolism and accumulation. In fact, despite the above-mentioned increased production of ethylene during a response to S deficiency in Nicotiana tabacum (Moniuszko et al., 2013) and Solanum lycopersicum (Zuchi et al., 2009), the transcription of only a fraction of ethylene responsive genes was affected. Similar results could be extracted from microarray studies on Arabidopsis (Hirai et al., 2003; Nikiforova et al., 2003).
Consistent lack of correlation of the transcriptomics data with ethylene measurements suggests an association of S deficit with ethylene signaling machinery rather than with ethylene production. Moreover, recent reports put forward the possible occurrence of the cross talk between Sulfur LIMitation 1 transcription factor (SLIM1, described in the next chapter) and ethylene receptors. The re-analysis of the Arabidopsis microarray data showed that silver nitrate mimics the signal for perception of sulfur deficiency in plants at the transcriptome level (Moniuszko, 2015). The author identified 20 genes that were similarly regulated under S deficit and AgNO3 treatment. Noteworthy, all 20 are considered S deficiency markers, and three of them (LSU1, LSU2, and SULTR1;2) are candidates for regulators of responses to S deficiency (Moniuszko et al., 2013; Zhang et al., 2014). Only two of them (APR2 and APR3) cannot be linked with SLIM1 during the plant's early response to S deficiency. The analysis also showed that the similarity between S deficit and AgNO3 treatment is rather linked to the silver nitrate action on ethylene receptors than to other AgNO3 effects (Moniuszko, 2015).
This mostly theory driven conclusion is supported by previously overlooked studies. It has been shown that Eruca sativa proteomic response to Ag+ ions is related to S metabolism (Vannini et al., 2013). The observed changes in S metabolites of E. sativa due two Ag+ exposures strongly suggest SLIM1 involvement. In addition, the heterologous expression of the Arabidopsis ethylene receptor gene, etr1-1 (which encodes mutated ETR1 protein unable to relay ethylene signal after hormone binding), in N. attenuata resulted in impaired sulfate uptake and S metabolism (Meldau et al., 2013). Abnormal phenotypes of such seedlings under optimal sulfate supply (similar to plants grown under S deficit) suggest a defect in SLIM1 action as a result of changes in ethylene signaling at the receptor level. Apparently, the etr1-1 receptor, despite (and in addition to) its inability to properly function in a classic linear ethylene-signaling pathway, was mimicking the signal of S deficiency.
On the other hand, proper ethylene signaling was found to be necessary for increased GSH accumulation after ozone treatment. In the ein2 Arabidopsis mutant plants, 6 h after ozone exposure, the increment of GSH level was much lower than in the control plants (Yoshida et al., 2009). Research involving the extrapolation of such regulation on different stresses falls way behind. Presently, the cross talk between GSH biosynthesis and ethylene signaling has been proposed only for Cd and drought (Masood et al., 2012; Saithong et al., 2015). Both cases have been discussed above regarding the S nutrition effect on ethylene production. However, we want to emphasize here that in the case of Cd treated mustard, the effects of additional S supply were reversed by the ethylene biosynthesis inhibitor, aminoethoxyvinylglycine (AVG), and that similar effects were achieved by additional S supply and ethephon treatment (Masood et al., 2012). Thus, the authors suggested a prominent role of ethylene (possibly on GSH biosynthesis) in S-induced alleviation of Cd stress. However, this might be the reflection of a switch between the ethylene receptors' role in S status sensing and linear ethylene signaling, as discussed in a recently proposed model (Moniuszko, 2015). Nevertheless, further studies are needed to clarify the exact molecular mechanism behind the observed effects of ethylene and ethylene signaling on sulfur metabolism and its regulation.
Possible Regulatory Mechanisms Responsible for Coupling Sulfur and Ethylene Signaling and Metabolism
The transcriptional control of gene expression very often serves to reprogram plant metabolism in order to cope with environmental challenges. So far the only described transcription factor exclusively assigned to affect gene expression during S deficiency is SLIM1 from Arabidopsis (Maruyama-Nakashita et al., 2006). Certainly, attracting attention in the perspective of this review is the fact that SLIM1 belongs to the same plant protein family as EIN3, the main transcription factor controlling the expression of ethylene-responsive genes. It was initially identified as ETHYLENE-INSENSITIVE-LIKE 3 (EIL3) coding for a putative transcription factor of unknown function (Guo and Ecker, 2004). Analyses of the knockout mutants revealed that SLIM1 affects the expression of various genes facilitating the increased flux through the sulfate assimilation pathway and translocation of sulfate to the shoot, but it also controls the degradation of glucosinolates under sulfur deficient conditions (Maruyama-Nakashita et al., 2006). The functional complementation of the slim1 mutant was only successful with SLIM1 and not any other protein member of EIL family, pointing out its specificity. Moreover, the treatment of plants with the precursor of ethylene, ACC, does not affect the transcription of any of SLIM1-dependent genes (Maruyama-Nakashita et al., 2006). It is tempting to speculate that the C-terminal part of the EIL proteins is responsible for that functional separation since all of them are highly homologous to one another, mainly in their N-terminal half of around 300 amino acid residues. All six members of the Arabidopsis EIL family share highly acidic N-terminal amino acids, five small clusters of basic amino acids scattered mostly in the first half of the protein and a proline-rich domain (Chao et al., 1997). SLIM1 served as a template to model the unique DNA-binding domain of the EIL family, consisting of five alpha helices, packed together into a globular shape as a whole (Yamasaki et al., 2005). The DNA-binding abilities of EIN3, EIL1, and EIL2 proteins have been demonstrated with ethylene response DNA elements, which are 28-nt imperfect palindromes, using an electro-mobility shift assay (Solano et al., 1998). The interaction of SLIM1 with those sequences is very unstable and is only detectable with surface plasmon resonance (Yamasaki et al., 2005), demonstrating the binding preferences between EIL family members. SLIM1 strongly binds to 20-nt consensus, called the UPE-box, which is only present in the promoters of eight genes that are strongly induced by S deficiency in Arabidospis (Wawrzynska et al., 2010). Yet three of these genes encode proteins from the LSU family, homologs of tobacco UP9C protein (Sirko et al., 2014). Silencing of UP9C expression in tobacco led to disturbances of the ethylene signaling and synthesis pathways during conditions of S deficiency (Moniuszko et al., 2013).
In contrast to EIN3, not much is known about SLIM1 posttranslational modifications or its interaction with other proteins (Wawrzynska and Sirko, 2014). Its transcription level is not modulated by the changes of S conditions (Maruyama-Nakashita et al., 2006); however a strong elevation is observed in root tissue during Fe deficiency (Garcia et al., 2010). SLIM1 can bind with MYB72, which together with MYB10 induce the nicotianamine synthase gene NAS4 governing proper homeostasis of Fe during its deficiency. However, this also triggers jasmonate/ethylene-dependent systemic resistance (Van Der Ent et al., 2008; Palmer et al., 2013). On the other hand, MYB72 is a direct target of FIT, a central regulator of Fe assimilation in roots (Sivitz et al., 2012). FIT abundance is controlled by interaction with EIN3, which reduces FIT proteasomal degradation leading to a higher level of expression of the genes involved in Fe acquisition (Lingam et al., 2011). Both SLIM1 and EIN3, therefore, seem to tune up Fe homeostasis when plants meet the conditions of deficiency.
Despite the possible cross talk between ethylene and S deficiency signals on the level of EIN3 and SLIM1 transcriptional factors, the regulation on the level of stability of enzymes involved in ethylene synthesis might be also envisaged. Such possibilities might be deduced from the reported interaction of the above-mentioned UP9C protein with ACO in tobacco (Moniuszko et al., 2013). Interestingly, many members of the LSU family are induced during S starvation and it is tempting to speculate that the interaction of these proteins with ACO serves some regulatory reason because of the lack of S-deficiency induced elevation of ethylene level in tobacco plants with lowered expression of UP9C (Moniuszko et al., 2013). Notably, the posttranslational regulation of ACS is a well-known phenomenon; however information about such regulation of ACO is thus far limited. Nevertheless, this possibility is supported by the transcriptomic-based kinetic model for ethylene synthesis in tomato fruits that indicates the existence of potential posttrancriptional regulation of ACO (Van De Poel et al., 2014).
Moreover, the small (five amino acids) peptide, phytosulfokine (PSK), a growth factor containing sulfated tyrosine might be an additional player in this complex signaling and regulatory network. PSK is produced from an 80-amino-acid-long precursor (there exist six PSK genes in Arabidopsis) via tyrosine sulfation and proteolytic processing (Matsubayashi, 2014; Sauter, 2015). Recent analysis of the Arabidopsis tpst-2 mutant defective in tyrosylprotein sulfotransferase revealed that PSK suppresses ethylene production (Wu et al., 2015).
The hypothetical model explaining possible co-regulation of sulfur and ethylene signaling in plants is shown in Figure 2. According to the model, S availability modulates ethylene sensitivity due to a switch of ethylene receptor function. S deficiency might also affect ethylene production by stabilizing ACO level or activity. On the other hand, ethylene production is negatively affected by the sulfated phytohormone, PSK. The functional ethylene pathway is necessary for increased level of GSH in some stresses. Moreover, ethylene seems to stimulate the activity of several enzymes involved in S assimilation.
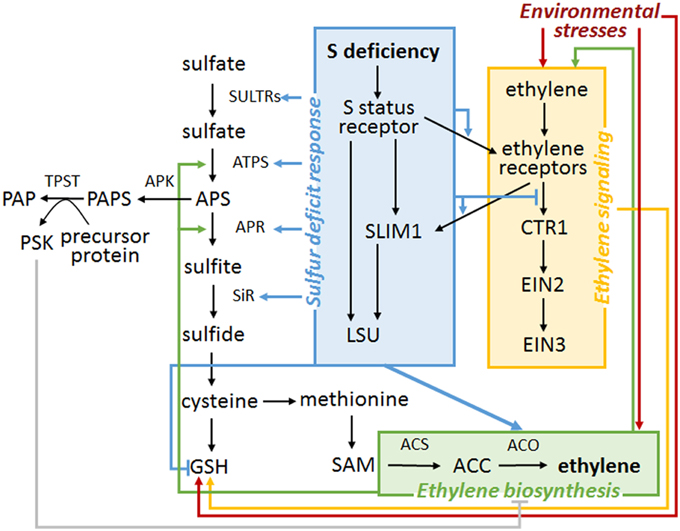
Figure 2. A hypothetical model of regulatory links between S− and ethylene sensing and signaling. Only the selected metabolites, enzymes and other players are presented. The black arrow represents one-step or multiple-step signaling or metabolic pathway progress. Colored arrows (gray, red, blue, green, orange) represent regulatory mechanisms reported in the published studies. At the current stage, most of these mechanisms are obscurely documented and need further research. Additionally, the S status sensor is elusive.
Conclusions
Ethylene production and sulfur assimilation pathways have close boundaries and share some metabolites. Thus, they might have also common regulatory elements. Although numerous observations suggest that these two pathways might indeed share some sensing or signaling elements, the molecular details are still obscure. Additional experiments are required to clarify and explain some contradicting and imprecise data. Answers to the following questions might help to elucidate the molecular basis of the postulated cross-talk of both signaling pathways: What is the S deficiency signal? What molecules function as the S status receptors? What factors are directly involved in linking these two pathways?
Author Contributions
AS drafted the manuscript. All authors were involved in the writing process and preparing the final version.
Funding
This work was supported by Narodowe Centrum Nauki (National Science Center), Grants 2014/15/B/NZ3/04854 and 2014/15/B/NZ1/01887.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Amir, R. (2010). Current understanding of the factors regulating methionine content in vegetative tissues of higher plants. Amino Acids 39, 917–931. doi: 10.1007/s00726-010-0482-x
Asgher, M., Khan, N. A., Khan, M. I., Fatma, M., and Masood, A. (2014). Ethylene production is associated with alleviation of cadmium-induced oxidative stress by sulfur in mustard types differing in ethylene sensitivity. Ecotoxicol. Environ. Saf. 106, 54–61. doi: 10.1016/j.ecoenv.2014.04.017
Bürstenbinder, K., Rzewuski, G., Wirtz, M., Hell, R., and Sauter, M. (2007). The role of methionine recycling for ethylene synthesis in Arabidopsis. Plant J. 49, 238–249. doi: 10.1111/j.1365-313X.2006.02942.x
Chan, K. X., Wirtz, M., Phua, S. Y., Estavillo, G. M., and Pogson, B. J. (2013). Balancing metabolites in drought: the sulfur assimilation conundrum. Trends Plant Sci. 18, 18–29. doi: 10.1016/j.tplants.2012.07.005
Chao, Q., Rothenberg, M., Solano, R., Roman, G., Terzaghi, W., and Ecker, J. R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144. doi: 10.1016/S0092-8674(00)80300-1
D'hooghe, P., Escamez, S., Trouverie, J., and Avice, J. C. (2013). Sulphur limitation provokes physiological and leaf proteome changes in oilseed rape that lead to perturbation of sulphur, carbon and oxidative metabolisms. BMC Plant Biol. 13:23. doi: 10.1186/1471-2229-13-23
Fontecave, M., Atta, M., and Mulliez, E. (2004). S-adenosylmethionine: nothing goes to waste. Trends Biochem. Sci. 29, 243–249. doi: 10.1016/j.tibs.2004.03.007
Forieri, I., Wirtz, M., and Hell, R. (2013). Toward new perspectives on the interaction of iron and sulfur metabolism in plants. Front. Plant Sci. 4:357. doi: 10.3389/fpls.2013.00357
García, M. J., Lucena, C., Romera, F. J., Alcántara, E., and Pérez-Vicente, R. (2010). Ethylene and nitric oxide involvement in the up-regulation of key genes related to iron acquisition and homeostasis in Arabidopsis. J. Exp. Bot. 61, 3885–3899. doi: 10.1093/jxb/erq203
Giraud, E., Ivanova, A., Gordon, C. S., Whelan, J., and Considine, M. J. (2012). Sulphur dioxide evokes a large scale reprogramming of the grape berry transcriptome associated with oxidative signalling and biotic defence responses. Plant Cell Environ. 35, 405–417. doi: 10.1111/j.1365-3040.2011.02379.x
Guo, H., and Ecker, J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7, 40–49. doi: 10.1016/j.pbi.2003.11.011
Hirai, M. Y., Fujiwara, T., Awazuhara, M., Kimura, T., Noji, M., and Saito, K. (2003). Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 33, 651–663. doi: 10.1046/j.1365-313X.2003.01658.x
Iqbal, N., Khan, N. A., Nazar, R., and Silva, J. A. T.D. (2012). Ethylene-stimulated photosynthesis results from increased nitrogen and sulfur assimilation in mustard types that differ in photosynthetic capacity. Environ. Exp. Bot. 78, 84–90. doi: 10.1016/j.envexpbot.2011.12.025
Iqbal, N., Masood, A., Khan, M. I., Asgher, M., Fatma, M., and Khan, N. A. (2013). Cross-talk between sulfur assimilation and ethylene signaling in plants. Plant Signal. Behav. 8:e22478. doi: 10.4161/psb.22478
Ivanov, R., Brumbarova, T., and Bauer, P. (2012). Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 5, 27–42. doi: 10.1093/mp/ssr065
Katz, Y. S., Galili, G., and Amir, R. (2006). Regulatory role of cystathionine-gamma-synthase and de novo synthesis of methionine in ethylene production during tomato fruit ripening. Plant Mol. Biol. 61, 255–268. doi: 10.1007/s11103-006-0009-8
Khan, M. I. R., Nazir, F., Asgher, M., Per, T. S., and Khan, N. A. (2015). Selenium and sulfur influence ethylene formation and alleviate cadmium-induced oxidative stress by improving proline and glutathione production in wheat. J. Plant Physiol. 173, 9–18. doi: 10.1016/j.jplph.2014.09.011
Koprivova, A., North, K. A., and Kopriva, S. (2008). Complex signaling network in regulation of adenosine 5′-phosphosulfate reductase by salt stress in Arabidopsis roots. Plant Physiol. 146, 1408–1420. doi: 10.1104/pp.107.113175
Larrainzar, E., Molenaar, J. A., Wienkoop, S., Gil-Quintana, E., Alibert, B., Limami, A. M., et al. (2014). Drought stress provokes the down-regulation of methionine and ethylene biosynthesis pathways in Medicago truncatula roots and nodules. Plant Cell Environ. 37, 2051–2063. doi: 10.1111/pce.12285
Lewandowska, M., Wawrzynska, A., Moniuszko, G., Lukomska, J., Zientara, K., Piecho, M., et al. (2010). A contribution to identification of novel regulators of plant response to sulfur deficiency: characteristics of a tobacco gene UP9C, its protein product and the effects of UP9C silencing. Mol. Plant 3, 347–360. doi: 10.1093/mp/ssq007
Lingam, S., Mohrbacher, J., Brumbarova, T., Potuschak, T., Fink-Straube, C., Blondet, E., et al. (2011). Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23, 1815–1829. doi: 10.1105/tpc.111.084715
Mao, D., Yu, F., Li, J., VAN DE Poel, B., Tan, D., Li, J., et al. (2015). FERONIA receptor kinase interacts with S-adenosylmethionine synthetase and suppresses S-adenosylmethionine production and ethylene biosynthesis in Arabidopsis. Plant Cell Environ. doi: 10.1111/pce.12570. [Epub ahead of print].
Maruyama-Nakashita, A., Nakamura, Y., Tohge, T., Saito, K., and Takahashi, H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18, 3235–3251. doi: 10.1105/tpc.106.046458
Masood, A., Iqbal, N., and Khan, N. A. (2012). Role of ethylene in alleviation of cadmium-induced photosynthetic capacity inhibition by sulphur in mustard. Plant Cell Environ. 35, 524–533. doi: 10.1111/j.1365-3040.2011.02432.x
Matsubayashi, Y. (2014). Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 65, 385–413. doi: 10.1146/annurev-arplant-050312-120122
Meldau, D. G., Meldau, S., Hoang, L. H., Underberg, S., Wünsche, H., and Baldwin, I. T. (2013). Dimethyl disulfide produced by the naturally associated bacterium bacillus sp B55 promotes Nicotiana attenuata growth by enhancing sulfur nutrition. Plant Cell 25, 2731–2747. doi: 10.1105/tpc.113.114744
Moniuszko, G. (2015). Ethylene signaling pathway is not linear, however its' lateral part is responsible for sensing and signaling of sulfur status in plants. Plant Signal. Behav. doi: 10.1080/15592324.2015.1067742. [Epub ahead of print].
Moniuszko, G., Skoneczny, M., Zientara-Rytter, K., Wawrzynska, A., Glów, D., Cristescu, S. M., et al. (2013). Tobacco LSU-like protein couples sulphur-deficiency response with ethylene signalling pathway. J. Exp. Bot. 64, 5173–5182. doi: 10.1093/jxb/ert309
Muneer, S., Lee, B. R., Kim, K. Y., Park, S. H., Zhang, Q., and Kim, T. H. (2014). Involvement of sulphur nutrition in modulating iron deficiency responses in photosynthetic organelles of oilseed rape (Brassica napus L.). Photosyn. Res. 119, 319–329. doi: 10.1007/s11120-013-9953-8
Nazar, R., Khan, M. I., Iqbal, N., Masood, A., and Khan, N. A. (2014). Involvement of ethylene in reversal of salt-inhibited photosynthesis by sulfur in mustard. Physiol. Plant. 152, 331–344. doi: 10.1111/ppl.12173
Nikiforova, V., Freitag, J., Kempa, S., Adamik, M., Hesse, H., and Hoefgen, R. (2003). Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J. 33, 633–650. doi: 10.1046/j.1365-313X.2003.01657.x
Palmer, C. M., Hindt, M. N., Schmidt, H., Clemens, S., and Guerinot, M. L. (2013). MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 9:e1003953. doi: 10.1371/journal.pgen.1003953
Paolacci, A. R., Celletti, S., Catarcione, G., Hawkesford, M. J., Astolfi, S., and Ciaffi, M. (2014). Iron deprivation results in a rapid but not sustained increase of the expression of genes involved in iron metabolism and sulfate uptake in tomato (Solanum lycopersicum L.) seedlings. J. Integr. Plant Biol. 56, 88–100. doi: 10.1111/jipb.12110
Roje, S. (2006). S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry 67, 1686–1698. doi: 10.1016/j.phytochem.2006.04.019
Saithong, T., Saerue, S., Kalapanulak, S., Sojikul, P., Narangajavana, J., and Bhumiratana, S. (2015). Gene co-expression analysis inferring the crosstalk of ethylene and gibberellin in modulating the transcriptional acclimation of cassava root growth in different seasons. PLoS ONE 10:e0137602. doi: 10.1371/journal.pone.0137602
Sauter, M. (2015). Phytosulfokine peptide signalling. J. Exp. Bot. 66, 5161–5169. doi: 10.1093/jxb/erv071
Sauter, M., Moffatt, B., Saechao, M. C., Hell, R., and Wirtz, M. (2013). Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 451, 145–154. doi: 10.1042/BJ20121744
Sirko, A., Wawrzynska, A., Rodríguez, M. C., and Sektas, P. (2014). The family of LSU-like proteins. Front. Plant Sci. 5:774. doi: 10.3389/fpls.2014.00774
Sivitz, A. B., Hermand, V., Curie, C., and Vert, G. (2012). Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 7:e44843. doi: 10.1371/journal.pone.0044843
Solano, R., Stepanova, A., Chao, Q., and Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Van De Poel, B., Bulens, I., Hertog, M. L., Nicolai, B. M., and Geeraerd, A. H. (2014). A transcriptomics-based kinetic model for ethylene biosynthesis in tomato (Solanum lycopersicum) fruit: development, validation and exploration of novel regulatory mechanisms. New Phytol. 202, 952–963. doi: 10.1111/nph.12685
Van De Poel, B., and Van Der Straeten, D. (2014). 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: more than just the precursor of ethylene! Front. Plant Sci. 5:640. doi: 10.3389/fpls.2014.00640
Van Der Ent, S., Verhagen, B. W., Van Doorn, R., Bakker, D., Verlaan, M. G., Pel, M. J., et al. (2008). MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 146, 1293–1304. doi: 10.1104/pp.107.113829
Vannini, C., Domingo, G., Onelli, E., Prinsi, B., Marsoni, M., Espen, L., et al. (2013). Morphological and proteomic responses of Eruca sativa exposed to silver nanoparticles or silver nitrate. PLoS ONE 8:e68752. doi: 10.1371/journal.pone.0068752
Wawrzynska, A., Lewandowska, M., and Sirko, A. (2010). Nicotiana tabacum EIL2 directly regulates expression of at least one tobacco gene induced by sulphur starvation. J. Exp. Bot. 61, 889–900. doi: 10.1093/jxb/erp356
Wawrzynska, A., and Sirko, A. (2014). To control and to be controlled: understanding the Arabidopsis SLIM1 function in sulfur deficiency through comprehensive investigation of the EIL protein family. Front. Plant Sci. 5:575. doi: 10.3389/fpls.2014.00575
Wu, T., Kamiya, T., Yumoto, H., Sotta, N., Katsushi, Y., Shigenobu, S., et al. (2015). An Arabidopsis thaliana copper-sensitive mutant suggests a role of phytosulfokine in ethylene production. J. Exp. Bot. 66, 3657–3667. doi: 10.1093/jxb/erv105
Yamasaki, K., Kigawa, T., Inoue, M., Yamasaki, T., Yabuki, T., Aoki, M., et al. (2005). Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. J. Mol. Biol. 348, 253–264. doi: 10.1016/j.jmb.2005.02.065
Yoshida, S., Tamaoki, M., Ioki, M., Ogawa, D., Sato, Y., Aono, M., et al. (2009). Ethylene and salicylic acid control glutathione biosynthesis in ozone-exposed Arabidopsis thaliana. Physiol. Plant. 136, 284–298. doi: 10.1111/j.1399-3054.2009.01220.x
Zhang, B., Pasini, R., Dan, H., Joshi, N., Zhao, Y., Leustek, T., et al. (2014). Aberrant gene expression in the Arabidopsis SULTR1;2 mutants suggests a possible regulatory role for this sulfate transporter in response to sulfur nutrient status. Plant J. 77, 185–197. doi: 10.1111/tpj.12376
Keywords: abiotic stress, ethylene, sulfur nutrition, LSU, SLIM1, signaling
Citation: Wawrzynska A, Moniuszko G and Sirko A (2015) Links Between Ethylene and Sulfur Nutrition—A Regulatory Interplay or Just Metabolite Association? Front. Plant Sci. 6:1053. doi: 10.3389/fpls.2015.01053
Received: 29 September 2015; Accepted: 12 November 2015;
Published: 01 December 2015.
Edited by:
Francisco Javier Romera, University of Cordoba, SpainReviewed by:
Stanislav Kopriva, University of Cologne, GermanyElizabeth Pilon-Smits, Colorado State University, USA
Copyright © 2015 Wawrzynska, Moniuszko and Sirko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agnieszka Sirko, asirko@ibb.waw.pl
 Anna Wawrzynska
Anna Wawrzynska Grzegorz Moniuszko
Grzegorz Moniuszko Agnieszka Sirko
Agnieszka Sirko