- 1Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD, Australia
- 2Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, VIC, Australia
The backbone cyclic and disulfide bridged sunflower trypsin inhibitor-1 (SFTI-1) peptide is a proven effective scaffold for a range of peptide therapeutics. For production at laboratory scale, solid phase peptide synthesis techniques are widely used, but these synthetic approaches are costly and environmentally taxing at large scale. Here, we developed a plant-based approach for the recombinant production of SFTI-1-based peptide drugs. We show that transient expression in Nicotiana benthamiana allows for rapid peptide production, provided that asparaginyl endopeptidase enzymes with peptide-ligase functionality are co-expressed with the substrate peptide gene. Without co-expression, no target cyclic peptides are detected, reflecting rapid in planta degradation of non-cyclized substrate. We test this recombinant production system by expressing a SFTI-1-based therapeutic candidate that displays potent and selective inhibition of human plasmin. By using an innovative multi-unit peptide expression cassette, we show that in planta yields reach ~60 μg/g dry weight at 6 days post leaf infiltration. Using nuclear magnetic resonance structural analysis and functional in vitro assays, we demonstrate the equivalence of plant and synthetically derived plasmin inhibitor peptide. The methods and insights gained in this study provide opportunities for the large scale, cost effective production of SFTI-1-based therapeutics.
Introduction
Beyond its biological role as a plant defense peptide, the 14 amino acid sunflower trypsin inhibitor-1 (SFTI-1) has attracted significant interest in the drug development field (Craik et al., 2006; Lesner et al., 2011). This interest largely stems from the cyclic backbone of SFTI-1, which together with a bridging disulfide bond imparts exceptional stability and conformational rigidity to the peptide (Korsinczky et al., 2001). Furthermore, SFTI-1 is readily tolerant of residue substitutions, exemplified by a wide range of combinatorial and rational design variants that have been applied to convert SFTI-1 into potent and stable inhibitors of therapeutically relevant proteases, including matriptases (Quimbar et al., 2013; Fittler et al., 2014; Gitlin et al., 2015), kallikreins (Shariff et al., 2014; Chen et al., 2016; de Veer et al., 2016; Jendrny and Beck-Sickinger, 2016), chymotrypsin (Swedberg et al., 2017), and furin (Fittler et al., 2015). SFTI-1 has also proven to be a useful scaffold for presenting and stabilizing small bioactive epitopes that by themselves would be unstable and not effective as pharmaceuticals (Chan et al., 2011; Qiu et al., 2017; Durek et al., 2018). These engineered cyclic SFTI-1 analogues uniformly display significantly enhanced serum stability compared with their linear counterparts, overcoming a major limitation of peptide-based therapeutics (Wang and Craik, 2018).
Peptide drugs are primarily produced via solid phase peptide synthesis techniques, which in large scale have considerable economic and environmental costs (Andersson et al., 2000). For some peptides, recombinant production is a feasible alternative, with prokaryotes and lower eukaryotic hosts most commonly used (Demain and Vaishnav, 2009). In the case of SFTI-1, backbone cyclization is required for maximum potency (Colgrave et al., 2010), so recombinant production strategies that incorporate this post-translational modification are required. Recently, an intein-mediated protein splicing approach to SFTI-1 cyclization in E. coli was reported, with SFTI-1 yields estimated at 180 μg/L of bacterial culture (Li et al., 2016). Although promising, intein splicing efficiency is highly sensitive to the residues at the extein-intein junction, potentially reducing its broad applicability (Aboye and Camarero, 2012). As an alternative, and considering that SFTI-1 is naturally produced and cyclized in sunflower, a plant-based production system is appealing. However, small peptides have typically proven difficult to produce in planta, presumably due to the unintended effects of proteolysis either in planta or during extraction phases (Benchabane et al., 2008; Habibi et al., 2017). Strategies to overcome this limitation have included expressing peptides with stabilizing fusion partners (Yasuda et al., 2005; Sainsbury et al., 2013), down-regulating interfering plant proteases (Robert et al., 2015), and the development of subcellular targeting approaches (Jackson et al., 2010; Yang et al., 2017). Despite advances using these strategies, the yields from plant-produced peptides have generally been low, typically in the low μg g−1 fresh weight (FW) range (Lico et al., 2012; Viana et al., 2012). In contrast, some endogenous cyclic plant peptides are known to accumulate to very high levels [~1.8 mg g−1 dry weight (DW)], most notably exemplified by the class of cyclic peptides termed cyclotides (Craik et al., 1999; Seydel and Dornenburg, 2006). Thus, determining the in planta biosynthetic pathways that govern cyclotide synthesis and accumulation in plants will be of great benefit if translatable to the recombinant production of “designer” therapeutic peptides.
SFTI-1 is produced in sunflower seeds, where it is post-translationally processed from the precursor protein PawS1 (preproalbumin with sunflower trypsin inhibitor-1) (Mylne et al., 2011) (Figure 1A). Because sunflower transformation is inefficient, most PawS1 processing studies have been done in the model plant Arabidopsis (Mylne et al., 2011), in situ using sunflower seed extracts (Bernath-Levin et al., 2015), or in vitro with recombinant processing enzymes and synthetic or recombinant substrates (Bernath-Levin et al., 2015; Franke et al., 2017; Haywood et al., 2018). Together these studies have unequivocally demonstrated the involvement of vacuolar cysteine proteases termed asparaginyl endopeptidases (AEPs) for both the cleavage and subsequent cyclization of SFTI-1. Similarly, cyclotides are known to be backbone cyclized by AEPs (Bernath-Levin et al., 2015; Harris et al., 2015; Poon et al., 2017), where a detailed understanding of mechanisms and structural requirements has emerged (Jackson et al., 2018; James et al., 2018). These ligase competent AEPs not only represent useful biotechnological tools for in vitro peptide and protein engineering applications (Harris et al., 2015; Nguyen et al., 2015; Hemu et al., 2016) but also open up opportunities for their deployment in plant biofactory applications for the production of cyclic peptides (Poon et al., 2017).
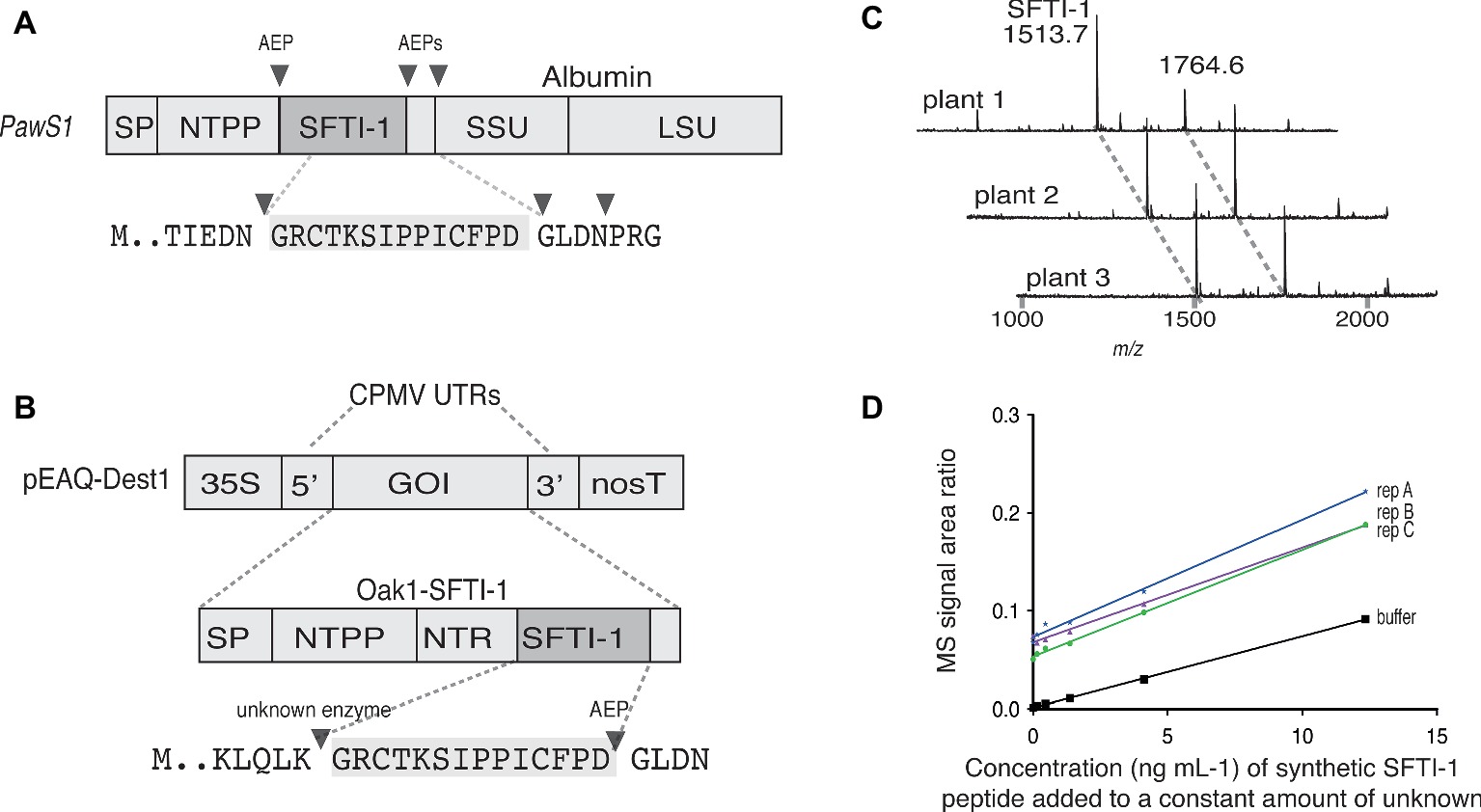
Figure 1. Transient expression analysis of SFTI-1 production in N. benthamiana. (A) Native SFTI-1 is produced in sunflower seed via expression of the PawS1 gene. SFTI-1 processing occurs via the concerted action of asparaginyl endopeptidases (AEPs), which have strict preference for asparagine or aspartic residues (shown as filled triangles). The 14-amino acid SFTI-1 peptide sequence and immediately flanking residues are displayed with the SFTI-1 sequence highlighted in grey. (B) For transient expression in N. benthamiana leaves, the pEAQ-Dest1 vector (Sainsbury et al., 2009) was used which provides high level transgene expression due the presence of the 35 s promoter and cowpea mosaic virus (CPMV) 5′ and 3′ UTRs. To produce SFTI-1, the Oak1 gene was engineered to include the SFTI-1 peptide encoding sequence, replacing that of kB1. Cleavage after an amino-terminal repeat (NTR) by an as yet unidentified protease is thought to occur first to liberate the N-terminal glycine required for AEP mediated backbone cyclization to the C-terminal aspartic residue. (C) MALDI-TOF MS analysis of peptides produced in N. benthamiana leaves upon co-expression of pEAQ-OaAEP1b with pEAQ-Oak1-SFTI-1. The mass for cyclic SFTI-1 (m/z 1513.7) was readily detected. An unrelated and endogenous peptide (m/z 1764.7) was also readily detected. (D) SFTI-1 peptides were quantified using the method of standard addition where a standard curve was built into each crude plant extract.
In this study, we evaluated a series of gene expression parameters for optimizing in planta SFTI-1 production using N. benthamiana as a biofactory host. We demonstrate that resultant yields are influenced by the choice of AEP ligase, the AEP recognition site used, and by using multi-unit peptide expression cassettes. We demonstrate the scalability and usefulness of this transient plant-based production system by producing and purifying a recently developed potent plasmin inhibitor therapeutic based on SFTI-1 (Swedberg et al., 2019).
Materials and Methods
Vector Construction
DNA encoding Oak1-SFTI-1_GLDN, Oak1-[D14N]SFTI-1_GLDN, Oak1_GLDN, Oak1-[N29D]_GLDN, Oak1-[T4Y,I7R]SFTI-1, and Oak1-[T4Y,I7R]SFTI-1_3R were synthesized by Integrated DNA Technologies (Singapore) as gene block fragments in preparation for in house cloning (Supplementary Figure S7). All peptide precursor genes and AEP genes were recombined into the plant expression vector pEAQ-DEST1 (Sainsbury et al., 2009) using Gateway™ LR Clonase™ technology (Invitrogen, Carlsbad CA). Sequence verified vectors were then transferred to Agrobacterium tumefaciens LBA4404 by electroporation.
Transient Expression in Nicotiana benthamiana
Nicotiana benthamiana plants were cultivated in Jiffy peat pellets in a plant growth room at 28°C under 160 μmol of LED illumination (AP67 spectra, Valoya Oy, Helsinki, Finland). Agrobacterium cultures harboring pEAQ-DEST1 expression cassettes were grown in Luria-Bertani media to stationary phase before centrifugation and resuspension in infiltration buffer (10 mM MES)(2-[N-morpholino]ethanesulfonic acid) (pH 5.6, 10 mM MgCl2, 100 uM acetosyringone). Separate cultures (OD600 of 1.0) harboring the AEP and peptide expression vectors were mixed at a ratio of 1:1 before vacuum infiltration of N. benthamiana plants at 5–6 weeks of age. For relative quantification experiments, a third Agrobacterium culture was added to the mix that contained an expression vector encoding a truncated kB6 peptide. This design which allowed only linear kB6 to be produced, irrespective of AEP expression, served as an internal control for normalizing SFTI-1 and kB1 levels.
Peptide Quantification
At 6 days post-infiltration, plant tissue was harvested for peptide extraction and analysis. To enable absolute quantification of SFTI-1 and [T4Y,I7R]SFTI-1, leaf tissue was lyophilized and ground using a Geno/Grinder® (SPEX Sample Prep) with homogenous subsamples used for MS-based quantification. Routinely, 5 mg of dry tissue was reconstituted in buffer [50% (v/v) acetonitrile, 0.1% (v/v) formic acid] with 0.05 μM codeine included as internal standard. Triplicate samples were spiked with a standard curve of concentrations of analyte before centrifugation. The analytes were quantified using targeted multiple reaction monitoring (MRM) analyses conducted on a SCIEX QTRAP 6500+ mass spectrometer interfaced with a SCIEX UPLC system. Studies were conducted using a Phenomenex Kinetex C18 UPLC column (150 mm × 2.0 mm, 1.7 μm particle size), maintained at 60°C with a linear acetonitrile gradient delivered at a flow rate of 0.4 ml min−1. Ion spray voltage was set at 5000 V, source temperature at 400°C, and MRM scans were conducted with unit resolution settings for both Q1 and Q3. MRM transition details for each analyte are provided in Supplementary Table S1. SCIEX MultiQuant (v 3.0.2) software was used to plot analyte signal intensities against concentrations added to the sample matrix and back-extrapolated to find negative X-axis intercepts, which were finally adjusted for dilution to calculate analyte concentrations in the sample.
For relative quantification, leaf disks were punched from infiltrated leaves and placed in microfuge tubes with ball bearings, before grinding to powder in liquid nitrogen using a Geno/Grinder® (SPEX Sample Prep). Peptides were extracted in 200 μl of aqueous [50% (v/v) acetonitrile, 1% (v/v) formic acid] with gentle mixing overnight. After centrifugation, the supernatant was diluted 1:5 with 1% formic acid before being desalted and concentrated using C18 ZipTips (Millipore). Samples were then mixed 1:1 with a-cyano-4-hydroxycinnamic acid [5 mg ml−1 in 50% acetonitrile, 0.1% TFA, 5 mM (NH4)H2PO4] before being spotted and dried onto a MALDI sample plate for matrix assisted laser desorption/ionization (MALDI)-time of flight (TOF) MS using an Applied Biosystems 4700 TOF-TOF Proteomics Analyzer. For relative quantification, the sum of the isotope cluster area corresponding to cyclic SFTI-1 or kB1 was normalized to the sum of the isotope cluster area of linear kB6 peptides.
Peptide Synthesis and Purification
All peptides were synthesized in house using established Fmoc solid-phase peptide synthesis methods (Cheneval et al., 2014). Peptides were isolated by RP-HPLC and characterized by high resolution MS and NMR spectroscopy.
In vitro Peptide Cyclization Assays
Recombinant OaAEP1b was prepared using the methods detailed in (Harris et al., 2015) with activated enzyme concentration estimated by BCA assay according to the manufacturer’s instructions. Linear target peptides (6.67 μM) were incubated with recombinant OaAEP1b (7.5 μg ml−1 final concentration) in activity buffer (50 mM sodium acetate, 50 mM NaCl, 1 mM EDTA, pH 5) for up to 40 h at room temperature. The reaction mixture (10 μl) was desalted using C18 ZipTips (Millipore) and mixed 1:1 with a-cyano-4-hydroxycinnamic acid [5 mg ml−1 in 50% acetonitrile, 0.1% TFA, 5 mM (NH4)H2PO4] before MALDI-TOF MS using an Applied Biosystems 4700 TOF-TOF Proteomics Analyzer.
Peptide Extraction and Purification
Harvested plant tissue at 6 days post-infiltration was lyophilized then homogenized using a Geno/Grinder® (SPEX Sample Prep) prior to solvent extraction with 50% (v/v) acetonitrile, 0.1% (v/v) formic acid. The supernatant collected was lyophilized, then redissolved with 10% (v/v) acetonitrile, 0.1% (v/v) formic acid before Solid-Phase Extraction (SPE) using a Phenomenex Strata C18-E SPE cartridge with 10 g resin capacity. The eluted fraction of 5–20% (v/v) acetonitrile, 0.1% (v/v) formic acid was then collected, lyophilized, and then reconstituted in 5% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid. The elution was then passed through a 0.45 μm filter before separation to homogeneity by HPLC on a semipreparative Phenomenex Jupiter C18 RP-HPLC column (250 mM × 10 mM, 5 μm particle size) followed by a preparative analytical Phenomenex Jupiter C18 RP-HPLC column (250 mm × 4.6 mm, 5 μm particle size). Fractions yielding homogeneous plant derived [T4Y,I7R]SFTI-1 were identified using MALDI-TOF as described above and lyophilized.
Structural Characterization of Plant Derived [T4Y,I7R]SFTI-1
High resolution MS comparison of synthetic and plant-produced peptides was conducted via UPLC-MS analysis on a SCIEX X500R mass spectrometer interfaced with a SCIEX UPLC. UPLC column details were identical to those used in the QTRAP analyses detailed earlier. A linear acetonitrile gradient was delivered over 15 min (flow rate 0.4 ml min−1) and monitored via positive ion TOF-MS acquisition (250 ms per scan, m/z 100–1,000). Retention times were determined from post-run XICs with a mass extraction width of 0.05 Da centered on monoisotopic peaks.
Prior to tandem MS, plant-derived [T4Y,I7R]SFTI-1 was redissolved in 100 mM NH4HCO3 (pH 8) for reduction, alkylation, and enzymatic digestion with bovine trypsin (Sigma T1426). Tandem MS of linear reduced and carboxyamidomethylated [T4Y,I7R]SFTI-1 fragment with sequence SRPPICFPDGR was collected on a SCIEX 5600 TripleTOF instrument interfaced with a Shimadzu UPLC. Sample was separated on a Agilent Zorbax 300SB-C18 column (100 mm × 2.1 mm, 1.8 μm particle size) and eluted with a linear acetonitrile gradient, and eluent was monitored using an information dependent acquisition experiment with a TOF-MS survey scan (mass range m/z 80–1,000 Da) triggering up to 20 MS/MS on precursor ions (mass range m/z 80–1,100 Da) with 50 ms scan times.
NMR Spectroscopy
The heterologously plant-produced and purified peptide [T4Y,I7R]SFTI-1 was dissolved in 90% H2O/10% D2O at a concentration of 80 μg ml−1. Spectra were recorded on a Bruker Avance III 600 MHz spectrometer equipped with a cryoprobe at 298 K. Phase-sensitive mode using time-proportional phase incrementation for quadrature detection in the t1 dimension was used for all two-dimensional spectra. Excitation sculpting with gradients was used to achieve water suppression. NMR experiments included TOCSY using a MLEV-17 spin lock sequence with an 80-ms mixing time, and NOESY with a 200-ms mixing time. Spectra were recorded with 4,096 data points in the F2 dimension and 512 increments in the F1 dimension. The t1 dimension was zero-filled to 1,024 real data points, and the F1 and F2 dimensions were multiplied by a sine-squared function before Fourier transformation. Chemical shifts were referenced to DSS. All spectra were processed using TopSpin (Bruker) and manually assigned with CCPNMR using the sequential assignment protocol (Wüthrich, 1986; Vranken et al., 2005).
Plasmin Inhibitory Assays
A serial dilution of plant-derived [T4Y,I7R]SFTI-1 was incubated with 1 nM native human plasmin (Sigma-Aldrich) for 30 min in assay buffer (0.1 M Tris-HCl, pH 8.0, 0.1 M NaCl, and 0.005% Triton X-100) in low binding 96-well plates (Corning). After addition of 100 μM of the colorimetric peptide substrate Acetyl-Arg-Met(sulphone)-Tyr-Arg-pNA (KM = 23.5 μM) to a final volume of 200 μl, the rate of substrate cleavage was monitored by the release of the pNA moiety at λ = 405 nm over 7 min. The inhibition constant (Ki) was determined from three independent assays by the Morrison equation and non-linear regression using GraphPad Prism 6.
Statistical Analysis
One-way ANOVA followed by Tukey’s multiple comparisons test was performed using GraphPad Prism version 7.0c for Mac OS X, GraphPad Software, La Jolla California USA, www.graphpad.com.
Accession Numbers
AEP gene sequences that have assigned GenBank accession numbers include OaAEP1b (KR259377), CtAEP1 (KF918345), PxAEP3b (MG720076), and HeAEP3 (MG720074).
Results
Nicotiana benthamiana Leaf-Based Transient Expression of Sunflower Trypsin Inhibitor-1
To express and cyclize SFTI-1 in planta, we used the pEAQ vector (Figure 1B) (Sainsbury et al., 2009) for recombinant production in N. benthamiana. Initially, to produce native SFTI-1, we tested peptide accumulation in leaves upon expression of a modified Oak1 gene [described in (Poon et al., 2017)] where the peptide sequence for SFTI-1 replaces the cyclotide peptide kalata B1 (kB1) (construct pEAQ-Oak-SFTI-1) (Figures 1A,B). In addition, we chose to replace the C-terminal GLPSLAA residues normally present in Oak1 with the residues GLDN that naturally flank SFTI-1 within the PawS1 precursor protein. As previously shown (Poon et al., 2017), cyclic SFTI-1 (m/z 1513.7) could only be detected in leaf extracts upon co-expression of the SFTI-1 precursor with the ligase-efficient AEP from O. affinis (OaAEP1b) (Figure 1C). To quantify the yield of SFTI-1 produced in N. benthamiana leaves, we used a quantitative mass spectroscopy (MS)-based approach (Bronsema et al., 2012) (Figure 1D). This approach, which requires a standard curve to be included in each replicate extraction, provides for a more accurate measurement by eliminating any effect that sample matrix might have on SFTI-1 signal intensity. Using this method, the yield of cyclic SFTI-1 extracted was determined to be 12.8 ± 3.0 μg g−1 DW (s.d., n = 3), which is substantially lower than the reported 199 μg g−1 DW yield obtained for the cyclotide kB1, using a similar expression strategy (Poon et al., 2017).
To determine if this lower in planta SFTI-1 yield correlates with a lower efficiency of OaAEP1b on SFTI-1 substrates, we set out to compare processing efficiencies between SFTI-1 and kB1 substrates in vitro (Figure 2). AEPs are known to have strict preference for either an Asn or Asp at the P1 position, thus we additionally wished to determine the effect that reciprocal Asn/Asp residue changes would have on processing efficiencies. For this, we directly compared recombinant OaAEP1b activity on the peptide substrates kB1_GLDN (Figure 2A), [N29D] kB1_GLDN (Figure 2B), SFTI-1_GLDN (Figure 2C), and [D14N]SFTI-1_GLDN (Figure 2D). Peptide cyclization assays were performed at pH 5.0 to simulate the low pH of leaf cell vacuoles, where AEP-mediated cyclization is predicted to occur (Jackson et al., 2007; Conlan et al., 2011). For all substrates, peptide cyclization was favored over hydrolysis by recombinant OaAEP1b with resulting MS signals for cyclic peptide dominating over linear peptide. For both kB1 substrates, the precursor was essentially quantitatively converted to cyclic kB1/[N29D] kB1 within 30 min (Figures 2A,B), while for the SFTI-1_GLDN and [D14N]SFTI-1_GLDN precursor peptides, unprocessed peptides were still detectable after 18 h (Figures 2C–E). Further analysis after 40 h of incubation revealed that processing was essentially complete with resulting cyclic to linear peptide MS signal ratios calculated at 96.86 ± 0.66% (s.d., n = 6) and 92.19 ± 1.89% (s.d., n = 6) for SFTI-1_GLDN and [D14N]SFTI-1_GLDN, respectively (Figure 2F). These findings indicate that SFTI-1 precursor peptides, irrespective of containing Asp or Asn at the AEP processing site, are although amenable to enzymatic cyclization, relatively poor substrates for OaAEP1b with significantly slower processing when compared to kB1 cyclotide precursors. Thus, we reasoned that the lower in planta yields observed for SFTI-1 over kB1 may be caused by OaAEP1b being outcompeted for SFTI-1 substrates by endogenous AEPs, which lack the ability to stabilize SFTI-1 by way of backbone cyclisation. Optimization or discovery of AEP ligases more conducive to SFTI-1 substrates, or downregulation of interfering endogenous AEP machinery are thus two approaches to increase the in planta yield of SFTI-1.
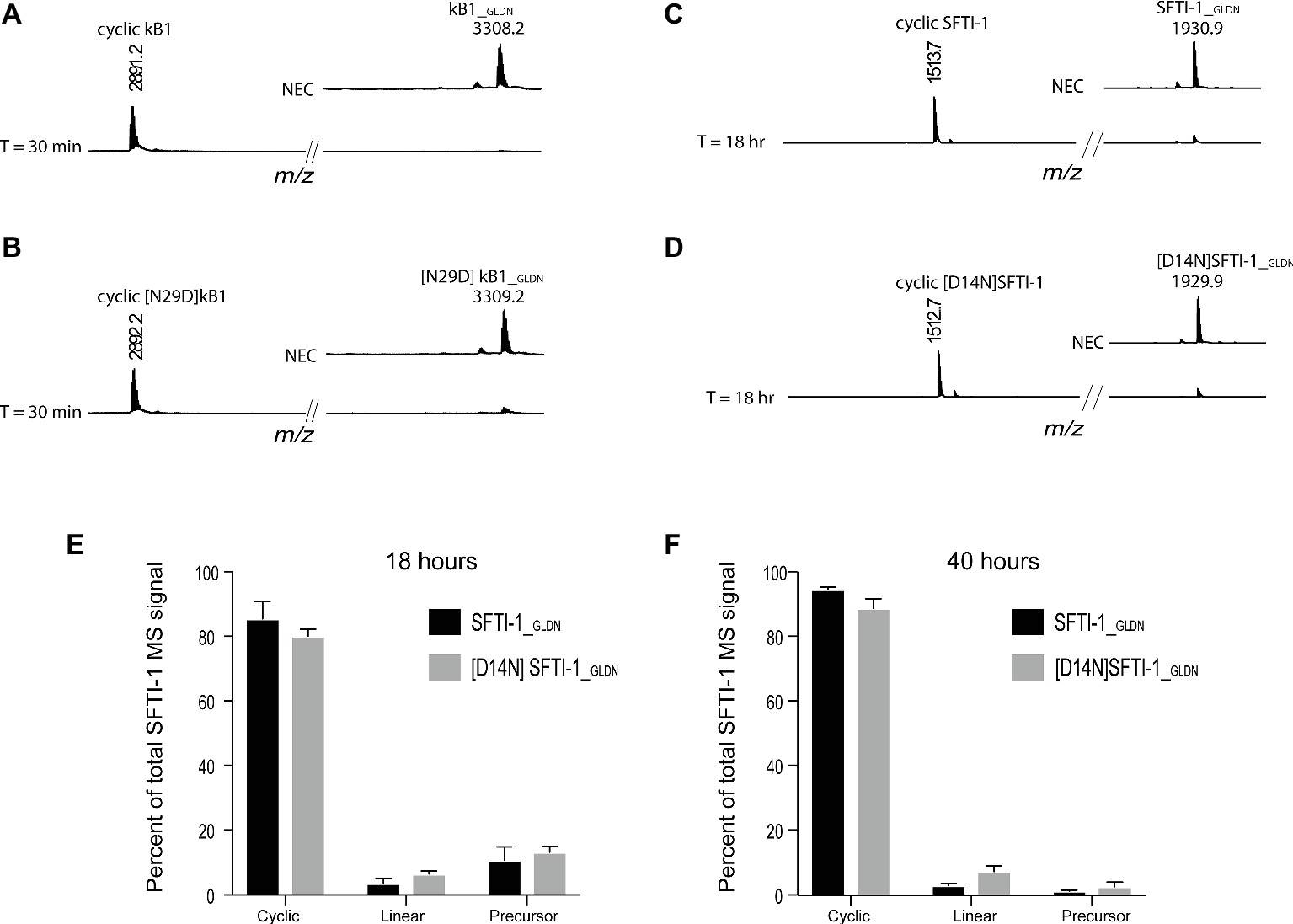
Figure 2. In vitro AEP cyclization efficiency. Representative MALDI-TOF-MS traces of the processing products of precursors (A) kB1_GLDN, (B) [N29D]kB1_GLDN, (C) SFTI-1_GLDN, and (D) [D14N]SFTI-1_GLDN. Both kB1 substrates were essentially quantitatively converted to cyclic forms within 30 min, while SFTI-1 substrates were still detectable after 18 h of reaction time. NEC, no enzyme control. Percent of total SFTI-1 and [D14N]SFTI-1 MS signal for cyclic, linear, and precursor form after 18 (E) and 40 h (F) of incubation (n = 6). Peptide precursor (6.67 μM final) was incubated at pH 5.0 with recombinant OaAEP1b enzyme at a final concentration 7.5 μg ml−1.
Assaying Diverse Asparaginyl Endopeptidases for in planta Cyclization of Sunflower Trypsin Inhibitor-1
In addition to OaAEP1b from O. affinis (Harris et al., 2015), several other AEP ligases have recently been characterized, including HeAEP3 (Hybanthus enneaspermus) (Jackson et al., 2018), PxAEP3b (petunia) (Jackson et al., 2018), and CtAEP1 (butelase-1; Clitoria ternatea) (Nguyen et al., 2015). To determine if any of these newly discovered AEP ligases have improved in planta processing ability for SFTI-1 over OaAEP1b, we set up an in planta assay wherein the precursor gene Oak1-SFTI-1, the AEP gene in question, and a C-terminally truncated kB6 peptide precursor gene were co-expressed (Figures 3A,B). Expression of the latter construct resulted in the production of a kB6 peptide precursor without the required C-terminal residues for AEP-mediated cyclization. Thus, the accumulation level of linear kB6 could serve as an internal control to normalize for differences in infiltration efficiencies between N. benthamiana leaves. Importantly, this approach produced very similar results to that obtained using normalization with a spiked peptide on a per dry weight basis (Supplementary Figure S1). Of the AEPs tested, OaAEP1b from O. affinis proved to be the best performing peptide ligase for SFTI-1 with a ~3 fold increase in relative abundance, when compared to the next best performing ligases, HeAEP3 and PxAEP3b, which yielded similar SFTI-1 levels (Figures 3A,B). CtAEP1 (butelase-1) produced only minimal cyclic SFTI-1 in agreement with the in vitro characterization of this enzyme as inefficient for cyclisation at Asp residues (Nguyen et al., 2015).
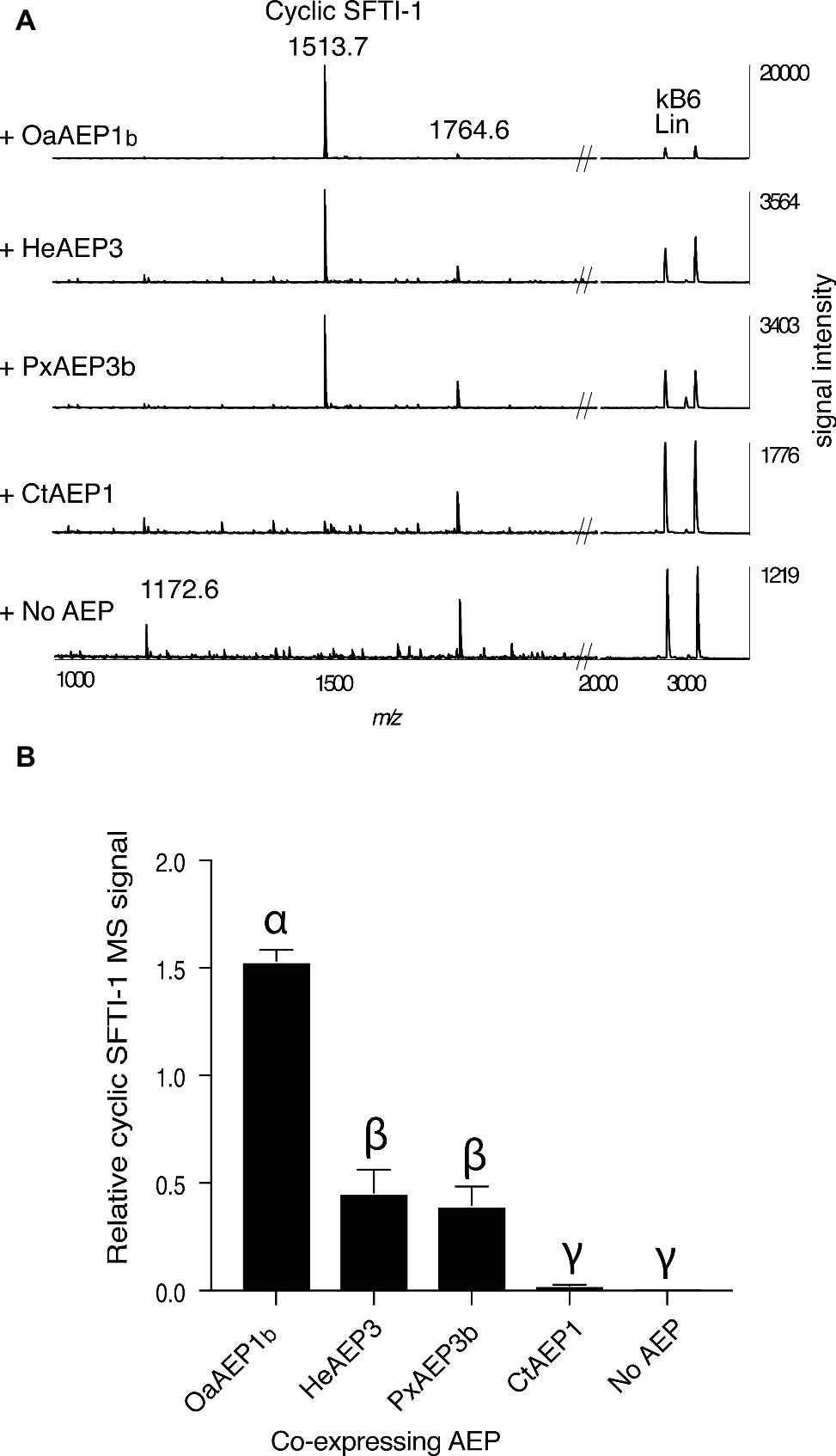
Figure 3. Comparison of AEP ligases for the in planta SFTI-1 peptide cyclization. (A) MALDI-TOF MS analysis of representative (n = 3) peptide extracts from N. benthamiana leaves which were co-infiltrated with pEAQ-Oak1-SFTI1_GLDN and pEAQ-Oak6_trun with or without pEAQ driven AEP ligase genes. Without AEP transgene expression, no cyclic or full length linear SFTI-1 related peptides were detectable. Smaller masses, however, were observed, with low signal strengths, and likely represent truncated SFTI-1 peptides (e.g., m/z 1172.6 consistent with linear oxidized GRCTKSIPPIC. By co-expressing AEP ligase genes from O. affinis (OaAEP1b), H. enneaspermus (HeAEP3) and Petunia “Mitchell” (PxAEP3b) cyclic SFTI-1 was readily detected which contrasted to expression of CtAEP1 which failed to produce any cyclic SFTI-1. For all infiltrations, the expression of pEAQ-Oak6_trun served as an internal control where MS signal intensities for linear kB6 were used to normalize SFTI-1 MS signals for relative quantification. (B) Relative MS signal intensities for cyclic SFTI-1 among co-expressed ligase capable AEPs (n = 3). Treatments carrying unique Greek lettering are significantly different (p < 0.05) as determined by Tukey’s ANOVA. Error bars are s.e.m.
Co-expression of AEPs with Oak1 (that harbors an asparagine at the cyclization site of kB1) revealed that CtAEP1 produced the highest relative level of kB1 when compared to OaAEP1b, HeAEP3, and PxAEP3b (Supplementary Figures S2A,C). With this in mind, we wished to determine if cyclization levels of SFTI-1 could be simply improved by combining CtAEP1 expression with a modified SFTI-1 where the cyclization residue was changed from the native aspartic acid to an asparagine (construct pEAQ-Oak1-[D14N]SFTI-1). Somewhat surprisingly however, co-expression of this modified peptide precursor, with any of the four AEPs tested, resulted in no detectable cyclic [D14N]SFTI-1. This result contrasts to our in vitro assessment of the [D14N]SFTI-1-GLDN substrate where recombinant OaAEP1b predominantly processed the substrate to cyclic [D14N]SFTI-1 (Figure 2D). These results suggest that in planta, [D14N]SFTI-1_GLDN is cyclizable but is prone to rapid degradation. Interestingly, this does not seem to be the case for kB1, where cyclic peptides containing either asparagine or aspartic acid residues at the kB1 cyclization point accumulate to high levels in N. benthamiana leaves (Supplementary Figure S2).
Plant Produced [T4Y,I7R] Sunflower Trypsin Inhibitor-1 and Synthetically Produced Peptide are Equivalent
The SFTI-1-based plasmin inhibitor [T4Y,I7R]SFTI-1 is the most potent inhibitor of plasmin developed to date (Swedberg et al., 2019). In addition to its high potency (Ki = 0.041 nM), the inhibitor displays a million-fold selectivity over other serine proteases found in blood and is a promising lead compound for certain antifibrinolytic treatments. As this inhibitor carries only two residue changes to SFTI-1 and retains an aspartic residue for AEP-mediated cyclization, we hypothesized that it is a good candidate for plant-based production. Similar to wild-type SFTI-1 peptide, [T4Y,I7R]SFTI-1 production required the co-expression of the O. affinis OaAEP1b ligase (Figure 4A) where yields of 12.3 ± 3.3 μg g−1 DW (s.d., n = 5) cyclic [T4Y,I7R]SFTI-1 were obtained (Figure 4B, Supplementary Figure S3). As an approach to further improve these yields, we then reengineered the Oak1 precursor to contain three tandem repeats of the [T4Y,I7R]SFTI-1 peptide (Figure 4A). Tandem repeats of kalata type peptides are commonly observed in cyclotide precursor genes (Craik and Malik, 2013) and, at least in part, may be responsible for the observed high yields. By making this change, we improved the in planta yield of [T4Y,I7R]SFTI-1 ~5-fold to 56.5 ± 10.8 μg g−1 DW (s.d., n = 7) (Figure 4B, Supplementary Figure S3).
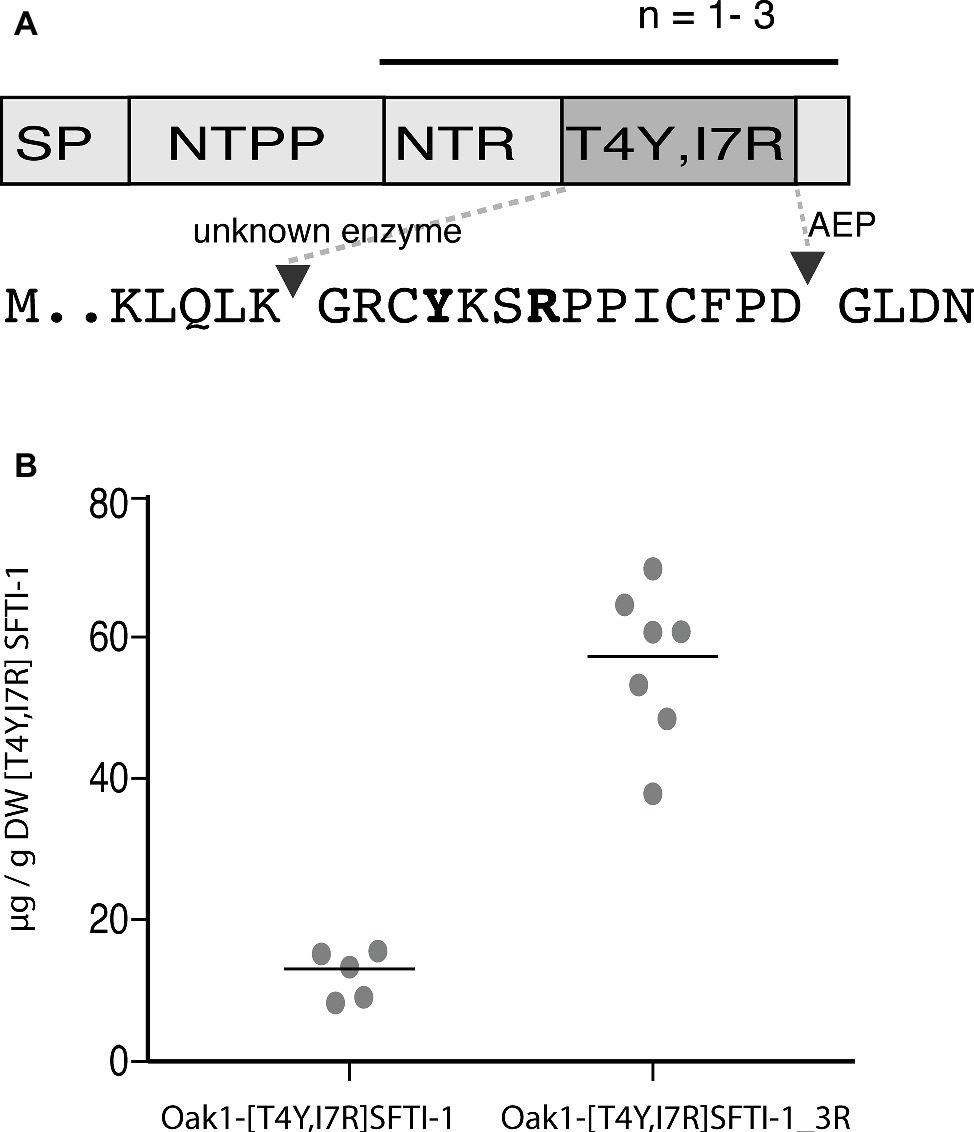
Figure 4. Transient expression analysis of [T4Y,I7R]SFTI-1 production in N. benthamiana. (A) Two constructs were prepared to produce [T4Y,I7R]SFTI-1 in planta. In the first, the kB1 domain normally present in the Oak1 precursor was substituted directly for [T4Y,I7R]SFTI-1. In the second, the amino terminal repeat (NTR), [T4Y,I7R]SFTI-1 and C-terminal tail domain were incorporated as three tandem repeats, adjacent the signal peptide (SP) and amino terminal propeptide (NTPP). (B) In planta yields (μg/g DW) were calculated using the method of standard addition (Supplementary Figure S3).
To validate the functional and structural equivalence of synthetic peptide vs. the in planta produced [T4Y,I7R]SFTI-1, we purified [T4Y,I7R]SFTI-1 peptide from lyophilized leaf tissue. Peptides were extracted, fractionated, and purified using C18 SPE with several rounds of reverse phase HLPC. Structural equivalence to synthetically produced [T4Y,I7R]SFTI-1 was demonstrated via LC-MS coelution (Supplementary Figure S4), MS-MS fragmentation patterns (Supplementary Figure S5), and NMR analysis (Figure 5A, Supplementary Figure S6). Functional equivalence of plant-produced [T4Y,I7R]SFTI-1 was demonstrated by determining the inhibitory constant of the purified peptide. The calculated Ki was 0.025 ± 0.004 nM (Figure 5B), which is comparable to the Ki 0.041 ± 0.005 nM previously calculated for synthetic peptide (Swedberg et al., 2019).
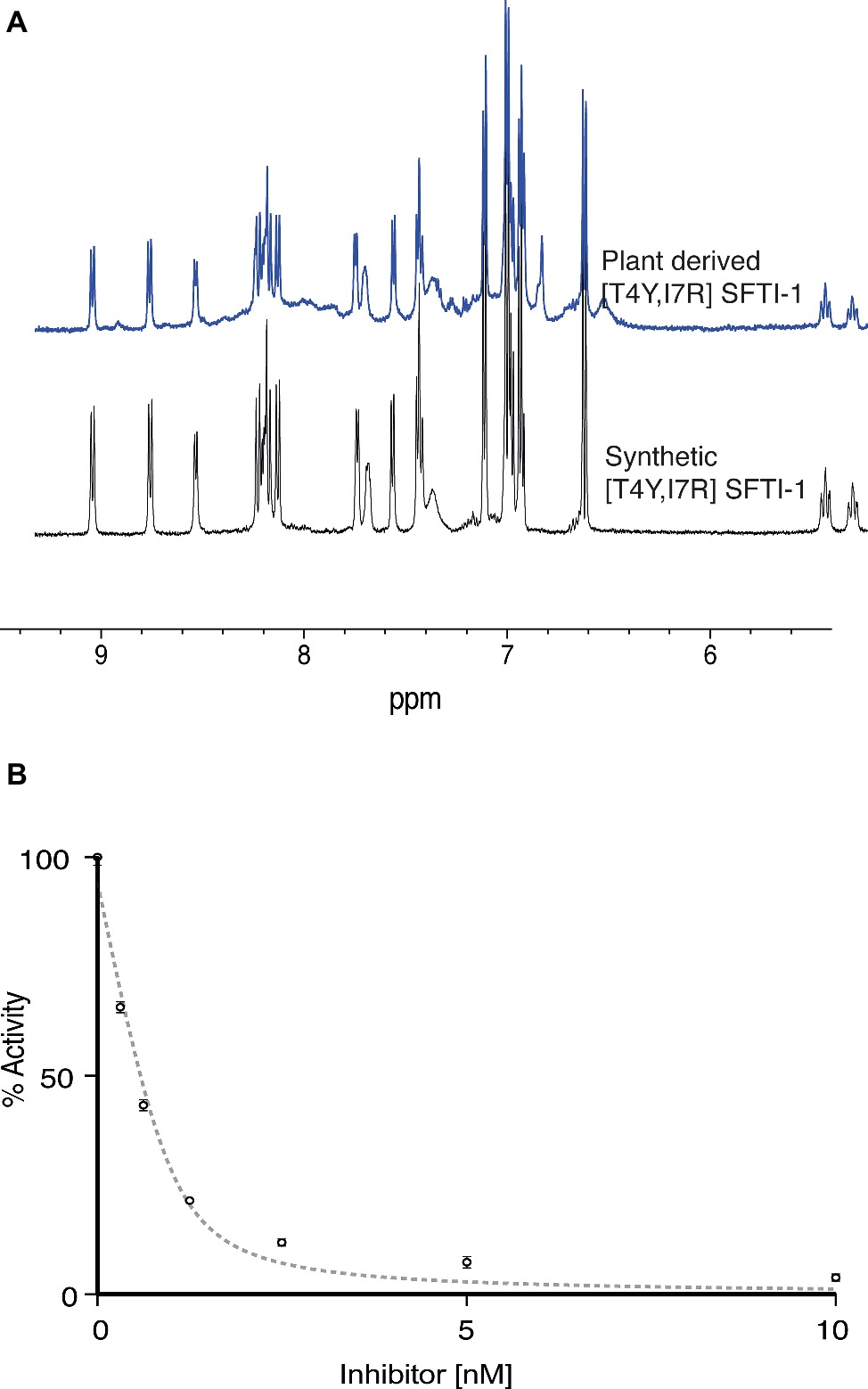
Figure 5. Structural and functional equivalence studies of plant derived vs. synthetic [T4Y,I7R]SFTI-1. (A) 1D 1H NMR fingerprint region of synthetic and in planta produced [T4Y,I7R]SFTI-1. (B) Plant-derived [T4Y,I7R]SFTI-1 inhibitor assay (n = 3).
Discussion
Peptides as therapeutics are posited to bridge the gap between traditional small molecule drugs and larger biologics, offering the potential of higher specificity, reduced off-target effects, and potentially lower production costs (Craik et al., 2013). However, one drawback is their poor stability, which directly affects efficacy due to shorter in vivo half-lives. To counter this limitation, much emphasis has been placed on developing strategies to stabilize peptides, of which backbone cyclization has shown great promise (Poth et al., 2013; Thapa et al., 2014). A recombinant production system that provides for post-translational backbone cyclization of peptides is thus highly desired. Here, we demonstrate a rapid, plant-based approach to produce and cyclize SFTI-1 peptide analogues in an environmentally friendly manner with capacity for scale-up.
In sunflower seed, SFTI-1 is processed from the PawS1 precursor, which additionally encodes for a seed storage albumin that is exclusively found in seed (Figure 1A) (Mylne et al., 2011). For expression in plant leaves, we chose to use the strategy described in (Poon et al., 2017) where the Oak1 gene was reengineered to include SFTI-1, replacing the kB1 domain (Figure 1B). This ensured that the vacuole targeting elements within Oak1 (Conlan et al., 2011) that work efficiently in N. benthamiana leaf cells would be sufficient to direct the engineered SFTI-1 precursor to the vacuole where functional AEP enzymes are believed to reside. An additional benefit of this approach was simplified biosynthesis, as it did not require the concerted action of multiple AEP isoforms, which are required for SFTI-1 maturation from the PawS1 precursor protein (Mylne et al., 2011). The success shown here for a N. benthamiana leaf-based SFTI-1 production provides hope that the “plug n play” type approach presented here could be extended to other bioactive peptides derived from seed, such as the trypsin inhibitor class of cyclic peptides derived from Momordica cochinchinensis seed (Hernandez et al., 2000).
By using transient gene expression technology, we were able to produce 12.8 ± 3.0 μg g−1 DW (s.d., n = 3) SFTI-1 upon 6 days of incubation (Figure 1D). Although this is an improvement on the reported natural SFTI-1 levels in mature sunflower seed (0.5 μg g−1 seed) (Bernath-Levin et al., 2015), it is lower then that previously reported for the production of the cyclotide kB1, using similar transient gene expression conditions (Poon et al., 2017). By comparing in vitro cyclization efficiencies, we demonstrated that this is due to the slower processing of SFTI-1 peptide precursors by OaAEP1b compared to kB1 precursor substrates. One strategy to increase efficiency would be to use the native AEP ligase from sunflower, which may be more efficient for SFTI-1 precursor processing, if expressed heterologously in N. benthamiana. However, so far no efficient ligase-type sunflower AEP has been reported (Bernath-Levin et al., 2015; Haywood et al., 2018). Attempts to improve the in planta yield of SFTI-1 by co-expressing other known AEP ligases were unsuccessful, with OaAEP1b remaining the superior ligase for SFTI-1. In contrast for kB1, the superior ligase for in planta activity is CtAEP1, with OaAEP1b being on par with HeAEP3 (Supplementary Figure S2). These results indicate that substrate preference plays a key role in the in planta performance of AEP ligases; however, differences in transcript stability, translational efficiency, enzyme maturation, and stability are also likely.
Plant AEPs are known to have a strict preference for processing at either asparagine or aspartic residues and thus it was surprising that expression of OaAEP1b in planta could only produce cyclic SFTI-1 and not [D14N]SFTI-1. We confirmed through in vitro cyclization experiments that this was not due to an inefficiency of the enzyme, but more likely due to instability of the precursor or cyclic [D14N]SFTI-1 in planta. As only the AEP processing residue was changed, we reasoned that the instability observed is likely governed by the pool of endogenous AEPs present in N. benthamiana leaf cells, which may either outcompete the transgene-derived AEP ligase or degrade any correctly cyclized [D14N]SFTI-1. Interestingly, this was not observed with kB1 cyclized with either an asparagine or aspartic residue at the cyclization site (Supplementary Figure S2) and suggests that the observed instability of [D14N]SFTI-1 may be specific to SFTI-1 peptides. This finding is particularly relevant in the case of SFTI-1 peptide analogues that require the D14N residue substitution for potency (Swedberg et al., 2011), in which case, developing strategies to downregulate or knock out endogenous interfering AEPs with emerging gene editing technologies (Puchta, 2017) would be beneficial.
SFTI-1 peptide analogues have been engineered for diverse therapeutic applications ranging from anti-cancer (Swedberg et al., 2009, 2011), anti-obesity (Durek et al., 2018), pro and anti-angiogenesis (Chan et al., 2011; Qiu et al., 2017) as well as for treatment of a range of skin conditions (Chen et al., 2016; Zhu et al., 2017). Here, we have shown that like SFTI-1, the engineered plasmin inhibitor [T4Y,I7R]SFTI-1 (Swedberg et al., 2019) is amenable to plant-based production with the resultant purified peptide displaying both structural and functional equivalence to synthetically produced material. Through expression of a multi peptide domain gene construct the yield of the 14 amino acid cyclic [T4Y,I7R]SFTI-1 reached ~60 μg/g DW, roughly equivalent to that obtained previously for kB1 (29 aa) on a molar basis. Importantly, this approach provides for economies of scale, with lower inputs and infrastructure costs than synthetic peptide synthesis. Although currently still an emerging industry, commercial facilities for plant-based recombinant production have begun to be established, primarily for vaccine production. One such facility operated by iBio Biotherapeutics has a reported capacity to process ~3,500 kg of plant material per week (Holtz et al., 2015). Although, the economics of scaling up a plant-based production approach for peptide therapeutic production must be considered on a case-by-case basis, backbone cyclized peptide scaffolds such as SFTI-1 represent a particularly suitable case, given their natural occurrence in plants.
Author Contributions
MJ, EG, TD, KH, DC, and MA conceived the experiments. MJ, SP, and HQ made gene constructs and performed transient assays. KY and AP performed MS analysis. KY and JS performed functional analysis. TD performed NMR analysis. KH and KY produced and assayed recombinant AEP. All authors contributed to the writing of the manuscript.
Funding
We acknowledge funding from the Australian Research Council (ARC Laureate Fellowship FL150100146 to DC, ARC grant DP150100443 to DC, EG, and TD). This research was also supported by the 2015 Ramaciotti Biomedical Research Award to DC and MA and from the Simon Axelsen Memorial Fund.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The pEAQ vectors were kindly provided by Prof. George Lomonossoff at the John Innes Centre and Plant Bioscience Ltd.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00602/full#supplementary-material
References
Aboye, T. L., and Camarero, J. A. (2012). Biological synthesis of circular polypeptides. J. Biol. Chem. 287, 27026–27032. doi: 10.1074/jbc.R111.305508
Andersson, L., Blomberg, L., Flegel, M., Lepsa, L., Nilsson, B., and Verlander, M. (2000). Large-scale synthesis of peptides. Biopolymers 55, 227–250. doi: 10.1002/1097-0282(2000)55:3<227::Aid-bip50>3.0.Co;2-7
Benchabane, M., Goulet, C., Rivard, D., Faye, L., Gomord, V., and Michaud, D. (2008). Preventing unintended proteolysis in plant protein biofactories. Plant Biotechnol. J. 6, 633–648. doi: 10.1111/j.1467-7652.2008.00344.x
Bernath-Levin, K., Nelson, C., Elliott, A. G., Jayasena, A. S., Millar, A. H., Craik, D. J., et al. (2015). Peptide macrocyclization by a bifunctional endoprotease. Chem. Biol. 22, 571–582. doi: 10.1016/j.chembiol.2015.04.010
Bronsema, K. J., Bischoff, R., and van de Merbel, N. C. (2012). Internal standards in the quantitative determination of protein biopharmaceuticals using liquid chromatography coupled to mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 893, 1–14. doi: 10.1016/j.jchromb.2012.02.021
Chan, L. Y., Gunasekera, S., Henriques, S. T., Worth, N. F., Le, S. J., Clark, R. J., et al. (2011). Engineering pro-angiogenic peptides using stable, disulfide-rich cyclic scaffolds. Blood 118, 6709–6717. doi: 10.1182/blood-2011-06-359141
Chen, W. J., Kinsler, V. A., Macmillan, D., and Di, W. L. (2016). Tissue kallikrein inhibitors based on the sunflower trypsin inhibitor scaffold - a potential therapeutic intervention for skin diseases. PLoS One 11:e0166268. doi: 10.1371/journal.pone.0166268
Cheneval, O., Schroeder, C. I., Durek, T., Walsh, P., Huang, Y. H., Liras, S., et al. (2014). Fmoc-based synthesis of disulfide-rich cyclic peptides. J. Org. Chem. 79, 5538–5544. doi: 10.1021/jo500699m
Colgrave, M. L., Korsinczky, M. J. L., Clark, R. J., Foley, F., and Craik, D. J. (2010). Sunflower trypsin inhibitor-1, proteolytic studies on a trypsin inhibitor peptide and its analogs. Biopolymers 94, 665–672. doi: 10.1002/bip.21415
Conlan, B. F., Gillon, A. D., Barbeta, B. L., and Anderson, M. A. (2011). Subcellular targeting and biosynthesis of cyclotides in plant cells. Am. J. Bot. 98, 2018–2026. doi: 10.3732/ajb.1100382
Craik, D. J., Cemazar, M., and Daly, N. L. (2006). The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discov. Devel. 9, 251–260. doi: 10.1007/s00018-009-0159-3
Craik, D. J., Daly, N. L., Bond, T., and Waine, C. (1999). Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294, 1327–1336. doi: 10.1006/jmbi.1999.3383
Craik, D. J., Fairlie, D. P., Liras, S., and Price, D. (2013). The future of peptide-based drugs. Chem. Biol. Drug Des. 81, 136–147. doi: 10.1111/cbdd.12055
Craik, D. J., and Malik, U. (2013). Cyclotide biosynthesis. Curr. Opin. Chem. Biol. 17, 546–554. doi: 10.1016/j.cbpa.2013.05.033
de Veer, S. J., Swedberg, J. E., Brattsand, M., Clements, J. A., and Harris, J. M. (2016). Exploring the active site binding specificity of kallikrein-related peptidase 5 (KLK5) guides the design of new peptide substrates and inhibitors. Biol. Chem. 397, 1237–1249. doi: 10.1515/hsz-2016-0112
Demain, A. L., and Vaishnav, P. (2009). Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 27, 297–306. doi: 10.1016/j.biotechadv.2009.01.008
Durek, T., Cromm, P. M., White, A. M., Schroeder, C. I., Kaas, Q., Weidmann, J., et al. (2018). Development of novel melanocortin receptor agonists based on the cyclic peptide framework of sunflower trypsin Inhibitor-1. J. Med. Chem. 61, 3674–3684. doi: 10.1021/acs.jmedchem.8b00170
Fittler, H., Avrutina, O., Empting, M., and Kolmar, H. (2014). Potent inhibitors of human matriptase-1 based on the scaffold of sunflower trypsin inhibitor. J. Pept. Sci. 20, 415–420. doi: 10.1002/psc.2629
Fittler, H., Depp, A., Avrutina, O., Dahms, S. O., Than, M. E., Empting, M., et al. (2015). Engineering a constrained peptidic scaffold towards potent and selective furin inhibitors. Chembiochem 16, 2441–2444. doi: 10.1002/cbic.201500447
Franke, B., James, A. M., Mobli, M., Colgrave, M. L., Mylne, J. S., and Rosengren, K. J. (2017). Two proteins for the price of one: structural studies of the dual-destiny protein preproalbumin with sunflower trypsin inhibitor-1. J. Biol. Chem. 292, 12398–12411. doi: 10.1074/jbc.M117.776955
Gitlin, A., Debowski, D., Karna, N., Legowska, A., Stirnberg, M., Gutschow, M., et al. (2015). Inhibitors of matriptase-2 based on the trypsin inhibitor SFTI-1. Chembiochem 16, 1601–1607. doi: 10.1002/cbic.201500200
Habibi, P., Prado, G. S., Pelegrini, P. B., Hefferon, K. L., Soccol, C. R., and Grossi-de-Sa, M. F. (2017). Optimization of inside and outside factors to improve recombinant protein yield in plant. Plant Cell Tissue Organ Cult. 130, 449–467. doi: 10.1007/s11240-017-1240-5
Harris, K. S., Durek, T., Kaas, Q., Poth, A. G., Gilding, E. K., Conlan, B. F., et al. (2015). Efficient backbone cyclization of linear peptides by a recombinant asparaginyl endopeptidase. Nat. Commun. 6:10199. doi: 10.1038/ncomms10199
Haywood, J., Schmidberger, J. W., James, A. M., Nonis, S. G., Sukhoverkov, K. V., Elias, M., et al. (2018). Structural basis of ribosomal peptide macrocyclization in plants. elife 7:e32955. doi: 10.7554/eLife.32955
Hemu, X., Qiu, Y. B., Nguyen, G. K. T., and Tam, J. P. (2016). Total synthesis of circular bacteriocins by butelase 1. J. Am. Chem. Soc. 138, 6968–6971. doi: 10.1021/jacs.6b04310
Hernandez, J. F., Gagnon, J., Chiche, L., Nguyen, T. M., Andrieu, J. P., Heitz, A., et al. (2000). Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 39, 5722–5730. doi: 10.1021/bi9929756
Holtz, B. R., Berquist, B. R., Bennett, L. D., Kommineni, V. J. M., Munigunti, R. K., White, E. L., et al. (2015). Commercial-scale biotherapeutics manufacturing facility for plant-made pharmaceuticals. Plant Biotechnol. J. 13, 1180–1190. doi: 10.1111/pbi.12469
Jackson, M. A., Gilding, E. K., Shafee, T., Harris, K. S., Kaas, Q., Poon, S., et al. (2018). Molecular basis for the production of cyclic peptides by plant asparaginyl endopeptidases. Nat. Commun. 9:2411. doi: 10.1038/s41467-018-04669-9
Jackson, M. A., Nutt, K. A., Hassall, R., and Rae, A. L. (2010). Comparative efficiency of subcellular targeting signals for expression of a toxic protein in sugarcane. Funct. Plant Biol. 37, 785–793. doi: 10.1071/FP09243
Jackson, M. A., Rae, A. L., Casu, R. E., Grof, C. P. L., Bonnett, G. D., and Maclean, D. J. (2007). A bioinformatic approach to the identification of a conserved domain in a sugarcane legumain that directs GFP to the lytic vacuole. Funct. Plant Biol. 34, 633–644. doi: 10.1071/FP07024
James, A. M., Haywood, J., and Mylne, J. S. (2018). Macrocyclization by asparaginyl endopeptidases. New Phytol. 218, 923–928. doi: 10.1111/nph.14511
Jendrny, C., and Beck-Sickinger, A. G. (2016). Inhibition of kallikrein-related peptidases 7 and 5 by grafting serpin reactive-center loop sequences onto sunflower trypsin inhibitor-1 (SFTI-1). Chembiochem 17, 719–726. doi: 10.1002/cbic.201500539
Korsinczky, M. L. J., Schirra, H. J., Rosengren, K. J., West, J., Condie, B. A., Otvos, L., et al. (2001). Solution structures by H-1 NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant. J. Mol. Biol. 311, 579–591. doi: 10.1006/jmbi.2001.4887
Lesner, A., Legowska, A., Wysocka, M., and Rolka, K. (2011). Sunflower trypsin inhibitor 1 as a molecular scaffold for drug discovery. Curr. Pharm. Des. 17, 4308–4317. doi: 10.2174/138161211798999393
Li, Y. L., Aboye, T., Breindel, L., Shekhtman, A., and Camarero, J. A. (2016). Efficient recombinant expression of SFTI-1 in bacterial cells using intein-mediated protein trans-splicing. Biopolymers 106, 818–824. doi: 10.1002/bip.22875
Lico, C., Santi, L., Twyman, R. M., Pezzotti, M., and Avesani, L. (2012). The use of plants for the production of therapeutic human peptides. Plant Cell Rep. 31, 439–451. doi: 10.1007/s00299-011-1215-7
Mylne, J. S., Colgrave, M. L., Daly, N. L., Chanson, A. H., Elliott, A. G., McCallum, E. J., et al. (2011). Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat. Chem. Biol. 7, 257–259. doi: 10.1038/nchembio.542
Nguyen, G. K. T., Kam, A., Loo, S., Jansson, A. E., Pan, L. X., and Tam, J. P. (2015). Butelase 1: a versatile ligase for peptide and protein macrocyclization. J. Am. Chem. Soc. 137, 15398–15401. doi: 10.1021/jacs.5b11014
Poon, S., Harris, K. S., Jackson, M. A., McCorkelle, O. C., Gilding, E. K., Durek, T., et al. (2017). Co-expression of a cyclizing asparaginyl endopeptidase enables efficient production of cyclic peptides in planta. J. Exp. Bot. 69, 633–641. doi: 10.1093/jxb/erx422
Poth, A. G., Chan, L. Y., and Craik, D. J. (2013). Cyclotides as grafting frameworks for protein engineering and drug design applications. Biopolymers 100, 480–491. doi: 10.1002/bip.22284
Puchta, H. (2017). Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr. Opin. Plant Biol. 36, 1–8. doi: 10.1016/j.pbi.2016.11.011
Qiu, Y. B., Taichi, M., Wei, N., Yang, H., Luo, K. Q., and Tam, J. P. (2017). An orally active bradykinin B-1 receptor antagonist engineered as a bifunctional chimera of sunflower trypsin inhibitor. J. Med. Chem. 60, 504–510. doi: 10.1021/acs.jmedchem.6b01011
Quimbar, P., Malik, U., Sommerhoff, C. P., Kaas, Q., Chan, L. Y., Huang, Y. H., et al. (2013). High-affinity cyclic peptide matriptase inhibitors. J. Biol. Chem. 288, 13885–13896. doi: 10.1074/jbc.M113.460030
Robert, S., Goulet, M. C., D’Aoust, M. A., Sainsbury, F., and Michaud, D. (2015). Leaf proteome rebalancing in Nicotiana benthamiana for upstream enrichment of a transiently expressed recombinant protein. Plant Biotechnol. J. 13, 1169–1179. doi: 10.1111/pbi.12452
Sainsbury, F., Thuenemann, E. C., and Lomonossoff, G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693. doi: 10.1111/j.1467-7652.2009.00434.x
Sainsbury, F., Varennes-Jutras, P., Goulet, M. C., D’Aoust, M. A., and Michaud, D. (2013). Tomato cystatin SlCYS8 as a stabilizing fusion partner for human serpin expression in plants. Plant Biotechnol. J. 11, 1058–1068. doi: 10.1111/pbi.12098
Seydel, P., and Dornenburg, H. (2006). Establishment of in vitro plants, cell and tissue cultures from Oldenlandia affinis for the production of cyclic peptides. Plant Cell Tissue Organ Cult. 85, 247–255. doi: 10.1007/s11240-005-9056-0
Shariff, L., Zhu, Y. N., Cowper, B., Di, W. L., and Macmillan, D. (2014). Sunflower trypsin inhibitor (SFTI-1) analogues of synthetic and biological origin via N -> S acyl transfer: potential inhibitors of human Kallikrein-5 (KLK5). Tetrahedron 70, 7675–7680. doi: 10.1016/j.tet.2014.06.059
Swedberg, J. E., de Veer, S. J., Sit, K. C., Reboul, C. F., Buckle, A. M., and Harris, J. M. (2011). Mastering the canonical loop of serine protease inhibitors: enhancing potency by optimising the internal hydrogen bond network. PLoS One 6:e19302. doi: 10.1371/journal.pone.0019302
Swedberg, J. E., Li, C. Y., de Veer, S. J., Wang, C. K., and Craik, D. J. (2017). Design of potent and selective cathepsin G inhibitors based on the sunflower trypsini inhibitor-1 scaffold. J. Med. Chem. 60, 658–667. doi: 10.1021/acs.jmedchem.6b01509
Swedberg, J. E., Nigon, L. V., Reid, J. C., de Veer, S. J., Walpole, C. M., Stephens, C. R., et al. (2009). Substrate-guided design of a potent and selective kallikrein-related peptidase inhibitor for kallikrein 4. Chem. Biol. 16, 633–643. doi: 10.1016/j.chembiol.2009.05.008
Swedberg, J. E., Wu, G., Mahatmanto, T., Durek, T., Caradoc-Davies, T., Whisstock, J. C., et al. (2019). Highly potent and selective plasmin inhibitors based on the sunflower trypsin inhibitor-1 scaffold attenuate fibrinolysis in plasma. J. Med. Chem. 62, 552–560. doi: 10.1021/acs.jmedchem.8b01139
Thapa, P., Espiritu, M. J., Cabalteja, C., and Bingham, J. P. (2014). The emergence of cyclic peptides: the potential of bioengineered peptide drugs. Int. J. Pept. Res. Ther. 20, 545–551. doi: 10.1007/s10989-014-9421-0
Viana, A. A. B., Pelegrini, P. B., and Grossi-de-Sa, M. F. (2012). Plant biofarming: novel insights for peptide expression in heterologous systems. Biopolymers 98, 416–427. doi: 10.1002/bip.22089
Vranken, W. F., Boucher, W., Stevens, T. J., Fogh, R. H., Pajon, A., Llinas, P., et al. (2005). The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696. doi: 10.1002/prot.20449
Wang, C. K., and Craik, D. J. (2018). Designing macrocyclic disulfide-rich peptides for biotechnological applications. Nat. Chem. Biol. 14, 417–427. doi: 10.1038/s41589-018-0039-y
Yang, R. L., Wong, Y. H., Nguyen, G. K. T., Tam, J. P., Lescar, J., and Wu, B. (2017). Engineering a catalytically efficient recombinant protein ligase. J. Am. Chem. Soc. 139, 5351–5358. doi: 10.1021/jacs.6b12637
Yasuda, H., Tada, Y., Hayashi, Y., Jomori, T., and Takaiwa, F. (2005). Expression of the small peptide GLP-1 in transgenic plants. Transgenic Res. 14, 677–684. doi: 10.1007/s11248-005-6631-4
Keywords: peptide, therapeutic, asparaginyl endopeptidase, cyclyzation, stability, biofactory, Nicotiana benthamiana, sunflower trypsin inhibitor
Citation: Jackson MA, Yap K, Poth AG, Gilding EK, Swedberg JE, Poon S, Qu H, Durek T, Harris K, Anderson MA and Craik DJ (2019) Rapid and Scalable Plant-Based Production of a Potent Plasmin Inhibitor Peptide. Front. Plant Sci. 10:602. doi: 10.3389/fpls.2019.00602
Edited by:
Suvi Tuulikki Häkkinen, VTT Technical Research Centre of Finland Ltd, FinlandReviewed by:
Joshua Lee Fuqua, University of Louisville, United StatesSomen Nandi, University of California, United States
Copyright © 2019 Jackson, Yap, Poth, Gilding, Swedberg, Poon, Qu, Durek, Harris, Anderson and Craik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David J. Craik, d.craik@imb.uq.edu.au
 Mark A. Jackson
Mark A. Jackson Kuok Yap
Kuok Yap Aaron G. Poth1
Aaron G. Poth1 Edward K. Gilding
Edward K. Gilding Thomas Durek
Thomas Durek Marilyn A. Anderson
Marilyn A. Anderson David J. Craik
David J. Craik