- 1Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Bundoora, VIC, Australia
- 2Bio21 Institute, University of Melbourne, Parkville, VIC, Australia
Extracellular vesicles (EVs) represent a system for the coordinated secretion of a variety of molecular cargo including proteins, lipids, nucleic acids, and metabolites. They have an essential role in intercellular communication in multicellular organisms and have more recently been implicated in host-pathogen interactions. Study of the role for EVs in fungal biology has focused on pathogenic yeasts that are major pathogens in humans. In this study we have expanded the investigation of fungal EVs to plant pathogens, specifically the major cotton pathogen Fusarium oxysporum f. sp. vasinfectum. EVs isolated from F. oxysporum f. sp. vasinfectum culture medium have a morphology and size distribution similar to EVs from yeasts such as Candida albicans and Cryptococcus neoformans. A unique feature of the EVs from F. oxysporum f. sp. vasinfectum is their purple color, which is predicted to arise from a napthoquinone pigment being packaged into the EVs. Proteomic analysis of F. oxysporum f. sp. vasinfectum EVs revealed that they are enriched in proteins that function in synthesis of polyketides as well as proteases and proteins that function in basic cellular processes. Infiltration of F. oxysporum f. sp. vasinfectum EVs into the leaves of cotton or N. benthamiana plants led to a phytotoxic response. These observations lead to the hypothesis that F. oxysporum f. sp. vasinfectum EVs are likely to play a crucial role in the infection process.
Introduction
Extracellular vesicles (EVs) are involved in intercellular communications in all kingdoms of life (Pathirana and Kaparakis-Liaskos, 2016; Maas et al., 2017; Bielska et al., 2018; Rybak and Robatzek, 2019). These small, lipid bilayer encapsulated particles provide a mechanism for the coordinated transport of heterogeneous cargo. This is not limited to transport between cells within an organism but also extends to cross kingdom interactions such as those between microbial pathogens and their hosts (Schorey et al., 2015; Rutter and Innes, 2018). The best characterized EVs are from mammalian cells; their cargo includes nucleic acids, proteins, lipids, and metabolites (Kalra et al., 2012; Maas et al., 2017). Uptake of EVs by target cells induces changes in gene/protein expression that consequently lead to altered physiology and other phenotypes (Greening and Simpson, 2018). Our understanding of EVs at the host-pathogen interface comes largely from the study of outer membrane vesicles (OMVs) produced by Gram negative bacteria that infect humans (Pathirana and Kaparakis-Liaskos, 2016). OMVs contribute to the host–pathogen interaction through their cargo, which can induce immune responses (Ellis et al., 2010), contribute to adhesion to mucosal surfaces (Olofsson et al., 2010), act as virulence factors (Bomberger et al., 2009) or toxins (Horstman and Kuehn, 2000), degrade antibiotics (Kim et al., 2018), acquire trace nutrients (Prados-Rosales et al., 2014), and function in inter-microbe competition (Vasilyeva et al., 2008).
Investigation of fungal EVs is in its relative infancy compared to mammalian EVs or bacterial OMVs with most studies on pathogenic yeasts that infect humans (reviewed in Bleackley et al., 2019). Like mammalian EVs and bacterial OMVs, fungal EV cargo consists of proteins, lipids, and nucleic acids. Polysaccharides are also transported by fungal EVs (Griffith et al., 2004; da Silva et al., 2015). Proteomic characterization of the cargo in EVs from pathogenic yeast has led to predicted roles in stress responses, and metabolism of various cellular building blocks including proteins, lipids, and carbohydrates. Functional analysis of EVs in vitro supports several roles for these vesicles in host–pathogen interactions. For example, EVs from a number of pathogenic yeast induce responses when applied to cultured innate immune cells, indicating EVs are probably sensed by the host during infection (Oliveira et al., 2010; Wolf et al., 2015; da Silva et al., 2016). Antifungal drug resistance is also linked to EV production; EVs from biofilms produced by the major human fungal pathogen Candida albicans contribute to resistance to antifungal azoles (Zarnowski et al., 2018) and S. cerevisiae EVs have a protective function against both caspofungin and antifungal peptides(Zhao et al., 2019). Virulence has also been linked to EV production. Hypervirulence of a Cryptococcus gattii strain is facilitated by intra-cellular communication between fungal cells mediated by EVs (Bielska et al., 2018).
Little is known about the role of EVs in plant–fungal interactions, although there has been speculation based on the study of mammalian yeast pathogens (Samuel et al., 2015; Boevink, 2017). Experimental investigation into the role for EVs in the plant–fungal interaction has focused on EVs produced by the plant, defense related cargo, and the effect they have on fungi (Rutter and Innes, 2018). Multivesicular bodies, which are the site of biogenesis for many EVs, accumulate at plasmodesmata during fungal infections leading to a potential role for vesicles in the callose deposition that isolates dying cells from living cells (An et al., 2006; Bohlenius et al., 2010; Li et al., 2018). Plant EVs may also deliver antifungal molecules to the pathogen. For example, EVs from sunflower seedlings inhibit the growth of Sclerotinia sclerotiorum spores (Regente et al., 2017). Plant EVs are also proposed to transport antimicrobial glucosinylates (Rutter and Innes, 2017). Very little is known about EVs produced by filamentous fungi, with data limited to a handful of studies on Alternaria infectoria (Silva et al., 2013), Trichophyton rubrum (Bitencourt et al., 2018), and Rhizopus delemar (Liu et al., 2018). The EVs from the filamentous fungi in these studies are similar to those produced by yeast with respect to morphology, cargo, and recognition by innate immune cells.
To address the lack of knowledge on how EVs contribute to fungal pathogenesis in plants, we have isolated and characterized EVs from the cotton pathogen Fusarium oxysporum f. sp. vasinfectum. The F. oxysporum species complex causes vascular wilt disease in many economically relevant crops and are considered hemibiotrophic pathogens because the infection consists of both a biotrophic and necrotrophic phase (Gordon 2017). The EVs had a similar morphology to those from yeasts and contained protein cargo that was functionally related to those described for human pathogens. EVs from F. oxysporum were deep purple in color and were phytotoxic when applied to plant leaves. This supports a role for fungal EVs in the host–pathogen interaction with plants.
Materials and Methods
Strains and Growth Conditions
F. oxysporum f. sp. vasinfectum, Australian isolate VCG01111 isolated from cotton, (gift from Wayne O’Neill, Farming Systems Institute, Department of Primary Industries (DPI), Queensland, Australia), was maintained on half strength potato dextrose agar (1/2 PDA) at 25°C. Liquid cultures were grown in half-strength potato dextrose broth (1/2 PDB) at room temperature with shaking at 90 rpm. S. cerevisiae BY4741 were maintained on 1% yeast extract, 2% peptone, 2% dextrose, 2% agar (YPD-agar) at 30°C. Liquid cultures were grown in YPD at 30°C with shaking at 125 rpm.
Isolation of EVs
EVs were isolated using a procedure modified from Rodrigues et al. (2007). Briefly, F. oxysporum f. sp. vasinfectum mycelium was cultured in ½ PDB for 72 h at room temperature. Mycelium was removed from the culture medium by filtration through sterile Miracloth. Spores and cell debris were removed from the culture medium by centrifugation at 4,000 x g for 15 min in a Heraeus Multifuge X3 centrifuge (Thermo Fisher) followed by 15,000 x g for 30 min in an Avanti J-E centrifuge (Beckman Coulter). Supernatant was further centrifuged at 100,000 x g using an SW32 rotor in an Optima L-100 XP ultracentrifuge (Beckman Coulter). EV pellets were resuspended in sterile phosphate-buffered saline (PBS). Protein content of EVs was initially assessed using a Qubit fluorimeter (Thermo Fisher) and then separated on SDS-PAGE gels and stained with SYPRO-Ruby (Invitrogen), and protein content was determined according to the manufacturer’s instructions. When needed, EVs were concentrated by pelleting using a TL-100 ultracentrifuge with a TLA100.3 rotor (Beckman Coulter); the supernatant was removed and the pellet resuspended in an appropriate volume to yield the desired concentration.
Transmission Electron Microscopy
EV samples (5 µl at 0.05 µg/ml protein) were deposited onto carbon-coated 400-mesh copper grids (ProSciTech) that had been glow discharged for 1 min in a K950X turbo evaporator coupled to a K350 glow discharge unit (Quorum Technologies Ltd), incubated for 1 min then washed with ultrapure water. Grids were then stained three times with 4 µl of 2% (v/v) uranyl acetate (Agar Scientific). Excess solution was blotted off and the grids were dried overnight. Images were captured using a JEM 2100 electron microscope (JEOL Ltd) operated at 200 kV.
Nanoparticle Tracking Analysis
Particle size and number of purified EVs were determined using a Nanosight NS300 with a 405 nm (blue) laser (Malvern Instruments). Samples (0.1 µg/ml) were diluted 1:1,000 with sterile PBS and injected using a syringe pump with a flow rate of 50. Three technical replicates were performed per sample with 1 min videos recorded for analysis. All samples were measured in triplicate and data were analyzed using NTA 3.2 Dev Build 3.2.16 with the auto-analysis settings.
Proteomic Analysis
Three separate pools of EVs (each pooled from two or three independent biological replicates) with 15 µg of total protein in each, were loaded onto precast NuPAGE® 4–12% Bis-Tris gels in 1x MES SDS running buffer. Gels were run at 150 V followed by visualization of proteins with Coomassie stain (Bio-Rad). Gel bands (10) were excised and subjected to in-gel reduction, alkylation, and trypsinization as described in Mathivanan et al. (2012). Briefly, gel bands were reduced with 10 mM DTT (Bio-Rad), alkylated with 25 mM iodoacetamide (Sigma) and digested overnight at 37°C with 150 ng of sequencing grade trypsin (Promega). Tryptic peptides were extracted by 0.1% trifluoroacetic acid in 50% (w/v) acetonitrile and analyzed by LC-MS/MS using LTQ Orbitrap Elite (Thermo Scientific), fitted with nanoflow reversed-phase-HPLC (Ultimate 3000 RSLC, Dionex). The nano-HPLC system was equipped with an Acclaim Pepmap nano-trap column (Dionex—C18, 100 Å, 75 μm × 2 cm) and an Acclaim Pepmap RSLC analytical column (Dionex—C18, 100 Å, 75 μm × 50cm). Typically, for each LC-MS/MS experiment, 3 μl of the peptide mix was loaded onto the enrichment (trap) column at an isocratic flow of 5 μl/min of 3% CH3CN containing 0.1% formic acid for 5 min before the enrichment column is switched in-line with the analytical column. The eluents used for the LC were 0.1% v/v formic acid (solvent A) and 100% CH3CN/0.1% formic acid v/v. The gradient used was 3% B to 25% B for 23 min, 25% B to 40% B in 2 min, 40% B to 85% B in 2 min, and maintained at 85% B for 2 min before equilibration for 10 min at 3% B prior to the next injection. The Orbitrap Elite MS was operated in the data-dependent mode with a capillary temperature of 250°C, nano ESI spray voltage of +1.9 kv, and S-lens RF value of 60%. All spectra were acquired in positive mode with full scan MS spectra scanning from m/z 300–1,650 in the FT mode at 240,000 resolution after accumulating to a target value of 1.00e6 and maximum accumulation time of 200 ms. Lockmass of 445.12003 m/z was used. For MS/MS, the 20 most intense peptide ions with minimum target value of 2,000 and charge states ≥2 were isolated with isolation window of 1.6 m/z and fragmented by low energy CID with normalized collision energy of 30 and activation Q of 0.25. A whole cell lysate (WCL) sample, prepared by lysing F. oxysporum f. sp. vasinfectum mycelia using glass beads in a Tissue lyser (Qiagen) and clarifying via centrifugation, was also analyzed using the same method for comparison.
Bioinformatic Analysis of Proteomic Data Set
Mascot Generic File Format (MGF) files were generated using MSConvert with the parameter of peak picking set. X!Tandem VENGEANCE (2015.12.15) was then used to search the MGF files against a target and decoy databases. A predicted protein set for F. oxysporum f. sp. vasinfectum was downloaded from Ensembl Fungi (FO_Cotton_V1, GCA_000260175) and used to generate the target database. Search parameters used were: fixed modification (carboamidomethylation of cysteine; +57 Da), variable modifications (oxidation of methionine; +16 Da and N-terminal acetylation; +42 Da), three missed tryptic cleavages, 20 ppm peptide mass tolerance, and 0.6 Da fragment ion mass tolerance. Protein identifications were shortlisted to obtain a master list with less than 1% false discovery rate. Proteins that were identified by at least two unique peptides in two of three samples were classified as EV proteins.
Predicted functions of F. oxysporum f. sp. vasinfectum ORFs were assigned by homology searching using Blast2Go v4.19 with default parameters (Conesa et al., 2005). Functional enrichment analysis was also performed using Blast2Go with Fisher’s exact test. Statistically significant GO terms that were enriched in EV proteins were condensed further to minimize overlap using the reduced GO terms function in Blast2Go.
Absorbance Spectroscopy of EVs
Absorbance spectroscopy was performed using a SpectraMax m2 spectrophotometer. EVs fractions were loaded into 1 cm pathlength 70 µl UV-Cuvette micro (BRAND, Germany) cuvettes and the absorbance spectra were measured in 10 nm intervals. Data were analyzed using Microsoft Excel.
Leaf Infiltration Assays
Coker 315 cotton seeds were germinated at 25°C in an Adaptis (Conviron) growth cabinet with 16 h light and 8 h dark for 10 days with 5 seeds per squat pot in standard pine park potting mix. Nicotiana benthamiana were grown in an Adaptis (Conviron) growth cabinet with 16 h light and 8 h dark for 4–6 weeks. Infiltrations were performed on the cotton cotyledons or N. benthamiana leaves using a protocol modified from Poon et al. (2018) used for infiltration of Agrobacterium into the leaves of N. benthamiana for transient protein expression. EVs were prepared to a protein concentration of 0.1 µg/ml in 1 ml PBS and infiltrated into the undersides of the cotyledons using a non-luer lock 1 ml syringe (Terumo). Four infiltrations were performed on each leaf. All infiltrations were paired with a PBS control on the second cotyledon of the plant for cotton or on the other half of the leaf for N. benthamiana. Cotton cotyledons were also infiltrated with a solution of hyphae and spores that had been removed from the culture supernatant during EV isolation. Hyphae and spores were washed, resuspended in PBS, and diluted until the solution was slightly turbid (OD 600 nm ~ 0.5) and still flowed easily to facilitate infiltration. After infiltration plants were returned to the growth cabinet for 5 days. After 5 days the cotyledons/leaves were removed from the plants and images of lesions on cotyledons were taken using a Nex-7 camera (Sony).
Optiprep Density Gradient Analysis of EVs
Density gradient ultracentrifugation was performed as described previously with modifications (Kalra et al., 2013). A discontinuous iodixanol gradient consisting of 40% w/v, 20% w/v, 10% w/v, and 5% w/v solutions of iodixanol was prepared by diluting a stock solution of OptiPrep [60% w/v aqueous iodixanol (Sigma)] in 0.25 M sucrose/10 mM Tris, pH 7.5. The crude EVs isolated from the F. oxysporum f. sp. vasinfectum culture by differential centrifugation coupled with ultracentrifugation, were resuspended in the OptiPrep solution, and overlaid onto the top layer. A control tube consisting 3 ml of each 40%, 20%, 10%, and 5% solutions was also prepared. Both tubes were simultaneously subjected to ultracentrifugation at 100,000 × g for 18 h at 4°C (Beckman Coulter: SW-28 rotor). The 12 fractions were collected separately for further analysis. EVs were pelleted by ultracentrifugation at 100,000 × g for 1 h at 4°C (Beckman Coulter: TLA-55 rotor). Pellets were then washed with 1 ml of PBS and the supernatant was removed with two successive ultracentrifugations at 100,000 × g for 1 h at 4°C (Beckman Coulter: TLA-55 rotor) and resuspended in 30 µl before being stored at -80°C.
Results
F. oxysporum f. sp. vasinfectum Produces EVs
EVs were isolated from the supernatant from F. oxysporum f. sp. vasinfectum suspension cultures grown in ½ PDB using differential centrifugation and ultracentrifugation. Unexpectedly the EV fraction had a deep purple color and dual absorbance maximum at 550 and 590 nm (Figure 1A). Nanoparticle tracking analysis was used to determine the number of particles and the distribution of particle diameters (Figure 1B). F. oxysporum f. sp. vasinfectum produced 1.0 x 1012 (st dev 2.8 x 10 11) EVs per mL of culture medium with an average mean particle diameter of 155.1 nm (st dev 3.5 nm) and an average mode of 150.0 nm (st dev 5.1 nm). The average protein concentration of the EV fraction measured by Qubit was 0.05 µg/µl. TEM revealed that the particles had the characteristic cup-like morphology of EVs (Figure 1C). In addition to the cup-shaped particles, there were an equivalent number of multi-lobed, rosette-shaped particles observed in the sample.
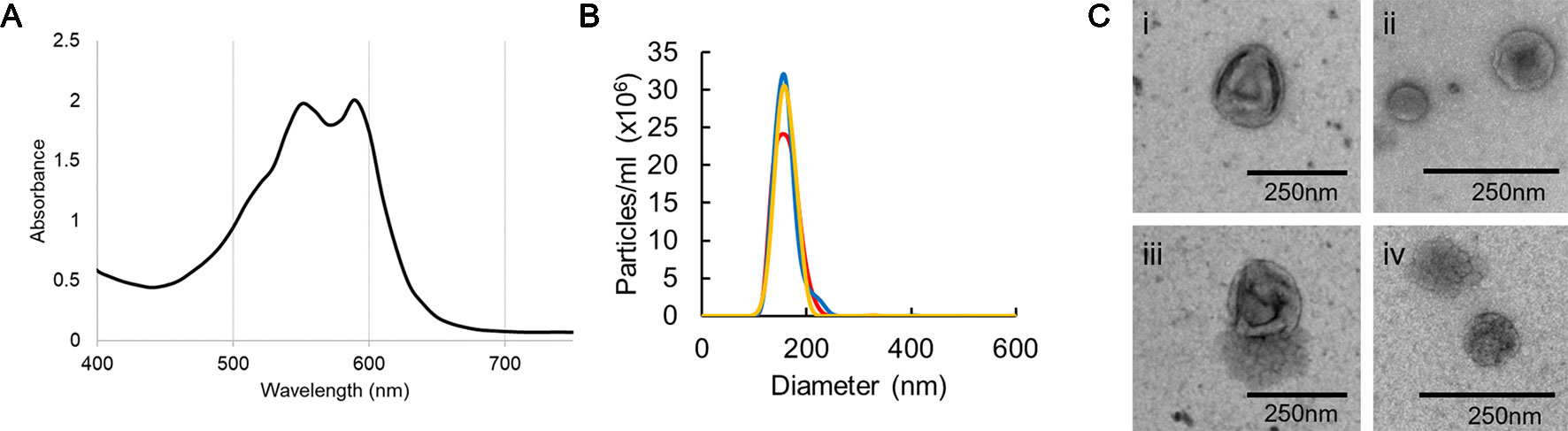
Figure 1 Characterization of EVs isolated from F. oxysporum f. sp. vasinfectum. EVs were isolated using differential centrifugation followed by ultracentrifugation. (A) Absorption spectrum of the F. oxysporum f. sp. vasinfectum EV preparation had dual maxima at 550 and 590 nm. Data are representative of three independent replicates. (B) The number and size distribution of the isolated EVs were determined using nanoparticle tracking analysis. The data from the three technical replicates from one biological sample are representative of all samples used in this study. The average quantity and size of EVs from three independent isolations of EVs from F. oxysporum f. sp. vasinfectum were 1.0 x 1012 ± 2.8 x 1011 particles/ml of culture, mean diameter 155.1 nm ± 3.5 nm, mode diameter 150.0 nm ± 5.1 nm. Values are presented ± standard deviation of three biological replicates. (C) TEM confirmed that most particles had the characteristic cup like morphology of EVs (i–iii); some particles had a multi-lobed rosette morphology as seen in (iv).
F. oxysporum f. sp. vasinfectum EVs Contain Proteins That Function in a Variety of Cellular Processes
The protein cargo of F. oxysporum f. sp. vasinfectum EVs was characterized by tryptic digest followed by LC-MS/MS using an LTQ Orbitrap Elite mass spectrometer. The criteria for positive identification of a protein from the mass spectrometry data set were the detection of at least two unique peptides in at least two of three samples. As a result, we identified list of 482 proteins (Table S1). The most abundant proteins (Table 1) in the EVs included three proteases (FOTG_10608, FOTG_12971, FOTG_06834), two frequency/period clock proteins (FOTG_02405, FOTG_15292), an HSP70 like protein (FOTG_06291), and a polyketide synthase (FOTG_13424) (Table 1). An additional polyketide synthase (FOTG_01390) and a fusarubin cluster-esterase (encoded by FOTG_16162) were also identified but at lower abundance.
Functional enrichment analysis of the EV proteins identified 287 GO terms that were enriched with an adjusted p-value cut-off of 0.05 (Table S2). The enriched GO terms were condensed to remove overlapping terms and give a broader picture of the annotated functions, cellular locations, and biological processes that were overrepresented in the F. oxysporum EVs (Figure 2). A similar enrichment analysis was performed using a proteomic data set from WCL. Basic cellular processes including protein and nucleotide metabolism, lipid biosynthesis, and cell structure were overrepresented EVs. Both the EVs and WCL were enriched for proteins that function in carbohydrate metabolism, the unfolded protein binding, GTP binding, GTPase activity, and ribosomes. Functions such as oxidoreductase activity, oxidative stress response, and vesicle mediated transport were only enriched in EVs. ATP binding was underrepresented in EVs but overrepresented in WCL, whereas DNA-binding transcription factor activity was overrepresented in EVs and underrepresented in WCL.
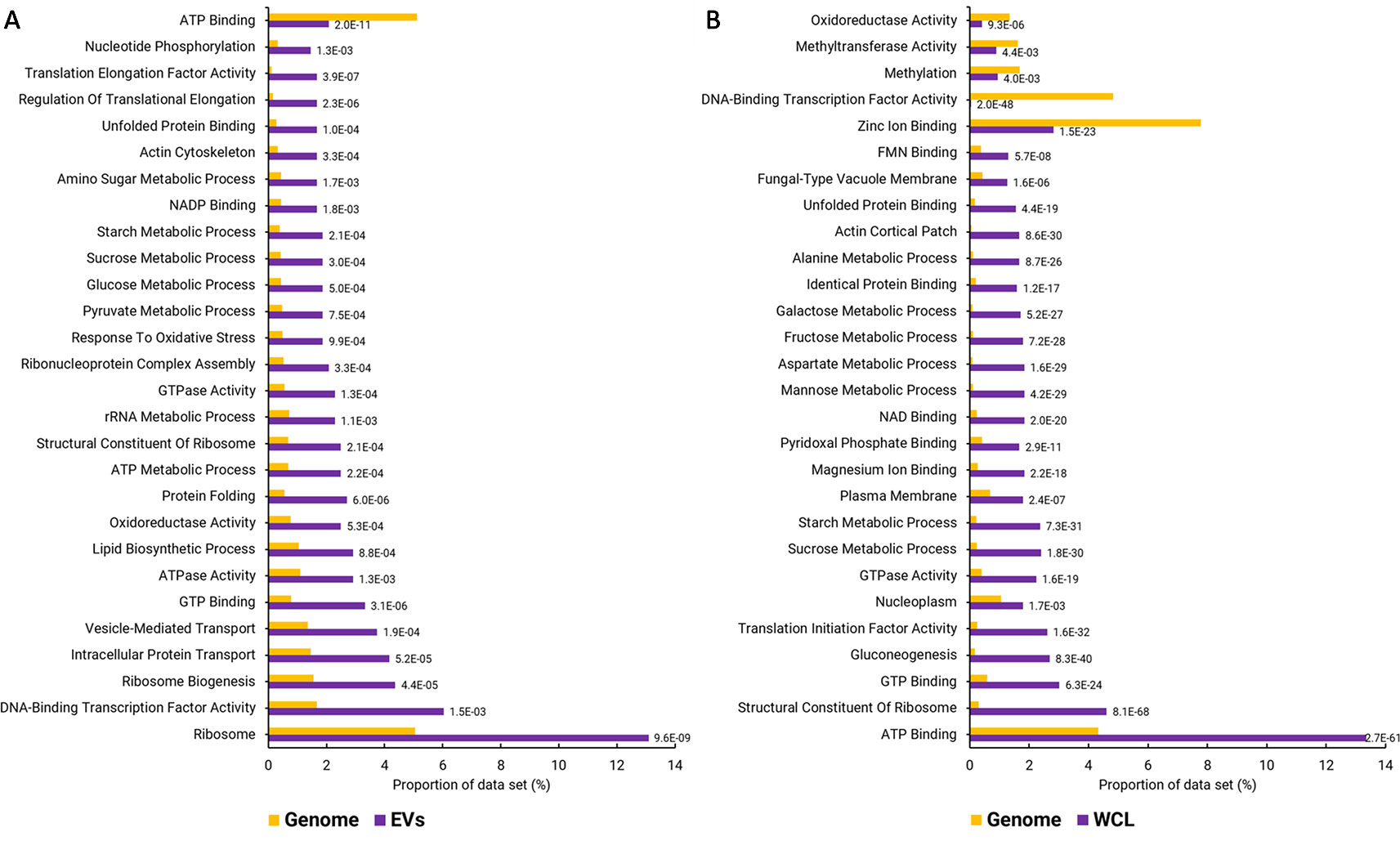
Figure 2 GO terms over- and underrepresented in F. oxysporum f. sp. vasinfectum EV proteins. (A) GO terms that are over- or underrepresented in the set of EV proteins. EV proteins were identified by LC-MS/MS with the peptides aligned to the predicted F. oxysporum f. sp. vasinfectum proteome. A protein was listed as an EV protein if there were at least two unique peptides from the protein identified in at least two of three EV pools. (B) Under- and overrepresented GO terms identified in the list of proteins identified in F. oxysporum f. sp. vasinfectum whole cell lysate. These lists are the top 30 terms; full lists can be found in Tables S2 and S3. GO terms were assigned to transcripts based on homology searches using Blast2Go. GO term enrichment was also performed using Blast2GO. The number next to each bar is the p-value for the GO term.
F. oxysporum f. sp. vasinfectum EVs Are Phytotoxic to Plant Leaves
To determine whether EVs produced by F. oxysporum f. sp. vasinfectum had any effect on plant tissue, EVs were infiltrated into the underside of the cotyledons of cotton seedlings or young leaves of N. benthamiana. After 5 days the F. oxysporum f. sp. vasinfectum EVs induced the formation of a discolored area on the cotyledon or leaf corresponding to the infiltration area, which is indicative of phytotoxicity (Figure 3A). In contrast, there was no discoloration in the infiltration areas of any of the cotyledons or leaves that were infiltrated with sterile PBS demonstrating that the infiltration procedure itself was not responsible for the discolored region. Infiltration of resuspended spores and hyphae led to the formation of discolored spots that were much smaller than those observed for the EVs. To determine whether the toxicity observed in the cotyledon was merely a response to foreign material or if it was specific to components of the F. oxysporum f. sp. vasinfectum EVs, the infiltration experiment was repeated with EVs isolated from the non-pathogenic fungus S. cerevisiae. No discoloration was observed in the regions infiltrated with S. cerevisiae EVs; indeed it was difficult to discern between the S. cerevisiae EV infiltrated and the paired PBS infiltrated cotyledons (Figure 3B). As F. oxysporum f. sp. vasinfectum is a specific pathogen of cotton, the infiltration experiment was repeated on N. benthamiana to determine if the phytotoxic effect was host specific. Infiltration of EVs from F. oxysporum f. sp. vasinfectum was also phytotoxic on N. benthamiana leaves (Figure 3C), indicating that the phytotoxic effect is not host specific. Consistent with observations in cotton, S. cerevisiae EVs did not elicit a response in N. benthamiana leaves (Figure 3D). Infiltration of cotton cotyledons with resuspended spores and hyphae led to a much-subdued phytotoxic effect (Figure 3E), indicating that it is not just a response to fungal material that causes the phytotoxic response in plants but something specific to EVs.
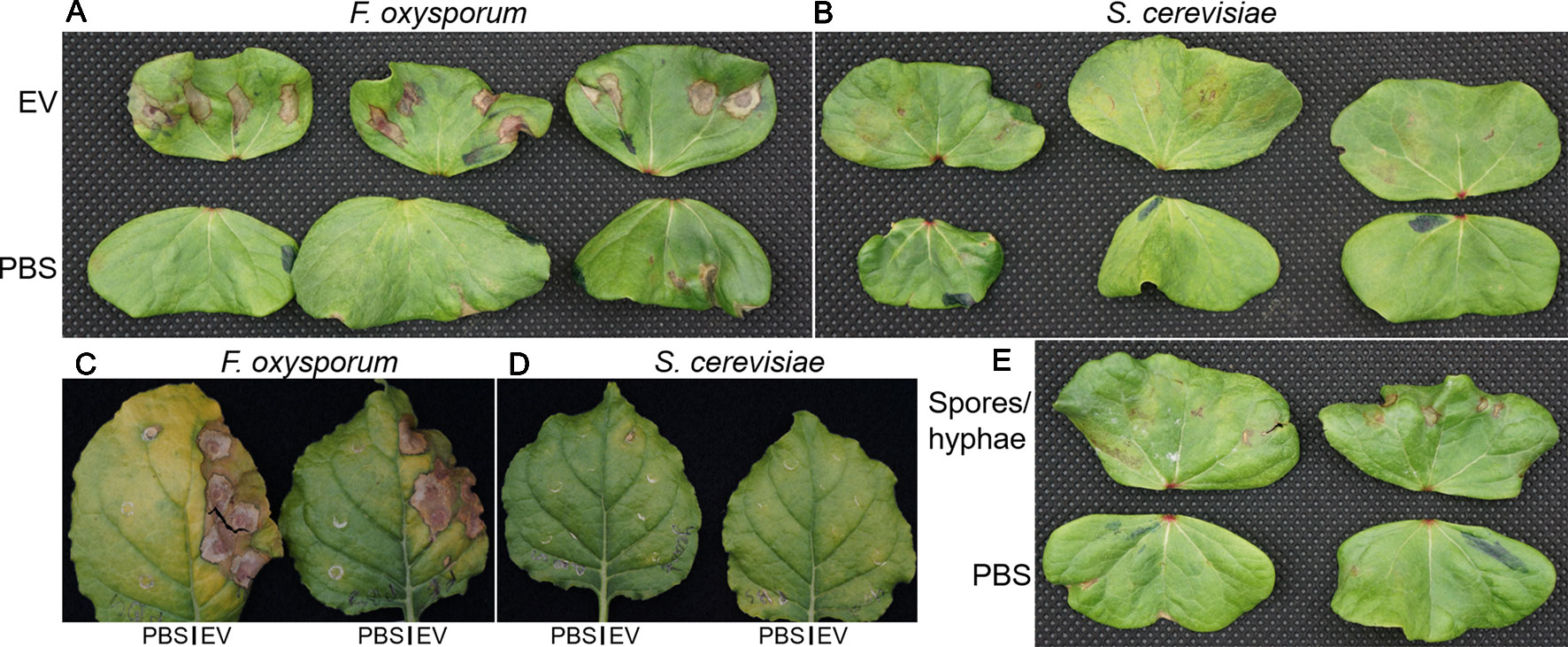
Figure 3 The effect of fungal EVs on cotton cotyledons. EVs were purified from F. oxysporum f. sp. vasinfectum and S. cerevisiae and diluted to a protein concentration of 0.1 µg/ml in PBS. (A, B) Paired cotyledons were infiltrated at four points with the EV solution and PBS control. After 5 days incubation, the cotyledons were removed from the plants and imaged. Cotyledons infiltrated with EVs from F. oxysporum f. sp. vasinfectum (A) had discolored/brown spots around the sites of infiltration, whereas the cotyledons infiltrated with EVs from S. cerevisiae (B) only had mild discoloration or no response. Cotyledons infiltrated with PBS had no observable effect (the brown spot on the third cotyledon in the F. oxysporum panel (A) is due to a pinch point as a result of a fold in the leaf and not infiltration). (C, D) N. benthamiana leaves were infiltrated with EVs at multiple points on the right half and PBS on the left. After 5 days the leaves were removed from the plants and imaged. Leaf halves infiltrated with EVs from F. oxysporum f. sp. vasinfectum (C) showed substantial discoloration around the sites of infiltration, whereas the half infiltrated with PBS had no discoloration. Leaf halves infiltrated with EVs from S. cerevisiae (D) appeared similar to the corresponding PBS infiltrated leave halves. Infiltration of cotton cotyledons with resuspended F. oxysporum f. sp. vasinfectum spores and hyphae (E) led to the formation of very small discolored spots at the sites of infiltration, but they were much smaller than those observed with EVs. Images are representative of three independent experiments.
As there are several different molecules produced by fungi that can induce a cell death response in plants further purification of the F. oxysporum f. sp. vasinfectum EVs by Optiprep density gradient centrifugation was performed. The purple pigment separated into two sections of the Optiprep gradient. The first section spanned fractions 4–7 and a second more condensed section in fraction 10 (Figure 4A). After removal of the iodixanol, a selection of these fractions were infiltrated into cotton cotyledons. Fractions 5, 6, and 7 induced the strongest response and also contained an enrichment of EVs (Figures 4B–D). Co-purification of the pigment with the EVs on the optiprep gradient supports the notion that the pigment is packaged in the EVs. The phytotoxicity of these fractions on cotton plants indicated that EVs and/or the pigment are responsible for initiating the response.
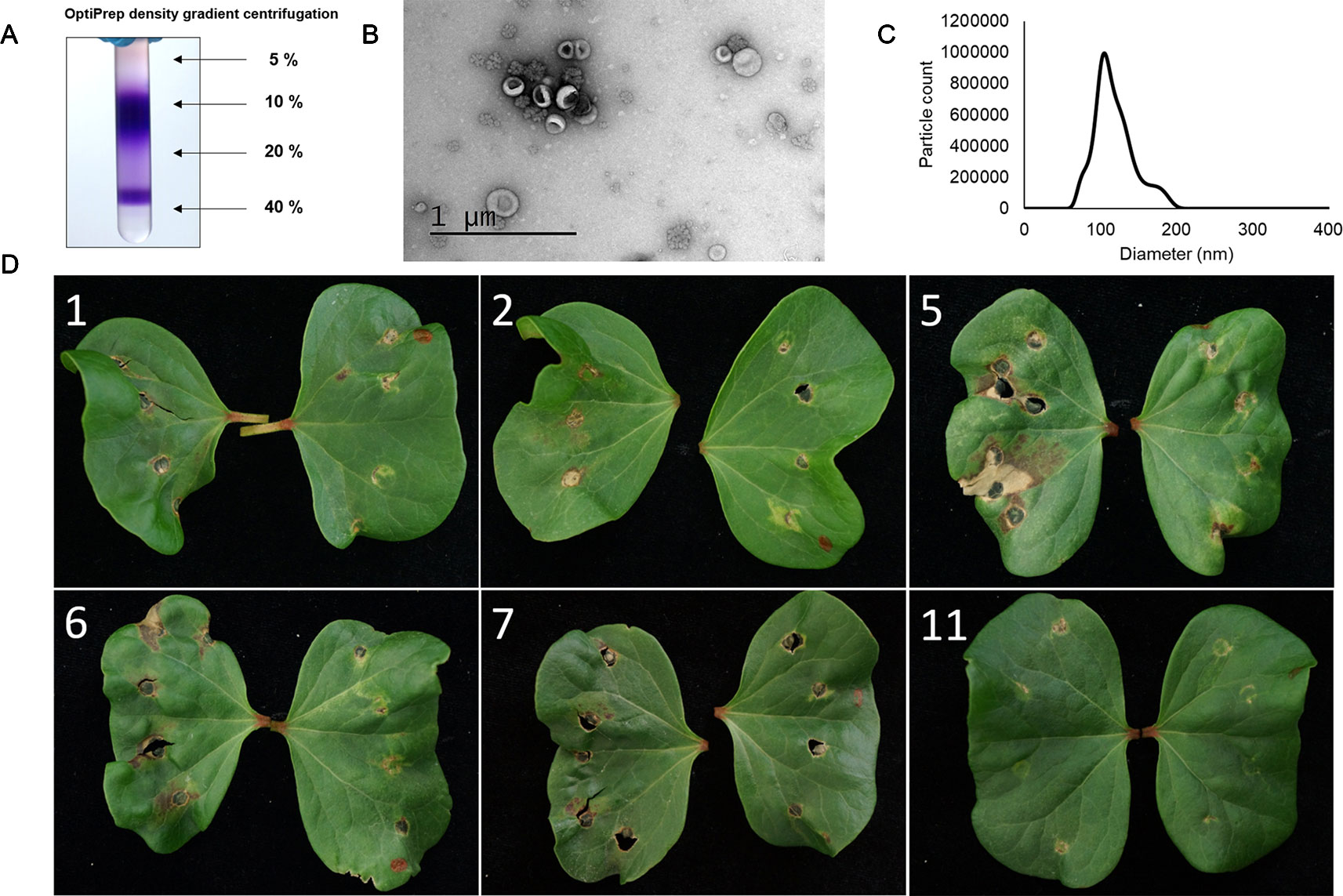
Figure 4 The optiprep fractions containing both EVs and the purple pigment are responsible for the hypersensitive response induced by the crude EV fraction in plants. (A) Optiprep gradient centrifugation of the crude EV fraction revealed that the purple pigment concentrates in two regions, one centered at 10% and a second close to 40% iodixanol. The purple pigment was concentrated in fractions 5–7. (B) Representative TEM of fractions 5–7. Particles with characteristic cup-like morphology of EVs were detected in these fractions as well as some rosette-shaped particles. (C) Representative NTA of fractions 5–7. The mode particle diameter was 104.5 nm; no particles were detected below 46.5 nm or above 226.5 nm. (D) Application of select fractions (numbered in white) to the cotyledons of a cotton plant. Fractions (left cotyledon) were all paired with a PBS control (right cotyledon). Fraction 5 induced the strongest response with fractions 6 and 7 inducing a lesser response. Fractions 1, 2, and 11 did not induce a response beyond that observed for the PBS control. The brown spots and holes on the control cotyledons are due to wounding of the leaf during infiltration.
Discussion
We have isolated EVs from the plant pathogen F. oxysporum f. sp. vasinfectum, characterized their protein cargo, and discovered that they have a phytotoxic effect when applied to the leaves of cotton and N. benthamiana. A unique feature of the EVs isolated from F. oxysporum f. sp. vasinfectum was their deep purple color, which has not been reported for EVs from any other species.
There are no marker proteins for fungal EVs so confirmation that the particles we had isolated via ultracentrifugation were indeed EVs depended on physical characterization. TEM confirmed that the EVs isolated from F. oxysporum f. sp. vasinfectum had the characteristic morphology of EVs from other organisms. NTA confirmed that the population of EVs had a similar size distribution to those reported for C. albicans and C. neoformans that also used NTA for size analysis (Bielska and May, 2019). Larger vesicles (diameter greater than 450 nm) have been detected in C. neoformans, C. albicans, and Malassezia sympodialis EV preparations (Bielska and May, 2019) but were not detected in this analysis. The rosette-shaped particles have been described previously in an isolation of EVs from S. cerevisiae, but their origin and identity remain unknown (Giardina et al., 2014).
Proteomic characterization of EVs from F. oxysporum f. sp. vasinfectum was conducted to elucidate potential functions of these particles in the host–pathogen interaction. Proteomic analysis F. oxysporum f. sp. vasinfectum EVs identified 482 proteins, which is greater than the 205 (or fewer) proteins identified in the early studies on yeast EVs (Rodrigues et al., 2008; Vallejo et al., 2012; Gil-Bona et al., 2014; Wolf et al., 2014; Vargas et al., 2015; Wolf et al., 2015) and lower than the 600 (or greater) reported from S. cerevisiae and C. albicans EVs obtained using similar methods of isolation and proteomic analysis (Zhao et al., 2019; unpublished data from the Anderson lab). This may have arisen from the lower yield of EVs from F. oxysporum f. sp. vasinfectum compared to yeast cultures. Functional enrichment analysis of the F. oxysporum f. sp. vasinfectum EV cargo proteins mostly identified basic cellular metabolic and biosynthetic processes. In addition, there were many proteins with unknown function. Yeast EVs are also enriched for proteins that function in these basal cellular processes as well as proteins that function in pathogenesis, stress responses, and cell wall dynamics (Bleackley et al., 2019). The latter protein groups were not identified in the F. oxysporum f. sp. vasinfectum EVs but this is probably due to the relatively poor annotation of the genome of F. oxysporum f. sp. vasinfectum. It is likely that a subset of the uncharacterized proteins, as well as proteins that have been incorrectly annotated based on sequence homology, have a function in F. oxysporum f. sp. vasinfectum pathogenesis.
An Hsp70 like protein was also found in F. oxysporum f. sp. vasinfectum EVs. Hsp70 was one of the few identified in all previously published proteomic data sets for C. albicans and C. neoformans EVs (reviewed in Bleackley et al., 2019) and is also found in mammalian exosomes (Lancaster and Febbraio, 2005). Hsp70 proteins are molecular chaperones that assist in protein folding and stability (Mayer and Bukau, 2005) that have been proposed as molecules that communicate intercellular stresses through packaging into EVs (De Maio, 2011). HSP70 may prove to be a valuable marker for fungal EVs. Marker proteins in mammalian EVs facilitate analysis of EV purity, that is, from potential contamination from organelle fragments (Lötvall et al., 2014), and the lack of markers for fungal EVs means that this is not currently possible.
One of the most striking observations in this study was the deep purple color of the EVs isolated from F. oxysporum f. sp. vasinfectum. The pigment not only co-purified with the EVs during ultracentrifugation but also co-purified with the EVs during the Optiprep gradient analysis. Packaging of pigments into EVs could represent a conserved secretion mechanism across fungi because melanin, a pigment that has a role in the virulence of C. neoformans, is also packaged into a subset of EVs (Rodrigues et al., 2008). Proteomic analysis of EVs isolated from A. infectoria, which also contain melanin, identified a polyketide synthase involved in biosynthesis of melanin and other pigments as EV cargo (Silva et al., 2013). Fusarium spp., like most fungi, produce a wide variety of secondary metabolites, which include mycotoxins that negatively impact agricultural production as well as molecules that have served as a starting point for development of pharmaceuticals (Brown et al., 2012). The source of the purple color was initially suspected to be a polyketide secondary metabolite similar to purpurfusarin, which was identified from a F. graminearum strain over expressing PGL1 (Frandsen et al., 2016). Purple pigments have since been isolated from F. oxysporum cultures grown on defined minimal dextrose broth (Lebeau et al., 2019). The absorbance spectra of an ethanol extract of lyophilized culture medium was consistent with the dual maxima at 550 and 590 nm of the EVs. These purple pigments were further refined and had typical UV-Vis absorbance profiles for napthoquinones, but specific peaks in the UV spectra indicated that they are not purpurfusarin (Frandsen et al., 2016) or 8-O-methyl anhydrofusarubin (Studt et al., 2012), which are known purple pigments from Fusarium. Bikaverin, a polyketide with antibiotic properties (Limón et al., 2010), was also detected in the extracts containing the pigments (Lebeau et al., 2019); it follows that perhaps bikaverin is also packaged into EVs. Analysis of the small molecule content of EVs is required to further enhance understanding of potential role of EVs in the secretion of polyketides and other fungal metabolites. Identification of the two polyketide synthases and one fusarubin cluster-esterase as EV cargo in this study could point towards EVs as a site of biosynthesis for pigments and/or toxins.
We speculated that F. oxysporum f. sp. vasinfectum EVs had a role in the fungal–plant interaction because the role of EVs in bacterial infections is well described in mammalian systems (Pathirana and Kaparakis-Liaskos, 2016). Furthermore, pathogenic yeasts such as C. albicans, C. neoformans, and Paracoccidioides brasiliensis are able to activate innate immune cells such as macrophages (Oliveira et al., 2010) and dendritic cells (Vargas et al., 2015; Wolf et al., 2015). EVs are also a contributing factor to the hypervirulence phenotype of a strain of C. gattii (Bielska et al., 2018). This led to the hypothesis that EVs would play a role in fungal pathogenesis in plants. EVs from F. oxysporum f. sp. vasinfectum that were infiltrated into cotyledons of cotton or leaves of N. benthamiana were phytotoxic. This toxicity was not merely a response to fungal cell wall polysaccharides or lipids as there was no phytotoxic response to EVs from S. cerevisiae or to intact fungal spores and hyphae from F. oxysporum f. sp. vasinfectum. EVs as the source of the phytotoxicity was further supported by the observation that after further fractionation of the initial EV prep using Optiprep gradient centrifugation, the EV enriched fractions retained phytotoxic activity whereas fractions that did not contain EVs did not. Although the leaf is not the normal route of infection for F. oxysporum f. sp. vasinfectum, the effect of EVs on leaf tissue demonstrates that the EVs do carry phytotoxic compounds. Furthermore, a similar phytotoxic effect was observed the F. oxysporum f. sp. vasinfectum EVs were infiltrated into N. benthamiana, a plant that is not a host for F. oxysporum f. sp. vasinfectum, demonstrates that the EVs contain a phytotoxic compound and it is not a host specific virulence factor that is eliciting this response. This could be related to the pigments and/or other polyketides that are associated with the EVs as these molecules can have a range of detrimental effects on plant cells (Möbius and Hertweck, 2009). Alternatively, EV proteins could be the cause of the phytotoxicity. Several proteases, a family of proteins that contribute to pathogenesis of various plant pathogens (Olivieri et al., 2002; Yike, 2011; Lowe et al., 2015), were packaged into EVs and could play a key role in contributing to the damage caused by the EVs. The phytotoxic effect of EVs during an infection is likely to be less extreme that the one observed here due to the amount of EVs used in these experiments compared to what would be present during an infection. EVs were isolated from a 1 L culture and resuspended in approximately 2 ml of PBS prior to infiltration and exceeds what would be expected to be produced by the fungus at any one point during an infection. However, a fungus would continuously be producing EVs during an infection so the cumulative exposure to EVs would increase over time. The high concentration, single dose of EVs was used to ensure that an effect was observed upon infiltration. Future investigation on the role of EVs in an infection will need to investigate physiologically relevant EV concentrations and use more sensitive techniques to measure the response in the plant.
In summary, F. oxysporum f. sp. vasinfectum produces EVs similar in morphology and size to EVs from other fungal species. These EVs carry protein cargo and pigments that may have a role in pathogenesis. Preliminary data demonstrated that EVs from F. oxysporum f. sp. vasinfectum are phytotoxic to leaves of both its host plant cotton and the unrelated plant N. benthamiana. This may be a response turned on by the plant in recognition of the fungal EVs to limit the spread of infection and may explain why F. oxysporum f. sp. vasinfectum enters the plant through the roots instead of the leaves. Alternatively, EV release could be downregulated during the biotrophic phase of the infection and then turned on when the fungus switches to necrotrophy. Further characterization of EVs from F. oxysporum f. sp. vasinfectum and other agricultural fungal pathogens will improve our understanding of the plant–pathogen interaction and may ultimately lead to identification of new targets to enhance disease protection in crop plants.
Data Availability Statement
All datasets for this study are included in the article/Supplementary Material.
Author Contributions
MB performed infiltration assays, analyzed proteomic data, and wrote the manuscript. MS performed EV isolations and characterization, prepared samples for mass spec, and performed infiltration assays. DG-C performed EV isolations and characterization, prepared samples for mass spec. JM aided in design of infiltration assays and collected images from infiltration assays. RL annotated the proteome and analyzed proteomic data. MP analyzed proteomic data. KZ isolated yeast EVs. C-SA performed proteomic data collection. SM conceived the project, oversaw the experimental work and data analysis. MA conceived the project, oversaw the experimental work and data analysis, and revised the manuscript.
Funding
This work was funded by Australian Research Council grants to MA (DP160100309) and SM (DP170102312 and DE150101777). JM is a Commonwealth Scientific and Industrial Research Organisation Science and Industry Endowment Fund STEM Fellow.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01610/full#supplementary-material
Table S1 | Full list of proteins in EVs.
Table S2 | EV enriched GO terms.
Table S3 | WCL enriched GO terms.
References
An, Q., Ehlers, K., Kogel, K. H., Van Bel, A. J., Hückelhoven, R. (2006). Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 172 (3), 563–576. doi: 10.1111/j.1469-8137.2006.01844.x
Bielska, E., May, R. C. (2019). Extracellular vesicles of human pathogenic fungi. Curr. Opin. Microbiol. 52, 90–99. doi: 10.1016/j.mib.2019.05.007
Bielska, E., Sisquella, M. A., Aldeieg, M., Birch, C., O’Donoghue, E. J., May, R. C. (2018). Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat. Commun. 9 (1), 1556. doi: 10.1038/s41467-018-03991-6
Bitencourt, T. A., Rezende, C. P., Quaresemin, N. R., Moreno, P., Hatanaka, O., Rossi, A., et al. (2018). Extracellular vesicles from the dermatophyte trichophyton interdigitale modulate macrophage and keratinocyte functions. Front. Immunol. 9, 2343. doi: 10.3389/fimmu.2018.02343
Bleackley, M. R., Dawson, C. S., Anderson, M. A. (2019). Fungal extracellular vesicles with a focus on proteomic analysis. Proteomics 19 (8), 1800232. doi: 10.1002/pmic.201800232
Boevink, P. C. (2017). Exchanging missives and missiles: the roles of extracellular vesicles in plant–pathogen interactions. J. Exp. Bot. 68 (20), 5411. doi: 10.1093/jxb/erx369
Bohlenius, H., Morch, S. M., Godfrey, D., Nielsen, M. E., Thordal-Christensen, H. (2010). The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell 22 (11), 3831–3844. doi: 10.1105/tpc.110.078063
Bomberger, J. M., Maceachran, D. P., Coutermarsh, B. A., Ye, S., O’Toole, G. A., Stanton, B. A. (2009). Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5 (4), e1000382. doi: 10.1371/journal.ppat.1000382
Brown, D. W., Butchko, R. A., Baker, S. E., Proctor, R. H. (2012). Phylogenomic and functional domain analysis of polyketide synthases in Fusarium. Fungal Biol. 116 (2), 318–331. doi: 10.1016/j.funbio.2011.12.005
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 (18), 3674–3676. doi: 10.1093/bioinformatics/bti610
da Silva, R. P., Heiss, C., Black, I., Azadi, P., Gerlach, J. Q., Travassos, L. R., et al. (2015). Extracellular vesicles from Paracoccidioides pathogenic species transport polysaccharide and expose ligands for DC-SIGN receptors. Sci. Rep. 5, 14213. doi: 10.1038/srep14213
da Silva, T. A., Roque-Barreira, M. C., Casadevall, A., Almeida, F. (2016). Extracellular vesicles from Paracoccidioides brasiliensis induced M1 polarization in vitro. Sci. Rep. 6, 35867. doi: 10.1038/srep35867
De Maio, A. (2011). Extracellular heat shock proteins, cellular export vesicles, and the Stress Observation System: a form of communication during injury, infection, and cell damage. It is never known how far a controversial finding will go! Dedicated to Ferruccio Ritossa. Cell Stress Chaperones 16 (3), 235–249. doi: 10.1007/s12192-010-0236-4
Ellis, T. N., Leiman, S. A., Kuehn, M. J. (2010). Naturally produced outer membrane vesicles from Pseudomonas aeruginosa Elicit a potent innate immune response via combined sensing of both Lipopolysaccharide and protein components. Infect. Immun. 78 (9), 3822–3831. doi: 10.1128/Iai.00433-10
Frandsen, R. J., Rasmussen, S. A., Knudsen, P. B., Uhlig, S., Petersen, D., Lysøe, E., et al. (2016). Black perithecial pigmentation in Fusarium species is due to the accumulation of 5-deoxybostrycoidin-based melanin. Sci. Rep. 6, 26206. doi: 10.1038/srep26206
Giardina, B. J., Stein, K., Chiang, H.-L. (2014). The endocytosis gene END3 is essential for the glucose-induced rapid decline of small vesicles in the extracellular fraction in Saccharomyces cerevisiae. J Extracell. Vesicles 3 (1), 23497. doi: 10.3402/jev.v3.23497
Gil-Bona, A., Llama-Palacios, A., Parra, C. M., Vivanco, F., Nombela, C., Monteoliva, L., et al. (2014). Proteomics unravels extracellular vesicles as carriers of classical cytoplasmic proteins in Candida albicans. J. Proteome Res. 14 (1), 142–153. doi: 10.1021/pr5007944
Gordon, T. R. (2017). Fusarium oxysporum and the Fusarium wilt syndrome. Ann. Rev. Phytopathol. 55, 23–39. doi: 10.1146/annurev-phyto-080615-095919
Greening, D. W., Simpson, R. J. (2018). Understanding extracellular vesicle diversity–current status. Expert Rev. Proteomic. 15 (11), 887–910. doi: 10.1080/14789450.2018
Griffith, C. L., Klutts, J. S., Zhang, L., Levery, S. B., Doering, T. L. (2004). UDP-glucose dehydrogenase plays multiple roles in the biology of the pathogenic fungus Cryptococcus neoformans. J Biol. Chem. 279 (49), 51669–51676. doi: 10.1074/jbc.M408889200
Horstman, A. L., Kuehn, M. J. (2000). Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275 (17), 12489–12496. doi: 10.1074/jbc.275.17.12489
Kalra, H., Simpson, R. J., Ji, H., Aikawa, E., Altevogt, P., Askenase, P., et al. (2012). Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 10 (12), e1001450. doi: 10.1371/journal.pbio.1001450
Kalra, H., Adda, C. G., Liem, M., Ang, C. S., Mechler, A., Simpson, R. J., et al. (2013). Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 13 (22), 3354–3364. doi: 10.1002/pmic.201300282
Kim, S. W., Park, S. B., Im, S. P., Lee, J. S., Jung, J. W., Gong, T. W., et al. (2018). Outer membrane vesicles from beta-lactam-resistant Escherichia coli enable the survival of beta-lactam-susceptible E. coli in the presence of beta-lactam antibiotics. Sci. Rep. 8 (1), 5402. doi: 10.1038/s41598-018-23656-0
Lancaster, G. I., Febbraio, M. A. (2005). Exosome-dependent trafficking of HSP70 A novel secretory pathway for cellular stress proteins. J Biol. Chem. 280 (24), 23349–23355. doi: 10.1074/jbc.M502017200
Lebeau, J., Petit, T., Clerc, P., Dufossé, L., Caro, Y. (2019). Isolation of two novel purple naphthoquinone pigments concomitant with the bioactive red bikaverin and derivates thereof produced by Fusarium oxysporum. Biotechnol. Prog. 35 (1), e2738. doi: 10.1002/btpr2738
Li, X., Bao, H., Wang, Z., Wang, M., Fan, B., Zhu, C., et al. (2018). Biogenesis and function of multivesicular bodies in plant immunity. Front. Plant Sci. 9, 979. doi: 10.3389/fpls.2018.00979
Limón, M. C., Rodríguez-Ortiz, R., Avalos, J. (2010). Bikaverin production and applications. Appl. Microbiol. Biotechnol. 87 (1), 21–29. doi: 10.1007/s00253-010-2551-1
Liu, M., Bruni, G. O., Taylor, C. M., Zhang, Z., Wang, P. (2018). Comparative genome-wide analysis of extracellular small RNAs from the mucormycosis pathogen Rhizopus delemar. Sci. Rep. 8 (1), 5243. doi: 10.1038/s41598-018-23611-z
Lötvall, J., Hill, A. F., Hochberg, F., Buzás, E. I., Di Vizio, D., Gardiner, C., et al. (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles, Taylor & Francis.
Lowe, R. G. T., McCorkelle, O., Bleackley, M., Collins, C., Faou, P., Mathivanan, S., et al. (2015). Extracellular peptidases of the cereal pathogen Fusarium graminearum. Front. Plant Sci. 6, 962. doi: 10.3389/fpls.2015.00962
Maas, S. L., Breakefield, X. O., Weaver, A. M. (2017). Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. 27 (3), 172–188. doi: 10.1016/j.tcb.2016.11.003
Mathivanan, S., Ji, H., Tauro, B. J., Chen, Y.-S., Simpson, R. J. (2012). Identifying mutated proteins secreted by colon cancer cell lines using mass spectrometry. J. Proteomics 76, 141–149. doi: 10.1016/j.jprot.2012.06.031
Mayer, M., Bukau, B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62 (6), 670. doi: 10.1007/s00018-004-4464-6
Möbius, N., Hertweck, C. (2009). Fungal phytotoxins as mediators of virulence. Curr. Opin. Plant Biol. 12 (4), 390–398. doi: 10.1016/j.pbi.2009.06.004
Oliveira, D. L., Freire-de-Lima, C. G., Nosanchuk, J. D., Casadevall, A., Rodrigues, M. L., Nimrichter, L. (2010). Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Infect. Immun. 78 (4), 1601–1609. doi: 10.1128/IAI.01171-09
Olivieri, F., Zanetti, M. E., Oliva, C. R., Covarrubias, A. A., Casalongué, C. A. (2002). Characterization of an extracellular serine protease of Fusarium eumartii and its action on pathogenesis related proteins. Eur. J. Plant Pathol. 108 (1), 63–72. doi: 10.1023/A:1013920929965
Olofsson, A., Vallstrom, A., Petzold, K., Tegtmeyer, N., Schleucher, J., Carlsson, S., et al. (2010). Biochemical and functional characterization of Helicobacter pylori vesicles. Mol. Microbiol. 77 (6), 1539–1555. doi: 10.1111/j.1365-2958.2010.07307.x
Pathirana, R. D., Kaparakis-Liaskos, M. (2016). Bacterial membrane vesicles: biogenesis, immune regulation and pathogenesis. Cell. Microbiol. 18 (11), 1518–1524. doi: 10.1111/cmi.12658
Poon, S., Harris, K. S., Jackson, M. A., McCorkelle, O. C., Gilding, E. K., Durek, T., et al. (2018). Co-expression of a cyclizing asparaginyl endopeptidase enables efficient production of cyclic peptides in planta. J. Exp. Bot. 69 (3), 633–641. doi: 10.1093/jxb/erx422
Prados-Rosales, R., Weinrick, B. C., Pique, D. G., Jacobs, W. R., Jr., Casadevall, A., Rodriguez, G. M. (2014). Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J. Bacteriol. 196 (6), 1250–1256. doi: 10.1128/JB.01090-13
Regente, M., Pinedo, M., San Clemente, H., Balliau, T., Jamet, E., de la Canal, L. (2017). Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J. Exp. Bot. 68 (20), 5485–5495. doi: 10.1093/jxb/erx355
Rodrigues, M. L., Nimrichter, L., Oliveira, D. L., Frases, S., Miranda, K., Zaragoza, O., et al. (2007). Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryotic Cell 6 (1), 48–59. doi: 10.1128/Ec.00318-06
Rodrigues, M. L., Nakayasu, E. S., Oliveira, D. L., Nimrichter, L., Nosanchuk, J. D., Almeida, I. C., et al. (2008). Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryotic Cell 7 (1), 58–67. doi: 10.1128/EC.00370-07
Rutter, B. D., Innes, R. W. (2017). Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173 (1), 728–741. doi: 10.1104/pp.16.01253
Rutter, B. D., Innes, R. W. (2018). Extracellular vesicles as key mediators of plant–microbe interactions. Curr. Opin. Plant Biol. 44, 16–22. doi: 10.1016/j.pbi.2018.01.008
Rybak, K., Robatzek, S. (2019). Functions of extracellular vesicles in immunity and virulence. Plant Physiol. 179 (4), 1236–1247. doi: 10.1104/pp.18.01557
Samuel, M., Bleackley, M., Anderson, M., Mathivanan, S. (2015). Extracellular vesicles including exosomes in cross kingdom regulation: a viewpoint from plant-fungal interactions. Front. Plant Sci. 6, 766. doi: 10.3389/fpls.2015.00766
Schorey, J. S., Cheng, Y., Singh, P. P., Smith, V. L. (2015). Exosomes and other extracellular vesicles in host–pathogen interactions. EMBO Rep. 16 (1), 24–43.
Silva, B. M., Prados-Rosales, R., Espadas-Moreno, J., Wolf, J. M., Luque-Garcia, J. L., Gonçalves, T., et al. (2013). Characterization of Alternaria infectoria extracellular vesicles. Sabouraudia 52 (2), 202–210. doi: 10.1093/mmy/myt003
Studt, L., Wiemann, P., Kleigrewe, K., Humpf, H.-U., Tudzynski, B. (2012). Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Appl. Environ. Microbiol. 78 (12), 4468–4480. doi: 10.1128/AEM.00823-12
Vallejo, M. C., Nakayasu, E. S., Matsuo, A. L., Sobreira, T. J., Longo, L. V., Ganiko, L., et al. (2012). Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: comparative analysis with other pathogenic fungi. J. Proteome Res. 11 (3), 1676–1685. doi: 10.1021/pr200872s
Vargas, G., Rocha, J. D., Oliveira, D. L., Albuquerque, P. C., Frases, S., Santos, S. S., et al. (2015). Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell. Microbiol. 17 (3), 389–407. doi: 10.1111/cmi.12374
Vasilyeva, N. V., Tsfasman, I. M., Suzina, N. E., Stepnaya, O. A., Kulaev, I. S. (2008). Secretion of bacteriolytic endopeptidase L5 of Lysobacter sp XL1 into the medium by means of outer membrane vesicles. Febs J. 275 (15), 3827–3835. doi: 10.1111/j.1742-4658.2008.06530.x
Wolf, J. M., Espadas-Moreno, J., Luque-Garcia, J. L., Casadevall, A. (2014). Interaction of Cryptococcus neoformans extracellular vesicles with the cell wall. Eukaryotic cell 13 (12), 1484–1493. doi: 10.1128/EC.00111-14
Wolf, J. M., Espadas, J., Luque-Garcia, J., Reynolds, T., Casadevall, A. (2015). Lipid biosynthetic genes affect Candida albicans extracellular vesicle morphology, cargo, and immunostimulatory properties. Eukaryotic Cell 14 (8), 745–754. doi: 10.1128/EC.00054-15
Yike, I. (2011). Fungal proteases and their pathophysiological effects. Mycopathologia 171 (5), 299–323. doi: 10.1007/s11046-010-9386-2
Zarnowski, R., Sanchez, H., Covelli, A. S., Dominguez, E., Jaromin, A., Berhardt, J., et al. (2018). Candida albicans biofilm–induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 16 (10), e2006872. doi: 10.1371/journal.pbio.2006872
Keywords: fungi, extracellular vesicle (EV), host-pathogen interaction, polyketide, pigment
Citation: Bleackley MR, Samuel M, Garcia-Ceron D, McKenna JA, Lowe RGT, Pathan M, Zhao K, Ang C-S, Mathivanan S and Anderson MA (2020) Extracellular Vesicles From the Cotton Pathogen Fusarium oxysporum f. sp. vasinfectum Induce a Phytotoxic Response in Plants. Front. Plant Sci. 10:1610. doi: 10.3389/fpls.2019.01610
Received: 31 July 2019; Accepted: 15 November 2019;
Published: 10 January 2020.
Edited by:
Jan Schirawski, Friedrich Schiller University Jena, GermanyReviewed by:
Wei-Hua Tang, Shanghai Institutes for Biological Sciences (CAS), ChinaLaurence Veronique Bindschedler, University of London, United Kingdom
Copyright © 2020 Bleackley, Samuel, Garcia-Ceron, McKenna, Lowe, Pathan, Zhao, Ang, Mathivanan and Anderson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marilyn A. Anderson, m.anderson@latrobe.edu.au
†Present address: Monisha Samuel, Division of Immunology and Allergy, Department of Medicine Solna, Karolinska University Hospital, Karolinska Institutet, Stockholm, Sweden
‡These authors have contributed equally to this work.
 Mark R. Bleackley
Mark R. Bleackley Monisha Samuel
Monisha Samuel Donovan Garcia-Ceron
Donovan Garcia-Ceron James A. McKenna
James A. McKenna Rohan G. T. Lowe
Rohan G. T. Lowe Mohashin Pathan1
Mohashin Pathan1 Suresh Mathivanan
Suresh Mathivanan Marilyn A. Anderson
Marilyn A. Anderson