- 1Sichuan Province Key Laboratory of Ecological Forestry Engineering on the Upper Reaches of the Yangtze River, College of Forestry, Sichuan Agricultural University, Chengdu, China
- 2College of Landscape Architecture, Sichuan Agricultural University, Chengdu, China
- 3Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, National Engineering Laboratory for Tree Breeding, College of Biological Sciences and Technology, Beijing Forestry University, Beijing, China
Chlorophyll (Chl) plays a crucial role in plant photosynthesis. The geranylgeraniol reductase gene (CHLP) participates in the terminal hydrogenation of chlorophyll biosynthesis. Although there are many studies related to the genome-wide analysis of Populus trichocarpa, little research has been conducted on CHLP family genes, especially those concerning growth and photosynthesis. In this study, three CHLP genes were identified in Populus. The evolutionary tree indicated that the CHLP family genes were divided into six groups. Moreover, one pair of genes was derived from segmental duplications in Populus. Many elements related to growth were detected by cis-acting element analysis of the promoters of diverse PtrCHLPs. Furthermore, PtrCHLPs exhibit different tissue expression patterns. In addition, PtrCHLP3 is preferentially expressed in the leaves and plays an important role in regulating chlorophyll biosynthesis. Silencing of PtrCHLP3 in poplar resulted in a decrease in chlorophyll synthesis in plants, thus blocking electron transport during photosynthesis. Furthermore, inhibition of PtrCHLP3 expression in poplar can inhibit plant growth through the downregulation of photosynthesis. Ultimately, PtrCHLP3 formed a co-expression network with photosynthesis and chlorophyll biosynthesis-related genes, which synergistically affected the growth and photosynthesis of poplars. Thus, this study provides genetic resources for the improved breeding of fast-growing tree traits.
Introduction
Chlorophyll plays a crucial role in photosynthetic organisms such as plants (Flood et al., 2011). Chlorophyll molecules are ubiquitous in these photosynthetic organisms and perform the process of capturing light energy in the antenna system by driving electron transport in the reaction center (Nagata et al., 2005). Chlorophyll a is the major electron donor in the reactive centers of PSI and PSII (Lin et al., 2014). With the development of molecular biology and protein structure, key members of the chlorophyll biosynthesis pathway have been fully explored and identified.
Chlorophyll biosynthesis is a complex process that begins with l-glutamyl-tRNA (Beale, 2005). Several enzymes and 27 genes are involved in this process (Beale, 2005; Nagata et al., 2005). Chlorophyll biosynthesis is a dynamic equilibrium process regulated by several genes, which can affect chlorophyll accumulation, photosynthetic capacity, chloroplast development, and leaf color (Mei et al., 2017; Ohmiya et al., 2019; Zhang et al., 2021). Many yellow-green leaf mutants are also closely related to genes involved in chlorophyll biosynthesis. In lGL1 mutants, the expression levels of some key genes involved in chlorophyll biosynthesis and photosynthesis are significantly changed in rice, such as ChlD, ChlI, Hema1, Ygl1, POR, Cab1R, Cab2R, PsaA, and rbcL (Mei et al., 2017). In rice, the YL-1 gene encodes magnesium protoporphyrin IX monomethyl ester cyclase, which is necessary for chlorophyll biosynthesis and chloroplast membrane stability (Sheng et al., 2017). In addition, it inhibits the expression of HrHEMA, HrPOR, and HrCAO genes in Chrysanthemum, resulting in leaf yellowing and chloroplast structural changes (Zhang et al., 2021).
The final stage of chlorophyll a (Chl a) biosynthesis is the esterification of the tetrapyrrole moiety and chlorophyllide (Havaux et al., 2003). Geranylgeranyl reductase (CHLP, EC: 1.3.1.111), a key enzyme in the final step of Chl a biosynthesis, participates in the terminal hydrogenation step of chlorophyll biosynthesis (Addlesee and Hunter, 2002; Zhang et al., 2015). In the last step of Chl a biosynthesis, chlorophyll synthase (CHL) catalyzes the esterification of chlorophyll (Chlide) with geranylgeranyl diphosphate (GGPP) or phytopyrophosphate (PPP) to form geranylgeranylated Chl a (Chl aGG) or phytylated Chl a, respectively (Shpilyov et al., 2005). In turn, CHLP catalyzes the reduction of free GGPP to PPP, and Chl aGG is reduced to phytyl-chlorophyll a, thus producing chlorophyll a (Addlesee and Hunter, 2002; Shpilyov et al., 2005).
CHLP genes have been identified in many species, including photosynthetic bacteria, algae, tobacco, rice, peaches, olives, and tomatoes (Addlesee and Hunter, 2002; Shpilyov et al., 2005; Deng et al., 2014; Zhang et al., 2015; Tan et al., 2019). Addlesee et al. (1996) discovered that the CHLP of the cyanobacterium Synechocystis sp. PCC 6803 can complement the function of the bchp mutant of R. sphaeroides. In 2005, Alexey studied a chlp mutant of the cyanobacterium Synechocystis sp. PCC 6803 and found that the cell chlorophyll and carotenoid content were decreased in the mutant, and the photosystem became unstable and could not grow autotrophically (Shpilyov et al., 2005). Plant growth was slow after CHLP gene silencing in tobacco, and the leaves showed a pale or mottled phenotype (Tanaka et al., 1999). Some chlp mutants have also been identified in Oryza sativa. The lyl1-1 mutant has a dynamic yellow-green leaf, with reduced chlorophyll content, inhibited chloroplast development, and sensitivity to light (Zhou et al., 2013). Similarly, the growth rate of the yellow-green leaf 502ys mutant was slowed, and the development of chlorophyll and chloroplasts was consistent with that of the lyl1-1 mutant (Wang et al., 2014). Moreover, it was revealed that an O. sativa mutant containing an inactivated CHLP gene could still produce tocopherols (Kimura et al., 2018). CHLP also plays an important role in the abiotic stress response. In tomatoes, CHLP overexpression improves seedling growth and tolerance to salt, osmotic, and oxidative stress by regulating the synthesis of chlorophyll a (Chl a) and tocopherol (TP) (Liu et al., 2015). However, the transcriptional regulatory mechanism of CHLP in plants has rarely been reported.
Recently, woody plants (such as Populus) have been proposed as models for the study of gene function due to their fast-growing properties and clear genomic information (Tuskan et al., 2006; He et al., 2021b,c). However, the role of the poplar CHLP family in plant growth and development has not been reported. Three putative CHLP proteins have been identified in the whole genome of P. trichocarpa. Comparative genomics, transcriptomics, and RT-qPCR were performed to investigate the poplar CHLP family and lay the foundation for studies on the features and functions of CHLPs in poplar growth and development. Moreover, inhibition of PtrCHLP3 expression in poplar can inhibit plant growth through the downregulation of photosynthesis. Furthermore, we investigated the role of PtrCHLP3 in the molecular regulatory networks of photosynthesis and growth. Therefore, this study provides genetic resources and a molecular basis for improved breeding of fast-growing tree traits.
Materials and Methods
Identification and Phylogenetic Analysis of the CHLP Gene Family
The identification and analysis of the CHLP gene family were based on the whole genome of P. trichocarpa. The relevant Populus genomic and protein sequences were downloaded from the Phytozome13 database1 (Goodstein et al., 2011). The other genomic data and protein sequences of Arabidopsis thaliana, Gossypium raimondii, Micromonas pusilla, Chlamydomonas reinhardtii, Triticum aestivum, Prunus persica, Volvox carteri, Oryza sativa, Arabidopsis lyrata, Trifolium pratense, Daucus carota, Phaseolus vulgaris, Populus deltoides, Sorghum bicoloa, Chromochloris zofingiensis, Lactuca sativa, Zea mays, Miscanthus sinensis, Setaria viridis, Botryococcus braunii, Panicum virgatum, Salix purpurea, Gossypium hirsutum, Dioscorea alata, Mimulus guttatus, Thuja plicata, Schrenkiella parvula, Spinacia oleracea, Phaseolus acutifolius, Diptychocarpus strictus, Cleome violacea, Panicum hallii, Zostera marina, Manihot esculenta, Paspalum vaginatum, Fragaria vesca, Glycine max, Solanum tuberosum, Quercus rubra, Solanum lycopersicum, and Hordeum vulgare were also downloaded from the Phytozome13 database. We used conserved domains GG-red-SF (TIGR02032) as seed sequence to search CHLP genes from the above species, the correct CHLP family genes were obtained using the CDD2 database (Lu et al., 2019).
To perform a more integral analysis of the CHLP gene family, all identified CHLP protein sequences were combined and compared using Clustal W, and a genealogical tree was constructed based on the neighbor-joining (NJ method, 1000 bootstraps) in MEGA-X (Hall, 2013). The results of the phylogenetic tree were drawn and modified using FigTree software (v1.4.3). Each CHLP protein was named according to the order in which it was located on a chromosome.
Structure, Conserved Motif, Gene Duplication, Cis-Element, and Expression Patterns Analysis of PtrCHLPs
All bioinformatics analyses of the CHLPs were performed as previously described (He et al., 2021a,b). Structure and conserved motif analysis of the CHLP gene family was performed using the Gene Structure Display Server (GSDS3) (Hu et al., 2014) and Multiple Em for Motif Elicitation (MEME4) online software (Bailey et al., 2015). Gene collinearity analysis of CHLP genes within and between species were performed using the Multiple Collinearity Scan toolkit (MCScanX). Moreover, cis-element analysis of PtrCHLPs were performed using the Plant Cis-Acting Regulatory Element (PlantCARE5) database (Lescot et al., 2002), and corresponding organization expression data was acquired from the Popgenie6 database (Sjodin et al., 2009), then graphics visualization was performed using Tbtools (Chen et al., 2020).
RNA Extraction and RT-qPCR Analysis
The RNA of 84 K poplar (P. alba × P. glandulosa) leaves, stems, roots, buds, phloem, and xylem was extracted using a plant RNA extraction kit (Accurate Biology, Hunan, China) and converted into cDNA using a reverse transcription kit (Accurate Biology, Hunan, China). Primers were designed by Premier 5.0 and synthesized by TSINGKE Biology Co., Ltd. (Chengdu, China). PtrUBQ and PtrActin were used as internal reference genes, and cDNA was used as a template for RT-qPCR amplification (He et al., 2018, 2019). Five biological replicates and four technical replicates were performed for each sample. PCR was performed on a Bio-Rad CFX96 instrument (Bio-Rad, Hercules, United States) and the 2–ΔΔ Ct algorithm was used to analyze the results (Livak and Schmittgen, 2001).
Genetic Transformation of Poplar
First, we cloned PtrCHLP3 from P. trichocarpa, which was subsequently cloned into the pEASY-T1 Kit vector and sequenced. The PtrCHLP3 fragment was amplified from the positive construct PtrCHLP3- pEASY. Double enzyme digestion (SacI/XbaI) was used to retrieve the pCambia2301-PS vector. The forward sequences of partial fragments of PtrCHLP3, sequences of RTM, and reverse sequences of partial fragments of PtrCHLP3 were sequentially recombined into the PCAMbiA2301-PS vector using the ClonExpress® II One Step Cloning Kit (Vazyme Biotech Co., Ltd, Nanjing, China). The correct clone, pCAMBIA2301-PtrCHLP3–RNAi, was introduced into Agrobacterium tumefaciens strain GV3101. The RNAi expression vector was transferred into poplar 84 K using Agrobacteria-mediated transformation methods (He et al., 2019). Transgenic poplar 84 K seedlings were selected in woody plant medium (WPM) containing 60 mg/L kanamycin, and the transformants were confirmed by PCR and stained with β-glucuronidase (GUS). Transgenic poplar lines were obtained and used in subsequent experiments. Related primers (Supplementary Table S1) were designed using Primer- Primer 6.0 tools.
Histochemical Staining of β-Glucuronidase
β-Glucuronidase tissue staining of the plants was performed as previously described (Yang et al., 2020). Histochemical staining of the leaves was performed using the M5 HiPer GUS staining kit (Mei5bio, Beijing, China). The prepared leaves were soaked in GUS dye solution overnight. The leaves were then treated with 70% ethanol 2–3 times until the negative control material was white.
Plant Materials and Growth Conditions
Wild type (WT) and transgenic poplar lines (chlp-3 and chlp-4) were the present study. Thirty-day-old plantlets were transplanted to pots and grown in a greenhouse (20–25°C; 70% humidity; 16 h light, 8 h dark) at Wenjiang, Chengdu, China (30°70’N, 103°85’E, 537.11 m above sea level) (He et al., 2020). All plants were cultivated in the same-sized pots with trays. After the plants grew normally in the greenhouse for 1 month, follow-up experiments were conducted.
Physiological and Biochemical Analysis
The plant height and stem elongation rate between WT and transgenic poplar were measured every 7 days, and the fresh weight and ground diameter of the plants were measured after 35 days of harvest. Measurement and calculation methods of plant height and stem, and stem elongation rates were based on published papers (Wang et al., 2016; He et al., 2018). Each sample was assayed in ten biological replicates. The ninth to eleventh leaves of the WT and transgenic poplar were measured using the LI-6800 Portable Photosynthesis System (LI-COR companies, Lincoln, United States). The light intensity and CO2 concentration were set as 800 μmol m–2 s–1 and 400 μmol mol–1, respectively, in the leaf chamber to measure the leaf net photosynthetic rate (Pn), transpiration rate (Tr), stomatal conductance (Gs), and instantaneous water use efficiency (WUE, Pn/Tr). Photosynthetic light response curves were determined at photosynthetically active radiation (PAR) levels of 1600, 1200, 1000, 800, 600, 400, 200, 100, 75, 50, 25, and 0 μmol m–2 s–1 to measure Pn (He et al., 2018). In addition, a non-rectangular hyperbolic model was used to calculate the Pn–light curve and obtain the initial quantum yield and Pmax (Thornley, 1976).
Chlorophyll content was detected in the terminal bud and leaves 1–8 (from top to bottom) of poplars using a portable chlorophyll meter (SPAD-502Plus, Konica Minolta, Osaka, Japan). Chlorophyll and carotenoids were extracted from fresh leaves using 80% acetone. The total chlorophyll (Chl), chlorophyll a (Chl a), chlorophyll b (Chl b), and carotenoid contents were measured using a spectrophotometer (UV-3800, Unico, Shanghai, China) and quantified according to the methods described by Arnon (1949). The parameters of the slow dynamic fluorescence curves, including the maximal PSII quantum yield (Fv/Fm), quantum yield of photochemical energy conversion in PSII [Y(II]), photochemical quenching parameter (qP), and non-photochemical quenching parameter (NPQ), were measured as previously described (He et al., 2018). Each parameter was measured in at least 40 replicates, including four technical repeats and ten biological repeats.
Tissue Section and Staining Analysis
Toluidine blue and phloroglucinol-HCl staining were performed as previously described (Gui et al., 2019), with slight modifications. Toluidine blue and phloroglucinol-HCl were used only to show the cell profile and the distribution and size of tissues in various parts of the poplar stem. The mature leaves of WT and transgenic poplars were located near the 8th internode. Transverse sections of the 8th internode of the WT and transgenic poplar were obtained using a semi-automatic frozen slicer. The cells were stained with 10% toluidine blue stain and 1% phloroglucinol (w/v) in 25% HCl and observed under a microscope (Olympus BX51, Tokyo, Japan), respectively. In addition, the number of cell layers and the proportion of xylem and bark in the cell cross-section were counted. Each parameter was repeated at least a hundred and twenty times, including three technical and 40 biological repeats.
Bioinformatics Analysis of Possible Upstream Transcription Factors and Co-expressed Gene of PtrCHLPs
The possible upstream transcription factors (PUTFs) for PtrCHLP3 were identified using the Plant Transcriptional Regulatory Map database (PlantRegMap7) (Tian et al., 2019). Moreover, the tissue expression of PUTF genes was obtained from the Popgenie database, and the co-expressed gene expression of PtrCHLP3 was obtained from Phytozome13 databases. Heat map expression patterns were drawn using TBTools (Chen et al., 2020).
Statistical Analyses
Microsoft Excel 2019 was used to calculate the average and standard error of all data. Moreover, Student’s t-test in SPSS software (v26.0) was applied to analyze the significance of the differences between control and treatment groups, and P-values were used to determine significant differences (*P < 0.05; **P < 0.01). Duncan’s multiple comparisons post hoc test was used to determine P-values among different poplar tissues. All the data were normalized and conformed to a normal distribution.
Results
Analysis of Phylogenetic Tree, Gene Structure, Protein Motif, and Conservative Domain of the CHLP Family
The 82 amino acid sequences of CHLP from 42 plants were used to establish a biogenetic tree to further investigate the evolution and discrepancy of CHLP family proteins between poplar and other plants (Figure 1A). All CHLP genes were divided into six groups (I–VI), and three CHLP genes were identified in poplar. PtrCHLP proteins were found in groups I and IV. The phylogenetic tree indicated that the CHLP genes of P. trichocarpa and S. purpurea were close to each other, indicating high homology. Statistical analysis of the CHLP genes in 42 plant species showed that 85.7% of the plants had no more than two CHLP genes (Figure 1B and Supplementary Table S2). Surprisingly, only one CHLP gene was found in the model plant Arabidopsis, whereas a maximum of five CHLP genes were found in L. sativa. Generally, the CHLP gene family differentiates and multiplies slowly during plant evolution.
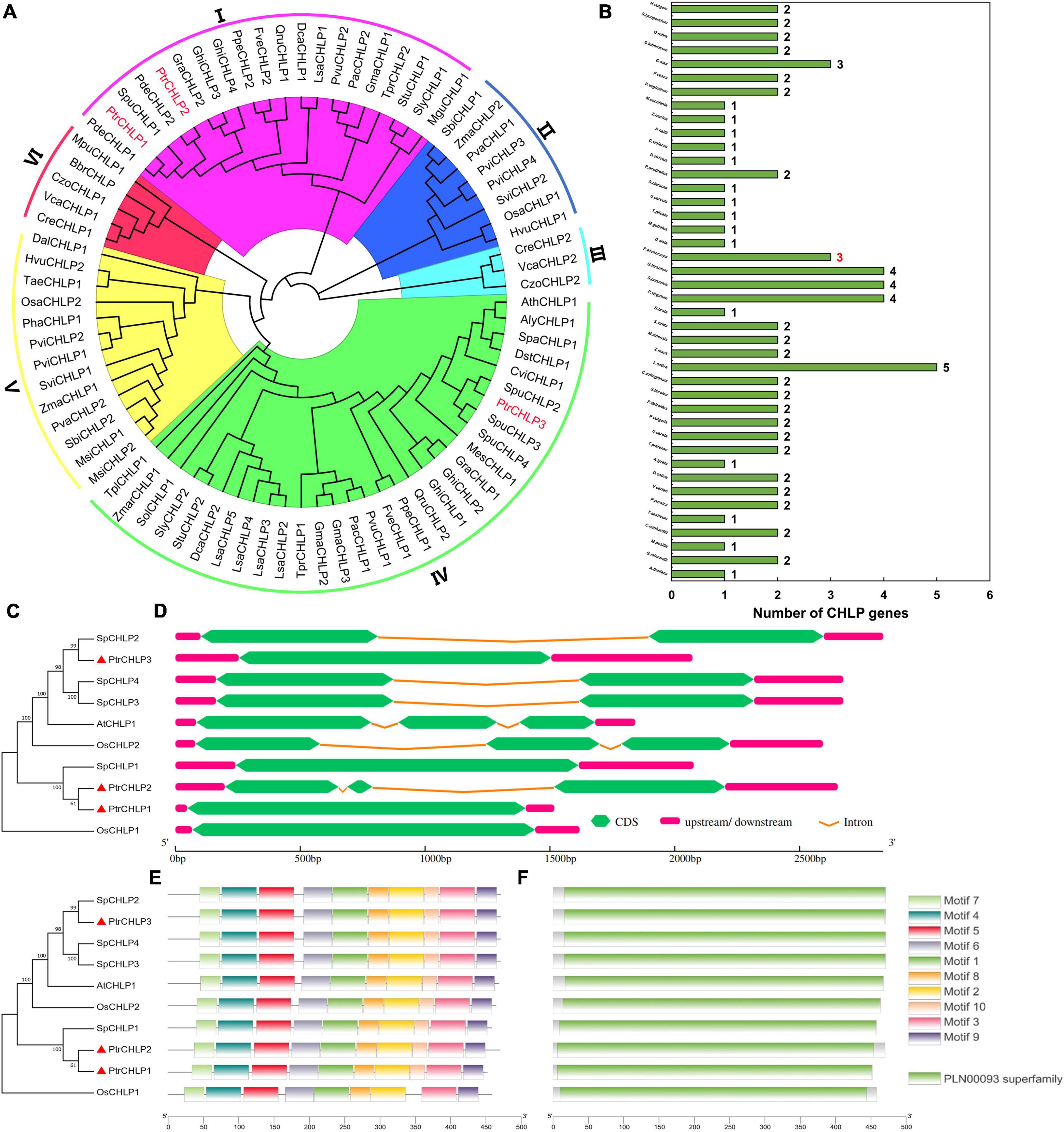
Figure 1. The phylogenetic tree and molecular structure analyses of PtrCHLP. (A) Evolutionary and phylogenetic analysis of the CHLP family among different plants. (B) Comparisons of CHLP protein numbers across 42 plant species. The accession numbers and gene names are all shown in Supplementary Table S2. The phylogenetic tree (C), gene structure (D), conserved motif (E), and conserved protein structure (F) analyses of CHLP genes from A. thaliana, O. sativa, and P. trichocarpa. The length of each model is exhibited in proportion.
To further analyze the phylogenetic relationships among the diverse members of the CHLP family, an evolutionary tree was constructed based on ten CHLP proteins from A. thaliana, O. sativa, S. purpurea, and P. trichocarpa (Figure 1C); and their gene structures, motifs, domains, and transmembrane domains were predicted. The phylogenetic tree showed that PtrCHLP1 and PtrCHLP2 clustered on the same branch, but were far from PtrCHLP3, suggesting that there may be functional differentiation between PtrCHLP1/2 and PtrCHLP3. In addition, PtrCHLP3, SpCHLP2/3/4, OsCHLP2, and AtCHLP1 were clustered in the same clade, suggesting that they may have similar biological functions. Most CHLP genes had one or two introns, whereas PtrCHLP1/3, SpCHLP1, and OsCHLP1 had no introns (Figure 1D). Moreover, one geranylgeranyl diphosphate reductase domain (PLAN00093 Superfamily) was discovered in the PtrCHLP protein (Figure 1F and Supplementary Table S3) and was made up of motif (1–10) components (Figure 1E and Supplementary Table S4). However, the C-terminus of OsCHLP1 lacks Motif10. The CHLP family is highly evolutionarily conserved.
Analysis of Chromosomal Location and Gene Duplication
The chromosomal position, collinearity, and evolution of CHLP genes in poplar were investigated to explore the phylogenetic relationship between CHLP family genes. The PtrCHLPs gene was located on chromosomes 4, 9, and 12 (Figure 2A and Supplementary Table S5). Chromosome regions of less than 200 kb of two or more genes were deemed tandem doubling events (Xie et al., 2018). The PtrCHLP gene family does not form tandem repeat regions on poplar chromosomes. However, two genes were represented by complex segmental duplication events (PtrCHLP1/2) using the MCScanX method (Singh et al., 2017). In summary, duplicated gene pairs (PtrCHLP1/2) were highly homologous, implying that they may jointly regulate the same biological pathways.
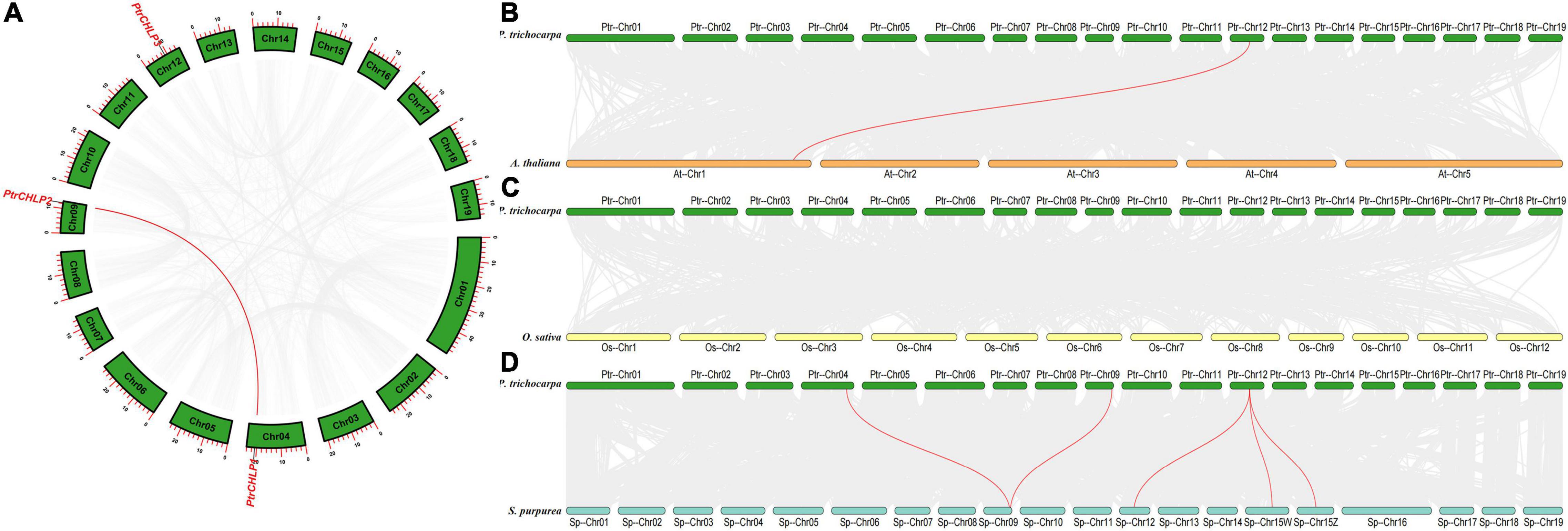
Figure 2. Synteny analysis of poplar CHLP gene family. (A) The synteny analysis of CHLP genes in P. trichocarpa. (B–D) Synteny analysis of CHLP genes between poplar and the other three typical plant species (A. thaliana, O. sativa, and S. purpurea). The synteny blocks in poplar and the other species are shown in gray lines, whereas the collinearity of CHLP gene pairs is emphasized in red lines from different species.
We compared collinear graphs among P. trichocarpa, A. thaliana, O. sativa, and S. purpurea to detect the evolutionary relationship of the CHLP gene in these species (Figures 2B–D). The collinear graphs showed that five pairs of isogenous genes were found between poplar and willow, and one pair of isogenous genes (PtrCHLP3 vs. AtCHLP1) was identified between poplar and A. thaliana (Figure 2B and Supplementary Table S5). The PtrCHLP3 genes were found to have a relationship with three isogenous gene (SpCHLP2/3/4) pairs in willow, suggesting that this gene has undergone a lot of differentiation in willows. Nevertheless, there are no CHLP homologous genes between poplar and rice, implying that many CHLP genes may be newly differentiated in dicotyledons (Figure 2C).
Cis-Element Analysis and Expression Patterns of PtrCHLPs in Poplar
The upstream sequences (∼2,000 bp) of the promoter were obtained from the genomic sequence of poplar to confirm the expression features of PtrCHLP1/2/3. The PlantCARE service was explored for cis-elements in the PtrCHLP promoter (Figure 3A). The detailed effects of these motifs (cis-elements) are listed in Supplementary Table S6. These elements are involved in plant growth and development, phytohormone responsiveness, and abiotic and biotic stress responses. All kinds of cis-acting elements related to development were explored, including the auxin responsiveness element, endosperm expression, gibberellin responsiveness element, and seed-specific regulation, all of which indicated the crucial role of cis-acting elements in poplar development of CHLP.
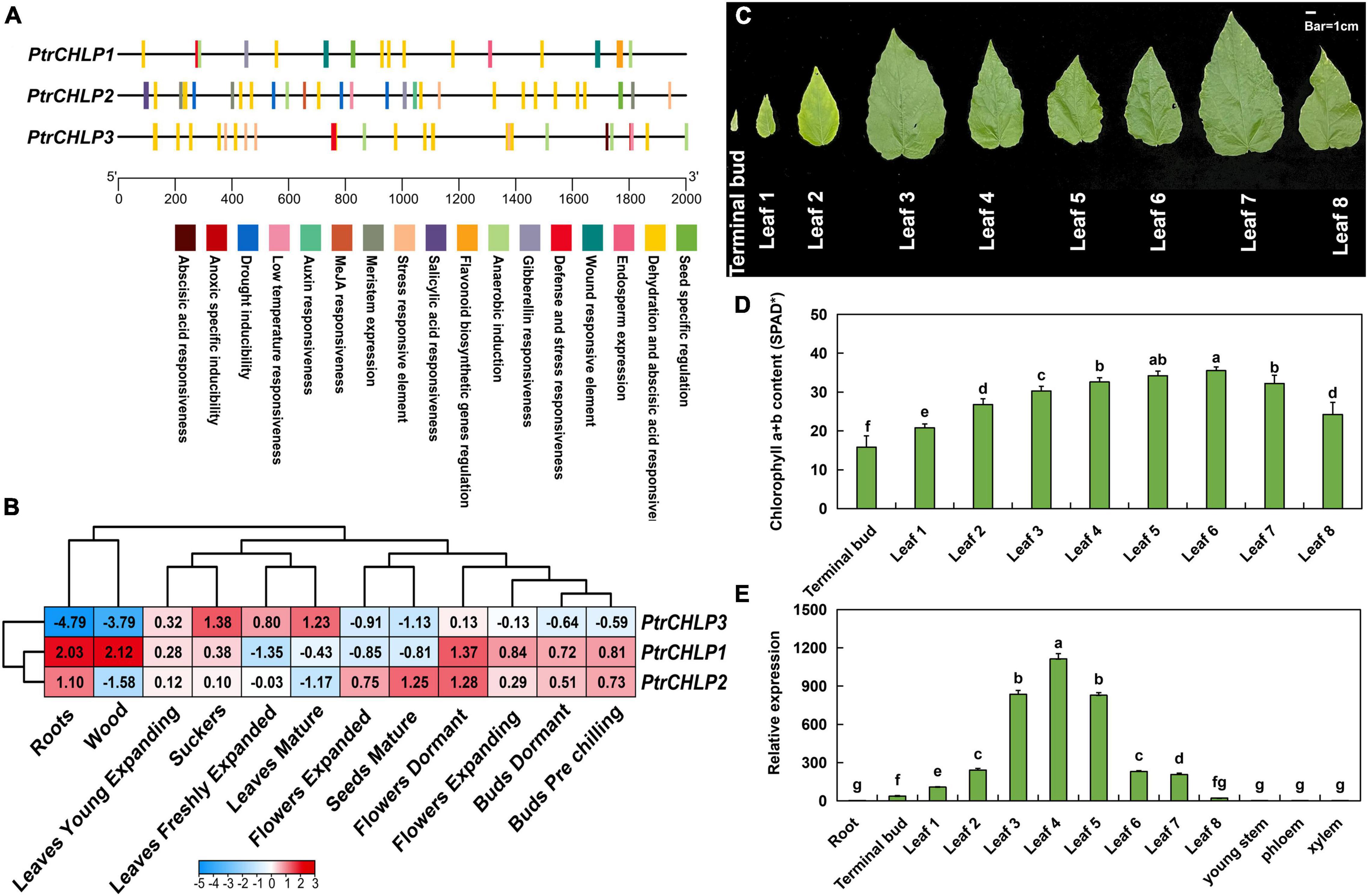
Figure 3. Analysis of cis-acting regulatory elements and expression patterns of CHLPs in Populus. (A) Analysis of cis-acting regulatory elements of PtrCHLPs. The corresponding positions of cis-elements correlated with stress and growth in PtrCHLP promoter regions. Diverse colors indicate various cis-acting elements, and their regions conform to the homologous region of the promoter. (B) Heat maps represent the expression of PtrCHLP1-3 in different plant tissues at various growth stages. The color bars indicate the range of fold change in expression. The phenotype (C) and chlorophyll content (D) analysis of 30-day-old poplar leaves from top to base. (E) RT-qPCR analysis of the expression features of PtrCHLPs in different tissues. The bars indicate mean ± SD (n = 20) from five independent trials. Different letters above the bars indicate statistically significant differences (adjusted P < 0.05, one-way ANOVA).
To further study the function of CHLP genes in growth and development, we investigated the tissue expression profiles of PtrCHLP1/2/3 in transcriptome data (Figure 3B and Supplementary Table S7). PtrCHLP1/2 showed highly similar expression patterns in tissues other than wood, expanded flowers, and mature seeds. However, the expression pattern of PtrCHLP3 was the opposite to that of PtrCHLP1/2, which was upregulated in photosynthetic organs (leaves) but downregulated in non-photosynthetic organs, especially in roots and xylem. Therefore, we hypothesized that PtrCHLP3 might be involved in chlorophyll biosynthesis in poplars. To better determine the function of these genes, we compared the absolute expression levels of three PtrCHLP1/2/3 genes in different tissues. The results showed that the absolute expression level of PtrCHLP3 was the highest in mature leaves, and the tissue expression model was similar to the relative expression pattern (Supplementary Figure S3).
One-month-old poplar leaves were analyzed from top to bottom to determine the phenotype and chlorophyll content. From top to bottom, the leaves changed color from pale green to green, and then to yellow (Figure 3C). Similarly, chlorophyll content first increased and then decreased, reaching the highest content in the sixth leaf (Figure 3D). Furthermore, the tissue expression pattern of poplar was analyzed using RT-qPCR (Figure 3E), and the relative expression of PtrCHLP3 in the photosynthetic organs (buds and leaves) of poplar was relatively high. In contrast, its expression levels were lower in the non-photosynthetic tissues. Overall, this tissue-specific expression suggests that PtrCHLP3 may be involved in chlorophyll biosynthesis in poplars.
Acquisition and Phenotypic Analysis of Transgenic Poplar
To investigate whether PtrCHLP3 affects chlorophyll biosynthesis in plants, the correct recombinant pCAMBIA2301-PtrCHLP3–RNAi vector was inserted into the poplar 84 K genome (Figure 4A). At least nine independently regenerated kanamycin-resistant lines were acquired, and nine transgenic lines (L1–L9) were detected using PCR analysis. We detected the predicted 500 bp band in the nine transgenic lines (Figure 4B). Furthermore, transgenic poplars were verified by GUS staining (Figure 4C). The relative expression levels of PtrCHLP3 in these transgenic lines were significantly lower than those in the WT, indicating that the pCAMBIA2301-PtrCHLP3–RNAi vector inhibited the expression of PtrCHLP3 in poplar 84 K. Because the transcript abundance of two independent transgenic lines (L3 and L4) was lower than that of the other lines (Figure 4D), chlp3 and chlp4 were selected for subsequent experiments.
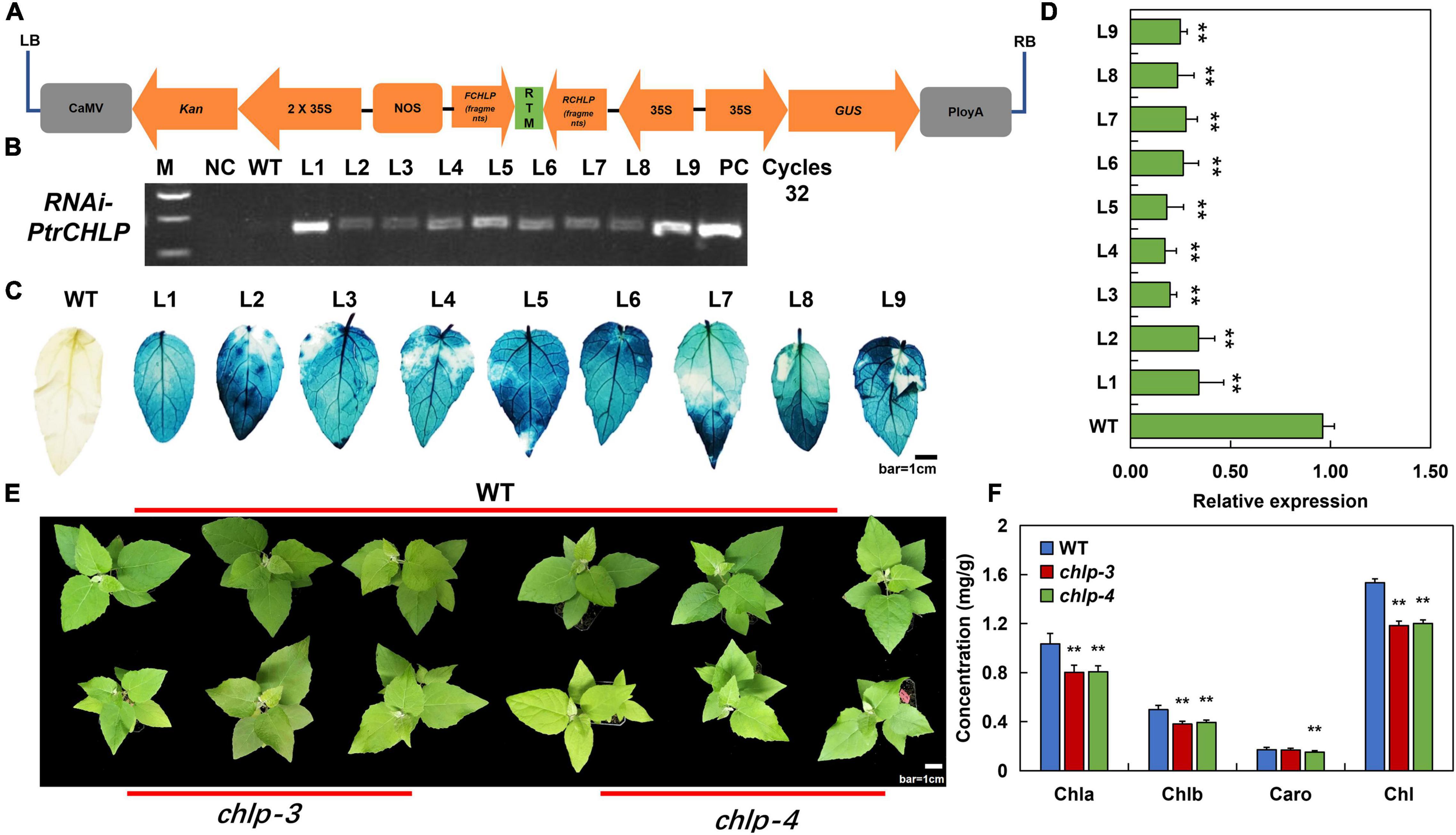
Figure 4. Analysis of the transgenic poplar plants suppressing PtrCHLP3. (A) Diagrammatic drawing of pCAMBIA2301-PtrCHLP3–RNAi vector. Expression of dsRNA-PtrCHLP3 is activated by the double 35S promoter. (B) PCR analysis of transgenic poplar: M, DNA molecular weight marker; PC, p2301-PtrCHLP–RNAi positive control vector; WT, wild-type; NC, negative control; L1-9, PCR products with genomic DNA from regenerated kanamycin-resistant poplar 84 K leaves as a template. (C) Histochemical GUS staining of different transgenic lines. Bar = 1 cm. (D) RT-qPCR analysis of PtrCHLP3 expression levels in different transgenic lines (chlp1–9). The phenotype (E) and chlorophyll content (F) of WT and PtrCHLP3–RNAi lines (chlp3 and chlp4). Bar = 1 cm. The value represents the mean ± standard deviation (n = 20). Asterisks indicate a significant difference at the same level: ** indicates P < 0.01.
In addition, phenotypic analysis showed that the leaf color of chlp3 and chlp4 was significantly yellow compared to that of the WT (Figure 4E). Meanwhile, chlorophyll content analysis indicated that the contents of chlorophyll a, chlorophyll b, and total chlorophyll in the leaves of transgenic plants were significantly lower than those in the leaves of WT (Figure 4F). In summary, the suppression of PtrCHLP3 expression may lead to the downregulation of chlorophyll levels in plants
PtrCHLP3 Involved in Poplar Growth
To further study the effects of PtrCHLP3 on the growth and development of poplar, WT and transgenic poplars were observed for 35 days under normal conditions. Phenotypic analysis showed that the height of the aboveground part and root length of transgenic poplar were significantly inhibited compared with those of WT (Figure 5A). With the time of growth, the plant height and stem elongation rate of WT plants were significantly higher than those of the transgenic plants (Figures 5B,D). Furthermore, the fresh weights of the transgenic poplar roots, stems, and leaves were significantly lower than those of the WT, and correspondingly decreased by 30–75%, 42–84%, and 46–82%, respectively (Figure 5C). In contrast to the WT, the stem diameter of the transgenic plants was significantly smaller (Figure 5E).
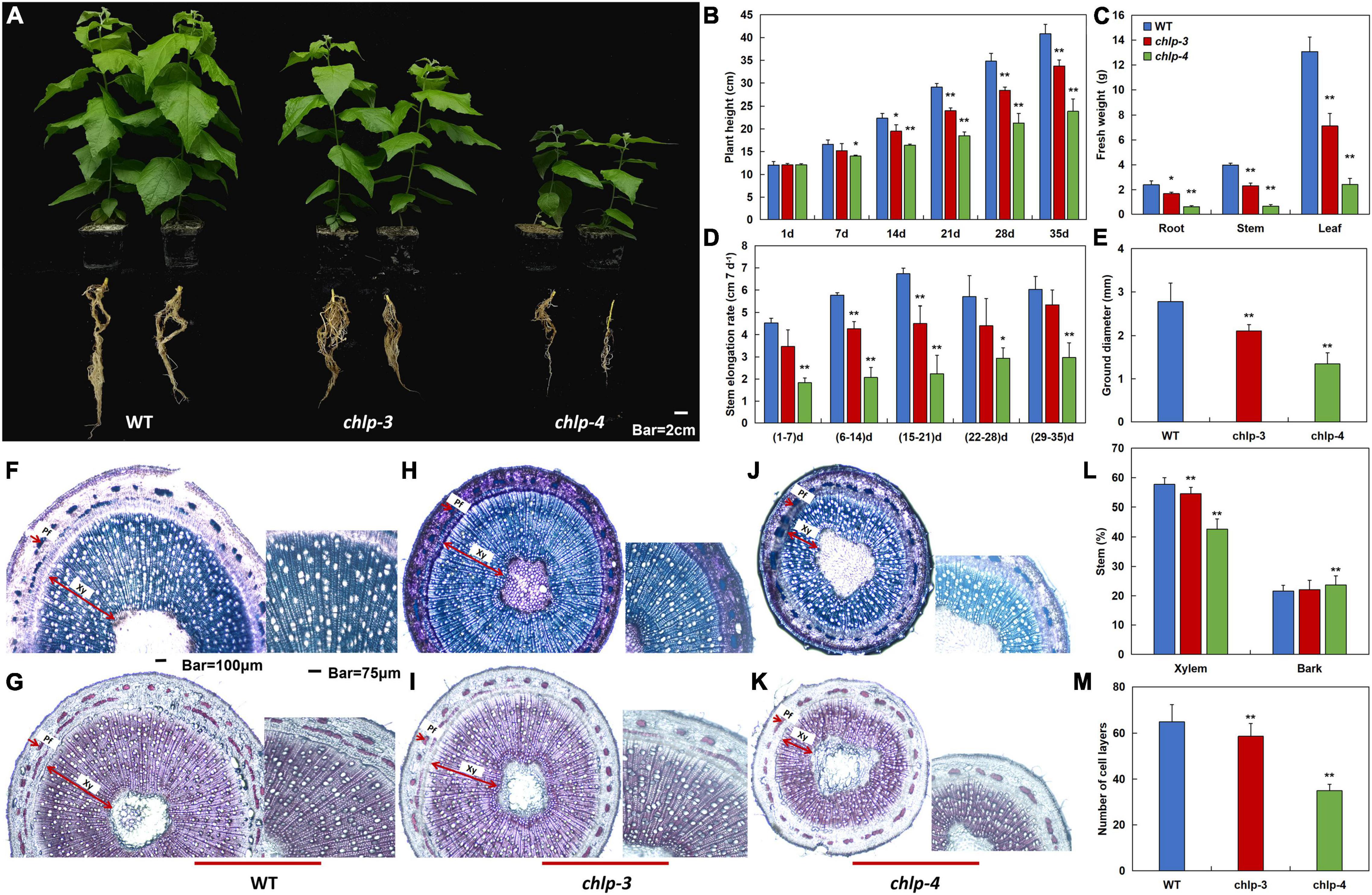
Figure 5. PtrCHLP3 positively regulates poplar growth. (A) Scrubby appearance of 35-day-old plants of independent PtrCHLP3-RNAi transgenic lines (chlp3 and 4). (B–E) The values of plant height (B), fresh weight (C), stem elongation rate (D), and stem diameter (E) of WT and chlp plants. (F–K) Transection and staining via toluidine blue (F,H,J) and phloroglucinol-HCl (G,I,K) of the 8th internode of WT and chlp plants. Pf, phloem fibers; Xy, xylem. (L) Secondary xylem (%) and bark (%) in stem between WT and chlp poplars. The scope of secondary xylem, bark, and total stem was counted using ImageJ in toluidine blue- and phloroglucinol-stained anatomical segments of the 8th internode of WT and chlp poplars. (M) Quantification of secondary xylem cell layers in WT and chlp poplars. The amount of secondary xylem cell layers was calculated in toluidine blue- and phloroglucinol-stained anatomical segments of the 8th internode of WT and chlp plants. The value represents the mean ± standard deviation (n = 120). Asterisks indicate a significant difference at the same level: ** indicates P < 0.01.
To further understand how PtrCHLP3 affects the thickness of poplar stems, toluidine blue and phloroglucinol-HCl were applied to dye a cross-section of the 8th internode of WT and chlp plants (Figures 5F–K). Phloroglucinol-HCl staining was performed to visualize lignin in the cell walls (red violet). In wild-type and transgenic plants, we observed no difference in the deposition of lignin in xylem, phloem fiber, and pith cells (Figures 5G,I,K). The widths of the phloem and xylem of the transgenic poplar were significantly thinner than those of the WT. In addition, compared to WT, the proportion of transgenic xylem in the stems was significantly reduced (Figure 5L). However, there was no significant difference in the bark ratio between transgenic and WT stems. Consistent with previous results, quantification of secondary xylem cell layers showed that xylem cell layers of transgenic plants were significantly smaller than those of WT plants (Figure 5M). These results suggest that the suppression of PtrCHLP3 expression can inhibit xylem expansion in poplars. In conclusion, PtrCHLP3 plays an important role in regulating poplar growth.
PtrCHLP3 Actively Regulates Photosynthesis in Poplar
To thoroughly study how PtrCHLP3 promotes plant growth, the photosynthetic index (Pn, Gs, Tr, and WUE) was compared between WT and transgenic poplars. The result showed that the Tr and Gs in transgenic plants were significantly larger than those in WT poplar (Figures 6A,B). However, the net photosynthesis (Pn) and water use efficiency (WUE) of transgenic plants were significantly lower than those of the WT plants (Figures 6C,D). Through simulation of photosynthesis-light response curves of transgenic and WT 84 K poplar, we found that when the light intensity was higher than 400 μmol m–2 s–1, the Pn of the transgenic plants was significantly lower than that of the WT (Figure 6E). However, there was no significant difference in the initial quantum yield between WT and transgenic poplars (Figure 6F). Among them, the Pmax of wild-type poplar 84 K was 22.202 μmol m–2 s–1, and those of the transgenic plants chlp3 and chlp4 were 20.064 μmol m–2 s–1 and 18.165 μmol m–2 s–1, respectively (Figure 6G). These results suggest that PtrCHLP3 influences the photosynthetic rate of plants through non-stomatal factors.
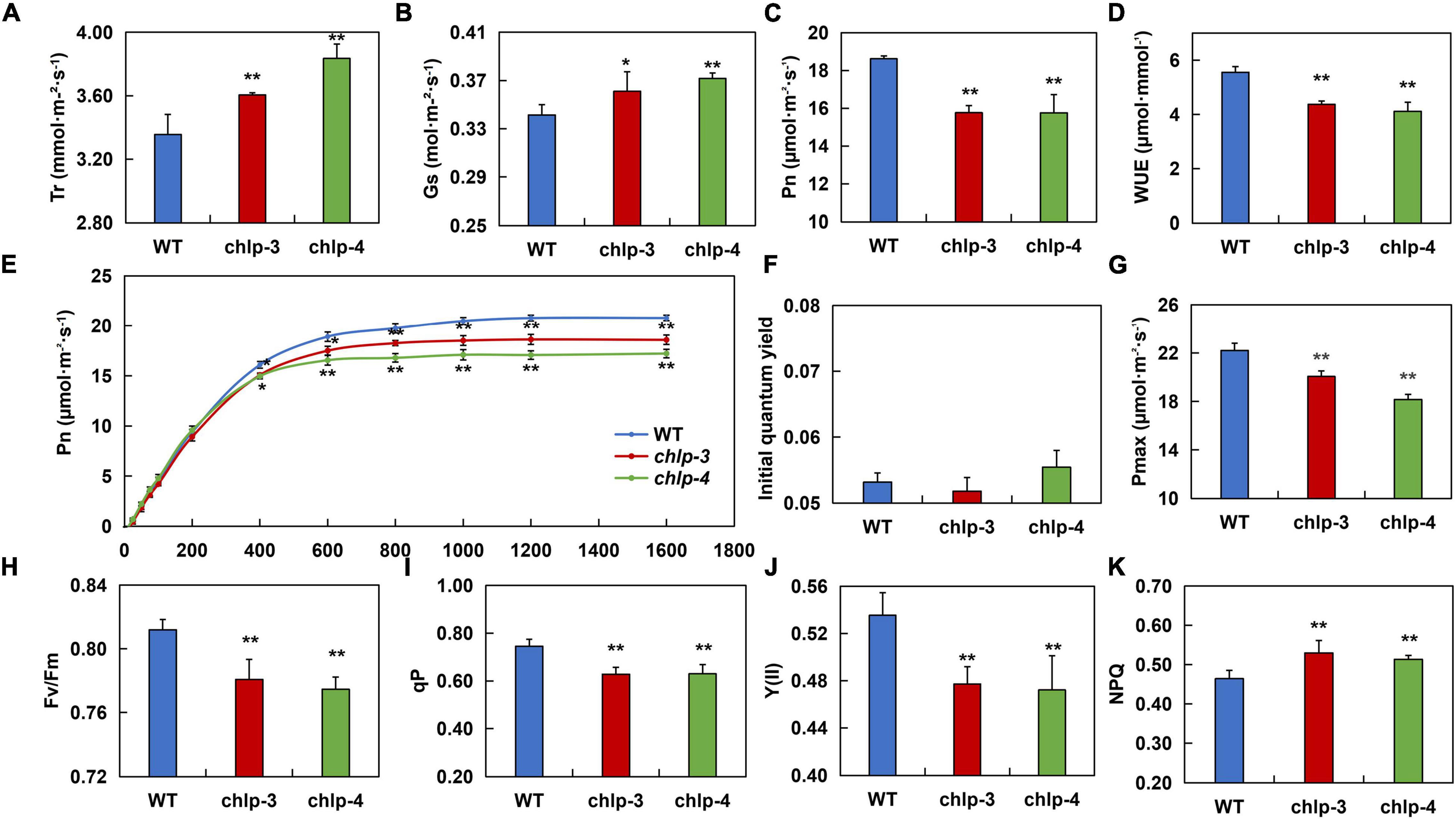
Figure 6. PtrCHLP3 actively regulates photosynthesis. (A–D) The photosynthetic parameters of chlp plants relative to those of WT plants. Tr, transpiration rate; Gs, stomatal conductance; Pn, net photosynthetic rate; WUE, instantaneous water-use efficiency; (E–G) Pn–light curve, initial quantum yield, and Pmax were measured in WT and chlp plants. (H–K) The value [Fv/Fm, Y (II), qP, and NPQ] of the slow dynamic fluorescence induction curve were measured in chlp and WT plants. The value represents the mean ± standard deviation (n = 40). Asterisks indicate a significant difference at the same level: * indicates P < 0.05; ** indicates P < 0.01, respectively.
To thoroughly determine how PtrCHLP3 affects electron transport during photosynthesis, fluorescence parameters [Fv/Fm, Y (II), qP, and NPQ] were measured between transgenic and WT plants. The results showed that the maximal Fv/Fm, qP, and Y (II) of transgenic plants were lower than those of WT plants (Figures 6H–J). In addition, NPQ was significantly higher in the transgenic plants than in WT (Figure 6K). In general, the downregulation of PtrCHLP3 levels in plants reduces electron transport rates during photosynthesis, thus weakening photosynthesis.
Molecular Network of PtrCHLP3 in Regulating Photosynthesis and Chlorophyll Biosynthesis in Poplar
To further elucidate the molecular mechanism by which PtrCHLP3 regulates photosynthesis and growth of poplar, the potential upstream transcription factors (PUTFs) of PtrCHLP3 were analyzed. The PUTFs of PtrCHLP3 were identified through bioinformatics, and their functions and expression levels in various tissues were analyzed (Figure 7B and Supplementary Table S9). Surprisingly, the PUTFs (Potri.012G060300, Potri.012G073900, Potri.010G240800, Potri.008G191800, Potri.002G228700, and Potri.001G155300) of PtrCHLP3 are MYB TF (Figure 7A and Supplementary Table S8). Potri.012G060300, Potri.012G073900, and Potri.002G228700 had similar expression patterns to PtrCHLP3, which were upregulated in mature leaves but inhibited in roots and stems. In contrast to the expression pattern of PtrCHLP3, Potri.010G240800 and Potri.001G155300 were downregulated in leaves but activated in roots and stems. In addition, the expression pattern of Potri.008G191800 is complex, and it is upregulated in both roots and mature leaves but downregulated in young leaves and stems.
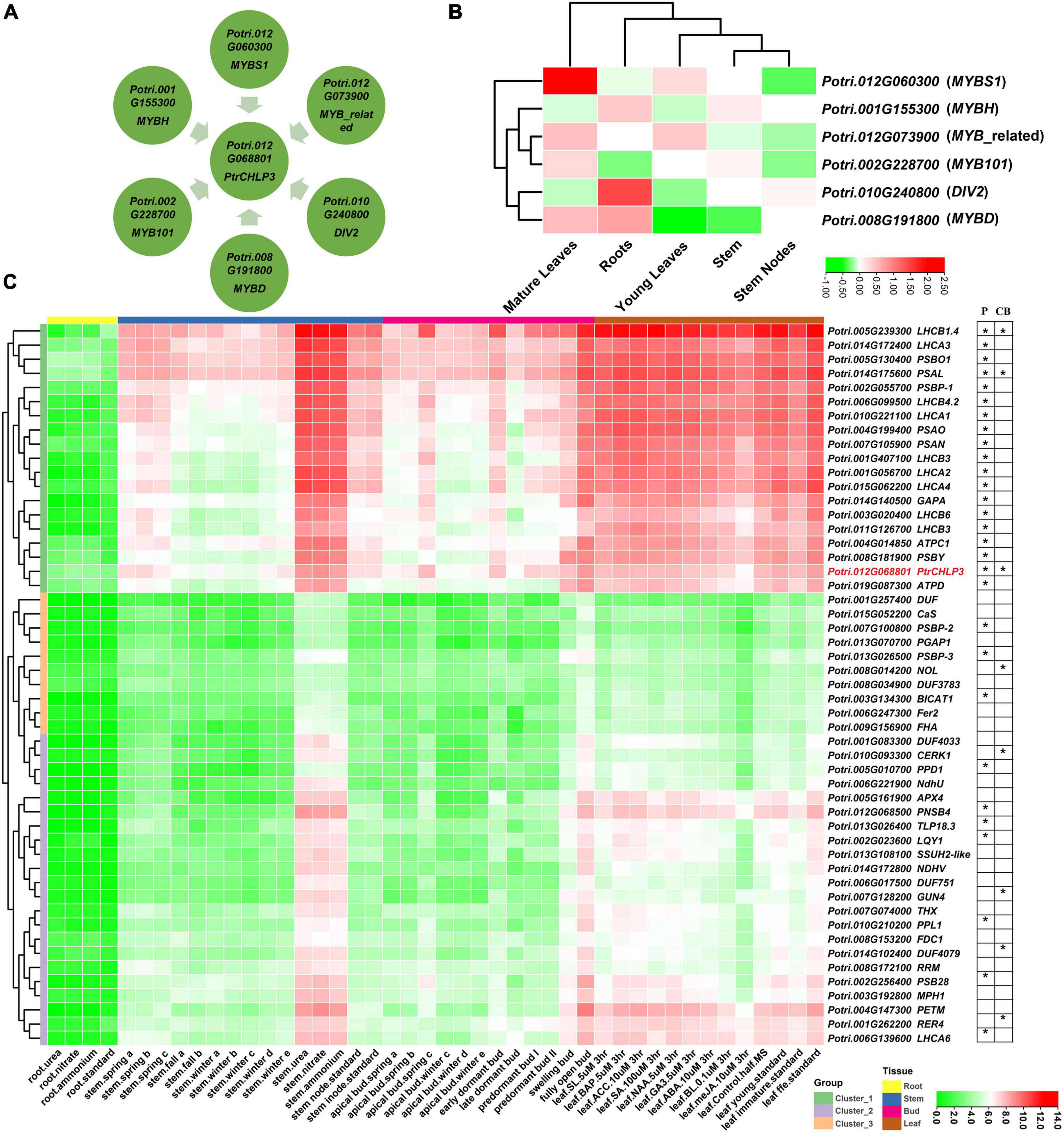
Figure 7. Construction of PtrCHLP3 molecular regulatory network. Bioinformatic analysis of TFs of PtrCHLP3. (A) Interaction network analysis of TFs-PtrCAX3. The blue rectangle represents TFs upstream of PtrCHLP3. (B) Heat maps showing the transcriptional abundance of upstream transcription factors of PtrCHLP3 under different plant tissues. The range of fold change in expression in the heat map is indicated by the color bar. (C) Cluster heat map analysis of genes co-expressed with PtrCHLP3 in different plant tissues and treatments. P, Photosynthesis; CB, Chlorophyll biosynthesis. The asterisk indicates that the gene is involved in the process of P or CB.
In addition, we constructed cluster heat maps co-expressed with PtrCHLP3 in different tissues (Figure 7C), which could be divided into three clusters (1–3). All genes in Cluster 1 were downregulated in roots, most were upregulated in stems and buds, and all were significantly upregulated in leaves. However, the gene expression level of Cluster 2 in each poplar tissue was almost suppressed. Furthermore, all genes in Cluster 3 were inhibited in roots, most were downregulated in stems and buds, and some were activated in leaves. A large number of genes involved in photosynthesis and chlorophyll biosynthesis were found to form a co-expression network with PtrCHLP3. Moreover, the following genes with similar expression patterns to PtrCHLP3 are located in Cluster 1: LIGHT HARVESTING COMPLEX PHOTOSYSTEM II SUBUNIT (LHCB1.4/3/4.2/4/6), PHOTOSYSTEM I LIGHT HARVESTING COMPLEX GENE (LHCA1/2/3/4), PHOTOSYSTEM II SUBUNIT O (PSBO1), PHOTOSYSTEM I SUBUNIT L (PSAL), PHOTOSYSTEM II SUBUNIT P (PSBP1), PHOTOSYSTEM I SUBUNIT O (PSAO), GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT (GAPA), ATPC1, ATP synthase D chain (ATPD), PSAN, and PHOTOSYSTEM II BY (PSBY); and they are all involved in plant photosynthesis. In addition, 25.755% of the genes were found to play a role in chloroplasts by GO cell component analysis. At the same time, GO biological process analysis found 14.085% genes involved in plant photosynthesis (Supplementary Figure S2). In conclusion, PtrCHLP3 and its co-expressed genes play important roles in regulating photosynthesis and chlorophyll biosynthesis in poplars.
Discussion
Chlorophyll is the most abundant pigment in photosynthetic organs and plays an irreplaceable role in chloroplast formation, homeostasis, and photosynthesis (Lin et al., 2014; Zhang et al., 2021). The final stage of Chl a synthesis is the esterification of the tetrapyrrole moiety and chlorophyllide. CHLP, a key enzyme in the final step of Chl a biosynthesis, participates in the terminal hydrogenation step of chlorophyll synthesis (Addlesee and Hunter, 2002). The CHLP family has been defined and explored in many plants, such as Arabidopsis, ralgae, tobacco, rice, peach, olive, and tomato (Addlesee and Hunter, 2002; Shpilyov et al., 2005; Deng et al., 2014; Zhang et al., 2015; Tan et al., 2019). However, there has been no related work on its distribution and function in poplar. We used comparative genomics, transcriptomics, and molecular biology to identify CHLP family genes in poplars and analyze their effects on plant photosynthesis and growth.
Phylogenesis, Molecular Characteristics, and Expansion of the CHLP Gene Family
We established a genealogical tree using the protein sequences of 42 plants and divided them into six groups to explore the evolution and phylogeny of CHLP genes (Figure 1A). PtrCHLP1, PtrCHLP2, and PtrCHLP3 were located on different branches (Figures 1A,C), indicating that CHLP genes may be functionally differentiated in poplar trees. Macromolecules (genes or proteins) with similar gene and protein structures may have similar biological functions (He et al., 2021b). Moreover, AtCHLP1 and PtrCHLP3 have similar conserved domains and motifs, and their clustering on the same branch indicates that they may be used for similar biological functions. However, motif10 was absent from the C-terminus of OsCHLP1 (Figure 1C), resulting in a single branch (Figure 1E).
In addition, the phylogenetic analysis of family genes can explain the evolution of genes (Zheng et al., 2021). Salix and poplar are closer because they belong to the same genus (Xiao et al., 2018; He et al., 2021a). Poplar and Salix are relatively tall dicotyledonous trees, and their CHLP1 genes clustered together (Figures 1A,C). Tandem and segmental replication play critical roles in the evolution of species (Kaur et al., 2017; Xie et al., 2018). One pair of genes (PtrCHLP1/2) that appeared as segmental duplications in Populus suggested that gene duplications played a role in the evolution of PtrCHLP genes in the genus (Figure 2A). Similar to the results of the evolutionary trees, only gene pairs formed between AtCHLP1 and PtrCHLP3 in Arabidopsis and poplar (Figure 2B), which ensured that their functions were similar and conserved. However, many gene pairs were detected between Populus and S. purpurea (Figure 2D), implying that conspicuous variation and duplication of CHLP genes could have been generated during the evolution of dicotyledons of the same genus.
Expression Pattern Analysis of CHLP in Poplar
The level of gene expression is very important for plant growth, development, and external responses (Khatun et al., 2017; He et al., 2021c). With the differentiation and amplification of genes, the expression profiles of different members of a gene family vary greatly (Mao et al., 2021). According to the transcriptomic data of poplars, the expression of PtrCHLP1/2/3 fluctuated slightly in each poplar tissue (Figure 3B). In addition, the tissue expression patterns of PtrCHLP1/2 and PtrCHLP3 were the opposite, which may be closely related to cis-acting elements on the gene promoter (Figure 3A). The cis-acting elements of promoters play an important role in the regulation of gene transcription (He et al., 2019). The promoter of PtrCHLP1/2 mainly contains elements that respond to hormones and stress, whereas the promoter of PtrCHLP3 contains many elements that are correlated with growth and development compared with PtrCHLP1/2 (Figure 3A and Supplementary Figure S1). CHLP has been reported to play a critical role in chlorophyll synthesis and is mainly expressed in the leaves (Shpilyov et al., 2005; Zhang et al., 2015). Consistent with the above results, PtrCHLP3 was significantly upregulated in leaves but inhibited in non-photosynthetic organs (Figure 3B). In addition, the expression levels of PtrCHLP3 in different leaves were different (Figure 3E). Although CHLP is involved in the last step of chlorophyll a synthesis, its expression level in leaves is not completely synchronous with chlorophyll content (Figures 3D,E). Mature leaves play a leading role in the photosynthetic contribution of plants, whereas young leaves need to synthesize a large amount of chlorophyll in the process of growing to mature leaves (Luo et al., 2019; Shen et al., 2021). This may be the main reason the expression of PtrCHLP3 reaches its peak before chlorophyll content in leaves; after all, the response rate of genes is faster than the phenotype.
PtrCHLP3 Is Involved in Plant Growth and Development Through Photosynthesis
In the present study, gene-silenced transgenic poplars (RNAi-PtrCHLP3/chlp3 and chlp4) were obtained and used for subsequent functional verification experiments (Figure 4). Compared with WT plants, the leaves of gene-silenced poplar plants were yellow due to the inhibition of chlorophyll synthesis (Figures 4E,F). Consistent with previous results, downregulation of PtrCHLP3 resulted in decreased chlorophyll content in plants (Figure 4D). In antisense N. tabacum transformants of CHL I, chlorophyll content in leaves was lower, and the leaf phenotype was light green or with white and green patches (Jutta Papenbrock et al., 2000; Havaux et al., 2003). The inhibition or reduction of chlorophyll synthesis results in the inhibition of plant growth and development (An et al., 2020; He et al., 2020). The plant height, stem diameter, and biomass of chlp3/4 were significantly lower than those of WT poplars (Figures 5A–E), which may be caused by severe chlorosis of the plant. It has been reported that GATA deletion in poplars inhibits chlorophyll synthesis, leading to yellowing and biomass reduction (An et al., 2020). This obstruction of chlorophyll synthesis may also be caused by variations in the chloroplast structure (An et al., 2020). However, the decrease in chlorophyll synthesis in chlp3/4 was mainly attributed to downregulation of the geranylgeraniol reductase (CHLP) gene (Figures 4D,F).
However, this reduction in chlorophyll or inhibition of chlorophyll synthesis in plants is itself a natural stress, such as plant senescence (Wang et al., 2021). Most of the reasons leading to inadequate plant productivity are attributed to the obvious weakening of plant photosynthetic capacity (An et al., 2020; Shen et al., 2021). Compared with WT plants, the Pn and Pmax of transgenic poplar were significantly reduced (Figures 6C,E,G). At the same time, the decrease in photosynthesis in transgenic plants resulted in a decrease in xylem cell layers, which slowed down the lateral growth of stems (Figures 5F–M). However, the Tr and Gs of transgenic plants were significantly higher than those of WT plants (Figures 6A,B), suggesting that downregulation of PtrCHLP3 in plants did not inhibit photosynthesis by affecting the dark reaction process. Plant photosynthesis mainly consists of light and dark reactions, involving light absorption, electron transfer, photophosphorylation, carbon assimilation, and other important reaction steps, which are of great significance in realizing energy conversion in nature and maintaining the carbon-oxygen balance in the atmosphere (Flood et al., 2011). The photoreaction phase converts light energy into chemical energy and produces ATP, which provides energy for dark reactions. It has been reported that Fv/Fm and Y (II) reflect the maximum and actual conversion efficiencies of plant photosystem II, respectively (He et al., 2019). However, downregulation of PtrCHLP3 in plants resulted in significantly lower Fv/Fm and Y (II) than in the WT (Figures 6H,J), which may be caused by reduced photosynthetic pigment synthesis (chlorophyll) in transgenic plants. In addition, the proportion of energy absorbed by photosystem II for photochemical reactions (qP) in transgenic plants was significantly lower than that in WT plants (Figure 6I). Concurrently, because of the low chlorophyll content of transgenic plants, they cannot carry out photosynthesis at high light intensities (Figure 6K), leading to the dissipation of excess light energy in the form of heat energy to prevent plant damage (Murchie and Niyogi, 2011). Therefore, PtrCHLP3 mediates plant growth and development by regulating photosynthesis.
Transcriptional Regulatory Networks Mediated by PtrCHLP3 in Photosynthesis and Chlorophyll Biosynthesis of Poplars
As a core enzyme in chlorophyll A synthesis, CHLP plays a key role in plant photosynthesis and chlorophyll biosynthesis (Shpilyov et al., 2005; Lin et al., 2014). However, there are few reports on how CHLP functions via transcriptional regulation. Therefore, it is essential to identify the PUTFs of PtrCHLP3. In addition, transcriptional regulation plays a crucial role in all aspects of the plant life cycle (Zhang et al., 2022). The PUTFs of PtrCHLP3 were identified using bioinformatic analysis (Figure 7A). Consistent with previous results (Figure 3B), the PUTFs (MYB) of PtrCHLP3 had a tissue expression pattern similar to that of PtrCHLP3, and only had a high expression level in the leaves (Figure 7B). For example, the MYB TF of poplar, LTF1, promotes plant growth and xylem formation by regulating downstream target genes (Gui et al., 2019). Moreover, the trichome density and resistibility of insect pests improve due to MYB186 overexpression in poplars, which affects plant growth (Plett et al., 2010). In short, several TFs may bind to elements such as MYB binding sites in the CHLP promoter, thereby regulating CHLP gene expression.
In plants, photosynthesis is a complex process in which a large number of genes work together to convert light energy into stable chemical energy (Flood et al., 2011). By constructing a co-expression network with the PtrCHLP3 gene in each plant tissue (Figure 7C), a large number of genes involved in photosynthesis have been identified, including LHCB, LHCA, PSBO, PSAL, PSBP1, PSAO, GAPA, ATPC, ATPD, PSAN, and PSBY (Haijun Liu et al., 2007; Yi et al., 2007, 2009; Bricker and Frankel, 2008; Peng et al., 2009; Seok et al., 2014; Otani et al., 2017; Ilikova et al., 2021). For example, LHCB3 and LHCB6 encode light-harvesting proteins involved in photosynthesis in Arabidopsis, and mutations in the LHCB3/6 in Arabidopsis lead to reduced plant chlorophyll content and growth inhibition (Damkjaer et al., 2009; Ilikova et al., 2021). Moreover, the three mutant (lhcb4/5/6) exhibited decreased energy-transfer efficiency from the LHCII (light-harvesting complex II) to the PSII reaction center (Chen et al., 2018). Similar to the phenotype of chlp3/4 (Figures 4F, 6H), their chlorophyll content and Fv/Fm in Mutant (lhcb4/5/6) were also lower than the wild type. PsbO1 is a thylakoid lumen-localized extrinsic subunit protein of PSII and plays a crucial role in the oxygen-evolving complex by stabilizing the catalytic manganese cluster (Suorsa and Aro, 2007). In addition, downregulation of PSBP in plants showed that PS II reaction centers decreased with downregulation of PSBP composition (Yi et al., 2009). Silencing the PtrCHLP3 gene in poplar resulted in the inhibition of chlorophyll synthesis and impaired photosynthetic ability (Figure 6). Furthermore, LHCB1.4/3/4.2/4/6, LHCA1/2/3/4, PSBO1, PSAL, PSBP1, PSAO, GAPA, ATPC1, ATPD, PSAN, and PSBY had similar expression patterns to PtrCHLP3 and were significantly activated in leaves and involved in photosynthesis. Functional enrichment analysis showed that most of these genes, co-expressed with PtrCHLP3, functioned in chloroplasts and participated in plant photosynthesis (Supplementary Figure S2). In general, PtrCHLP3 and co-expressed genes synergistically mediate photosynthesis and chlorophyll biosynthesis in poplars.
Conclusion
The data obtained in this study were used to construct a working frame to investigate whether PtrCHLP3 regulates poplar growth (Figure 8). In summary, a whole-genome analysis of the CHLP family of P. trichocarpa identified three PtrCHLP genes that play pivotal roles in poplar growth and photosynthesis. The gene structure, evolution, phylogenetics, chromosomal position, gene doubling, cis-elements, and expression profiles of CHLP were determined using genomics and bioinformatics. In addition, PtrCHLP3 is preferentially expressed in the leaves and plays an important role in regulating chlorophyll biosynthesis. Furthermore, inhibition of PtrCHLP3 expression in poplar can inhibit plant growth through the downregulation of photosynthesis. Ultimately, PtrCHLP3 formed a co-expression network with photosynthesis-related genes, which synergistically affected the growth and photosynthesis of the poplar trees. Thus, this study provides genetic resources for the improved breeding of fast-growing tree traits.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
FH, Y-JS, QC, J-LL, M-XN, C-HF, M-ML, Q-LL, F-FT, FZ, T-TL, L-HC, and X-QW conceived and performed the original research project. FH, Y-JS, and J-LL performed the experiments. FH, Y-JS, FZ, and X-QW designed the experiments and analyzed the data. FH refined the project and wrote the manuscript with contributions from all authors. T-TL, FZ, and X-QW supervised the experiments and revised the writing. X-QW and FH obtained the funding for the research project. All the authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32101481 to FH; 31870645 to X-QW), and supported by Sichuan Science and Technology Program (2021YFYZ0032 to X-QW), and by the Project of Science and Technology Department of Sichuan Province (22NSFSC3282 to FH; 2021YJ0301 to FZ). Furthermore, we appreciate the Project of Tianfu Emei of Sichuan Province and the China Postdoctoral Foundation (2021M692331 to FH) for supporting this research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.870970/full#supplementary-material
Footnotes
- ^ https://phytozome-next.jgi.doe.gov/
- ^ https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi?
- ^ http://gsds.gao-lab.org/
- ^ https://meme-suite.org/meme/tools/meme
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ https://popgenie.org/
- ^ http://plantregmap.gao-lab.org/network.php
References
Addlesee, H. A., Gibson, L. C. D., Jensen, P. E., and Hunter, C. N. (1996). Cloning, sequencing and functional assignment of the chlorophyll biosynthesis gene, chlP, of Synechocystis sp. PCC 6803. FEBS Lett. 389, 126–130. doi: 10.1016/0014-5793(96)00549-2
Addlesee, H. A., and Hunter, C. N. (2002). Rhodospirillum rubrum possesses a variant of the bchP gene, encoding geranylgeranyl-bacteriopheophytin reductase. J. Bacteriol. 184, 1578–1586. doi: 10.1128/JB.184.6.1578-1586.2002
An, Y., Zhou, Y., Han, X., Shen, C., Wang, S., Liu, C., et al. (2020). The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 71, 1969–1984. doi: 10.1093/jxb/erz564
Arnon, D. I. (1949). Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME Suite. Nucleic Acids Res. 43, W39–W49.
Beale, S. I. (2005). Green genes gleaned. Trends Plant Sci. 10, 309–312. doi: 10.1016/j.tplants.2005.05.005
Bricker, T. M., and Frankel, L. K. (2008). The psbo1 mutant of Arabidopsis cannot efficiently use calcium in support of oxygen evolution by photosystem II. J. Biol. Chem. 283, 29022–29027. doi: 10.1074/jbc.M805122200
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y. E., Ma, J., Wu, N., Su, Y. Q., Zhang, Z. W., Yuan, M., et al. (2018). The roles of Arabidopsis proteins of Lhcb4, Lhcb5 and Lhcb6 in oxidative stress under natural light conditions. Plant Physiol. Biochem. 130, 267–276. doi: 10.1016/j.plaphy.2018.07.014
Damkjaer, J. T., Kereiche, S., Johnson, M. P., Kovacs, L., Kiss, A. Z., Boekema, E. J., et al. (2009). The photosystem II light-harvesting protein Lhcb3 affects the macrostructure of photosystem II and the rate of state transitions in Arabidopsis. Plant Cell 21, 3245–3256. doi: 10.1105/tpc.108.064006
Deng, X. J., Zhang, H. Q., Wang, Y., He, F., Liu, J. L., Xiao, X., et al. (2014). Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLoS One 9:e99564. doi: 10.1371/journal.pone.0099564
Flood, P. J., Harbinson, J., and Aarts, M. G. (2011). Natural genetic variation in plant photosynthesis. Trends Plant Sci. 16, 327–335. doi: 10.1016/j.tplants.2011.02.005
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2011). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Gui, J., Luo, L., Zhong, Y., Sun, J., Umezawa, T., and Li, L. (2019). Phosphorylation of LTF1, an MYB Transcription Factor in Populus, Acts as a Sensory Switch Regulating Lignin Biosynthesis in Wood Cells. Mol. Plant 12, 1325–1337. doi: 10.1016/j.molp.2019.05.008
Haijun Liu, L. K. F., And Terry, M., and Bricker. (2007). Functional Analysis of Photosystem II in a PsbO-1-Deficient Mutant in Arabidopsis. Biochemistry 46, 7607–7613. doi: 10.1021/bi700107w
Hall, B. G. (2013). Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 30, 1229–1235. doi: 10.1093/molbev/mst012
Havaux, M., Lutz, C., and Grimm, B. (2003). Chloroplast membrane photostability in chlP transgenic tobacco plants deficient in tocopherols. Plant Physiol. 132, 300–310. doi: 10.1104/pp.102.017178
He, F., Li, H.-G., Wang, J.-J., Su, Y., Wang, H.-L., Feng, C.-H., et al. (2019). PeSTZ1, a C2H2-type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2. Plant Biotechnol. J. 17, 2169–2183. doi: 10.1111/pbi.13130
He, F., Niu, M. X., Feng, C. H., Li, H. G., Su, Y., Su, W. L., et al. (2020). PeSTZ1 confers salt stress tolerance by scavenging the accumulation of ROS through regulating the expression of PeZAT12 and PeAPX2 in Populus. Tree Physiol. 40, 1292–1311. doi: 10.1093/treephys/tpaa050
He, F., Shi, Y.-J., Mi, J.-X., Zhao, K.-J., Cui, X.-L., Chen, L.-H., et al. (2021a). Genome-Wide Investigation of the NF-X1 Gene Family in Populus trichocarpa Expression Profiles during Development and Stress. Int. J. Mol. Sci. 22:4664. doi: 10.3390/ijms22094664
He, F., Shi, Y. J., Zhao, Q., Zhao, K. J., Cui, X. L., Chen, L. H., et al. (2021b). Genome-wide investigation and expression profiling of polyphenol oxidase (PPO) family genes uncover likely functions in organ development and stress responses in Populus trichocarpa. BMC Genom. 22:731. doi: 10.1186/s12864-021-08028-9
He, F., Zhao, Q., Huang, J.-L., Niu, M.-X., Feng, H.-C., Shi, Y.-J., et al. (2021c). External application of nitrogen alleviates toxicity of cadmium on poplars via starch and sucrose metabolism. Tree Physiol. 41, 2126–2141. doi: 10.1093/treephys/tpab065
He, F., Wang, H.-L., Li, H.-G., Su, Y., Li, S., Yang, Y., et al. (2018). PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA-induced stomatal closure by ROS production in Populus. Plant Biotechnol. J. 16, 1514–1528. doi: 10.1111/pbi.12893
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Ilikova, I., Ilik, P., Opatikova, M., Arshad, R., Nosek, L., Karlicky, V., et al. (2021). Towards spruce-type photosystem II: consequences of the loss of light-harvesting proteins LHCB3 and LHCB6 in Arabidopsis. Plant Physiol. 187, 2691–2715. doi: 10.1093/plphys/kiab396
Jutta Papenbrock, E. P., and Hans-Peter Mock, and Bernhard Grimm. (2000). Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J. 22, 155–164. doi: 10.1046/j.1365-313x.2000.00724.x
Kaur, S., Dhugga, K. S., Beech, R., and Singh, J. (2017). Genome-wide analysis of the cellulose synthase-like (Csl) gene family in bread wheat (Triticum aestivum L.). BMC Plant Biol. 17:193. doi: 10.1186/s12870-017-1142-z
Khatun, K., Nath, U. K., Robin, A. H. K., Park, J. I., Lee, D. J., Kim, M. B., et al. (2017). Genome-wide analysis and expression profiling of zinc finger homeodomain (ZHD) family genes reveal likely roles in organ development and stress responses in tomato. BMC Genom. 18:695. doi: 10.1186/s12864-017-4082-y
Kimura, E., Abe, T., Murata, K., Kimura, T., Otoki, Y., Yoshida, T., et al. (2018). Identification of OsGGR2, a second geranylgeranyl reductase involved in α-tocopherol synthesis in rice. Sci. Rep. 8:1870. doi: 10.1038/s41598-018-19527-3
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Lin, Y. P., Lee, T. Y., Tanaka, A., and Charng, Y. Y. (2014). Analysis of an Arabidopsis heat-sensitive mutant reveals that chlorophyll synthase is involved in reutilization of chlorophyllide during chlorophyll turnover. Plant J. 80, 14–26. doi: 10.1111/tpj.12611
Liu, H., Liu, J., Zhao, M. M., and Chen, J. S. (2015). Overexpression of ShCHL P in tomato improves seedling growth and increases tolerance to salt, osmotic, and oxidative stresses. Plant Grow. Regul. 77, 211–221. doi: 10.1007/s10725-015-0054-x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M., Geer, R., Gonzales, N., et al. (2019). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Luo, X. Z., Croft, H., Chen, J. M., He, L. M., and Keenan, T. F. (2019). Improved estimates of global terrestrial photosynthesis using information on leaf chlorophyll content. Glob. Chang. Biol. 25, 2499–2514. doi: 10.1111/gcb.14624
Mao, K., Yang, J., Wang, M., Liu, H., Guo, X., Zhao, S., et al. (2021). Genome-wide analysis of the apple CaCA superfamily reveals that MdCAX proteins are involved in the abiotic stress response as calcium transporters. BMC Plant Biol. 21:81. doi: 10.1186/s12870-021-02866-1
Mei, J., Li, F., Liu, X., Hu, G., Fu, Y., and Liu, W. (2017). Newly identified CSP41b gene localized in chloroplasts affects leaf color in rice. Plant Sci. 256, 39–45. doi: 10.1016/j.plantsci.2016.12.005
Murchie, E. H., and Niyogi, K. K. (2011). Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol. 155, 86–92. doi: 10.1104/pp.110.168831
Nagata, N., Tanaka, R., Satoh, S., and Tanaka, A. (2005). Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17, 233–240. doi: 10.1105/tpc.104.027276
Ohmiya, A., Oda-Yamamizo, C., and Kishimoto, S. (2019). Overexpression of CONSTANS-like 16 enhances chlorophyll accumulation in petunia corollas. Plant Sci. 280, 90–96. doi: 10.1016/j.plantsci.2018.11.013
Otani, T., Yamamoto, H., and Shikanai, T. (2017). Stromal Loop of Lhca6 is Responsible for the Linker Function Required for the NDH-PSI Supercomplex Formation. Plant Cell Physiol. 58, 851–861. doi: 10.1093/pcp/pcx009
Peng, L., Fukao, Y., Fujiwara, M., Takami, T., and Shikanai, T. (2009). Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21, 3623–3640. doi: 10.1105/tpc.109.068791
Plett, J. M., Wilkins, O., Campbell, M. M., Ralph, S. G., and Regan, S. (2010). Endogenous overexpression of Populus MYB186 increases trichome density, improves insect pest resistance, and impacts plant growth. Plant J. 64, 419–432. doi: 10.1111/j.1365-313X.2010.04343.x
Seok, M. S., You, Y. N., Park, H. J., Lee, S. S., Aigen, F., Luan, S., et al. (2014). AtFKBP16-1, a chloroplast lumenal immunophilin, mediates response to photosynthetic stress by regulating PsaL stability. Physiol. Plant 150, 620–631. doi: 10.1111/ppl.12116
Shen, C., Zhang, Y., Li, Q., Liu, S., He, F., An, Y., et al. (2021). PdGNC confers drought tolerance by mediating stomatal closure resulting from NO and H2O2 production via the direct regulation of PdHXK1 expression in Populus. N. Phytol. 230, 1868–1882. doi: 10.1111/nph.17301
Sheng, Z., Lv, Y., Li, W., Luo, R., Wei, X., Xie, L., et al. (2017). Yellow-Leaf 1 encodes a magnesium-protoporphyrin IX monomethyl ester cyclase, involved in chlorophyll biosynthesis in rice (Oryza sativa L.). PLoS One 12:e0177989. doi: 10.1371/journal.pone.0177989.
Shpilyov, A. V., Zinchenko, V. V., Shestakov, S. V., Grimm, B., and Lokstein, H. (2005). Inactivation of the geranylgeranyl reductase (ChlP) gene in the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys Acta. 1706, 195–203. doi: 10.1016/j.bbabio.2004.11.001
Singh, V. K., Rajkumar, M. S., Garg, R., and Jain, M. (2017). Genome-wide identification and co-expression network analysis provide insights into the roles of auxin response factor gene family in chickpea. Sci. Rep. 7:10895. doi: 10.1038/s41598-017-11327-5
Sjodin, A., Street, N. R., Sandberg, G., Gustafsson, P., and Jansson, S. (2009). The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. N. Phytol. 182, 1013–1025. doi: 10.1111/j.1469-8137.2009.02807.x
Suorsa, M., and Aro, E.-M. (2007). Expression, assembly and auxiliary functions of photosystem II oxygen-evolving proteins in higher plants. Photosynt. Res. 93, 89–100. doi: 10.1007/s11120-007-9154-4
Tan, J., Zhang, T., Xia, S., Yan, M., Li, F., Sang, X., et al. (2019). Fine mapping of a novel yellow-green leaf 14 (ygl14) mutant in rice. Euphytica 215:100.
Tanaka, R., Oster, U., Kruse, E., Rudiger, W., and Grimm, B. (1999). Reduced activity of geranylgeranyl reductase leads to loss of chlorophyll and tocopherol and to partially geranylgeranylated chlorophyll in transgenic tobacco plants expressing antisense RNA for geranylgeranyl reductase. Plant Physiol. 120, 695–704. doi: 10.1104/pp.120.3.695
Thornley, J. H. M. (1976). Mathematical Models in Plant Physiology : a Quantitative Approach to Problems in Plant and Crop Physiology. New York: Academic Press.
Tian, F., Yang, D.-C., Meng, Y.-Q., Jin, J., and Gao, G. (2019). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz1020
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604.
Wang, C., Liu, S., Dong, Y., Zhao, Y., Geng, A., Xia, X., et al. (2016). PdEPF1 regulates water-use efficiency and drought tolerance by modulating stomatal density in poplar. Plant Biotechnol. J. 14, 849–860. doi: 10.1111/pbi.12434
Wang, H.-L., Zhang, Y., Wang, T., Yang, Q., Yang, Y., Li, Z., et al. (2021). An alternative splicing variant of PtRD26 delays leaf senescence by regulating multiple NAC transcription factors in Populus. Plant Cell 33, 1594–1614. doi: 10.1093/plcell/koab046
Wang, P., Li, C., Wang, Y., Huang, R., Sun, C., Xu, Z., et al. (2014). Identification of a Geranylgeranyl reductase gene for chlorophyll synthesis in rice. SpringerPlus 3:201. doi: 10.1186/2193-1801-3-201
Xiao, G., He, P., Zhao, P., Liu, H., Zhang, L., Pang, C., et al. (2018). Genome-wide identification of the GhARF gene family reveals that GhARF2 and GhARF18 are involved in cotton fibre cell initiation. J. Exp. Bot. 69, 4323–4337. doi: 10.1093/jxb/ery219
Xie, T., Chen, C., Li, C., Liu, J., Liu, C., and He, Y. (2018). Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genom. 19:490. doi: 10.1186/s12864-018-4880-x
Yang, Y., Li, H.-G., Wang, J., Wang, H.-L., He, F., Su, Y., et al. (2020). ABF3 enhances drought tolerance via promoting ABA-induced stomatal closure by directly regulating ADF5 in Populus euphratica. J. Exp. Bot. 71, 7270–7285. doi: 10.1093/jxb/eraa383
Yi, X., Hargett, S. R., Frankel, L. K., and Bricker, T. M. (2009). The PsbP protein, but not the PsbQ protein, is required for normal thylakoid architecture in Arabidopsis thaliana. FEBS Lett. 583, 2142–2147. doi: 10.1016/j.febslet.2009.05.048
Yi, X., Hargett, S. R., Liu, H., Frankel, L. K., and Bricker, T. M. (2007). The PsbP protein is required for photosystem II complex assembly/stability and photoautotrophy in Arabidopsis thaliana. J. Biol. Chem. 282, 24833–24841. doi: 10.1074/jbc.m705011200
Zhang, C., Zhang, W., Ren, G., Li, D., Cahoon, R. E., Chen, M., et al. (2015). Chlorophyll Synthase under Epigenetic Surveillance Is Critical for Vitamin E Synthesis, and Altered Expression Affects Tocopherol Levels in Arabidopsis. Plant Physiol. 168, 1503–1511. doi: 10.1104/pp.15.00594
Zhang, H., Zhu, J., Gong, Z., and Zhu, J.-K. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119.
Zhang, J., Sui, C., Liu, H., Chen, J., Han, Z., Yan, Q., et al. (2021). Effect of chlorophyll biosynthesis-related genes on the leaf color in Hosta (Hosta plantaginea Aschers) and tobacco (Nicotiana tabacum L.). BMC Plant Biol. 21:45. doi: 10.1186/s12870-020-02805-6
Zheng, Y., Wang, L. B., Sun, S. F., Liu, S. Y., Liu, M. J., Lin, J., et al. (2021). Phylogenetic and ion-response analyses reveal a relationship between gene expansion and functional divergence in the Ca(2+)/cation antiporter family in Angiosperms. Plant Mol. Biol. 105, 303–320. doi: 10.1007/s11103-020-01088-7
Keywords: poplar, CHLP, chlorophyll biosynthesis, photosynthesis, plant growth
Citation: He F, Shi Y-J, Chen Q, Li J-L, Niu M-X, Feng C-H, Lu M-M, Tian F-F, Zhang F, Lin T-T, Chen L-H, Liu Q-l and Wan X-Q (2022) Genome-Wide Investigation of the PtrCHLP Family Reveals That PtrCHLP3 Actively Mediates Poplar Growth and Development by Regulating Photosynthesis. Front. Plant Sci. 13:870970. doi: 10.3389/fpls.2022.870970
Received: 07 February 2022; Accepted: 21 April 2022;
Published: 10 May 2022.
Edited by:
Els J. M. Van Damme, Ghent University, BelgiumReviewed by:
Guanjun Liu, Northeast Forestry University, ChinaYi An, Zhejiang A&F University, China
Copyright © 2022 He, Shi, Chen, Li, Niu, Feng, Lu, Tian, Zhang, Lin, Chen, Liu and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue-Qin Wan, w-xue@163.com
†These authors have contributed equally to this work
 Fang He1†
Fang He1† Fan Zhang
Fan Zhang Tian-Tian Lin
Tian-Tian Lin Xue-Qin Wan
Xue-Qin Wan