- 1Department of Agronomy, College of Agriculture and Life Sciences, Chungnam National University, Daejeon, South Korea
- 2Department of Southern Area Crop Science, Rural Development Administration, Miryang, South Korea
- 3Crop Breeding Division, National Institute of Crop Science, Wanju-Gun, South Korea
- 4LG Chem., Ltd., Seoul, South Korea
- 5USDA-ARS Crops Pathology and Genetics Research Unit, Davis, CA, United States
- 6Department of Plant Sciences, University of California, Davis, Davis, CA, United States
Four near-isogenic lines (NILs) with different allele combinations of the starch branching enzyme 3 (SBE3) and granule-bound starch synthase 1 (GBSS1) were developed by crossing the japonica rice cultivars “Dodamssal” and “Hwayeong.” The associations between sequence variations in SBE3 and GBSS1, and starch-related traits were investigated. These sequence variations led to changes in seed morphology, starch structure, starch crystallinity, amylopectin chain length distribution, digestibility, apparent amylose content (AAC), and resistant starch content (RS). SBE3 and GBSS1 showed genetic interaction in regulating AAC and RS. Gene expression profiling of panicle tissues revealed significant differences in expression levels of GBSS1, SBE3, and other starch-related genes among the four NILs, indicating that variations in GBSS1 and SBE3 changed the expression level of starch-related genes. These variations contributed to the changes observed in AAC, RS, and physico-chemical characteristics of the rice starch from the NILs.
Introduction
Rice (Oryza sativa L.), one of the most important cereal crops in the world, is high in starch. Starch is the main energy-reserve polysaccharide in cereal crops, and is the chief source of carbohydrates in the human diet (Amagliani et al., 2016). Starch also has many technological, medical, and industrial uses (Miura et al., 2021). Rice starch is mainly composed of two biopolymers: amylose and amylopectin. Amylose is a linear molecule which is consisted of α (1–4) glucosidic chains, while amylopectin is highly branched glucan with α (1–6) glucosidic chains for a connection of linear chains (Yang et al., 2012).
Over the last few years, higher amylose content in staple crops has gained considerable interest due to its positive correlation with resistant starch (RS; Kim et al., 2018). RS is resistant to enzymatic hydrolysis (i.e., digestion) in the small intestine and is fermented by microorganism in the large intestine to produce short chain fatty acid (Englyst et al., 1982; Yang et al., 2016). RS-enriched cereals offer potential benefits for human health. RS consumption could lead to reduced glycemic and insulin responses and could lower the risk of developing type-II diabetes mellitus, obesity, and cardiovascular diseases (Zhou et al., 2016; Wang et al., 2017). High-amylose starches also exhibit different physico-chemical properties. Thus, understanding how these physico-chemical traits are controlled will be of practical value in many industrial food and non-food industries (Wang et al., 2017; Kim et al., 2018).
As the potential health benefits of RS-enriched cereals become more apparent, higher amylose rice mutants and varieties have been developed to address consumer demands. Several genes controlling amylose content have been cloned and characterized. The amylose in rice endosperm is mainly synthesized by granule-bound starch synthase I (GBSS1), which is encoded by the Waxy (Wx) gene on chromosome 6 (Sano, 1984). Three major Wx alleles have been identified: wx, Wxa, and Wxb. In the 5′ splicing site of intron 1, the Wxa allele harbors a G-SNP (Chr 6: 1,765,761) and is mainly found in indica rice whereas the Wxb allele harbors a T-SNP at this position and typically occurs in japonica rice (Cai et al., 1998; Yamanaka et al., 2004). The T/G SNP is associated with a variation of amylose content with the G allele (Wxa) corresponding to a higher expression level of GBSS1 and increased amylose content (Sano et al., 1986). In addition, a non-synonymous SNP in Ex10-115 (C/T) is associated with gel consistency and a low rapid viscosity analysis (RVA) phenotype with most indica (Wxa) and japonica (Wxb) rices having a T (Serine) and a C (Proline), respectively (Chen et al., 2008; Zhang et al., 2019). Recently, the ancestral allele of Waxy, Wxlv (allele responsible for low viscosity and high amylose content), was isolated using a map-based cloning approach (Zhang et al., 2019). Although both Wxlv and Wxa possess a G SNP at 5′ splicing site of intron 1, Wxa harbors a T SNP at Ex10-115 whereas Wxlv carries a C SNP, which is mainly found in japonica rices (Wxb; Zhang et al., 2019). Wxlv allele expression level was higher than Wxb and was similar to Wxa due to the G-SNP at the 5′ splicing site in intron 1. Although the expression level of Wxa and Wxlv was similar, their eating and cooking qualities were significantly different due to the SNP at Ex10-115.
Amylopectin is a branched glucose polymer and is mainly synthesized via concerted reactions catalyzed by three types of biosynthetic enzymes: starch synthases (SSs), starch branch enzymes (SBEs), and debranching enzymes (DBEs; Wang et al., 2017). Among these enzymes, mutations in the SBE isozymes showed significant association with amylose content variation (Nishi et al., 2001; Satoh et al., 2003; Butardo et al., 2011; Yang et al., 2012; Kim et al., 2018; Adeva et al., 2020; Miura et al., 2021). In rice, three isoforms of SBE are present: BEI (SBE1 or BE1), BEIIa (SBE4 or BE2a), and BEIIb (SBE3 or BE2b). Among these isozymes, SBE3 plays a major role in amylopectin synthesis, and a mutation in SBE3 has been found to increase the amylose and RS content in the rice endosperm (Nishi et al., 2001; Yang et al., 2012; Adeva et al., 2020). Yang et al. (2012) identified an SNP (T/C) in the SBE3 coding region resulting in a missense mutation (Leu-599-Pro) that was possibly responsible for high RS. This was confirmed by complementation (Yang et al., 2016). The same T/C SNP was also identified in SBE3 of high amylose variety Dodamssal with high amylose and RS content (Adeva et al., 2020).
Recently, double mutant lines with mutations in or downregulation of amylose or amylopectin biosynthesis genes have been reported. The beI/beIIb double knockdown transgenic lines showed drastic increase in apparent amylose content (AAC) in an indica genetic background (Zhu et al., 2012). Asai et al. (2014) crossed two japonica rice mutants EM10 (be2b) and e1 (ss3a) to generate a ss3a/be2b double null mutant line with 45% AAC. A higher amylose line (be1be2b) with an ultra-high level of RS was developed by crossing BEI and BE2b mutant lines (Miura et al., 2021). The be1be2b mutant had 51.7% AAC and 35.1% RS content. These reports showed that SBE3 (BE2b) plays a crucial role in regulating amylose and amylopectin content. Understanding comprehensively the genetic interaction between starch synthase genes controlling amylose content will greatly contribute to breeding higher amylose rice varieties.
In this study, we developed four near-isogenic lines (NILs) with different allele combinations of SBE3 and GBSS1 derived from the cross between a Korean elite line Hwayeong and a high-amylose cultivar Dodamssal. The four NILs and two parental lines were evaluated for physico-chemical characteristics including seed morphology and starch structure, X-ray diffraction, starch viscosity, and amylopectin chain length distribution. Also, we analyzed the expression of the starch biosynthesis genes in the NILs to understand the interaction among genes with respect to control of amylose content. Our results indicate that SBE3 and GBSS1 alleles of Dodamssal synergistically contributed to an increase in the AAC and altered various physico-chemical characteristics. In addition, SBE3 directly or indirectly regulates expression of the starch synthase and ADP-glucose pyrophosphorylase (AGPase) genes and interacts with GBSS1 in the transcription and posttranslational pathways. Based on these results, the Dodamssal alleles of SBE3 and GBSS1 may be useful in rice breeding for fine-tuning amylose content and producing starch suited for industrial applications.
Materials and methods
Generation of the four near-isogenic lines
In our previous study, a recombinant inbred line (RIL) population (F6:7) was developed from a cross between Hwayeong (SBE3/Wxb) and Dodamssal (sbe3/Wxlv; Adeva et al., 2020). Among the RIL population, two lines (CR2121 and CR2138) harboring the Dodamssal alleles of sbe3 and Wxlv, were crossed with Hwayeong to generate F1 seeds and the F2 plants were used for genetic analysis (Adeva et al., 2020). Among 210 F2 plants, four homozygous genotypes (hereafter referred to as the NIL 1, NIL 2, NIL 3, and NIL 4) were selected using two gene-specific CAPS markers (CS02_001 for SBE3 T/C SNP and CS06_001 for GBSS1 T/G SNP; Supplementary Figure 1; Supplementary Table 1). NIL 1 was homozygous for Hwayeong at SBE3 (SBE3) and GBSS1 (Wxb) whereas NIL 2 was homozygous for Hwayeong at SBE3 (SBE3) and Dodamssal at GBSS1 (Wxlv). NIL 3 was homozygous for Dodamssal at SBE3 (sbe3) and Hwayeong at GBSS1 (Wxb) and NIL 4 has a genotype of homozygous for Dodamssal at SBE3 (sbe3) and GBSS1 (Wxlv). For NIL 1, NIL 2, NIL 3, and NIL 4, 18, 11, four, and nine F2 plants were selected, respectively, to account for possible background effects. Selected F2 plants of NIL 1–4 were grown as lines (20 plants per line) during the F3:4 generation and F5 seeds harvested from F4 plants were used in this study. Plant materials used in this study were grown in the experimental field of Chungnam National University, Daejeon, South Korea in 2020 and seeds were harvested from five plants per line at 45 days after flowering and dried in the greenhouse for 2 weeks.
DNA extraction and genotype analysis
Fresh leaves were collected from each plant and genomic DNA extraction was performed using the CTAB method as described by Causse et al. (1994) with minor modifications. PCR was conducted as described by Shim et al. (2019) with minor modifications in the amplification profile: 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 55–58°C for 30 s, and 72°C for 30 s, and 5 min at 72°C of final extension. PCR amplicons were separated on a 3% agarose gel stained with StaySafe Nucleic Acid Gel Stain (RBC, New Taipei City, Taiwan). Restriction enzymes AccI and SpeI (NEB, MA, United States) were used according to manufacturer’s instructions. Sanger sequencing of PCR products was conducted by SolGent sequencing service (SolGent Co. Ltd., Daejeon, Korea).
Evaluation of seed morphological traits
Grain weight was measured from three plants per line with 100 randomly selected seeds per plant. Endosperm microstructure was observed using scanning electron microscopy (SEM). Transverse section of the middle of the fully matured seeds was made using a razor blade (DORCO, Seoul, Korea) and coated with platinum (Pt). Endosperms of each sample were observed using CLARA (TESCAN, Czech Republic) at 10 kV with magnification of 1,000 and 3,000x in the Center for Research Facilities (Chungnam National University, Daejeon, Korea).
Starch isolation
Rice starch was isolated from polished rice of Dodamssal, Hwayeong, and the four NILs following the method described by Kim et al. (2020) with some modifications. Polished rice samples (10 g) were treated with 20 ml of 0.2% sodium hydroxide solution. After 2 h of shaking at room temperature at 200 rpm, samples were ground with a mortar and pestle. Thirty milliliter of 0.2% sodium hydroxide solution was added and samples were shaken for another 2 h at room temperature at 200 rpm. The resultant slurry was then filtered through 100- and 325-mesh sieves. The filtrate was centrifuged at 3,000 g for 20 min. The sediment was washed with distilled water (50 ml) and centrifuged at 3,000 g for 15 min, and these steps were repeated twice. After discarding the supernatant, the dark tailings layer atop the starch was carefully scraped and discarded. The starch was washed with water and centrifuged at 3,000 g for 15 min. These steps were repeated until the tailings fraction became negligible. The starch was washed with absolute ethanol and centrifuged at 3,000 g for 15 min and then dried at 40°C for 2 days before X-ray diffraction analysis, chain length distribution analysis, and in vitro starch digestibility assay.
Chain length distribution of amylopectin in rice starch
Chain length distribution of amylopectin in rice starch was determined using a High-Performance Anion-Exchange Chromatography Coupled with Pulsed Electrochemical Detection (HPAEC-PAD) system. Sample preparation and analysis were conducted following the method described by Park et al. (2020) with minor modifications. Starch samples (10 mg) were dispersed in 2 ml 90% dimethyl sulfoxide DMSO and boiled with continuous stirring for 20 min. The solubilized starch samples were precipitated with 6 ml of absolute ethanol, and centrifuged at 2,700 rpm for 12 min. The precipitates were dissolved in 2 ml of 50 mM sodium acetate buffer (pH 3.5) and heated in a boiling water bath with continuous stirring for 20 min. After the solution was equilibrated at 37°C, isoamylase (E-ISAMY, 4.16 μl, 240 U/mg, Megazyme Co., Wicklow, Ireland) was added and the starch solution was incubated at 37°C with continuous stirring for 24 h. The enzyme was inactivated by boiling for 10 min. An aliquot (200 μl) of the debranched starch was diluted with 2 ml of 150 mM NaOH. The sample was filtered through a 0.45-μm nylon syringe filter and injected into the HPAEC-PAD on a Dionex ICS-5000 system (Dionex Co., CA, United States) consisting of a CarboPac PA200 column (3 × 250 mm). Separation was achieved using a gradient eluent with 150 mM NaOH and 0.6 M sodium acetate in 150 mM NaOH at a flow rate of 0.5 ml/min.
X-ray diffraction
X-ray diffraction (XRD) analysis was performed using the isolated starch with X-ray diffractometer D8 ADVANCE (Bruker, Bremen, Germany) operated at 40 kV and 40 mA in the Center for Research Facilities (Chungnam National University, Daejeon, Korea). Diffractograms were obtained from 4 to 50° (2θ) at a scan rate of 2 s/step.
Pasting characteristics
The pasting characteristics of rice flour were evaluated using rapid viscosity analysis RVA Super 4 (Newport Scientific, Sydney, Australia). Fine rice flours (3 g) of each sample were placed in an aluminum container and dispersed in 25 ml of deionized water. Samples were kept at 50°C for 1 min, and the temperature was increased from 50 to 95°C for 3.48 min and maintained at 95°C for 2.05 min. After cooling at 50°C for 3.48 min viscosity characteristics (peak, trough, breakdown, final, and setback) of the samples were evaluated. The analysis was carried out in the Department of Southern Area Crop Science, Rural Development Administration, Miryang, Korea.
In vitro starch digestibility assay
In vitro starch digestibility assay was conducted following the methods described by Englyst et al. (1992) and Chung et al. (2009) with modifications. 450 mg of porcine pancreatic alpha-amylase (P-7545; Sigma-Aldrich Co., MO, United States) was dispersed in 4 ml of distilled water in the 50 ml of conical tube (SPL Life Science, Pocheon, Korea) and centrifuged at 1,500 g for 10 min. A 2.7 ml aliquot of the supernatant was transferred to a new 50-ml tube and 0.3 ml of amyloglucosidase (A-9913; Sigma-Aldrich Co., MO, United States) and 0.2 ml of invertase (E-INVPD; Megazyme Co., Wicklow, Ireland; 250 mg/ml) were added to the solution. Starch (100 mg) and 4 ml of 0.5 M sodium acetate buffer (pH 5.2) were added to each glass test tube (15 mm × 120 mm). One milliliter of enzyme solution and five glass beads (4 mm diameter) were added to each glass tube which were then incubated in a shaking water bath (37°C, 200 rpm). Aliquots (0.1 ml) were collected at each six time points (0, 15, 30, 60, 120, and 240 min) and 1 ml of 80% ethanol was added to stop the reactions. Tubes were centrifuged for 10 min with 2,000 rpm, and the glucose content was measured by supernatant of each sample. GOPOD kit (K-GLUC; Megazyme Co., Wicklow, Ireland) was used for determination of glucose content.
Amylose and resistant starch content
All measurements and calculations in estimating the AC and RS content were performed by strictly following the Megazyme assay kit procedures (Megazyme Co., Wicklow, Ireland). For estimation of amylose content, finely crushed polished rice samples (25 mg) were completely dispersed by heating in 1 ml of dimethyl sulfoxide (DMSO). Ethanol (95%) precipitation of the starch was performed to remove lipids. The amylopectin fraction was removed by precipitation with concanavalin A followed by centrifugation. The amylose and total starch (in a separate aliquot of the acetate/salt solution) were both enzymatically hydrolyzed to D-glucose. The amylose and total starch were measured using a spectrophotometer and the absorbance was read at 510 nm against a reagent blank. For estimation of RS content, each sample (100 mg) was incubated in a shaking 37°C water bath and digested with pancreatic α-amylase and amyloglucosidase (AMG) for 16 h. The reaction was terminated by adding an equal volume of absolute ethanol and, after the centrifugation, the RS was recovered as a pellet. The RS pellets were dissolved in 2 ml of 2 M KOH with vigorous mixing in an ice-water bath with a magnetic stirrer followed by neutralization with acetate buffer. AMG was added to the solutions to hydrolyze the starch to glucose which was measured with glucose oxidase/peroxidase (GOPOD). The solution served as the measure of the RS content of each sample. The absorbance was read at 510 nm against a reagent blank.
RNA extraction and qRT-PCR
Total RNA was extracted from panicle samples of the four NILs and two parental lines at 15 days after flowering as described by Jeon et al. (2021). Panicle samples at 15 days after flowering were used as starch-related genes are actively expressed at this stage. cDNA was synthesized from the total RNA samples and amplified using a kit (SMART GENE, Daejeon, Republic of Korea), and quantitative real-time PCR (qRT–PCR) was performed using a CFX real-time PCR system with SYBR Green Master mix (SMARTGENE, Daejeon, Korea). The rice Ubiquitin5 (UBQ5) gene was used for normalization. Primers for starch-related genes developed from previous studies were used and are listed in Supplementary Table 1 (Zeng et al., 2007; Zhou et al., 2016; Baysal et al., 2020).
Statistical analysis
One-way ANOVA, Tukey’s test, and regression analysis were conducted using Minitab19 software (https://www.minitab.co.kr/; accessed on February 3, 2022). Genetic interaction was determined by multiple regression model with two genes as independent variables and interaction term.
Results
Comparison of SBE3 and GBSS1 sequence in parental lines
In our previous study, nucleotide sequences of SBE3 and GBSS1 were compared using whole-genome sequencing (WGS) between Hwayeong and Dodamssal (Adeva et al., 2020). In this study, we confirmed these sequence variations using Sanger sequencing. For SBE3 gene, one T/C SNP (T for Hwayeong and C for Dodamssal) was detected in 16th exon, which led to change of amino acid from leucine (Hwayeong) to proline (Dodamssal; Supplementary Figure 2). For GBSS1, Hwayeong has a Wxb allele whereas Dodamssal has a Waxy ancestral allele Wxlv derived from a Korean weedy rice “Gimcheonaengmi” (Adeva et al., 2020).
SBE3 variation changed seed morphology and starch structure in rice endosperm
Seeds were compared between the four NILs and parental lines (Hwayeong and Dodamssal; Figure 1). Grains of Dodamssal, NIL 3, and NIL 4, all of which harbor sbe3 alleles, exhibited an opaque and floury endosperm, while a translucent endosperm was observed in grains of Hwayeong, NIL 1, and NIL 2, which carry SBE3 alleles. This observation suggests that variation in SBE3 was associated with change in endosperm morphology.

Figure 1. Comparison of seed morphology of two parental lines and four near-isogenic lines (NILs). (A) Dodamssal, (B) Hwayeong, (C) NIL 1 (SBE3/Wxb), (D) NIL 2 (SBE3/Wxlv), (E) NIL 3 (sbe3/Wxb), and (F) NIL 4 (sbe3/Wxlv).
To observe the starch granule structure of the grains, SEM analysis was conducted (Figure 2). Hwayeong, NIL 1, and NIL 2 showed polygonal starch granules with sharp edges, smooth flat surfaces, and compound starch granules (Figure 2). However, the endosperms of Dodamssal, NIL 3, and NIL 4 were filled with rounded, irregularly shaped starch granules with air spaces between granules (Figure 2). No other obvious differences were observed between NIL 1 and NIL 2, and NIL 3 and NIL 4, indicating that the variation in SBE3 might be a major determinant of the difference in seed morphology and granule structure between Hwayeong and Dodamssal.
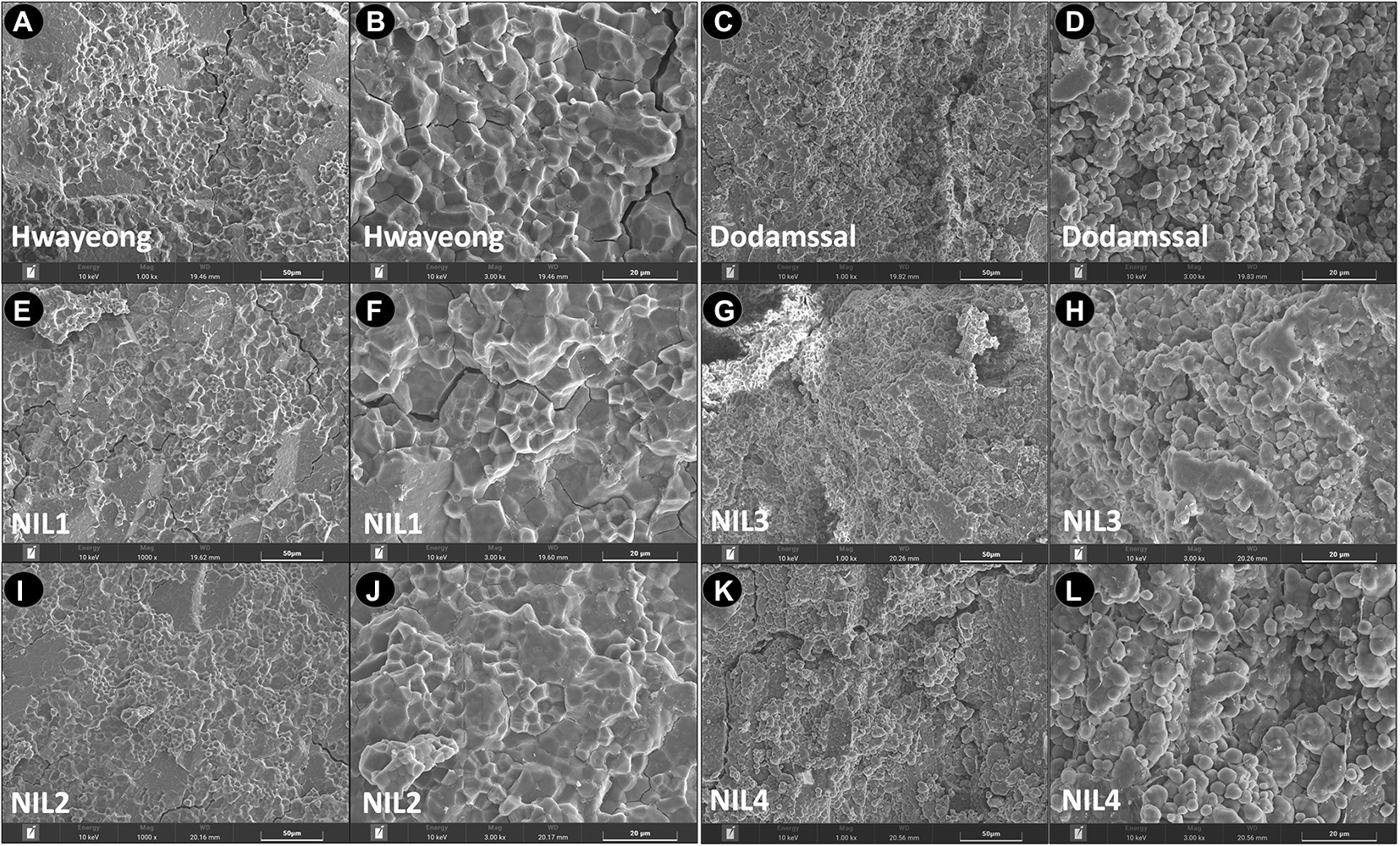
Figure 2. Ultrastructure of endosperm surface structure observed by scanning electron microscope in two parental lines and four NILs. Hwayeong (A,B), Dodamssal (C,D), NIL 1 (E,F), NIL 2 (I,J), NIL 3 (G,H), and NIL 4 (K,L) with two different magnifications (x 1,000 and x 3,000).
One hundred grain weight (100-GW) was also measured, and NIL 1, NIL 2, NIL 3, and NIL 4 showed 2.48, 2.62, 2.15, and 2.49 g 100-GW, respectively (Supplementary Figure 3A). NIL 2 had a significantly higher 100-GW among genotypes while NIL 3 had a significantly lower 100-GW than the other genotypes. Although NIL 4 has an sbe3 allele, 100-GW of NIL 4 was significantly higher than NIL 3. This could be explained by the presence of the Wxlv in NIL 4. The four NILs did not show significant differences in other agronomic traits, such as grain fertility, plant height, panicle number, and panicle length (Supplementary Figures 3B–E). These results suggest that the progenies from two japonica lines are similar in agronomic traits except 100-GW and starch-related traits. It is also possible that the background effects were neutralized by selecting 4–18 F2 plants per each NIL for evaluation.
SBE3 Dodamssal allele led to increase in long amylopectin chains
Chain length distribution of amylopectin was examined to understand the difference in amylopectin structure (Figure 3A). The six genotypes were classified into two groups based on their patterns of chain length distribution. Hwayeong, NIL 1, and NIL 2 composed one group, and the other group included Dodamssal, NIL 3, and NIL 4. This result indicates that the sequence variation in SBE3 greatly affected amylopectin chain length distribution, but variation in GBSS1 did not. Despite Dodamssal and NIL 4 having higher AAC than NIL 3, the amylopectin chain length distribution patterns of the three genotypes were similar. To observe the detailed differences in chain length distribution, each value of four NILs was subtracted showing two major different distribution patterns and Δ (Delta) normalized peak area (Figure 3B). For example, NIL 2-1 is the value obtained by subtracting NIL 1 from NIL 2. NIL 3-1, NIL 4-1, NIL 3-2, and NIL 4-2 showed similar distribution pattern while NIL 2-1 and NIL 4-3 had different distribution patterns overlapping to each other. Comparison of NIL 3-1, NIL 4-1, NIL 3-2, and NIL 4-2 indicated the effect of SBE3 variation and the subtraction curve of this group showed a decrease in the proportion of shorter chains, degree of polymerization (DP) 6-16 with an increase in the fraction of longer chains (DP 17–81). Both NIL 2-1 and NIL 4-3 displayed an increase in DP 6–12, but the difference was insignificant. Amylopectin chains were classified into four groups (A, B1, B2, and B3) based on the amylopectin cluster model (Hanashiro et al., 1996). Dodamssal, NIL 3, and NIL 4 exhibited significantly higher average chain length and distribution of B2 and B3 chains, while Hwayeong, NIL 1, and NIL 2 showed significantly increased A chains (Supplementary Table 2). These results indicate that the Dodamssal sbe3 allele conferred a change in the amylopectin structure.
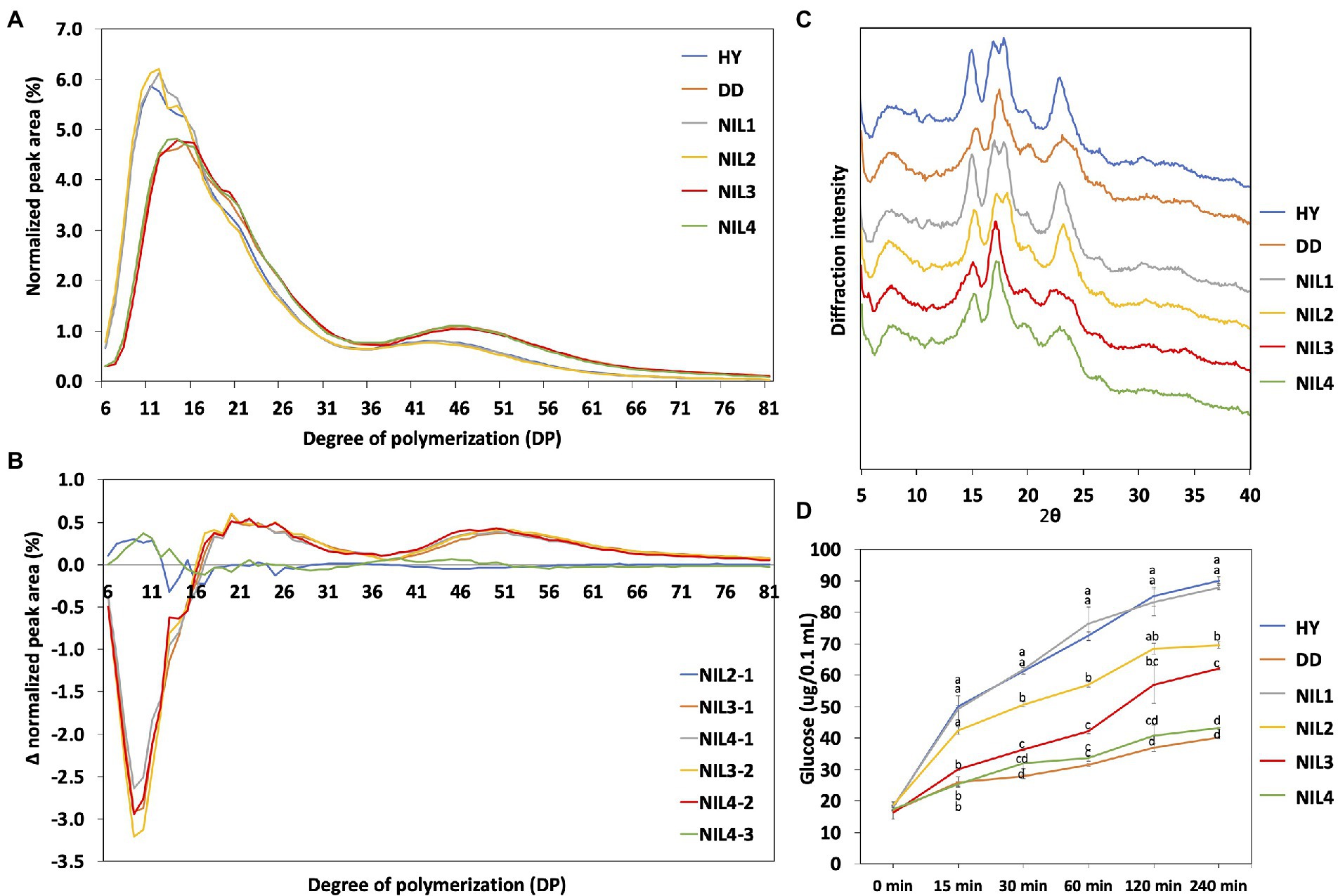
Figure 3. Physico-chemical characteristics of four NILs. (A) Chain length distribution of the two parental lines and four NILs. (B) Differences in the molecular structure of amylopectin between the four NILs. Differences are shown as Δ (Delta) normalized peak area. (C) X-ray diffraction pattern of rice starch samples. (D) Starch digestibility analysis of the two parental lines and four NILs. Glucose content was determined by GOPOD kit (Megazyme Co., Wicklow, Ireland) from the digestion of each starch sample. The same letter is not significantly different at p = 0.05 based on Tukey’s test. HY, Hwayeong; DD, Dodamssal.
Starch crystallinity of four NILs
To examine the crystal structure of the rice starch in parental lines and the four NILs, XRD analysis was conducted (Figure 3C). Starch crystallinity is classified into three types: A-type, B-type, and C-type, a hybrid of A-type and B-type based on its XRD pattern (Park et al., 2020). Hwayeong, NIL 1, and NIL 2 showed continuous double diffraction peaks at 17 and 18° 2θ, and strong diffraction peak at 15 and 23° 2θ. Based on these results, these three genotypes were classified as having an A-type starch. However, a strong single diffraction peak was detected at near 17° 2θ in Dodamssal, NIL 3, and NIL 4, and relatively weak peaks were found at 15 and 23° 2θ, indicating that these lines could be classified as having B-type starch crystals. These results indicated that SBE3 plays an important role in rice starch crystal structure.
Different starch viscosity properties in four NILs
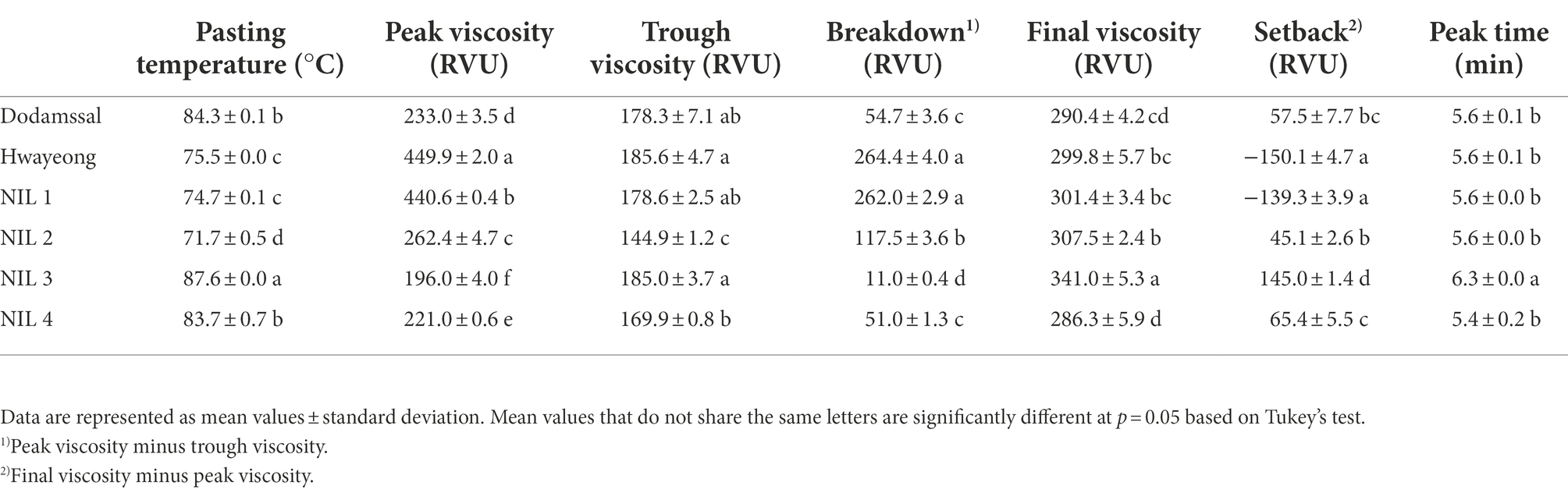
To investigate the viscosity properties of rice starch, a Rapid Visco-Analyser (RVA) was employed (Table 1). Significant differences were observed in all properties. NIL 3 showed the highest pasting temperature, final viscosity, and peak time. Dodamssal and NIL 4 had similar starch viscosity properties, and pasting temperature of Dodamssal and NIL 4 were higher than those of Hwayeong, NIL 1, and NIL 2. Hwayeong and NIL 1 displayed the highest peak viscosity. Peak viscosity of NIL 3 with the sbe3 Dodamssal allele was lower than the other three NILs. Although NIL 2 and NIL 3 had similar amylose content, their starch viscosity traits were considerably different suggesting that the variation in SBE3 and GBSS1 alleles affects the starch viscosity properties.
High amylose and resistant starch in Dodamssal is associated with low starch digestibility
To investigate the effect of variation in SBE3 and GBSS1 on starch digestibility, digestibility assays were performed as shown in Figure 3D. The glucose content of the digested starch samples was measured at six time intervals each (0, 15, 30, 60, 120, and 240 min). Hwayeong and NIL 1 had the highest increase in glucose content. NIL 2 was lower than that of Hwayeong and NIL 1, but higher than that of NIL 3. Although NIL 2 and NIL 3 had similar AAC, the starch of NIL 2 was more easily digested than NIL 3. This result might be due to different RS contents between NIL 2 and NIL 3 (see below). NIL 4 and Dodamssal showed the lowest starch digestibility due to high amylose content and RS content. These data suggest that the combination of two alleles sbe3 and Wxlv would be valuable for non-digestible starch rice breeding program.
Genetic interaction of SBE3 and GBSS1 in amylose and resistant starch content
Apparent amylose content of Hwayeong, Dodamssal, and the four NILs were compared (Figure 4). Dodamssal and Hwayeong showed about 33 and 15% AAC, respectively (Figure 4A). AAC values about 15, 21, 21, and 35% were observed in NIL 1, NIL 2, NIL 3, and NIL 4, respectively, indicating that the Dodamssal alleles of SBE3 and GBSS1 (sbe3 and Wxlv) significantly enhanced the AAC (Figure 4B). Replacement of Wxb in NIL 1 with Wxlv NIL 2 at GBSS1 corresponded to a 5% increase of AAC, while the difference in AAC between NIL 3 and NIL 4 was about 14%. To determine the genetic interaction between SBE3 and GBSS1, a multiple regression model was employed. Significant interaction was found between two genes (p < 0.001, Supplementary Table 3), and the Dodamssal alleles of SBE3 and GBSS1 synergistically increased the AAC in the Hwayeong genetic background, which was consistent with our previous observation (Adeva et al., 2020).
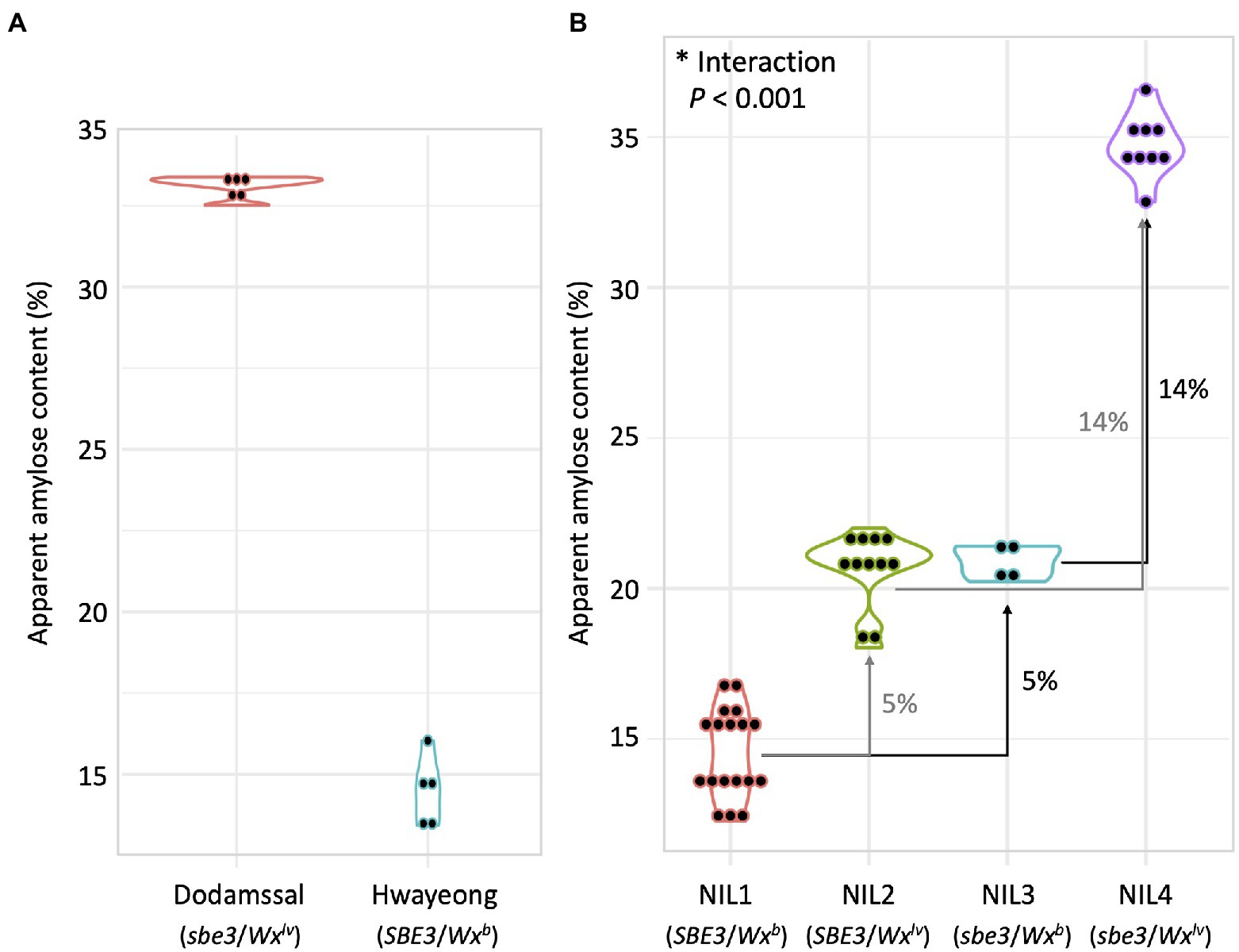
Figure 4. Comparison of apparent amylose content (AAC) between (A) Dodamssal and Hwayeong, (B) four NILs. Violin plots were used to show distribution of data points. The numbers beside the arrows indicate increased AAC.
Resistant starch content was also measured from rice flours of Hwayeong, Dodamssal, and the four NILs. Hwayeong, NIL 1, and NIL 2 had 0.07, 0.07, and 0.30% RS content, respectively (Supplementary Figure 4). While the RS content of NIL 2 was higher than those of Hwayeong and NIL 1, the difference was not significant. Dodamssal and NIL 4 exhibited similar RS content of 8.17 and 7.55%, respectively. NIL 3 had 11.51% RS content and was significantly higher than Dodamssal and NIL 4 (p < 0.001). Despite NIL 3 and NIL 4 both carrying the Dodamssal sbe3 allele, the RS content of NIL 3 was higher than that of NIL 4 suggesting an interaction between SBE3 and GBSS1. Multiple regression analysis showed a significant interaction between SBE3 and GBSS1 in regulating RS (p < 0.001, Supplementary Table 4). Further study is needed to understand the association between SBE3 and GBSS1 in the regulation of the RS content.
Dynamic changes in starch-related genes expression
To understand the genetic interaction of two genes (GBSS1 and SBE3) on AAC and the physico-chemical properties of the four NILs, gene expression profiling was carried out using panicle samples at 15 days after flowering (Figure 5; Supplementary Figure 5). Significant differences were observed in expression level of GBSS1 between the NILs. NIL 4 showed the highest gene expression consistent with AAC and the GBSS1 expression level in NIL 4 was 6.7, 1.3, and 3.5 times higher than that observed for NIL 1, NIL 2, and NIL 3, respectively. Although NIL 2 and NIL 3 displayed similar AAC, expression of GBSS1 in NIL 2 was 2.7 times higher than NIL 3. SBE3 expression in NIL 3 was significantly higher (about 2-fold) than the other genotypes. Paralogous isoforms of GBSS1 and SBE3 expression were also examined (Supplementary Figure 5A). The expression level of GBSS2 and SBE1 was upregulated in NIL 3 and NIL 4 compared to NIL 1 and NIL 2. NIL 4 showed the highest SBE4 expression level, which was significantly different from the other NILs.
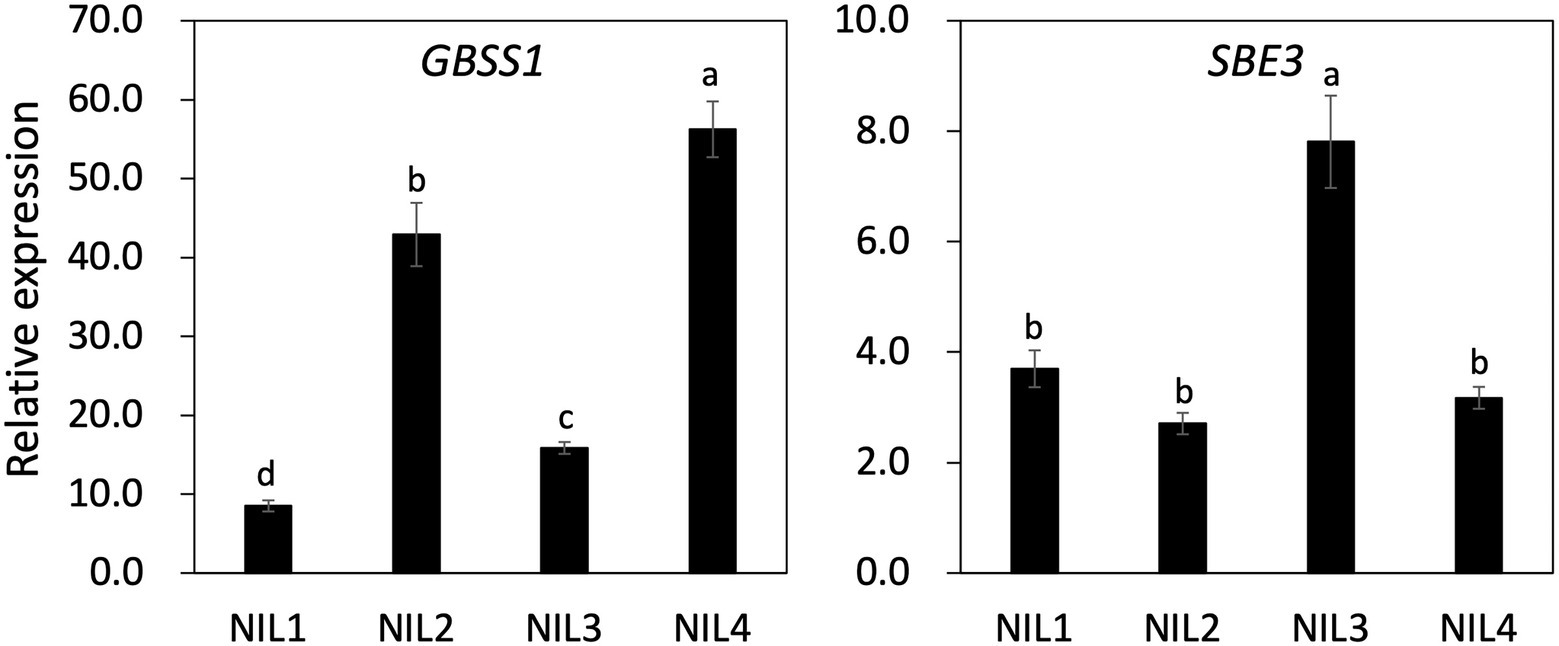
Figure 5. Gene expression of GBSS1 and SBE3 in four NILs using panicles samples from 15 days after flowering. UBQ5 was used for gene normalization. The same letter on the bar is not significantly different at p < 0.05 based on Tukey’s test, respectively.
The expression of other starch-related genes including ADP-glucose pyrophosphorylase (AGP; Supplementary Figure 5B), soluble starch synthase (SS; Supplementary Figure 5C), starch debranching enzyme [Pullulanase (PUL) and Isoamylase (ISA); Supplementary Figure 5D], and starch phosphorylase (PHOL and PHOH; Supplementary Figure 5E) was examined. NIL 3 and NIL 4 or NIL 3 alone showed significantly higher expression of the soluble starch synthase isoforms compared to NIL 1 and NIL 2. Moreover, NIL 3 had higher expression of SSI, SSIIa, SSIIc, SSIIIa, SSIIIb, and SSIVb than NIL 4. Similar gene expression patterns were also observed for the AGPase genes and starch debranching enzyme genes. The transcript levels of AGPS1, AGPS2a, AGPS2b, and AGPL2 were higher in NIL 3 than NIL 1 and NIL 2.
The transcript level of GBSS1 and SBE3 was examined in Hwayeong and Dodamssal (Supplementary Figure 6). Dodamssal showed significantly higher expression level of GBSS1, while SBE3 expression was not significantly different between two parental lines. Dodamssal also displayed significantly higher expression in two major starch biosynthesis genes, SSI and SS3a, than Hwayeong. For SBE paralogous genes, no significant difference between two lines was not observed in SBE1 whereas Hwayeong showed higher SBE4 level than Dodamssal.
Transcript profiling indicates that the different alleles and allele combinations of GBSS1 and SBE3 changed the expression level of not only GBSS1 and SBE3 but also other starch-related genes that might be responsible for the increased AAC and altered physico-chemical characteristics of the rice starch in the NILs.
Discussion
The functional rice variety “Dodamssal” with high amylose and resistant starch content (RS) is widely used for products such as rice noodles (Cho et al., 2019; Park et al., 2020). However, Dodamssal has weaknesses such as lacks of disease and insect pest resistances and low-yield. The program to develop new versions of Dodamssal with high-yield potential and resistance to diseases is underway using the marker assisted selection (Cho et al., 2019). In our previous study, we determined that two major starch biosynthesis genes, SBE3 and GBSS1 are responsible for the high amylose and RS in Dodamssal (Adeva et al., 2020). However, the molecular mechanism how these two genes interact at the molecular level and alter the physico-chemical properties of rice starch remained to be determined. In this study, we developed four near-isogenic lines carrying different allelic combinations of two major starch biosynthesis genes, SBE3 and GBSS1, to understand the genetic interaction and how these two genes alter the physico-chemical properties of rice starch.
Analysis of the seed morphology and starch granule structure of the NILs and parental lines confirmed the association of these traits with the variation in SBE3 (Figures 1, 2). These results are consistent with previously reported SBE3 mutants, EM10 and Jiangtangdao 1, which also showed a similar floury endosperm phenotype (Yang et al., 2012). In addition to differences in their endosperms, grain weight (100-GW) variation was observed among the four NILs with NIL 3 showing significantly lower 100-GW than the other three NILs (Supplementary Figure 3A). Nishi et al. (2001) found that SBE3 (Ae) gene dosage was associated with grain weight and size. The ae allele dosage in the endosperm decreased grain weight and grain size with a decrease in the relative BE2b protein content. Yang et al. (2016) also showed that SBE3 overexpression of transgenic T1 plants generated in the Jiangtangdao 1 (sbe3) genetic background had significantly higher 1,000-seed weight than control plants. The significantly higher 100-GW of NIL 2 than NIL 1 appears to be due to the presence of Wxlv in NIL 2. This result is consistent with that of the previous study that the Wxlv allele explained 14.6% (p < 0.001) of the phenotypic variation of the grain weight in the RIL population (Adeva et al., 2020). Although these results suggest the association of Wxlv with grain weight, it remains to be determined whether Wxlv or a tightly linked QTL contributed to an increase in grain weight.
Variations in SBE3 and GBSS1 significantly altered the physico-chemical properties of rice starch. Of these two genes, variation in SBE3 strongly affected the starch properties. For X-ray diffraction analysis, Hwayeong, NIL 1, and NIL 2, harboring an SBE3 allele, showed an A-type diffraction pattern, while Dodamssal, NIL 3, and NIL 4, harboring an sbe3 allele, displayed a B-type XRD pattern. Park et al. (2020) reported that Dodamssal displayed a C-type X-ray diffraction pattern with a predominant B-type (CB-type) based on the broad peak at 22–23° 2θ without shoulder peak at 18° 2θ. Similar diffraction patterns were observed in Dodamssal, NIL 3, and NIL 4. However, the diffraction peak of NIL 3 at 22–23° 2θ was slightly broader than Dodamssal and NIL 4. The SBE3 (BEIIb) mutant EM10 and the be1/be2b double mutant were classified as having B-type crystal structure (Miura et al., 2021). Jiangtangdao1, which harbors the same T/C mutation in SBE3 as NIL 3, also had a B-type diffraction pattern (Yang et al., 2016). The donor of the sbe3 allele, “Goami2” was reported as a B-type starch (Yoon et al., 2013). These studies consistently confirmed that mutation in SBE3 led to a B-type or a CB-type crystal structure.
With regard to starch viscosity properties, NIL 3 had the highest pasting temperature, final viscosity, and peak time values although NIL 4 harbors the same sbe3 allele and has higher AAC than NIL 3. NIL 3 requires more energy and time for gelatinization, which may be associated with a higher RS and the amylose-lipid complexes found in resistant starch. When the thermal properties of starch in the be1, be2b, and be1be2b mutants were analyzed, the onset, peak, and conclusion gelatinization temperature were associated with resistant starch and amylose contents (Miura et al., 2021). It was previously demonstrated that Wxlv affected eating and cooking quality of rice grains by modulating the size of amylose molecules (Zhang et al., 2019). Thus, presence of Wxlv in NIL 4 may explain the difference in starch viscosity traits compared to NIL 3.
Variations in GBSS1 and SBE3 also changed the amylopectin chain length distribution (Figure 3B). The comparison of amylopectin chain length distribution between NIL 2–1 and NIL 4–3 revealed that Wxlv was associated with an increase in amylopectin short chains (DP 6–12), consistent with a previous observation (Crofts et al., 2012). Moreover, a T/C SNP in SBE3 was associated with a decrease in short chains (DP 6–16) and an increase in long chains (DP 16–81). In particular, the distribution patterns of NIL 3-1, NIL 4-1, NIL 3-2, and NIL 4-2 were consistent with those of previous reports on the distribution pattern between EM10 and wild type (Nishi et al., 2001; Miura et al., 2021).
Deficiencies in starch synthases or starch branching enzymes can significantly increase the AAC. Two SS3a mutant lines, a Tos17 insertion and T65 MNU mutant, displayed an increased GBSS1 protein expression level in the developing seeds, and GBSS1 mRNA was upregulated in Tos17 insertion mutant compared to the wild type (Fujita et al., 2007). The GBSS1 protein level of ss3a, be2b, and ss3a/be2b deficiency mutants slightly increased compared to their wild type counterparts (Asai et al., 2014). In addition, a significant interaction was reported between ss3a and Wx allele in starch synthesis, and the ss3a mutant in the indica (Wxa) genetic background showed higher amylose content or resistant starch than the ss3a mutant in japonica (Wxb) rice (Crofts et al., 2012; Zhou et al., 2016). The relative GBSS1 content was enhanced in ss3a/Wxb than SS3a/Wxb genotype, indicating a loss-of-function of SS3a led to upregulation of the GBSS1 protein level (Crofts et al., 2012). Synergistic increase of the AAC was observed when the different genotype combinations of SS3a and GBSS1 were compared (Crofts et al., 2012). The ss3a/wxa line showed the highest AAC (41.3%), and two alleles ss3a and Wxa contributed to an increase in the AAC compared to their counterparts SS3a and Wxb, respectively. Interestingly, the difference in the GBSS1 protein expression between SS3a/Wxa and ss3a/wxa genotype was not significant and the synergistic effect was possibly due to the elevated AGPase activity (Crofts et al., 2012). The expression level of GBSS1 mRNA or protein in ss3a mutant developed by Zhou et al. (2016) was not significantly different with wild type, indicating that interaction possibly occurred at the posttranslational level (Zhou et al., 2016). It is apparent that the genetic interaction might exist between starch synthase genes and GBSS1 in regulating amylose content.
In the present study, the sbe3 allele led to significantly increased amylose content and GBSS1 gene expression (Figures 4, 5). NIL 3 (sbe3/Wxb) showed 21% AAC and 1.9 times higher GBSS1 transcript level than NIL 1 (SBE3/Wxb), and NIL 4 (sbe3/Wxlv) has 35% AAC with 6.6 times higher GBSS1 expression than NIL 1 (SBE3/Wxb). The data combined from this study and previous reports suggest that SBE3 directly or indirectly regulates the expression of starch synthase and AGPase genes including SS3a and interacted with GBSS1 in the transcriptional and posttranslational pathways. However, more specific mechanisms need to be examined to fully validate the possible genetic interactions among SBE3, SS3a, and GBSS1. Another genetic interaction between SBE3 and GBSS1 was observed in the regulation of RS content (Supplementary Figure 4; Supplementary Table 4). Wxlv decreased the RS content in the sbe3 genetic background [comparison between NIL 3 (sbe3/Wxb) and NIL 4 (sbe3/Wxlv)] while Wxlv did not significantly change the RS content in the SBE3 genetic background [comparison between NIL 1 (SBE3/Wxb) and NIL 2 (SBE3/Wxlv)]. A similar result was observed from a previous study. Goami2, one of the parental lines of Dodamssal, has the sbe3 and Wxb alleles which is of same genotype with NIL 3 (Adeva et al., unpublished data) and displayed 17.71% RS content (Cho et al., 2019). However, the RS content of Dodamssal was 13.62%, indicating the existence of genetic interaction between two genes in controlling RS content in rice (Cho et al., 2019). The interaction could be caused by substrate competition between amylose and RS biosynthesis. Further study is required to understand the genetic mechanisms in regulating starch content and their interactions.
Previously, studies have focused on identifying single genes controlling traits of agronomic importance. To better understand how the quantitative traits are regulated, QTL-QTL or gene–gene interactions need to be studied using genetically homogeneous populations, such as chromosome substitution, introgression, and near-isogenic lines to reduce background noises (Mackay, 2014). In the present study, we developed near-isogenic lines derived from a cross between two closely related japonica varieties. Because of their genetic closeness between two parental lines, the interaction between two genes, SBE3 and GBSS1, was clearly examined without interference of other genetic factors. NILs for various starch-related genes with different alleles will be necessary to better define the genetic relationships between starch-related genes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
K-CS, CA, and S-NA designed the experiments and wrote the manuscript. TT edited the manuscript and provided advice on the experiments. J-WK and J-HC performed physico-chemical trait evaluation. H-SL, HK, and NL conducted the agronomic traits investigation and qRT-PCR analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ015757)” Rural Development Administration, Republic of Korea.
Acknowledgments
We thank Hyun-Jung Chung, Chonnam National University, for chain length distribution analysis and Young-Sook Kim and Eun-Jung Choi for technical assistance.
Conflict of interest
HK was employed by the company LG Chem., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls. 2022.968795/full#supplementary-material
Abbreviations
SBE, Starch branching enzyme; GBSS1, Granule binding starch synthase 1; SS, Soluble starch synthase; DBE, Starch debranching enzyme; ISA, Isoamylase; PUL, Pullulanase; AGP, ADP-glucose pyrophosphorylase; NILs, Near-isogenic lines; QTL, Quantitative trait loci; SNP, Single nucleotide polymorphism; XRD, X-ray diffraction; AAC, Apparent amylose content; RS, Resistant starch.
References
Adeva, C. C., Lee, H.-S., Kim, S.-H., Jeon, Y.-A., Shim, K.-C., Luong, N. H., et al. (2020). Two complementary genes, SBE3 and GBSS1 contribute to high amylose content in japonica cultivar Dodamssal. Plant Breed. Biotechnol. 8, 354–367. doi: 10.9787/PBB.2020.8.4.354
Amagliani, L., O'regan, J., Kelly, A. L., and O’mahony, J. A. (2016). Chemistry, structure, functionality and applications of rice starch. J. Cereal Sci. 70, 291–300. doi: 10.1016/j.jcs.2016.06.014
Asai, H., Abe, N., Matsushima, R., Crofts, N., Oitome, N. F., Nakamura, Y., et al. (2014). Deficiencies in both starch synthase IIIa and branching enzyme IIb lead to a significant increase in amylose in SSIIa-inactive japonica rice seeds. J. Exp. Bot. 65, 5497–5507. doi: 10.1093/jxb/eru310
Baysal, C., He, W. S., Drapal, M., Villorbina, G., Medina, V., Capell, T., et al. (2020). Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc. Natl. Acad. Sci. U. S. A. 117, 26503–26512. doi: 10.1073/pnas.2014860117
Butardo, V. M., Fitzgerald, M. A., Bird, A. R., Gidley, M. J., Flanagan, B. M., Larroque, O., et al. (2011). Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing. J. Exp. Bot. 62, 4927–4941. doi: 10.1093/jxb/err188
Cai, X. L., Wang, Z. Y., Xing, Y. Y., Zhang, J. L., and Hong, M. M. (1998). Aberrant splicing of intron 1 leads to the heterogeneous 5 ' UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 14, 459–465. doi: 10.1046/j.1365-313X.1998.00126.x
Causse, M. A., Fulton, T. M., Cho, Y. G., Ahn, S. N., Chunwongse, J., Wu, K. S., et al. (1994). Saturated molecular map of the Rice genome based on an interspecific backcross population. Genetics 138, 1251–1274. doi: 10.1093/genetics/138.4.1251
Chen, M. H., Bergman, C. J., Pinson, S. R. M., and Fjellstrom, R. G. (2008). Waxy gene haplotypes: associations with pasting properties in an international rice germplasm collection. J. Cereal Sci. 48, 781–788. doi: 10.1016/j.jcs.2008.05.004
Cho, J. H., Song, Y. C., Lee, J. H., Lee, J. Y., Son, Y. B., Oh, S. H., et al. (2019). ‘Dodamssal (Milyang 261)’, functional Rice as a resistant starch with a high amylose content. Kor. Soc. Breed. Sci. 51, 515–522. doi: 10.9787/KJBS.2019.51.4.515
Chung, H. J., Liu, Q., and Hoover, R. (2009). Impact of annealing and heat-moisture treatment on rapidly digestible, slowly digestible and resistant starch levels in native and gelatinized corn, pea and lentil starches. Carbohydr. Polym. 75, 436–447. doi: 10.1016/j.carbpol.2008.08.006
Crofts, N., Abe, K., Aihara, S., Itoh, R., Nakamura, Y., Itoh, K., et al. (2012). Lack of starch synthase IIIa and high expression of granule-bound starch synthase I synergistically increase the apparent amylose content in rice endosperm. Plant Sci. 193-194, 62–69. doi: 10.1016/j.plantsci.2012.05.006
Englyst, H. N., Kingman, S. M., and Cummings, J. H. (1992). Classification and measurement of nutritionally important starch fractions. Eur. J. Clin. Nutr. 46, S33–S50.
Englyst, H., Wiggins, H. S., and Cummings, J. (1982). Determination of the non-starch polysaccharides in plant foods by gas-liquid chromatography of constituent sugars as alditol acetates. Analyst 107, 307–318. doi: 10.1039/an9820700307
Fujita, N., Yoshida, M., Kondo, T., Saito, K., Utsumi, Y., Tokunaga, T., et al. (2007). Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 144, 2009–2023. doi: 10.1104/pp.107.102533
Hanashiro, I., Abe, J., and Hizukuri, S. (1996). A periodic distribution of the chain length of amylopectin as revealed by high-performance anion-exchange chromatography. Carbohydr. Res. 283, 151–159. doi: 10.1016/0008-6215(95)00408-4
Jeon, Y. A., Lee, H. S., Kim, S. H., Shim, K. C., Kang, J. W., Kim, H. J., et al. (2021). Natural variation in rice ascorbate peroxidase gene APX9 is associated with a yield-enhancing QTL cluster. J. Exp. Bot. 72, 4254–4268. doi: 10.1093/jxb/erab155
Kim, H. J., Woo, K. S., Lee, H. U., Nam, S. S., Lee, B. W., Kim, M. Y., et al. (2020). Physicochemical characteristics of starch in sweet potato cultivars grown in Korea. Prev. Nutr. Food Sci. 25, 212–218. doi: 10.3746/pnf.2020.25.2.212
Kim, H., Yoon, M. R., Chun, A., and Tai, T. H. (2018). Identification of novel mutations in the rice starch branching enzyme I gene via TILLING by sequencing. Euphytica 214:94. doi: 10.1007/s10681-018-2174-7
Mackay, T. F. C. (2014). Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 15, 22–33. doi: 10.1038/nrg3627
Miura, S., Koyama, N., Crofts, N., Hosaka, Y., Abe, M., and Fujita, N. (2021). Generation and starch characterization of non-transgenic BEI and BEIIb double mutant Rice (Oryza sativa) with ultra-high level of resistant starch. Rice 14:3. doi: 10.1186/s12284-020-00441-0
Nishi, A., Nakamura, Y., Tanaka, N., and Satoh, H. (2001). Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127, 459–472. doi: 10.1104/pp.010127
Park, J., Oh, S. K., Chung, H. J., and Park, H. J. (2020). Structural and physicochemical properties of native starches and non-digestible starch residues from Korean rice cultivars with different amylose contents. Food Hydrocoll. 102:105544. doi: 10.1016/j.foodhyd.2019.105544
Sano, Y. (1984). Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 68, 467–473. doi: 10.1007/BF00254822
Sano, Y., Katsumata, M., and Okuno, K. (1986). Genetic studies of speciation in cultivated rice. 5. Inter-and intraspecific differentiation in the waxy gene expression of rice. Euphytica 35, 1–9. doi: 10.1007/BF00028534
Satoh, H., Nishi, A., Yamashita, K., Takemoto, Y., Tanaka, Y., Hosaka, Y., et al. (2003). Starch-branching enzyme I-deficient mutation specifically affects the structure and properties of starch in rice endosperm. Plant Physiol. 133, 1111–1121. doi: 10.1104/pp.103.021527
Shim, K.-C., Kim, S., Le, A. Q., Lee, H.-S., Adeva, C., Jeon, Y.-A., et al. (2019). Fine mapping of a low-temperature Germinability QTL qLTG1 using introgression lines derived from Oryza rufipogon. Plant Breed. Biotechnol. 7, 141–150. doi: 10.9787/PBB.2019.7.2.141
Wang, J., Hu, P., Chen, Z. C., Liu, Q. Q., and Wei, C. X. (2017). Progress in high-amylose cereal crops through inactivation of starch branching enzymes. Front. Plant Sci. 8:469. doi: 10.3389/fpls.2017.00469
Yamanaka, S., Nakamura, I., Watanabe, K. N., and Sato, Y. I. (2004). Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theor. Appl. Genet. 108, 1200–1204. doi: 10.1007/s00122-003-1564-x
Yang, R. F., Bai, J. J., Fang, J., Wang, Y., Lee, G. S., and Piao, Z. Z. (2016). A single amino acid mutation of OsSBEIIb contributes to resistant starch accumulation in rice. Breed. Sci. 66, 481–489. doi: 10.1270/jsbbs.16037
Yang, R., Sun, C., Bai, J., Luo, Z., Shi, B., Zhang, J., et al. (2012). A putative gene sbe3-rs for resistant starch mutated from SBE3 for starch branching enzyme in rice (Oryza sativa L.). PLoS One 7:e43026. doi: 10.1371/journal.pone.0043026
Yoon, M., Lee, J., Lee, J., Kwak, J., Chun, A., and Kim, B. (2013). Content and characteristics of resistant starch in high amylose mutant rice varieties derived from Ilpum. Kor. J. Breed. Sci. 45, 324–331. doi: 10.9787/KJBS.2013.45.4.324
Zeng, D., Yan, M. X., Wang, Y. H., Liu, X. F., Qian, Q., and Li, J. Y. (2007). Du1, encoding a novel Prp1 protein, regulates starch biosynthesis through affecting the splicing of Wx(b) supercript stop pre-mRNAs in rice (Oryza sativa L.). Plant Mol. Biol. 65, 501–509. doi: 10.1007/s11103-007-9186-3
Zhang, C. Q., Zhu, J. H., Chen, S. J., Fan, X. L., Li, Q. F., Lu, Y., et al. (2019). Wx(lv), the ancestral allele of Rice waxy gene. Mol. Plant 12, 1157–1166. doi: 10.1016/j.molp.2019.05.011
Zhou, H. J., Wang, L. J., Liu, G. F., Meng, X. B., Jing, Y. H., Shu, X. L., et al. (2016). Critical roles of soluble starch synthase SSIIIa and granule-bound starch synthase waxy in synthesizing resistant starch in rice. Proc. Natl. Acad. Sci. U. S. A. 113, 12844–12849. doi: 10.1073/pnas.1615104113
Keywords: amylose content, near-isogenic lines, physico-chemical trait, starch synthase gene, rice
Citation: Shim K-C, Adeva C, Kang J-W, Luong NH, Lee H-S, Cho J-H, Kim H, Tai TH and Ahn S-N (2022) Interaction of starch branching enzyme 3 and granule-bound starch synthase 1 alleles increases amylose content and alters physico-chemical properties in japonica rice (Oryza sativa L.). Front. Plant Sci. 13:968795. doi: 10.3389/fpls.2022.968795
Edited by:
Mallikarjuna Swamy, International Rice Research Institute (IRRI), PhilippinesReviewed by:
Zilhas Ahmed Jewel, Sejong University, South KoreaRajeev Ranjan, Purdue University, United States
Copyright © 2022 Shim, Adeva, Kang, Luong, Lee, Cho, Kim, Tai and Ahn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Nag Ahn, ahnsn@cnu.ac.kr
†These authors have contributed equally to this work
 Kyu-Chan Shim
Kyu-Chan Shim Cheryl Adeva
Cheryl Adeva Ju-Won Kang
Ju-Won Kang Ngoc Ha Luong
Ngoc Ha Luong Hyun-Sook Lee
Hyun-Sook Lee Jun-Hyeon Cho2
Jun-Hyeon Cho2 HyunJung Kim
HyunJung Kim Thomas H. Tai
Thomas H. Tai Sang-Nag Ahn
Sang-Nag Ahn