- Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland
In nature, all living organisms must continuously sense their surroundings and react to the occurring changes. In the cell, the information about these changes is transmitted to all cellular compartments, including the nucleus, by multiple phosphorylation cascades. Sucrose Non-Fermenting 1 Related Protein Kinases (SnRK2s) are plant-specific enzymes widely distributed across the plant kingdom and key players controlling abscisic acid (ABA)-dependent and ABA-independent signaling pathways in the plant response to osmotic stress and salinity. The main deleterious effects of salinity comprise water deficiency stress, disturbances in ion balance, and the accompanying appearance of oxidative stress. The reactive oxygen species (ROS) generated at the early stages of salt stress are involved in triggering intracellular signaling required for the fast stress response and modulation of gene expression. Here we established in Arabidopsis thaliana that salt stress or induction of ROS accumulation by treatment of plants with H2O2 or methyl viologen (MV) induces the expression of several genes encoding transcription factors (TFs) from the WRKY DNA-Binding Protein (WRKY) family. Their induction by salinity was dependent on SnRK2.10, an ABA non-activated kinase, as it was strongly reduced in snrk2.10 mutants. The effect of ROS was clearly dependent on their source. Following the H2O2 treatment, SnRK2.10 was activated in wild-type (wt) plants and the induction of the WRKY TFs expression was only moderate and was enhanced in snrk2.10 lines. In contrast, MV did not activate SnRK2.10 and the WRKY induction was very strong and was similar in wt and snrk2.10 plants. A bioinformatic analysis indicated that the WRKY33, WRKY40, WRKY46, and WRKY75 transcription factors have a similar target range comprising numerous stress-responsive protein kinases. Our results indicate that the stress-related functioning of SnRK2.10 is fine-tuned by the source and intracellular distribution of ROS and the co-occurrence of other stress factors.
Introduction
The environment is highly dynamic and undergoes constant changes. The fluctuating environmental conditions that have a negative impact on plant functioning are considered stress factors (Srivastava et al., 2021). Salinity is one of the most common environmental factors limiting plant productivity and affecting agricultural yield (Acosta-Motos et al., 2017). It is estimated that globally about 20% of irrigated land is affected by salinity and therefore unsuitable for agriculture, and by 2050 this fraction will increase to 50% (Srivastava et al., 2021). The main deleterious effects of salinity are the reduction of water potential and the appearance of water deficiency stress, and disturbances in ion balance. As a result, salt stress limits plant growth, inhibits photosynthesis, affects redox homeostasis, modulates antioxidant metabolism, the uptake and balance of mineral nutrients, and accumulation of osmolytes (for review see Yang and Guo, 2018; Hasanuzzaman et al., 2021; Srivastava et al., 2021). A proper recognition of the stress conditions, triggering adequate signaling pathways and metabolic adjustment are critical for the optimization of plant growth, reproduction, and survival under challenging conditions.
Stress signals are recognized and transmitted to diverse cellular compartments by specialized signaling pathways in which protein kinases and phosphatases are key components. Among the protein kinases involved in stress and ABA signal transduction, Sucrose non-fermenting 1-Related protein Kinases (SnRKs) play an important role. The SnRKs are classified into three subfamilies: SnRK1, SnRK2, and SnRK3. In this study, we focus on the SnRK2 subfamily, which are plant-specific Ser/Thr protein kinases. They have been identified in Arabidopsis thaliana (Boudsocq et al., 2004) and in other species such as rice, sorghum, maize, tobacco, wheat, soybean, fava bean, potato, and algae (for review see Kulik et al., 2011 and Shinozawa et al., 2019; Kamyiama et al., 2021). The SnRK2 subfamily has been divided into three groups, based on their reaction to the phytohormone abscisic acid (ABA): group 1 – kinases not activated by ABA, group 2 – weakly activated by this hormone, and group 3 - kinases strongly activated by ABA. All SnRK2s are activated rapidly in response to osmotic stress (salinity, desiccation) (for review see Kamyiama et al., 2021). The physiological role of SnRK2s from group 1 (SnRK2.1, SnRK2.4, SnRK2.4, SnRK2.9, and SnRK2.10) is relatively poorly understood. In 2012, McLoughlin et al. (2012) established a role of SnRK2.4 and SnRK2.10 in the response to salt stress. SnRK2.4 was found to stimulate primary root growth, SnRK2.10 lateral root density under salt stress, whereas SnRK2.1, SnRK2.5, and SnRK2.9 were shown to control root growth under non-stress conditions. It has also been shown recently that ABA non-activated SnRK2s phosphorylate VARICOSE (VCS), an mRNA decapping activator, and thus regulate mRNA decay under osmotic stress conditions and osmotic stress-dependent transcript accumulation (Soma et al., 2017; Kawa et al., 2020). Our recent phosphoproteomic study suggested that SnRK2.10 phosphorylates two dehydrin proteins, EARLY RESPONSIVE TO DEHYDRATION 10 and 14, in response to osmotic stress (Maszkowska et al., 2019). Moreover, SnRK2.10 conditions the plant tolerance to salinity by helping to maintain photosynthetic efficiency through the protection of the photosynthetic machinery from salinity-caused damage and diminution of ROS accumulation (Mazur et al., 2021). SnRK2.4 is also a negative regulator of root growth in the presence of cadmium ions and putatively influences iron homeostasis (Kulik et al., 2012). There is additional data indicating that the ABA non-activated SnRK2s are activated by salinity, drought, bacterial elicitors, and oxidative stress in other plant species and potentially could regulate their tolerance to those factors (Kulik et al., 2015; for review see Kulik et al., 2011 and Fàbregas et al., 2020).
Salinity induces the accumulation of ROS in plant cells, including hydrogen peroxide (H2O2), superoxide anion radical (O2·-), hydroxyl radical (OH.), and singlet oxygen (1O2). In response to different types of environmental stresses, ROS are mainly generated in chloroplasts, but also in mitochondria, peroxisomes, and on the outside of the cell membrane (Sewelam et al., 2014; Smirnoff and Arnaud, 2019; Liu et al., 2021). The ROS formed upon salinity have two distinct roles, both positive and negative. Those generated at the early stages of stress mostly serve as an early warning system triggering the intracellular signaling required for rapid stress response and modulation of gene expression (Allu et al., 2014; Mittler, 2017; Srivastava et al., 2021). Not all ROS have the same potential as a secondary signaling molecule. H2O2, being moderately long-lived in vivo (the half-life of milliseconds to seconds), can be transported and accumulated transiently in various cellular compartments and thus serve as a signal initiating distinct signaling pathways. H2O2 is involved in a cross-talk with many other signaling molecules such as other ROS and RNS (Reactive Nitrogen Species) or phytohormones, and it also modulates the activity of protein kinases (Smirnoff and Arnaud, 2019; Liu et al., 2021; Zentgraf et al., 2022). Its accumulation is controlled by scavenging enzymes, catalases and peroxidases, localized in different cellular compartments (Sewelam et al., 2016; Mittler, 2017; Zentgraf et al., 2022). When a stress is too strong the balance between ROS production and scavenging is disturbed and ROS accumulate uncontrollably causing damage to proteins, nucleic acids, and lipids, which leads to irreversible defects in cell functioning and eventually to its death (Hasanuzzaman et al., 2021; Srivastava et al., 2021). The extent of ROS accumulation and oxidative damage vary among distinct plant organs. Although an accumulation of ROS, in particular H2O2, has been observed under salinity in both roots and leaves (Szymańska et al., 2019; Mazur et al., 2021; for review see Hasanuzzaman et al., 2021), the roots suffer more damage than the rest of the plant, because they are usually the first to be exposed to salinity (Hasanuzzaman et al., 2021). The ability to cope with an excessive accumulation of salt-induced ROS is often correlated with the resistance of individual genotypes to salinity. This suggests that the type of ROS accumulated, their level and the site of the accumulation determine the overall response to salinity stress and are subject to subtle regulation (Murata et al., 2012; Zentgraf et al., 2022).
Transcription factors (TFs) are important constituents of plant signaling pathways that determine long-term (i.e., those involving changes in gene expression) responses to biotic and abiotic stimuli (Joshi et al., 2016). Large-scale transcriptomic analyses have revealed that the response to salinity is regulated in Arabidopsis by numerous transcription factors from distinct families, among others WRKY (Seki et al., 2002; Allu et al., 2014), which is a unique superfamily of TFs in higher plants and algae playing important roles in diverse life processes, and biotic and abiotic stress responses (Eulgem et al., 2000; Zhang and Wang, 2005; Li et al., 2020). Their functioning is regulated at the transcriptional, post-transcriptional, and post-translational levels (Giacomelli et al., 2012; Phukan et al., 2016). Expression of WRKY genes is controlled by TFs belonging to other families and by WRKYs. The cellular abundance, properties and activity of the WRKY TFs are modulated by ubiquitination (Yu et al., 2013), phosphorylation (Qiu et al., 2008a; Xie et al., 2010; Adachi et al., 2015), and interactions with other signaling components, including other WRKYs (by homo- and heterodimerization) (Chi et al., 2013). They can also be controlled through inter-organelle retrograde signaling; for example, the AtWRKY18-40-60 cluster is regulated by chloroplast-mediated retrograde signals (Shang et al., 2010).
Owing to their high plasticity and responsiveness to a broad range of environmental and intracellular signals, the WRKY TFs are an interesting but complex model to study the integration and differentiation of cellular signaling pathways and responses. Not all upstream factors modulating their abundance and activity have been identified, including the numerous SnRK2s. Here we focused on the poorly characterized SnRK2.10 and studied its effects on WRKY expression under salinity and oxidative stress. We monitored the expression levels of four WRKY genes, WRKY33, WRKY40, WRKY46, and WRKY75, upon salinity, H2O2 application, and oxidative stress evoked by treatment of plants with MV in wild-type and snrk2.10 insertion mutants. Putative targets of the WRKY33, WRKY40, WRKY46 and WRKY75 TFs were predicted by a bioinformatic analyses of data collected from numerous data bases. The four WRKY TFs showed highly similar target specificity, which included numerous genes encoding stress-responsive protein kinases.
Materials and methods
Plant lines and growth conditions
The Arabidopsis thaliana lines used in the study: wild-type Col-0, snrk2.10-1 (WiscDsLox233E9), and snrk2.10-3 (SAIL_698_105). The line expressing SnRK2.10-GFP was kindly provided by Prof. Christa Testerink (Wageningen University).
For salinity-dependent gene expression analysis plants were grown under short-day conditions in a hydroponic culture (Araponics system) as previously described (Mazur et al., 2021). For MV-dependent gene expression analysis plants were grown in soil for three weeks.
For aseptic hydroponic cultures, seeds were sterilized by gentle shaking in 70% ethanol for 2 min, then incubated in water: bleach solution (13:1, v:v) for 20 min and washed five times with sterile water. About 100 seeds were planted to glass flasks containing 100 mL of ½ Murashige and Skoog medium supplemented with ½ Murashige and Skoog vitamin solution, 500 mg/L MES, 10 g/L sucrose, pH 5.7 and imbibed at 4°C for 5 d. Seedlings were grown for the next 10 days under constant shaking at 22°C and long-day conditions.
Stress application
For salinity-dependent gene expression analysis and H2O2 visualization five-week-old plants were treated or not (control) with 150 mM NaCl for up to six days and whole rosettes were collected for further analysis as described (Mazur et al., 2021).
Treatment with H2O2 was performed on seedlings grown in aseptic hydroponic cultures. For gene expression analysis the seedlings were incubated with 10 mM H2O2 for 5 h, as described (Allu et al., 2014). For SnRK2.10 kinase activity analysis, the seedlings were treated with 2 mM H2O2 for up to 2 h, according to (Kulik et al., 2012), with 250 mM NaCl for 5 min, or with 50 µM MV for up to 2 h. The seedlings were collected immediately after the treatment and frozen in liquid nitrogen.
For MV-dependent gene expression analysis and H2O2 visualization, soil-grown three-week-old plants were sprayed on the abaxial and adaxial sides of leaves with 25 µM or 50 µM MV respectively, and incubated under illumination for 7 h as described (Benina et al., 2015). Only fully developed leaves were collected for further analysis.
Immunoprecipitation and Immunocomplex kinase activity assay
The procedure was performed as described in Mazur et al. (2021). In brief, for immunoprecipitation of GFP-fused proteins, 400 µg of crude protein extract from seedlings treated with H2O2 or MV was incubated with 10 µL of GFP-Trap®_A (Chromotek) for 2.5 h with gentle rocking. After intensive washing, agarose beads with bound immunocomplexes were suspended in 20 mM Tris–HCl, pH 7.5 supplemented with 150 mM NaCl and 4 µg of Myelin Basic Protein (Sigma-Aldrich) per sample. To each sample, ATP supplemented with 1 μCi of [γ32P]ATP in kinase buffer (25 mM Tris-HCl, pH 7.5, 5 mM EGTA, 1 mM DTT, 30 mM MgCl2) was added to 50 μM final concentration. After 15 min of incubation at 37°C samples were mixed with Laemmli sample buffer and incubated for 3 min at 95°C with vigorous shaking. Proteins were separated on 12% SDS polyacrylamide gel and signal was detected on Medical X-ray Blue/MXBE Film (Carestream).
Immunoblotting
Immunoblotting was performed as previously described (Mazur et al., 2021). PVDF membranes were stained with 0.2% Ponceaus S for protein loading visualization. For the detection of GFP-conjugated protein, HRP-conjugated anti-GFP antibody (Santa Cruz Biotechnology, USA) diluted 1:1000 was used according to the manufacturer’s protocol.
RT-qPCR
Rosettes were ground to a fine powder in liquid nitrogen. RNA was extracted with Trizol (Molecular Research Center) according to the manufacturer’s instructions and treated with DNase 1 (Thermo Scientific). Reverse transcription was performed on 1 µg of RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). The resulting cDNA was diluted tenfold with water and 1 μL of the sample was used for qPCR in a Step One Plus device (Applied Biosystems) using the GoTaq® qPCR Master Mix (Promega) and specific pairs of primers (Supplemental Table 1). Expression levels were calculated relative to the housekeeping genes PDF2 (At1g13320) and PEX4 (At5g25760) using the delta-delta Ct method.
Statistical analysis
Statistical analysis was performed by One Way ANOVA followed by post hoc Tukey’s test (p < 0.05).
Intracellular localization of H2O2
For detection of H2O2 localization in response to salinity plants were grown for 5 weeks in a hydroponic system (Mazur et al., 2021). For the experiment, we employed two ways of salt stress application. First, we added NaCl directly into the growing media (root application): 250 mM for 30 min or 150 mM NaCl for 3 days. The undetached leaves were stained with 50 µM BES-H2O2-Ac probe (Wako Chemicals) in 10 µM PIPES buffer, pH 6.8 for 30 min, cut from the rosette, washed 5 times for 1 min in the above buffer and observed immediately.
In the second attempt (direct treatment) the undetached leaves were firstly stained as described above and then cut from the plant and treated with 150 mM NaCl or 250 mM in 10uM PIPES buffer, pH 6.8 and immediately examined under the confocal microscope. Images were taken after 30 min of the salt treatment.
For monitoring of paraquat-triggered H2O2 accumulation in leaf cells, plants grown for 3 weeks in soil were sprayed with 50 µM MV and incubated on light for 2 to 7 h, and stained with BES-H2O2-Ac as described above.
The BES-H2O2-Ac fluorescence from leaf tissues was registered with the Nikon EZ-C1 confocal microscope using an excitation light at a wavelength of 488nm set at 1% of maximum power (20mW, Sapphire, Coherent, USA). The emission of BES-H2O2-Ac was collected with 515/30 emission filter and displayed in false green. Simultaneously the chlorophyll autofluorescence was detected by a 610 long pass filter and displayed in false magenta. The pinhole and exposure time were optimized and all settings of fluorescence detection were the same for experiments. Leaf samples have been imaged in the epidermis layer and first layer of spongy mesophyll cells using 20x oil immersion objective (Nikon, CFI Plan Fluor NA 0.75) and 60x oil immersion objective (Nikon, CFI Plan Apochromat NA 1.4) Single confocal sections and stacks were collected in Nikon EZ-C1 software. The images were digitally processed using FIJI software and the figures compiled in FigureJ plugin (NIH, Bethesda, MD, USA) (https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0240280).
Bioinformatic resources and analysis
Protein interactors of WRKY33, WRKY40, WRKY46, and WRKY75 were extracted from the following databases: BioGRID 3.5.187 (Oughtred et al., 2019), TAIR Interactome 2.0 (Berardini et al., 2015), STRING 11.0 (Szklarczyk et al., 2019), and Kihara Bioinformatics Laboratory’s resources (Ding and Kihara, 2019). Predicted target genes of WRKY33, WRKY40, WRKY46, and WRKY75 were extracted from Expresso (Aghamirzaie et al., 2017), PlantRegMap (Tian et al., 2020), Plant Cistrome Database (O’Malley et al., 2016), AthaMap (Hehl et al., 2016), and Chip-seq experimental data (Birkenbihl et al., 2017). For validation and GO term analysis, the predicted target genes were analyzed with TF2Network (Kulkarni et al., 2018). The Venny 2.1 online resource (Oliveros, 2007-2015) was used to create Venn diagrams. BioMart (Durinck et al., 2009) was used to enlist the information related to the obtained genes.
Results
SnRK2.10 controls the expression of several WRKY TFs under salinity
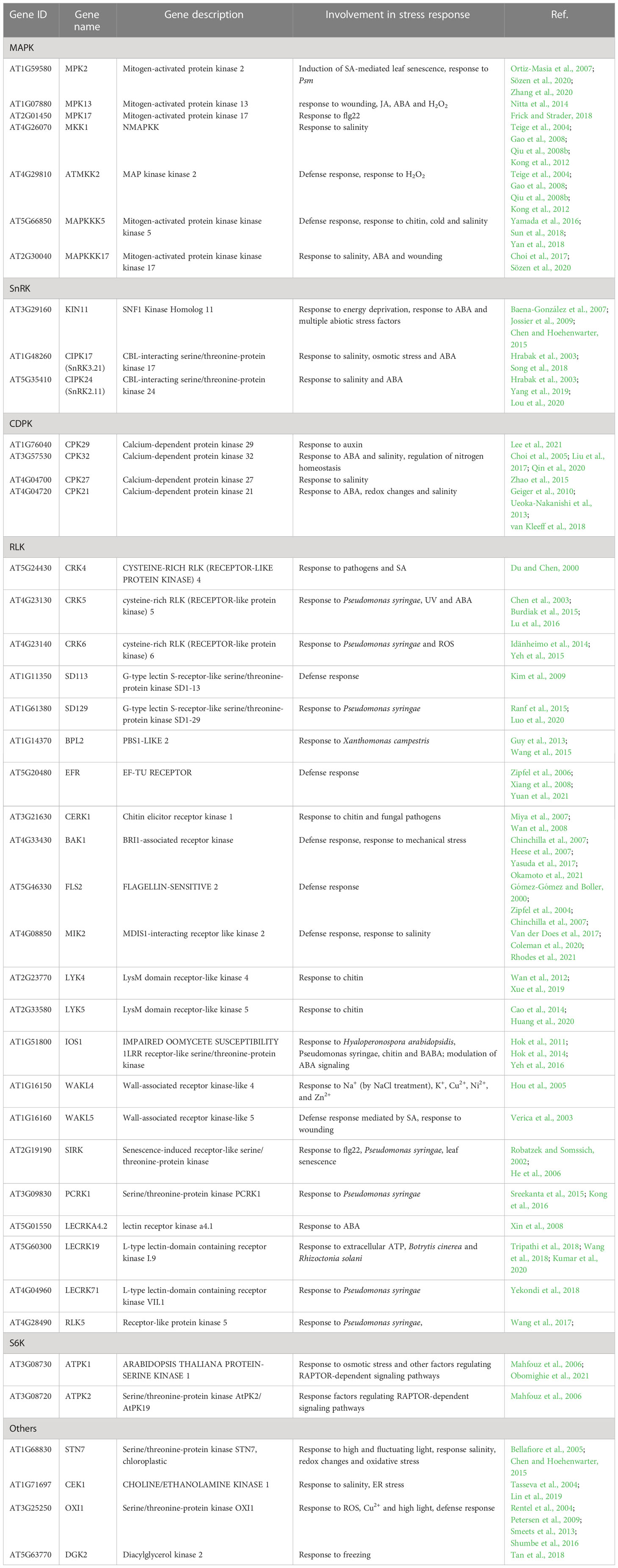
During the past twenty years, an extensive transcriptomic analysis of the action of ABA-activated SnRK2s (SnRK2.2, SnRK2.3, and SnRK2.6) under osmotic stress, salinity, and ABA treatment has identified numerous genes regulated by these kinases, including many encoding transcription factors, which helped us to understand the mechanisms by which the ABA-activated SnRK2s determine plant resistance to water deficiency (Fujita et al., 2009). However, the genes regulated by the ABA non-activated SnRK2s, and particularly SnRK2.10, were characterized only partially. To begin filling this gap we compared the expression of selected genes in wt Arabidopsis and snrk2.10 mutants exposed to salinity for up to six days. SnRK2.10 was found to play a major role in the induction of four genes from the WRKY family of transcription factors (Figure 1). Prior to salinity treatment, the basal expression of the genes in question was the same in wt plants and the mutants. Following exposure to salinity, the expression of WRKY33, WRKY40, and WRKY46 was induced slightly in the wt after three days of treatment and at day six reached maximum values of approximately 360-, 210-, and 140-fold induction, respectively. The expression of WRKY75 was induced already on day one of salinity and reached 240-fold induction on day six. Notably, in the both tested snrk2.10 mutants the induction of these genes on the sixth day of salinity was significantly lower than in wild-type plants. Next, we analyzed the expression of CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 13 (CYP71A13), which is a direct target gene of WRKY33 and WRKY40 (Birkenbihl et al., 2012; Birkenbihl et al., 2017), and of OSMOTIN 34 (OSM34), a target of WRKY33 (Zheng et al., 2006; Jiang et al., 2007). The expression of CYP71A13 and OSM34 increased in all plant lines, reaching about 700- and 30- fold induction, respectively, on day six of salinity, in the wild-type plants, and only ca. half of those values in the snrk2.10 mutants. These results indicate that the expression of WRKY33, WRKY40, WRKY46, and WRKY75 and of two of their targets is regulated by a SnRK2.10-dependent signaling pathway(s).
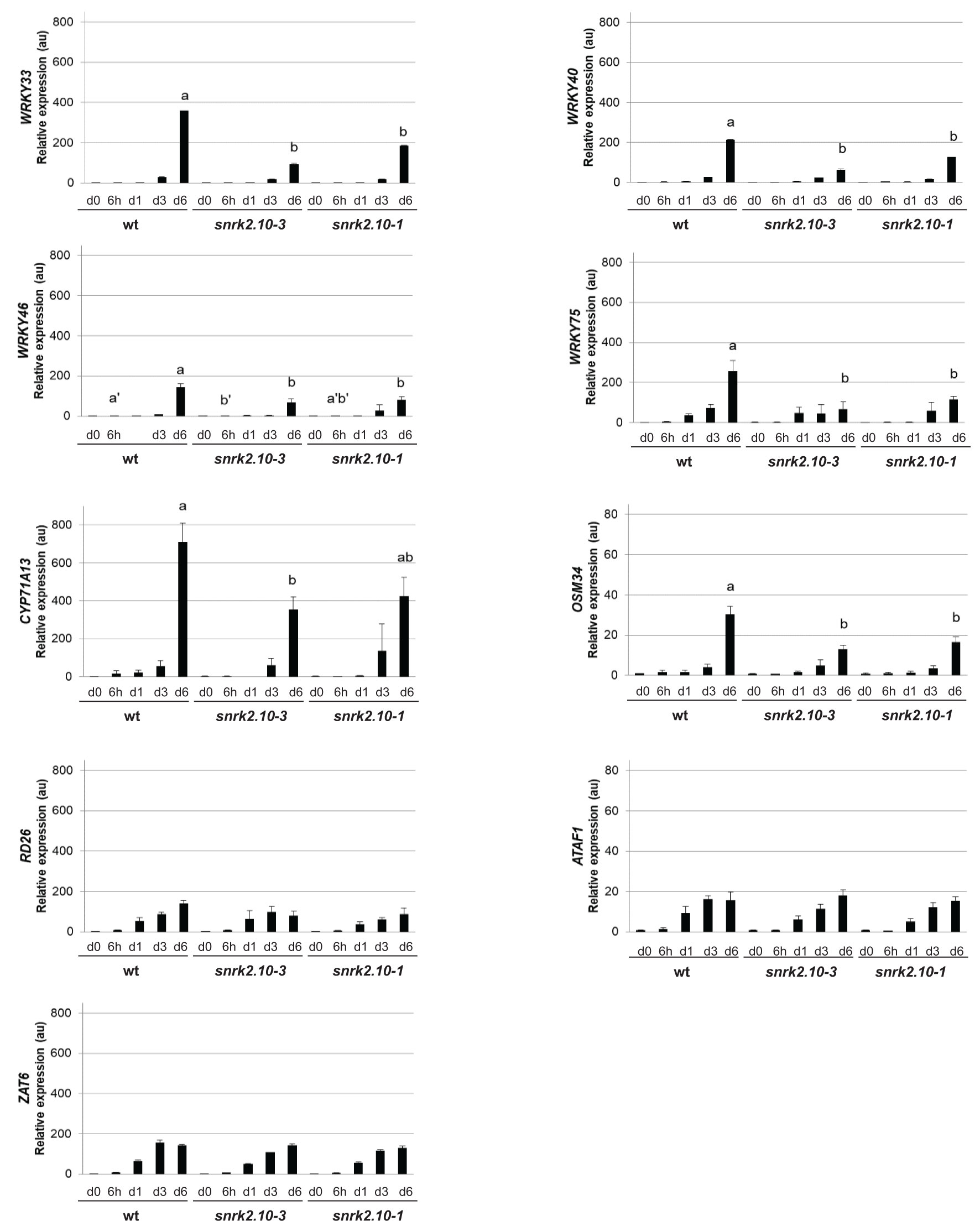
Figure 1 Effect of SnRK2.10 gene disruption on the expression of selected genes upon salinity. Transcript level was determined by RT-qPCR in rosettes of five-week-old plants not treated (day 0; d0) or treated with 150 mM NaCl for 6 hours (6h), 1, 3 and 6 days (d1, d3, d6), and normalized against PEX4 (At5g25760) gene. The expression of each gene is shown relative to that in non-treated wild-type plants (Col-0). Mean values from three independent biological replicates, each with 8-10 plants from each line, +/- SD are shown. Statistical significance of differences between groups was determined by ANOVA and Tukey post hoc test. The same letters denote values belonging to the same homogeneous group (p < 0.05).
Next, we analyzed the expression of transcription factors belonging to other families. The expression of RESPONSIVE TO DESSICATION26/ANAC072 (RD26) and ARABIDOPSIS TRANSCRIPTION ACTIVATOR FACTOR 1 (ATAF1), both genes belonging to the Arabidopsis NAC Domain Containing Protein (ANAC) family and involved in the maintenance of tolerance to salinity and water regime (Wu et al., 2009; Liu et al., 2016; Ye et al., 2017), was continuously rising in the wt plants and in both snrk2.10 mutants upon salinity. On day six the expression of RD26 was increased nearly 150-fold in the wt and ca. 100-fold in the mutant, and of ATAF1 over 15-fold in all three lines (Figure 1).
As a representative of the C2H2 zinc finger family of transcription factors we analyzed the expression of ZINC FINGER OF ARABIDOPSIS THALIANA 6 (ZAT6) known to positively regulate seed germination and plant tolerance to salinity (Liu et al., 2013; Shi et al., 2014; Tang and Luo, 2018). Its expression was rising continuously upon salinity and on day six it was 150 times higher than on day zero in all three lines (Figure 1).
A cross-talk between WRKY33, WRKY40, WRKY46, and WRKY75 gene targets and protein interactors
The transcription factors from the WRKY superfamily are involved in plant responses to diverse biotic and abiotic stresses (Phukan et al., 2016; Li et al., 2020; Wani et al., 2021). They often function as homo- or heterodimers, heterooligomers, or act redundantly to each other. Owing to those features, the genes regulated by individual WRKY TFs frequently overlap, which allows specific functional groups to be discerned within the WRKY family (Li et al., 2020; Wani et al., 2021). We, therefore, conducted a comparative bioinformatic analysis of putative target genes of WRKY33, WRKY40, WRKY46, and WRKY75 extracted from five databases (see Materials and methods for details). We found 4941, 4932, 6425, and 4796 putative target genes of WRKY33, WRKY40, WRKY46, and WRKY75, respectively (Figure 2A and Supplemental Data Sheet 1). About 351, 1593, 791, and 451 of those were specific for the respective TFs, while ca. 26% (2337) genes were ranked as regulated by all four TFs. These common WRKY target genes were subjected to a functional analysis using the Gene Ontology (GO) categorization. The most significantly enriched categories included genes encoding proteins having a kinase activity, mainly protein serine-threonine kinase activity (Figure S1). These kinases belong to very distinct families and are involved in plant responses to multiple environmental biotic and abiotic stress factors (Table 1).
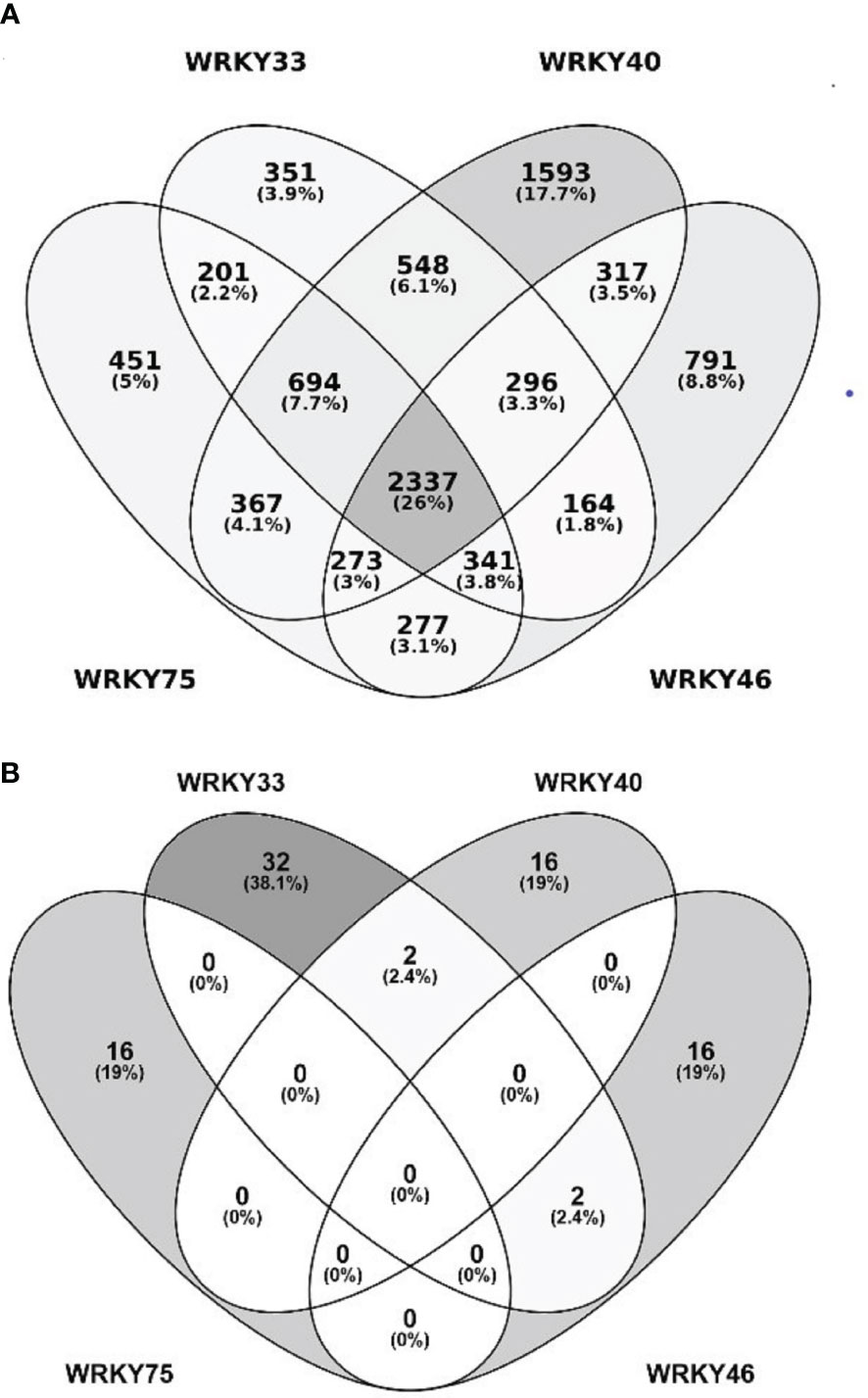
Figure 2 WRKY33, WRKY40, WRKY46, and WRKY75 target genes and interacting proteins. (A) Number of predicted target genes of WRKY33, WRKY40, WRKY46, and WRKY75. Color intensity reflects the number of genes. (B) Number of protein partners of WRKY33, WRKY40, WRKY46, and WRKY75.
In contrast to the highly overlapping gene target sets, the protein partners of WRKY33, WRKY40, WRKY46, and WRKY75 showed high specificity towards particular transcription factors (Figure 2B). Thirty-two proteins have previously been identified as interacting specifically with WRKY33, and 16 each with WRKY40, WRKY46, and WRKY75. In addition, four proteins interacted with two of the TFs. Among specific interactors, protein kinases involved in stress response and transcription factors were particularly abundant (Supplemental Data Sheet 2).
SnRK2.10 is activated under oxidative stress and controls the expression of several WRKY TFs
The ROS accumulated during the early response to biotic and abiotic stress factors function as secondary messengers and play a substantial role in triggering cellular signaling. To investigate the role of SnRK2.10 in the ROS-mediated signaling we focused on H2O2 because of its relatively long life and high mobility (Smirnoff and Arnaud, 2019; Liu et al., 2021). To investigate whether H2O2 can activate SnRK2.10 in the absence of salt stress, transgenic seedlings expressing SnRK2.10-GFP were treated with 2 mM hydrogen peroxide for up to two hours (Figure 3A). The SnRK2.10 activity was detected after fifteen minutes of the exposure, reached a maximum after 30 minutes, and returned to the control level after two hours. The activity triggered by H2O2 treatment was lower than that observed after 5 min of salinity. The level of the SnRK2.10-GFP protein did not change during the treatment.
Figure 3 Involvement of SnRK2.10 in plant response to H2O2. (A) SnRK2.10 activity. Seedlings of transgenic Arabidopsis line expressing SnRK2.10-GFP were grown in hydroponic culture for ten days and exposed or not to 2 mM H2O2 for up to 2 hours. Kinase activity was determined by in-gel kinase activity assay with γ-[32P] ATP and MBP (myelin basic protein) as substrates. SnRK2.10-GFP protein was quantified by western blotting with anti-GFP antibodies. A representative result of three independent repeats is shown. CBB – Coomassie Brilliant Blue. (B) Expression of selected genes. Transcript level was monitored by RT-qPCR in ten-day-old seedlings not treated (C) or treated with 10 mM H2O2 for 5 hours (H2O2), and normalized against PEX4 (At5g25760) gene. The expression of each gene is shown relative to that in non-treated wild-type plants (Col-0). Mean values from three independent biological replicates, each with 8-10 plants from each line, +/- SD are shown. Statistical significance of differences between groups was determined by ANOVA and Tukey post hoc test. The same letters denote values belonging to the same homogeneous group (p < 0.05).
According to Allu et al. (2014) WRKY33, WRKY40, and WRKY75 are upregulated in plants in response to ROS/H2O2 treatment. To determine whether SnRK2.10 contributes to this induction, ten-day-old seedlings of the wt and the snrk2.10 mutants were treated with 10 mM H2O2 as previously described (Allu et al., 2014). In all three lines the expression of the WRKY33, WRKY40, WRKY46, and WRKY75 genes was significantly increased compared to control levels (Figure 3B). Although, the induction of WRKYs expression in H2O2 treated samples was lower than after salinity stress (Figure 1). In wt plants the increase was, respectively, 8-, 17-, 7-, and 17-fold, while in the snrk2.10 mutants it was significantly higher: 10-, 25-, 16-, and 25-fold, respectively (Figure 3B). The activation of SnRK2.10 by salinity (Figure 1) had an opposite effect on WRKY expression to the activation in response to H2O2 (Figure 3), suggesting the existence of two distinct signaling pathways involving SnRK2.10 and triggering different stress-specific transcriptional responses.
SnRK2.10 is not activated by methyl viologen-generated ROS
Leaf chloroplasts are very sensitive to different environmental factors, and thus can rapidly perceive and transfer the stress signal to other cellular compartments e.g., by generation of stress-specific ROS (Liu et al., 2021; Zentgraf et al., 2022). A chloroplast-specific ROS formation can be triggered by the herbicide methyl viologen (MV; N,-N’-dimethyl 4, -4’-bipyrydinium dichloride, also known as paraquat). Methyl viologen has been in use in field agriculture at high concentrations for ca. 60 years (Baltazar et al., 2013). In experimental plant biology, it is commonly used in low concentrations (in the nM to µM range) to study ROS signaling and oxidative stress tolerance (for review see Nazish et al., 2022). In the chloroplasts, in the presence of light, MV competes with ferredoxin for electrons on the acceptor side of photosystem I (PSI) to produce monocationic MV radical (MV˙+) which reacts rapidly with oxygen to form superoxide radical (O2˙-) (Hassan, 1984; Dodge, 1989; Fuerst and Norman, 1991; Hartel et al., 1992). The highly reactive O2·- further generates other ROS, like H2O2 or OH˙, which may play signaling functions and damage the cell (Babbs et al., 1989). To determine whether SnRK2.10 can be activated by ROS generated in chloroplasts, we treated seedlings expressing SnRK2.10-GFP with 10 µM MV and analyzed the kinase activity by an in-gel assay. Only traces of SnRK2.10 activity were detected after up to 2 h of the treatment vis-a-vis a very strong signal observed following application of H2O2 (Figure 4A). Nevertheless, the MV treatment caused a nearly two-fold induction of the SnRK2.10 gene, suggesting a hitherto unknown role of this kinase in the plant response to paraquat (Figure 5). Despite the lack of a substantial SnRK2.10 activation, the WRKY33, WRKY40, WRKY46, and WRKY75 genes were induced by MV in wild-type Arabidopsis leaves approximately 60-, 420-, 38- and 240-fold, respectively, and to a similar extent also in the snrk2.10 mutants (Figure 4B). These results indicate that SnRK2.10 does not regulate their expression in the conditions studied.
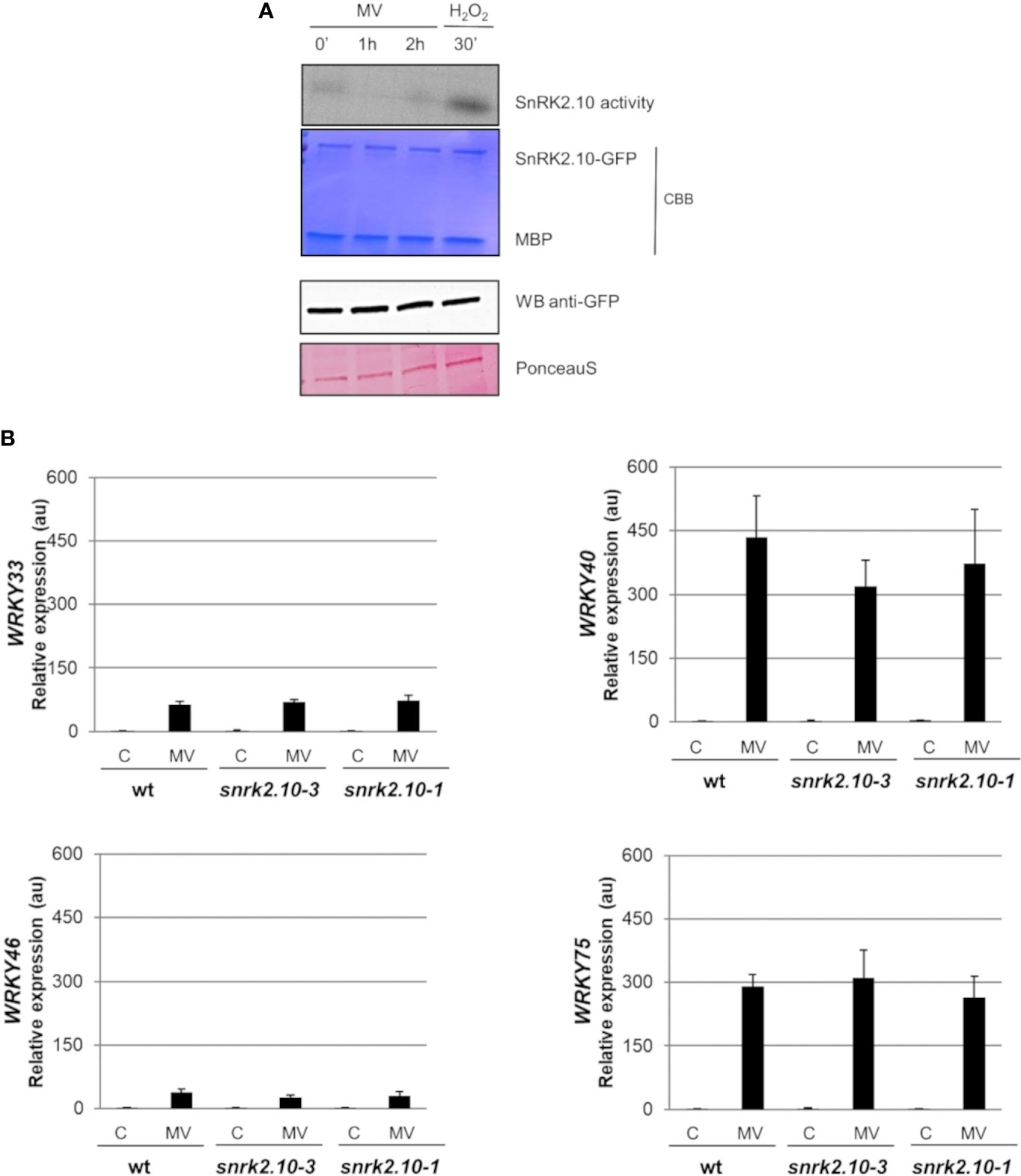
Figure 4 Involvement of SnRK2.10 in plant response to methyl viologen. (A) SnRK2.10 activity. Seedlings of transgenic Arabidopsis line expressing SnRK2.10-GFP were grown in hydroponic culture for ten days and exposed or not to 50 µM MV for up to 2 hours. Kinase activity was determined by in-gel kinase activity assay with γ-[32P] ATP and MBP (myelin basic protein) as substrates. SnRK2.10-GFP protein was quantified by western blotting with anti-GFP antibodies. A representative result of three independent repeats is shown. CBB – Coomassie Brilliant Blue (B) Expression of selected genes. Transcript level was monitored by RT-qPCR in rosettes of three-week-old plants not treated (C) or treated with 10 µM MV for up to 2 hours (MV) and normalized against PEX4 (At5g25760) gene. The expression of each gene is shown relative to that in non-treated wild-type plants (Col-0). Mean values from three independent biological replicates, each with 8-10 plants from each line, +/- SD are shown. Statistical significance of differences between groups was determined by ANOVA and Tukey post hoc test (p < 0.05).
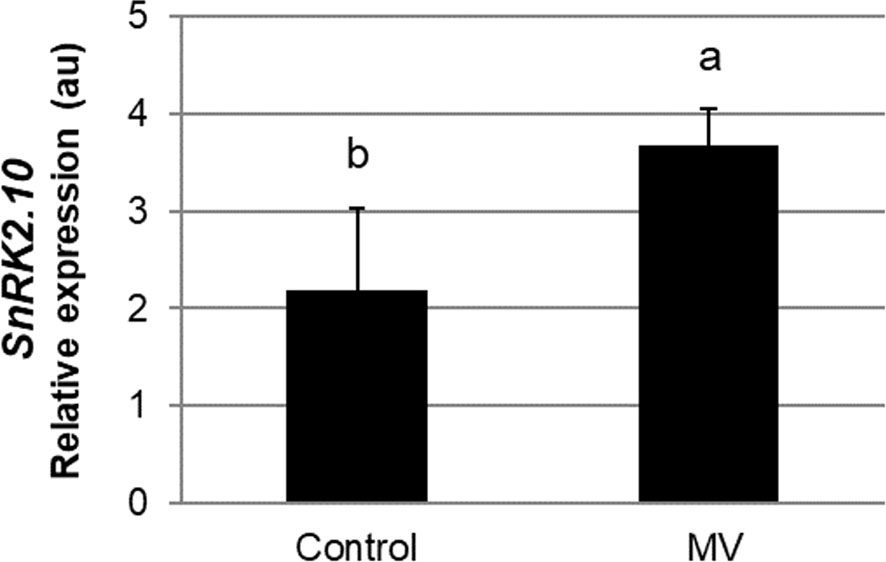
Figure 5 Effect of methyl viologen on SnRK2.10 gene expression. Transcript level was monitored by RT-qPCR in rosettes of three-weeks-old plants not treated (Control) or treated with 10 µM MV for up to 2 hours (MV) and normalized against PEX4 (At5g25760) gene. Mean values from three independent biological replicates, each with 8-10 plants from each line, +/- SD are shown. Statistical significance of differences between groups was determined by ANOVA and Tukey post hoc test (p < 0.05).
H2O2 accumulates in leaves in a site-specific manner under salinity and MV treatments
Generation of different types of ROS, and particularly H2O2, has been widely observed in plants challenged with salinity (Mittler, 2017; Srivastava et al., 2021). These observations are performed very frequently by the histochemical staining of leaves with 3,3’-diaminobenzidine (DAB) or by measuring the hydrogen peroxide concentration in the homogenized biological materials using diverse biochemical methods (Ben Rejeb et al., 2015; Nguyen et al., 2017; Mazur et al., 2021). However, to fulfill their specific signaling role salinity-triggered ROS production must occur in specific cellular compartments. To analyze the NaCl-specific intercellular localization of H2O2, we treated Arabidopsis plants grown hydroponically for five weeks with 150 mM NaCl for 3 days or 250 mM NaCl for 30 min, or we not treated them (control). We then stained the leaves with an H2O2-specific fluorescent probe BES-H2O2-Ac. This fluorescent probe was previously used for the observation of dynamic hydrogen peroxide localization in plant leaves (Zhuang et al., 2021; Shi et al., 2022) and roots (Tsukagoshi et al., 2010). In our control experimental conditions, fluorescence was observed in the cytoplasm and nucleus but not in chloroplasts (Figures 6I and 6II, A–D). This indicates that hydrogen peroxide is present mainly in the first two cellular compartments. After 30 min of salinity, NaCl applied to roots, the fluorescence signal was more pronounced and started to disperse more widely in the cytoplasm (Figures 6I and 6II, E–H). In leaves of hydroponically grown plants treated with 150 mM NaCl for 3 days, H2O2 accumulation was widely dispersed in the cytoplasm, present in the nucleus and in many, but still not all, chloroplasts (Figures 6I and 6II, I–L). It should be noted that, in leaves directly immersed in 150 or 250 mM NaCl for 30 min fluorescence was much stronger and the pattern was different. Hydrogen peroxide was present in the cytoplasm, nucleus, chloroplasts, and in numerous intensive cytoplasm-localized spots, cytoplasmic strands, and bubble-shaped structures (Figures 6I and 6II, M–T and Figure S2). This suggests that in leaves, the NaCl-dependent accumulation site of H2O2 strongly depends on the stress duration, intensity and way of salt application, NaCl applied indirectly into the roots or directly into leave tissue. This potentially may have an impact on downstream signaling events.
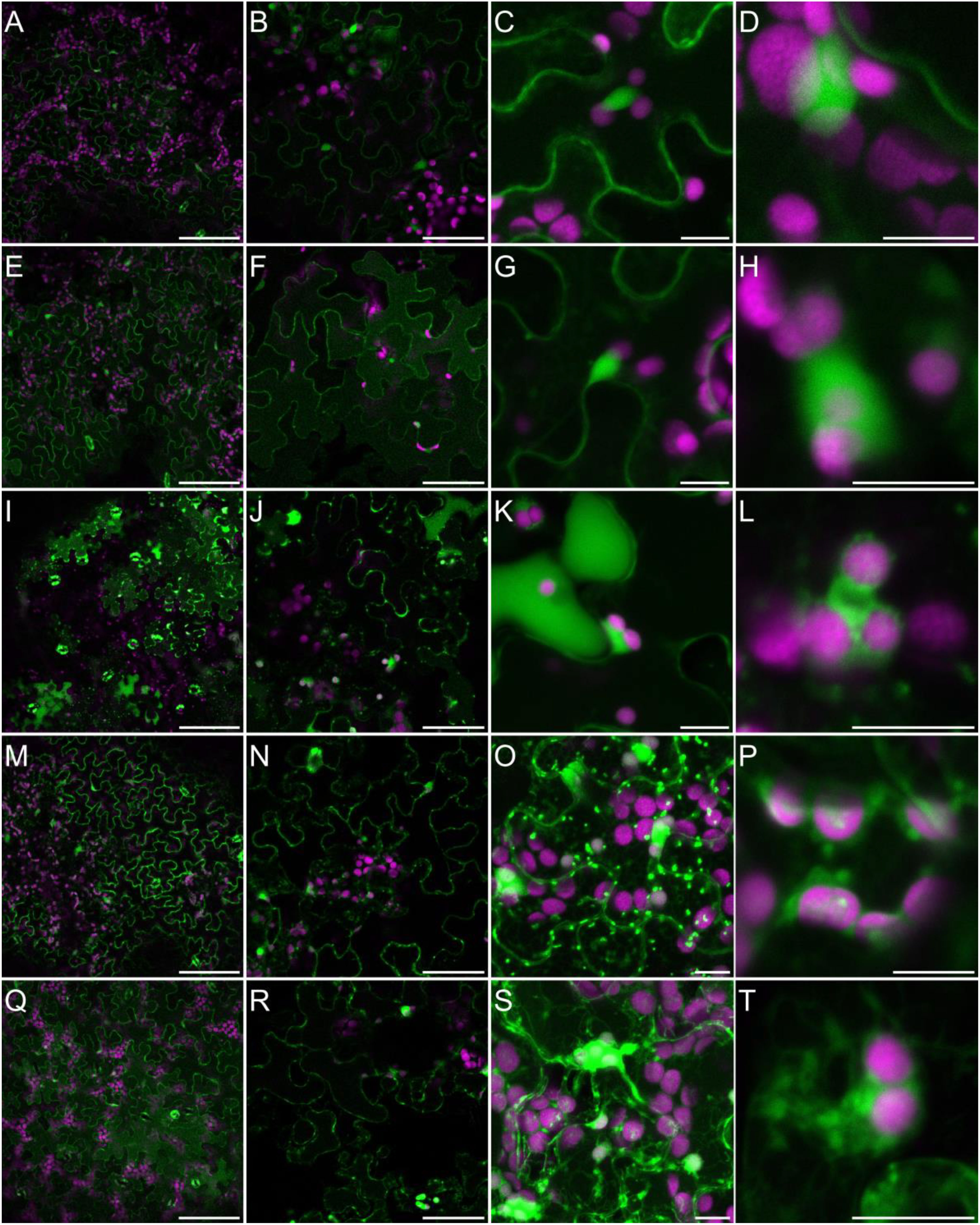
Figure 6I Localization of H2O2 accumulation in response to salinity. Staining of Arabidopsis leaves with an H2O2-specific BES- H2O2-Ac probe for intracellular hydrogen peroxide detection was carried out as described in Materials and Methods. Detection of BES- H2O2-Ac fluorescence is shown in false green and auto-fluorescence of chlorophyll is depicted in false magenta. The panel consists of images of leaves from control conditions (A–D); 30 min of 250 mM NaCl applied to roots (E–H); 3 days of 150 mM NaCl applied to roots (I–L); 30 min of 150 mM NaCl applied directly to leaves (M–P); 30 min of 250 mM NaCl applied directly to leaves (Q–T). Scale bars: 100 μm in the first two columns and 10 μm in the last three columns. Images were made as a single scan in the first two columns, in the remaining columns merged projections from 14-25 individual optical slices are presented. Six to eight leaves from different plants and from independent biological treatments were observed for each treatment. Images from multiple optical sections were collected from each leaf. The panel shows representative images.
It has been previously shown that in leaves exposed to MV, different ROS are accumulated, among them hydrogen peroxide (Bulgakov et al., 2012; Cui et al., 2019; Wang et al., 2019). Although, the sites of its accumulation are not well documented. Thus, for monitoring of paraquat-triggered H2O2 accumulation in leaf cells, plants grown in soil were sprayed with 50 µM MV and incubated on light for 2 to 7 h, and stained with BES-H2O2-Ac. The H2O2 accumulation was observed mainly in the cytoplasm and nucleus but not in chloroplasts in both, control and MV-treated leaves (Figure 6II). This indicates that in response to MV H2O2 accumulates in leaf cells in the same cellular compartments/organelles as during normal growth, but must probably to higher concentrations. Alternatively, hydrogen peroxide accumulation in some cellular spaces/organelles may be very weak and beyond our detection range, since Ugalde et al. (2021) reported MV-dependent H2O2 formation in chloroplasts. This problem needs further detailed investigation and application of more sophisticated methods.
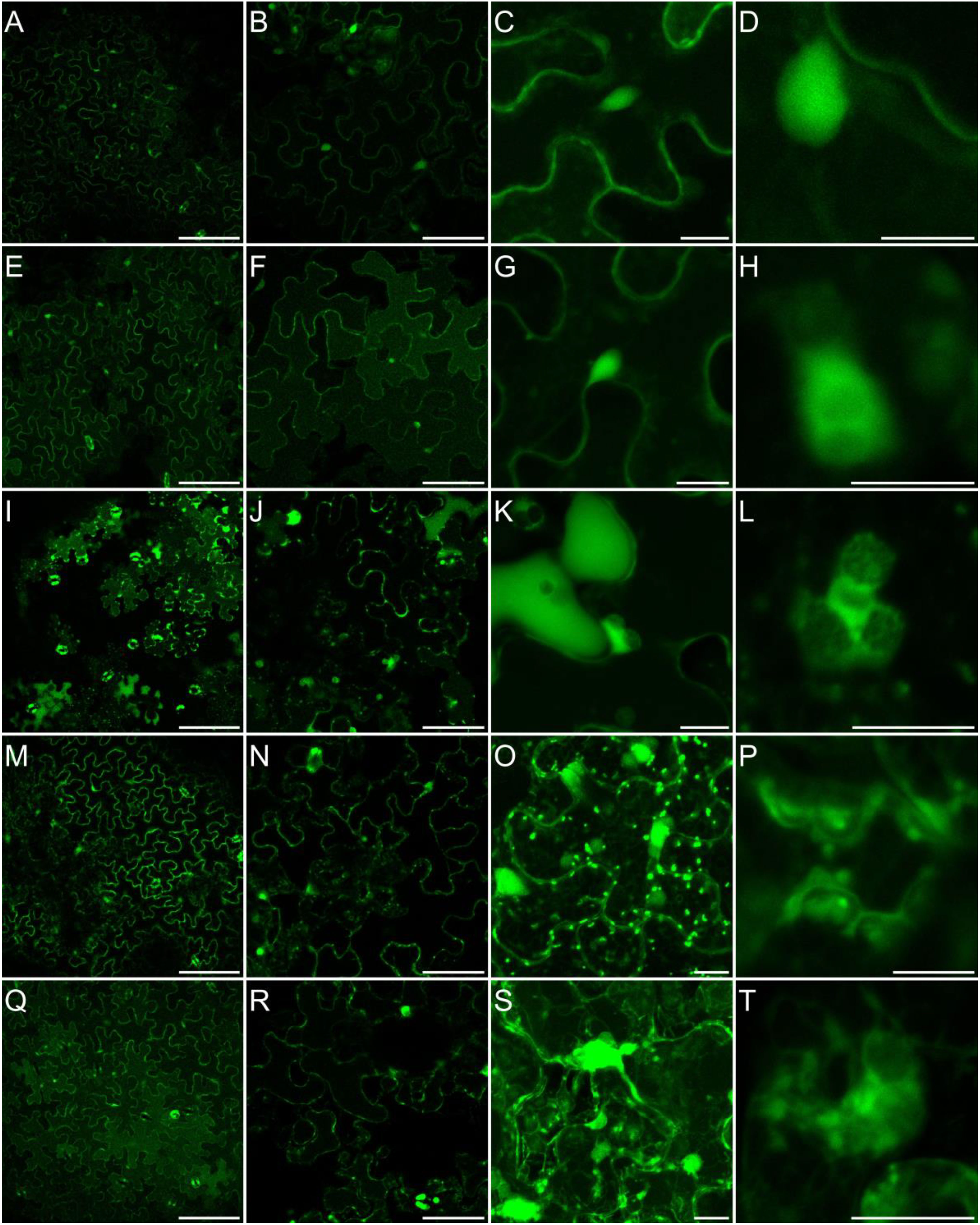
Figure 6II Localization of H2O2 accumulation in response to salinity. Staining of Arabidopsis leaves with an H2O2 -specific BES- H2O2-Ac probe for intracellular hydrogen peroxide detection was carried out as described in Materials and Methods. Detection of BES- H2O2 -Ac fluorescence is shown in false green. All figure captions correspond to the description of Figure 6I.
Discussion
Monitoring the activation of stress-responsive kinases upon different environmental stimuli and/or the induction of their expression is the first step toward identifying their involvement in the regulation of plant responses to particular conditions. In most cases, a signal must be perceived by specific receptors to initiate a highly structured sequence of down-stream events in which the information is transduced through a branched network of intracellular pathways leading to the final response. The transfer of information about a stress is a non-linear and extremely complex process resembling an extensive root system in structure (Sewelam et al., 2016). One of the key signaling second messengers in the cell are ROS, particularly hydrogen peroxide. Several H2O2-induced protein kinases have been identified, such as MITOGEN-ACTIVATED PROTEIN KINASE 6, CALCIUM-DEPENDENT PROTEIN KINASE 5 and OXIDATIVE SIGNAL-INDUCIBLE 1 (OXI1) (Rentel et al., 2004; Wang et al., 2010; Dubiella et al., 2013). Furthermore, H2O2-dependent oxidation of specific thiol groups in type 2C protein phosphatase HYPERSENSITIVE TO ABA 1 (HAB1) inhibits its catalytic activity and the ability to interact with SnRK2.6, an ABA-activated SnRK2. This, in turn, allows auto-phosphorylation and auto-activation of SnRK2.6 and phosphorylation of its downstream targets located mainly in guard cells (Sridharamurthy et al., 2014). In our laboratory, Nicotiana tabacum OSMOTIC STRESS-ACTIVATED PROTEIN KINASE (NtOSAK) has been identified as a hydrogen peroxide-activated kinase, activated also by cadmium ions in an H2O2-dependent manner (Kulik et al., 2012). NtOSAK belongs to ABA non-activated SnRK2s and is closely related to SnRK2.4 and SnRK2.10 from Arabidopsis (Kulik et al., 2011). In this study, we showed that SnRK2.10, like NtOSAK, is activated by hydrogen peroxide. Moreover, the kinase alters the expression of four H2O2-responsive WRKY genes. The phosphorylation of SnRK2.10 is an element of early response to salinity and H2O2 which has been indicated by several studies including a large-scale phosphoproteomic study (Chen and Hoehenwarter, 2015). The question of whether SnRK2.10 is activated during salinity in an H2O2–dependent manner remains open for further investigation.
An opposite regulation of WRKY TFs expression by SnRK2.10 under salinity and in response to H2O2 suggests the existence of two distinct signaling pathways involving SnRK2.10 and triggering different stress-specific transcriptional responses. A differentiation of the signaling in response to H2O2 vs. prolonged salinity has already been proposed (Allu et al., 2014). To understand the specificity and identify other components of the two putative signaling pathways, further study is required, e.g., the identification of respective SnRK2.10 partners and targets under salinity and H2O2 stresses. Under salinity, VARICOSE (VCS) has been identified as an ABA-non-activated SnRK2s target in Arabidopsis (Soma et al., 2017; Kawa et al., 2020). VCS is a scaffold protein for DCP1 and DCP2 proteins which together form a complex that catalyzes the decapping of 5’mRNA, which is followed by the degradation of mRNA by 5′–>3′ exoribonuclease XRN4 (Sorenson et al., 2018). Within VARICOSE, multiple SnRK2.10-related phosphorylation residues have been identified. Although to date, the consequences of VCS phosphorylation remain controversial (Soma et al., 2017; Kawa et al., 2020) and we cannot undoubtedly state whether SnRK2 protein kinases enhance or inhibit 5′ mRNA decay via phosphorylation of VCS. Mutant plants with disrupted expression of snrk2.4, snrk2.10, vcs or xrn4 show similar disturbances in the growth of the main root and lateral roots, respectively, under saline conditions (Kawa et al., 2020). It has been shown that transcripts of WRKY33, WRKY40, and WRKY46 are enhanced in vcs and xrn4 mutants (Basbouss-Serhal et al., 2017; Sorenson et al., 2018; Nagarajan et al., 2019; Carpentier et al., 2020). The effect of VCS-XRN4 5’mRNA decay module on WRKYs expression seems to be complex and needs further investigation, also under oxidative stress conditions. The post-transcriptional regulation mediated by the ‘subclass I SnRK2s–VARICOSE’ signaling module represents one of mechanisms of gene expression control under osmotic stress (Soma et al., 2017) and we cannot exclude other pathways by which SnRK2.10 controls WRKY33, WRKY40, WRKY46, and WRKY75 transcription under salinity and oxidative stress. For instance, phosphoproteomic study of Maszkowska et al. (2019) revealed several putative proteins involved in mRNA metabolism phosphorylated by SnRK2.10 in roots under salinity. The schematic putative functioning of SnRK2.10-depepndent regulation of WRKYs expression in response to salinity has been presented on Figure S3.
Chloroplasts play a key role in plant functioning as the site of photosynthesis. It should be noted that they are also very sensitive sensors of environmental stresses, including salinity, and the important source of ROS generated in plants under stress (for review see Smirnoff and Arnaud, 2019; Liu et al., 2021; Zentgraf et al., 2022). Our present results show that the ROS formed upon induction of an MV-dependent oxidative burst do not induce the activity of SnRK2.10. This is perhaps unsurprising since SnRK2.10 has never been observed in chloroplasts, although its abundance in the cellular structures linking physically and functionally chloroplasts and the nucleus remains to be determined. Recently, it has been demonstrated that ROS generated locally in intact Arabidopsis chloroplasts by methyl viologen treatment cause dynamic changes in H2O2 accumulation in the cytosol and in mitochondria (Ugalde et al., 2021). A simple diffusion of H2O2 across cellular membranes is strongly limited, and its efficient transport occurs only through aquaporins, which are present in the plasma membrane, tonoplast, and most likely in chloroplast membranes allowing for retrograde signaling involving H2O2 moving via cytosol (for review see Mullineaux et al., 2019; Smirnoff and Arnaud, 2019). A second route of H2O2 transportation outside the chloroplasts is through so-called plastid-nuclear complexes and stromules (stroma-filled tubular plastid extensions) that link chloroplasts physically with the nucleus and play a pivotal role in retrograde signaling (Mullineaux et al., 2019). Salinity, drought, and ABA are among the agents inducing stromule formation (Gray et al., 2012). According to Zentgraf et al. (2022), H2O2 signaling in plants involves not only simple accumulation of the molecule but also a modulation of the ratio of H2O2 concentrations between different compartments. For instance, it has been shown that H2O2 produced in chloroplasts or in peroxisomes induces two types of transcriptomic responses: one independent of the subcellular site of H2O2 production and another that is organelle-specific (Sewelam et al., 2014). Therefore, it seems plausible that the induction of SnRK2.10-dependent signaling pathways requires not only an overall ROS accumulation in the cell but also an appropriate ROS ratio between compartments and/or specific site of ROS accumulation. Our results clearly showed that the ROS accumulation following paraquat treatment does not affect SnRK2.10-signaling in Arabidopsis, although the moderate induction of SnRK2.10 expression in these conditions suggests its putative role in the plant response to the herbicide.
Identifying and establishing the roles of salt-responsive genes is key for understanding the mechanisms of the plant response to salinity as well as the molecular basis of their resistance. Transcription factors deserve special attention here because they often regulate a broad spectrum of responses to abiotic and biotic stresses. Proteins containing the WRKY domain comprise one of the largest families of transcription factors in plants and modulate numerous processes including senescence, seed development, dormancy, and germination, and diverse biotic and abiotic stress responses (Phukan et al., 2016; Birkenbihl et al., 2018).
It has been shown that WRKY33 is induced by chloroplast-derived hydrogen peroxide in Arabidopsis plants overexpressing glycolate oxidase and challenged with high light conditions (Schmidt et al., 2020). On the other hand, WRKY33 suppression leads to enhanced H2O2 accumulation (Sun et al., 2020). Jiang and Deyholos (2009) reported that WRKY33 expression was induced under salinity and the wrky33 null mutant showed only a moderately increased NaCl sensitivity when primary root length and ion leakage were monitored. Besides, several research groups have shown that WRKY33 strongly affects plant resistance to Botrytis cinerea through negative regulation of ABA biosynthesis and signaling, and by inducing synthesis of camalexin (Birkenbihl et al., 2012; Liu et al., 2015; Sham et al., 2017; Zhou et al., 2020). Further, WRKY33 together with SALT TOLERANCE ZINC FINGER (STZ, ZAT10) and ARABIDOPSIS TOXICOS EN LEVADURA 31 (ATL31) take part in a common transcriptional regulatory network inducing hypocotyl elongation downstream of the auxin perception module (Rigal et al., 2021).
Regarding WRKY40, recent research points to its role in responses to ABA, drought, and pathogens. It has been shown that the WRKY40 gene is induced by drought and salinity, and a wrky40 insertion mutant displays an ABA-hypersensitive phenotype in seed germination, green cotyledon formation, and primary root elongation tests (Chen et al., 2010; Rasheed et al., 2016; Ahmad et al., 2019; Wang et al., 2021; Gigli-Bisceglia et al., 2022). In cooperation with WRKY18 and WRKY60, WRKY40 modulates the plants response and susceptibility to the hemibiotrophic bacterial pathogen Pseudomonas syringae, and the necrotrophic fungal pathogen Botrytis cinerea (Xu et al., 2006; Birkenbihl et al., 2017; Abeysinghe et al., 2018).
WRKY46 has been reported to mediate leaf senescence and undergo induction in Arabidopsis plants under various stresses, e.g., drought, salinity, H2O2, and salicylic acid treatments; it confers resistance to drought and salinity by contributing to an inhibition of osmotic/salt stress-dependent formation of lateral roots via regulation of ABA signaling and auxin homeostasis (Ding et al., 2015; Ding et al., 2015; Chen et al., 2017; Zhang et al., 2021). The phenotypes exhibited by the wrky46 and snrk2.10 insertion mutant plants challenged by salinity are similar to some extent. It has been shown that wrky46 seedlings achieve smaller increments of dry mass accumulation and exhibit higher salt sensitivity than wild-type plants, whereas WRKY46 overexpressing lines are more resistant to salinity. Seedlings of wrky46 (Ding et al., 2014) and adult snrk2.10 plants exposed to salinity (Mazur et al., 2021) accumulate more ROS in the leaves, suggesting that the two respective proteins may function in the regulation of cellular redox homeostasis. And finally, WRKY46 and SnRK2.10 are both engaged in lateral root formation under osmotic and salt stress (McLoughlin et al., 2012; Ding et al., 2015). Thus, there are ample indicators of a functional similarity of the SnRK2.10 and WRKY46 signaling pathways during the response to salinity or even their partial overlapping, the confirmation of which, however, requires additional research.
The WRKY75 transcription factor takes part in the signaling pathways of diverse plant hormones and acts as a multilink of the response to abiotic and biotic stressors, e.g., phosphorus starvation (Devaiah et al., 2007; Rishmawi et al., 2014), Sclerotinia sclerotiorum infection (Chen et al., 2013), and treatment with flagellin (Birkenbihl et al., 2018). It has been also shown that WRKY75 is induced after 24 h of salinity, plays the role of a genuine regulator of the ER-stress cellular responses, and its overexpression confers plant resistance to salt stress (Hossain et al., 2016). Besides its role in stress response, WRKY75 also participates in the regulation of plant development. In particular, it plays a role in the formation of root architecture (Devaiah et al., 2007), promotes flowering via gibberellin-dependent pathways (Zhang et al., 2018), mediates ABA-dependent seed germination, and senescence, where a tripartite amplification loop involving WRKY75, salicylic acid, and ROS has been reported (Guo et al., 2017; Zhang et al., 2022).
Recent studies indicate that molecular dynamics, specific homo- and heterodimerizations, as well as modular flexibility and posttranslational modifications, determine the functional specificity of many TFs engaged in environmental adaptation (Golldack et al., 2011). Among the most intensively studied TFs are those belonging to the WRKY family, notably for their propensity to function as hubs in complex protein-protein networks. Numerous WRKY hubs have been identified in biotic and abiotic stress responses, including WRKY18, WRKY33, WRKY40, WRKY46, WRKY51, WRKY53, WRKY60, and WRKY70 (Friedel et al., 2012; Chi et al., 2013; Choura et al., 2015; Birkenbihl et al., 2017). There is a significant overlap between the sets of various WRKY target genes. One such gene is CYP71A13 encoding camalexin biosynthesis enzyme, and it is induced by NaCl treatment (Xu et al., 2008). However, the role of camalexin in the response to salinity remains unknown. The CYP71A13 promoter is a direct target of WRKY33, WRKY40, and WRKY18, and the former two also bind to the WRKY75 promoter in response to salinity or biotic stimuli (Birkenbihl et al., 2012; Birkenbihl et al., 2017). WRKY40 is a central node in abiotic stress response regulation in shoots, forming the regulatory network with other TFs, including WRKY33 and WRKY46. WRKY46 is a hub molecule in plant roots challenged with abiotic stress (Friedel et al., 2012). Furthermore, WRKY40 and WRKY46 act as hubs in the plant - pathogen interaction system (Biniaz et al., 2022). OSM34 analyzed by us in this report is also controlled by WRKY TFs, e.g., WRKY33 (Zheng et al., 2006; Jiang et al., 2007). We found that under salinity the expression of the WRKY33, WRKY40, WRKY75, CYP71A13, and OSM34 genes was reduced in snrk2.10 mutants compared to wt plants. This suggests that SnRK2.10 affects the expression of CYP71A13, OSM34, and WRKY75 by influencing their upstream regulators WRKY33 and WRKY40. Interestingly, WRKY33, WRKY40, WRKY75, and CYP71A13 all belong to clusters of genes upregulated in Arabidopsis rosettes by H2O2 treatment and during developmental- and NaCl-induced senescence (Allu et al., 2014). This is in agreement with the observation that high and prolonged salinity can induce genes involved in programmed cell death and senescence initiation (Golldack et al., 2011). Currently, it is considered that the function of SnRK2.10 is dedicated to the response to osmotic/salt and to some extent to Cd2+ -induced stress (Kulik et al., 2012; McLoughlin et al., 2012; Julkowska et al., 2015; Maszkowska et al., 2019; Szymańska et al., 2019). The present results suggest that SnRK2.10, and possibly also other ABA non-activated SnRK2s, could play a more general role in the regulation of the responses to abiotic and biotic stimuli, nutrients imbalance, induction of senescence, and other developmental events by influencing the hubs of transcription factor networks. Our analysis of putative WRKY33/40/46/75 targets revealed a high proportion of genes regulated by several of those TFs and coding for proteins involved in global stress responses.
In conclusion, we have shown that NaCl-activated SnRK2.10 kinase signaling is involved in the induction of four WRKY TFs, WRKY33, WRKY40, WRKY46, and WRKY75, in Arabidopsis leaves, whereas H2O2-induced activation of the kinase attenuates their expression. The activation of SnRK2.10 and the following transcriptional responses do not depend on the ROS accumulation per se, but rather are fine-tuned depending on the source of ROS and their intracellular distribution in different compartments, and the co-occurrence with other stress factors. This indicates a previously unanticipated plasticity and variable specificity of the SnRK2.10-dependent signaling pathways, most likely achieved through interactions with other, so far very poorly understood, components and regulators. The regulation of the hub WRKY transcription factors by SnRK2.10 indicates its pivotal role in the response to abiotic stress. Analyzed by us WRKY TFs regulate a large number of stress-related protein kinases, which suggests that SnRK2.10 could modulate the pleiotropic cellular responses to salinity and ROS, including both common and stress-specific responses. Its involvement in the response to other stress factors (e.g., nutrient imbalance, pathogens) and in the regulation of plant development (e.g., root architecture, germination, senescence), all controlled by the four WRKY TFs, should also be taken into consideration.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
AK designed and supervised the study. AK, JR, AA-M, MB and KS performed the experiments. AK interpreted results and wrote the manuscript. All authors revised the manuscript.
Funding
This work was supported by the National Science Centre, Poland, grant 2017/27/B/NZ3/01763 to AK.
Acknowledgments
We are grateful to Professor C. Testerink (the Wageningen University) for seeds of the SnRK2.10-GFP transgenic plants and to Professor G. Dobrowolska (Institute of Biochemistry and Biophysics, PAS) for stimulating discussions and insightful comments. Fluorescence microscopy was performed in the Fluorescence Microscopy Facility in IBB PAS, Poland.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1135240/full#supplementary-material
Supplementary Figure 1 | Selected Gene Ontology Terms for WRKY33, WRKY40, WRKY46, and WRKY75 enriched in the target gene set common to all four WRKY TFs. Bar lengths indicate -log10 -transformed False Discovery Rate (FDR). BP – Biological Process, MF – Molecular Function, CC – Cellular Component.
Supplementary Figure 2 | Detailed localization of H2O2 accumulation in the cytoplasm in response to salinity. Staining of Arabidopsis leaves with an H2O2-specific BES-H2O2-Ac probe for intracellular hydrogen peroxide detection was carried out as described in Materials and Methods. Detection of BES-H2O2-Ac fluorescence is shown in false green (A–F). The panel consists of images of leaves after 30 min of 150 mM NaCl applied directly to leaves (A–C) or 30 min of 250 mM NaCl applied directly to leaves (D–F). Scale bar: 10 µm. This panel shows three examples of individual optical sections from the projections shown in Figure 6II O, S.
Supplementary Figure 3 | Model presenting putative regulation of WRKYs expression by SnRK2.10 in response to salinity. Model is based on the results published by Sorenson et al. (2018); Soma et al. (2017); Kawa et al. (2020) and the present data. In response to salinity, SnRK2.10 is phosphorylated and interplays with cellular H2O2/ROS which triggers specific signaling pathways leading to the induction of expression of WRKY33, WRKY40, WRKY46, and WRKY75. Synthetized mRNA may be translated, which leads to the formation of WRKYs protein and specific regulation of stress response. Parallelly, SnRK2.10 may phosphorylate VCS and influence the 5’mRNA decay system, thus alternatively post-transcriptionally controlling WRKYs expression.
Supplementary Table 1 | Primers used in this study.
Supplementary Data Sheet 1 | Target genes of WRKY33, WRKY40, WRKY46, and WRKY75.
Supplementary Data Sheet 2 | Protein partners of WRKY33, WRKY40, WRKY46, and WRKY75.
References
Abeysinghe, J. K., Lam, K. M., Ng, D. W. (2018). Differential regulation and interaction of homoeologous WRKY18 and WRKY40 in arabidopsis allotetraploids and biotic stress responses. Plant J. 97 (2), 352–367. doi: 10.1111/tpj.14124
Acosta-Motos, J. R., Ortuño, M. F., Bernal-Vicente, A., Diaz-Vivancos, P., Sanchez-Blanco, M. J., Hernandez, J. A. (2017). Plant responses to salt stress: adaptive mechanisms. Agronomy 7, 18. doi: 10.3390/agronomy7010018
Adachi, H., Nakano, T., Miyagawa, N., Ishihama, N., Yoshioka, M., Katou, Y., et al. (2015). WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27, 2645–2663. doi: 10.1105/tpc.15.00213
Aghamirzaie, D., Velmurugan, K. R., Wu, S., Altarawy, D., Heath, L. S., Grene, R. (2017). Expresso: a database and web server for exploring the interaction of transcription factors and their target genes in arabidopsis thaliana using ChIP-seq peak data. F1000 Res. 6, 372. doi: 10.12688/f1000research
Ahmad, R., Liu, Y., Wang, T.-J., Meng, Q., Yin, H., Wang, X., et al. (2019). GOLDEN2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid. Plant Physiol. 179 (4), 1844–1860. doi: 10.1104/pp.18.01466
Allu, A. D., Soja, A. M., Wu, A., Szymanski, J., Balazadeh, S. (2014). Salt stress and senescence: identification of cross-talk regulatory components. J. Exp. Bot. 65 (14), 3993–4008. doi: 10.1093/jxb/eru173
Babbs, C. F., Pham, J. A., Coolbaugh, R. C. (1989). Lethal hydroxyl radical production in paraquat-treated plants. Plant Physiol. 90 (4), 1267–1270. doi: 10.1104/pp.90.4.1267
Baena-González, E., Rolland, F., Thevelein, J. M., Sheen, J. (2007). A central integrator of transcription netwo rks in plant stress and energy signaling. Nature 448, 938–942. doi: 10.1038/nature06069
Baltazar, T., Dinis-Oliveira, R. J., Duarte, J. A., de Lourdes Bastos, M., Carvalho, F. (2013). Paraquat research: do recent advances in limiting its toxicity make its use safer? Br. J. Pharmacol. 168 (1), 44–45. doi: 10.1111/j.1476-5381.2012.02017.x
Basbouss-Serhal, I., Pateyron, S., Cochet, F., Leymarie, J., Bailly, C. (2017). 5′ to 3′ mRNA decay contributes to the regulation of arabidopsis seed germination by dormancy. Plant Physiol. 173 (3), 1709–1723. doi: 10.1104/pp.16.01933
Bellafiore, S., Barneche, F., Peltier, G., Rochaix, J.-D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433, 892–895. doi: 10.1038/nature03286
Benina, M., Ribeiro, D. M., Gechev, T. S., Mueller-Roeber, B., Schippers, J. H. M. (2015). A cell type-specific view on the translation of mRNAs from ROS-responsive genes upon paraquat treatment of arabidopsis thaliana leaves. Plant Cell Environ. 38 (2), 349–363. doi: 10.1111/pce.12355
Ben Rejeb, K., Benzarti, M., Debez, A., Bailly, C., Savouré, A., Abdelly, C. (2015). NADPH oxidase-dependent H2O2 production is required for salt-induced antioxidant defense in arabidopsis thaliana. J. Plant Physiol. 174, 5–15. doi: 10.1016/j.jplph.2014.08.022
Berardini, T. Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The arabidopsis information resource: making and mining the "gold standard" annotated reference plant genome. Genesis 53, 474–485. doi: 10.1002/dvg.22877
Biniaz, Y., Tahmasebi, A., Tahmasebi, A., Albrectsen, B. R., Poczai, P., Afsharifar, A. (2022). Transcriptome meta-analysis identifies candidate hub genes and pathways of pathogen stress responses in Arabidopsis thaliana. Biol 11, 1155. doi: 10.3390/biology11081155
Birkenbihl, R. P., Diezel, C., Somssich, I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward botrytis cinerea infection. Plant Physiol. 159 (1), 266–285. doi: 10.1104/pp.111.192641
Birkenbihl, R. P., Kracher, B., Roccaro, M., Somssich, I. E. (2017). Induced genome-wide binding of three arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 29 (1), 20–38. doi: 10.1105/tpc.16.00681
Birkenbihl, R. P., Kracher, B., Ross, A., Kramer, K., Finkemeier, I., Sommish, I. E. (2018). Principles and characteristics of the arabidopsis WRKY regulatory network during early MAMP-triggered immunity. Plant J. 96, 487–502. doi: 10.1111/tpj.14043
Boudsocq, M., Barbier-Brygoo, H., Laurière, C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stress in Arabidopsis thaliana. J. Biol. Chem. 279, 41758–41766. doi: 10.1074/jbc.M405259200
Bulgakov, V. P., Gorpenchenko, T. Y., Veremeichik, G. N., Shkryl, Y. N., Tchernoded, G. K., Bulgakov, D. V., et al. (2012). The rolB gene suppresses reactive oxygen species in transformed plant cells through the sustained activation of antioxidant defense. Plant Physiol. 158, 1371–1381. doi: 10.1104/pp.111.191494
Burdiak, P., Rusaczonek, A., Witoń, D., Głów, D., Karpiński, S. (2015). Cysteine-rich receptor-like kinase CRK5 as a regulator of growth, development, and ultraviolet radiation responses in Arabidopsis thaliana. J. Exp. Bot. 66 (11), 3325–3337. doi: 10.1093/jxb/erv143
Cao, Y., Liang, Y., Tanaka, K., Nguyen, C. T., Jedrzejczak, R. P., Joachimiak, A., et al. (2014). The kinase LYK5 is a major chitin receptor in arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3, e03766. doi: 10.7554/eLife.03766
Carpentier, M. C., Deragon, J. M., Jean, V., Be, S. H. V., Bousquet-Antonelli, C., Merret, R. (2020). Monitoring of XRN4 targets reveals the importance of cotranslational decay during arabidopsis development. Plant Physiol. 184 (3), 1251–1262. doi: 10.1104/pp.20.00942
Chen, K., Du, L., Chen, Z. (2003). Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in arabidopsis. Plant Mol. Biol. 53, 61–74. doi: 10.1023/B:PLAN.0000009265.72567.58
Chen, Y., Hoehenwarter, W. (2015). Changes in the phosphoproteome and metabolome link early signaling events to rearrangement of photosynthesis and central metabolism in salinity and oxidative stress response in arabidopsis. Plant Physiol. 169, 3021–3033. doi: 10.1104/pp.15.01486
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., Xu, X. (2010). Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10, 281. doi: 10.1186/1471-2229-10-281
Chen, X., Liu, J., Lin, G., Wang, A., Wang, Z., Lu, G. (2013). Overexpression of AtWRKY28 and AtWRKY75 in arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Rep. 32, 1589–1599. doi: 10.1007/s00299-013-1469-3
Chen, J., Nolan, T. M., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29 (6), 1425–1439. doi: 10.1105/tpc.17.00364
Chi, Y., Yang, Y., Zhou, Y., Zhoua, J., Fan, B., Yu, J.-Q., et al. (2013). Protein–protein interactions in the regulation of WRKY transcription factors. Mol. Plant 6 (2), 287–300. doi: 10.1093/mp/sst026
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Choi, S.-W., Lee, S.-B., Na, Y.-J., Jeung, S.-G., Yo, S. (2017). Arabidopsis MAP3K16 and other salt-inducible MAP3Ks regulate ABA response redundantly. Mol. Cells 40 (3), 230–242. doi: 10.14348/molcells.2017.0002
Choi, H.-I., Park, H.-J., Park, J. H., Kim, S., Im, M.-Y., Seo, H.-H., et al. (2005). Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 139, 1750–1761. doi: 10.1104/pp.105.069757
Choura, M., Rebai, A., Masmoudi, K. (2015). Unraveling the WRKY transcription factors network in arabidopsis thaliana by integrative approach. Net. Biol. 5 (2), 55–61.
Coleman, A. D., Maroschek, J., Raasch, L., Takken, F. L. W., Ranf, S., Hüuckelhoven, R. (2020). The arabidopsis leucine-rich repeat receptor-like kinase MIK2 isa crucial component of early immune responses to a fungal-derived elicitor. New Phytol. 229, 3453–3466. doi: 10.1111/nph.17122
Cui, F., Brosché, M., Shapiguzov, A., He, X.-Q., Vainonen, J. P., Leppälä, J., et al. (2019). Interaction of methyl viologen-induced chloroplast and mitochondrial signalling in arabidopsis. Free Rad. Biol. Med. 134, 555–566. doi: 10.1016/j.freeradbiomed.2019.02.006
Devaiah, B. N., Karthikeyan, A. S., Raghothama, K. G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in arabidopsis. Plant Physiol. 143 (4), 1789–1801. doi: 10.1104/pp.106.093971
Ding, Z., Kihara, D. (2019). Computational identification of protein-protein interactions in model plant proteomes. Sci. Rep. 9, 8740. doi: 10.1038/s41598-019-45072-8
Ding, Z. J., Yan, J. Y., Li, C. X., Li, G. X., Wu, Y. R., Zheng, S. J. (2015). Transcription factor WRKY46 modulates the development of arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 84 (1), 56–69. doi: 10.1111/tpj.12958
Ding, Z. J., Yan, J. Y., Xu, X. Y., Yu, D. Q., Li, G. X., Zhang, S. Q., et al. (2014). Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in arabidopsis. Plant J. 79 (1), 13–27. doi: 10.1111/tpj.12538
Dodge, A. D. (1989). Herbicides interacting with photosystem IHerbicides and plant metabolism. Cambridge Univ. Press pp, 1–277. doi: 10.1017/CBO9780511752315.004
Du, L., Chen, Z. (2000). Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in arabidopsis. Plant J. 24 (6), 837–847. doi: 10.1111/j.1365-313X.2000.00923.x
Dubiella, U., Seybold, H., Durian, G., Komander, E., Lassig, R., Witte, C.-P., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U.S.A. 110 (21), 8744–8749. doi: 10.1073/pnas.1221294110
Durinck, S., Spellman, P., Birney, E., Huber, W. (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Prot. 4, 1184–1191. doi: 10.1038/nprot.2009.97
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Tren. Plant Sci. 5 (5), 199–206. doi: 10.1016/s1360-1385(00)01600-9
Fàbregas, N., Yoshida, T., Fernie, A. R. (2020). Role of raf-like kinases in SnRK2 activation and osmotic stress response in plants. Nat. Comm. 11, 6184. doi: 10.1038/s41467-020-19977-2
Frick, E. M., Strader, L. C. (2018). Kinase MPK17 and the peroxisome division factor PMD1 influence salt-induced peroxisome proliferation. Plant Physiol. 176, 340–351. doi: 10.1104/pp.17.01019
Friedel, S., Usadel, B., von Wiren, N., Sreenivasulu, N. (2012). Reverse engineering: a key component of systems biology to unravel global abiotic stress cross-talk. Front. Plant Sci. 3 (294). doi: 10.3389/fpls.2012.00294
Fuerst, E. P., Norman, M. A. (1991). Interactions of herbicides with photosynthetic electron transport. Weed Sci. 39, 458–464. doi: 10.1017/S0043174500073227
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in arabidopsis. Plant Cell Physiol. 50 (12), 2123–2132. doi: 10.1093/pcp/pcp147
Gao, M., Liu, J., Bi, D., Zhang, Z., Cheng, F., Chen, S., et al. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18, 1190–1198. doi: 10.1038/cr.2008.300
Geiger, D., Scherzer, S., Mumm, P., Marten, I., Ache, A., Matschi, S., et al. (2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. U.S.A. 107 (17), 8023–8028. doi: 10.1073/pnas.0912030107
Giacomelli, J., Weigel, D., Chan, R. L., Manavella, P. A. (2012). Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 195, 766–773. doi: 10.1111/j.1469-8137.2012.04259.x
Gigli-Bisceglia, N., van Zelm, E., Huo, W., Lamers, J., Testerink, C. (2022). Arabidopsis root responses to salinity depend on pectin modification and cell wall sensing. Develop 149, dev200363. doi: 10.1242/dev.200363
Golldack, D., Lüking, I., Yang, O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30 (8), 1391. doi: 10.1007/s00299-011-1068-0
Gómez-Gómez, L., Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/s1097-2765(00)80265-8
Gray, J. C., Hansen, M. R., Shaw, D. J., Graham, K., Dale, R., Smallman, P., et al. (2012). Plastid stromules are induced by stress treatments acting through abscisic acid. Plant J. 69, 387–398. doi: 10.1111/j.1365-313X.2011.04800.x
Guo, P., Li, Z., Huang, P., Li, B., Fang, S., Chu, J., et al. (2017). A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29 (11), 2854–2870. doi: 10.1105/tpc.17.00438
Guy, E., Lautier, M., Chabannes, M., Roux, B., Lauber, E., Arlat, M., et al. (2013). xopAC-triggered immunity against xanthomonas depends on arabidopsis receptor-like cytoplasmic kinase genes PBL2 and RIPK. PloS One 8 (8), e73469. doi: 10.1371/journal.pone.0073469
Hartel, H., Haseloff, R. F., Ebert, B. (1992). Free radical formation in chloroplasts: methyl viologen action. Photochem. Photobiol. 12, 375–387. doi: 10.1016/1011-1344(92)85042-S
Hasanuzzaman, M., Raihan, M. R. H., Masud, A. A. C., Rahman, K., Nowroz, F., Rahman, M., et al. (2021). Regulation of reactive oxyden species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 22, 9326. doi: 10.3390/ijms22179326
Hassan, H. M. (1984). Exacerbation of superoxide radical formation by paraquat. Methods Enzym. pp, 523–532. doi: 10.1016/S0076-6879(84)05072-2
He, P., Shan, L., Lin, N.-C., Martin, G. B., Kemmerling, B., Nürnberger, T., et al. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in arabidopsis innate immunity. Cell 125 (3), 563–575. doi: 10.1016/j.cell.2006.02.047
Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M. E., He, K., Li, J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. U.S.A. 104 (29), 12217–12222. doi: 10.1073/pnas.0705306104
Hehl, R., Norval, L., Romanov, A., Bülow, L. (2016). Boosting AthaMap database content with data from protein binding microarrays. Plant Cell Physiol. 57 (1), e4. doi: 10.1093/pcp/pcv156
Hok, S., Allasia, V., Andrio, E., Naessens, E., Ribes, E., Panabières, F., et al. (2014). The receptor kinase IMPAIRED OOMYCETE SUSCEPTIBILITY1 attenuates abscisic acid responses in arabidopsis. Plant Physiol. 166 (3), 1506–1518. doi: 10.1104/pp.114.248518
Hok, S., Danchin, E. G. J., Allasia, V., Panabieres, F., Attard, A., Keller, H. (2011). An arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ. 34, 1944–1957. doi: 10.1111/j.1365-3040.2011.02390.x
Hossain, M. A., Henríquez-Valencia, C., Gómez-Páez, M., Medina, J., Orellana, A., Vicente-Carbajosa, J. (2016). Identification of novel components of the unfolded protein response in arabidopsis. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00650
Hou, X., Tong, H., Selby, J., DeWitt, J., Peng, X., He, Z.-H. (2005). Involvement of a cell wall-associated kinase, WAKL4, in arabidopsis mineral responses. Plant Physiol. 139 (4), 1704–1716. doi: 10.1104/pp.105.066910
Hrabak, E. M., Chan, C. W. M., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N. (2003). The arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132 (2), 666–680. doi: 10.1104/pp.102.011999
Huang, C., Yan, Y., Zhao, H., Ye, Y., Cao, Y. (2020). Arabidopsis CPK5 phosphorylates the chitin receptor LYK5 to regulate plant innate immunity. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00702
Idänheimo, N., Gauthier, A., Salojärvi, J., Siligato, R., Brosché, M., Kollist, H., et al. (2014). The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Comm. 445 (2), 457–462. doi: 10.1016/j.bbrc.2014.02.013
Jiang, Y., Deyholos, M. K. (2009). Functional characterization of arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69 (1-2), 91–105. doi: 10.1007/s11103-008-9408-3
Jiang, Y.-Q., Yang, B., Harris, N. S., Deyholos, M. K. (2007). Comparative proteomic analysis of NaCl stress-responsive proteins in arabidopsis roots. J. Exp. Bot. 58, 3591–3607. doi: 10.1093/jxb/erm207
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01029
Jossier, M., Bouly, J.-P., Meimoun, P., Arjmand, A., Lessard, P., Hawley, S., et al. (2009). SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in arabidopsis thaliana. Plant J. 59, 316–328. doi: 10.1111/j.1365-313X.2009.03871.x
Julkowska, M. M., McLoughlin, F., Galvan-Ampudia, C. S., Rankenberg, J. M., Kawa, D., Klimecka, M., et al. (2015). Identification and functional characterization of the arabidopsis Snf1-related protein kinase SnRK2.4 phosphatidic acid-binding domain. Plant Cell Environ. 38 (3), 614–624. doi: 10.1111/pce.12421
Kamyiama, Y., Katagiri, S., Umezawa, T. (2021). Growth promotion or osmotic stress response: how SNF1-related protein kinase 2 (SnRK2) kinases are activated and manage intracellular signaling in plants. Plants 10 (7), 1443. doi: 10.3390/plants10071443
Kawa, D., Meyer, A. J., Dekker, H. L., Abd-El-Haliem, A. M., Gevaert, K., van de Slijke, E., et al. (2020). SnRK2 protein kinases and mRNA decapping machinery control root development and response to salt. Plant Physiol. 182, 361–377. doi: 10.1104/pp.19.00818
Kim, H. S., Jung, M. S., Lee, S. M., Kim, K. E., Byun, H., Choi, M. S. (2009). An s-locus receptor-like kinase plays a role as a negative regulator in plant defense responses. Biochem. Biophys. Res. Comm. 381 (3), 424–428. doi: 10.1016/j.bbrc.2009.02.050
Kong, Q., Qu, N., Gao, M., Zhang, Z., Ding, X., Yang, F., et al. (2012). The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in arabidopsis. Plant Cell 24, 2225–2236. doi: 10.1105/tpc.112.097253
Kong, Q., Sun, T., Qu, N., Ma, J., Li, M., Cheng, Y., et al. (2016). Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol. 171, 1344–1354. doi: 10.1104/pp.15.01954
Kulik, A., Anielska-Mazur, A., Bucholc, M., Koen, E., Szymańska, K., Zmieńko, A., et al. (2012). SNF1-related protein kinases type 2 are involved in plant responses to cadmium stress. Plant Physiol. 160, 868–883. doi: 10.1104/pp.112.194472
Kulik, A., Noirot, E., Grandperret, V., Bourque, S., Fromentin, J., Salloignon, P., et al. (2015). Interplays between nitric oxide and reactive oxygen species in cryptogein signalling. Plant Cell Environ. 38, 331–348. doi: 10.1111/pce.12295
Kulik, A., Wawer, I., Krzywińska, E., Bucholc, M., Dobrowolska, G. (2011). SnRK2 protein kinases–key regulators of plant response to abiotic stresses. OMICS 15, 859–872. doi: 10.1089/omi.2011.0091
Kulkarni, S. R., Vaneechoutte, D., Van de Velde, J., Vandepoele, K. (2018). TF2Network: predicting transcription factor regulators and gene regulatory networks in arabidopsis using publicly available binding site information. Nuc. Acids Res. 46 (6), e31. doi: 10.1101/173559
Kumar, S., Tripathi, D., Okubara, P. A., Tanaka, K. (2020). Purinoceptor P2K1/ DORN1 enhances plant resistance against a soilborne fungal pathogen, Rhizoctonia solani. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.572920
Lee, H., Ganguly, A., Baik, S., Cho, H.-T. (2021). Calcium-dependent protein kinase 29 modulates PIN-FORMED polarity and arabidopsis development via its own phosphorylation code. Plant Cell 33 (11), 3513–3531. doi: 10.1093/plcell/koab207
Li, W., Pang, S., Lu, Z., Jin, B. (2020). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 9, 1515. doi: 10.3390/plants9111515
Lin, Y.-C., Kanehara, K., Nakamura, Y. (2019). Arabidopsis CHOLINE/ETHANOLAMINE KINASE 1 (CEK1) is a primary choline kinase localized at the endoplasmic reticulum (ER) and involved in ER stress tolerance. New Phytol. 223 (4), 1904–1917. doi: 10.1111/nph.15915
Liu, J., Chengcheng, F., Guangjing, L., Khan, M. N., Wu, H. (2021). ROS homeostasis and plant salt tolerance: plant nanobiotechnology updates. Sustainability 13, 3552. doi: 10.3390/su13063552
Liu, S., Kracher, B., Ziegler, J., Birkenbihl, R. P., Somssich, I. E. (2015). Negative regulation of ABA signaling by WRKY33 is critical for arabidopsis immunity towards botrytis cinerea 2100. eLife 4, e07295. doi: 10.7554/eLife.07295
Liu, X. M., Nguyen, X. C., Kim, K. E., Han, H. J., Yoo, Y., Lee, K., et al. (2013). Phosphorylation of the zinc finger transcriptional regulator ZAT6 by MPK6 regulates arabidopsis seed germination under salt and osmotic stress. Biochem. Biophys. Res. Comm. 430 (3), 1054–1059. doi: 10.1016/j.bbrc.2012.12.039
Liu, K. H., Niu, Y., Konishi, M., Wu, Y., Du, H., Chung, H. S., et al. (2017). Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 545, 311–316. doi: 10.1038/nature22077
Liu, Y., Sun, J., Wu, Y. (2016). Arabidopsis ATAF1 enhances the tolerance to salt stress and ABA in transgenic rice. J. Plant Res. 129 (5), 955–962. doi: 10.1007/s10265-016-0833-0
Lou, J., Yu, F., Tian, M., Liu, G., Wu, Y., Wu, Y., et al. (2020). ESCRT-I component VPS23A sustains salt tolerance by strengthening the SOS module in arabidopsis. Mol. Plant 13 (8), 1134–1148. doi: 10.1016/j.molp.2020.05.010
Lu, K., Liang, S., Wu, Z., Bi, C., Yu, Y.-T., Wang, X.-F., et al. (2016). Overexpression of an arabidopsis cysteine-rich receptor-like protein kinase, CRK5, enhances abscisic acid sensitivity and confers drought tolerance. J. Exp. Bot. 67 (17), 5009–5027. doi: 10.1093/jxb/erw266
Luo, X., Wu, W., Liang, Y., Xu, X., Wang, Z., Zou, H., et al. (2020). Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J. 39, e102856. doi: 10.15252/embj.2019102856
Mahfouz, M. M., Kim, S., Delauney, A. J., Verma, D. P. S. (2006). Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18 (2), 477–490. doi: 10.1105/tpc.105.035931
Maszkowska, J., Dędski, J., Kulik, A., Kistowski, M., Bucholc, M., Lichocka, M., et al. (2019). Phosphoproteomic analysis reveals that dehydrins ERD10 and ERD14 are phosphorylated by SNF1-related protein kinase 2.10 in response to osmotic stress. Plant Cell Environ. 42 (3), 931–946. doi: 10.1111/pce.13465
Mazur, R., Maszkowska, J., Anielska-Mazur, A., Garstka, M., Polkowska-Kowalczyk, L., Czajkowska, A., et al. (2021). The SnRK2.10 kinase mitigates the adverse effects of salinity by protecting photosynthetic machinery. Plant Physiol. 187 (4), 2785–2802. doi: 10.1093/plphys/kiab438
McLoughlin, F., Galvan-Ampudia, C. S., Julkowska, M. M., Caarls, L., van der Does, D., Laurière, C., et al. (2012). The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 72, 436–449. doi: 10.1111/j.1365-313X.2012.05089.x
Mittler, R. (2017). ROS are good. Trends Plant Sci. 22 (1), 11–19. doi: 10.1016/j.tplants.2016.08.002
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104 (49), 19613–19618. doi: 10.1073/pnas.0705147104
Mullineaux, P. M., Exposito-Rodriquez, M., Laissue, P. P., Smirnoff, N., Park, E. (2019). Spatial chloroplast-to-nucleus signaling involving plastid-nuclear complexes and stromules. Phil. Trans. R Soc. B 375, 20190405. doi: 10.1098/rstb.2019.0405
Murata, T., Noshi, M., Tanouchi, A., Tamoi, M., Yabuta, Y., Yoshimura, K., et al. (2012). H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J. Biol. Chem. 287 (15), 11717–11729. doi: 10.1074/jbc.M111.292847
Nagarajan, V. K., Kukulich, P. M., Hagel, B., Green, P. J. (2019). RNA Degradomes reveal substrates and importance for dark and nitrogen stress responses of arabidopsis XRN4. Nuc. Acids Res. 47 (17), 9216–9230. doi: 10.1093/nar/gkz712
Nazish, T., Huang, Y.-J., Zhang, J., Xia, J.-Q., Alfatih, A., Luo, C., et al. (2022). Understanding paraquat resistance mechanisms in arabidopsis thaliana to facilitate the development of paraquat-resistant crops. Plant Comm. 3, 100321. doi: 10.1016/j.xplc.2022.100321
Nguyen, H. M., Sako, K., Matsui, A., Suzuki, Y., Mostofa, M. G., Ha, C. V., et al. (2017). Ethanol enhances high-salinity stress tolerance by detoxifying reactive oxygen species in arabidopsis thaliana and rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01001
Nitta, Y., Ding, P., Zhang, Y. (2014). Identification of additional MAP kinases activated upon PAMP treatment. Plant Sign Behav. 9 (11), e976155. doi: 10.4161/15592324.2014.976155
Obomighie, I., Lapenas, K., Murphy, B. E., Bowles, A. M. C., Bechtold, U., Prischi, F. (2021). The role of ribosomal protein S6 kinases in plant homeostasis. Front. Mol. Biosci. 8. doi: 10.3389/fmolb.2021.636560
Okamoto, T., Takatani, S., Motose, H., Iida, H., Takahashi, T. (2021). The root growth reduction in response to mechanical stress involves ethylene-mediated microtubule reorganization and transmembrane receptor-mediated signal transduction in arabidopsis. Plant Cell Rep. 40, 575–582. doi: 10.1007/s00299-020-02653-6
Oliveros, J. C. (2007-2015) Venny. an interactive tool for comparing lists with venn's diagrams. Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
O’Malley, R. C., Huang, S. C., Song, L., Lewsey, M. G., Bartlett, A., Joseph, R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165 (5), 1280–1292. doi: 10.1016/j.cell.2016.04.038
Ortiz-Masia, D., Perez-Amador, M. A., Carbonell, J., Marcote, M. J. (2007). Diverse stress signals activate the C1 subgroup MAP kinases of arabidopsis. FEBS Let. 581, 1834–1840. doi: 10.1016/j.febslet.2007.03.075
Oughtred, R., Stark, C., Breitkreutz, B. J., Rust, J., Boucher, L., Chang, C., et al. (2019). The BioGRID interaction database: 2019 update. Nucleic Acids Res. 8, D529–D541. doi: 10.1093/nar/gky1079
Petersen, L. N., Ingle, R. A., Knight, M. R., Denby, K. J. (2009). OXI1 protein kinase is required for plant immunity against Pseudomonas syringae in arabidopsis. J. Exp. Bot. 60 (13), 3727–3735. doi: 10.1093/jxb/erp219
Phukan, U. J., Jeena, G. S., Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00760
Qin, D. B., Liu, M. Y., Yuan, L., Zhu, Y., Li, X. D., Chen, L. M., et al. (2020). Calcium-dependent protein kinase 32-mediated phosphorylation is essential for the ammonium transport activity of AMT1;1 in arabidopsis roots. J. Exp. Bot. 71, 5087–5097. doi: 10.1093/jxb/eraa249
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008a). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221. doi: 10.1038/emboj.2008.147
Qiu, J. L., Zhou, L., Yun, B.-W., Nielsen, H. B., Katrine Fiil, B., Petersen, K., et al. (2008b). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS11. Plant Physiol. 148, 212–222. doi: 10.1104/pp.108.120006
Ranf, S., Gisch, N., Schäffer, M., Illig, T., Westphal, L., Knirel, Y. A., et al. (2015). A lectin s-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immun. 16, 426–433. doi: 10.1038/ni.3124
Rasheed, S., Bashir, K., Matsui, A., Tanaka, M., Seki, M. (2016). Transcriptomic analysis of soil-grown arabidopsis thaliana roots and shoots in response to a drought stress. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00180
Rentel, M. C., Lecourieux, D., Ouaked, F., Usher, S. L., Petersen, L., Okamoto, H., et al. (2004). OXI1 kinase is necessary for oxidative burst-mediated signalling in arabidopsis. Nature 427 (6977), 858–861. doi: 10.1038/nature02353
Rhodes, J., Yang, H., Moussu, S., Boutrot, F., Santiago, J., Zipfel, C. (2021). Perception of a divergent family of phytocytokines by the arabidopsis receptor kinase MIK2. Nat. Comm. 12, 705. doi: 10.1038/s41467-021-20932-y
Rigal, A., Doyle, S. M., Ritter, A., Raggi, S., Vain, T., O’Brien, J. A., et al. (2021). A network of stress-related genes regulate hypocotyl elongation downstream of selective auxin perception. Plant Physiol. 187 (1), 430–445. doi: 10.1093/plphys/kiab269
Rishmawi, L., Pesch, M., Juengst, C., Schauss, A. C., Schrader, A., Hülskamp, M. (2014). Non-Cell-Autonomous regulation of root hair patterning genes by WRKY75 in arabidopsis. Plant Physiol. 165 (1), 186–195. doi: 10.1104/pp.113.233775
Robatzek, S., Somssich, I. E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. doi: 10.1101/gad.222702
Schmidt, A., Mächtel, R., Ammon, A., Engelsdorf, T., Schmitz, J., Maurino, V. G., et al. (2020). Reactive oxygen species dosage in arabidopsis chloroplasts can improve resistance towards Colletotrichum higginsianum by the induction of WRKY33. New Phytol. 226, 1189–1204. doi: 10.1111/nph.16332
Seki, M., Narusaka, M., Ishida, J., Nanjo, T., Fujita, M., Oono, Y., et al. (2002). Monitoring the expression profiles of 7000 arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31 (3), 279–292. doi: 10.1046/j.1365-313x.2002.01359.x
Sewelam, N., Jaspert, N., van der Kelen, K., Tognetti, V. B., Schmitz, J., Frerigmann, H., et al. (2014). Spatial H2O2 signaling specificity: H2O2 from chloroplsts and peroxisomes modulates the plant transcriptome differentially. Mol. Plant 7, 1191–1210. doi: 10.1093/mp/ssu070
Sewelam, N., Kazan, K., Schenk, P. M. (2016). Global planr stress signaling: reactive oxygen species at the cross-road. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00187
Sham, A., Moustafa, K., Al-Shamisi, S., Alyan, S., Iratni, R., AbuQamar, S. (2017). Microarray analysis of arabidopsis WRKY33 mutants in response to the necrotrophic fungus Botrytis cinerea. PloS One 12 (2), e0172343. doi: 10.1371/journal.pone.0172343
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The mg-chelatase h subunit of arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shi, W., Wang, L., Yao, L., Hao, W., Han, C., Fan, M., et al. (2022). Spatially patterned hydrogen peroxide orchestrates stomatal development in arabidopsis. Nat. Comm. 13, 5040. doi: 10.1038/s41467-022-32770-7
Shi, H., Wang, X., Ye, T., Chen, F., Deng, J., Yang, P., et al. (2014). The Cysteine2/Histidine2-type transcription factor ZINC FINGER OF ARABIDOPSIS THALIANA6 modulates biotic and abiotic stress responses by activating salicylic acid-related genes and c-REPEAT-BINDING FACTOR genes in arabidopsis. Plant Physiol. 165 (3), 1367–1379. doi: 10.1104/pp.114.242404
Shinozawa, A., Otake, R., Takezawa, D., Umezawa, T., Komatsu, K., Tanaka, K., et al. (2019). SnRK2 protein kinases represent an ancient system in plants for adaptation to a terrestrial environment. Commun. Biol. 2, 30. doi: 10.1038/s42003-019-0281-1
Shumbe, L., Chevalier, A., Legeret, B., Taconnat, L., Monnet, F., Havaux, M. (2016). Singlet oxygen-induced cell death in arabidopsis under high-light stress is controlled by OXI1 kinase. Plant Physiol. 170 (3), 1757–1771. doi: 10.1104/pp.15.01546
Smeets, K., Opdenakker, K., Remans, T., Forzani, C., Hirt, H., Vangronsveld, J., et al. (2013). The role of the kinase OXI1 in cadmium- and copper-induced molecular responses in Arabidopsis thaliana. Plant Cell Environ. 36, 1228–1238. doi: 10.1111/pce.12056
Smirnoff, N., Arnaud, D. (2019). Hydrogen peroxide metabolism and functions in plants. New Phytol. 221, 1197–1214. doi: 10.1111/nph.15488
Soma, F., Mogami, J., Yoshida, T., Abekura, M., Takahashi, F., Kidokoro, S., et al. (2017). ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants 3, 16204. doi: 10.1038/nplants.2016.204
Song, S.-J., Feng, Q.-N., Li, C.-L., Li, E., Liu, Q., Kang, H., et al. (2018). A tonoplast-associated calcium-signaling module dampens ABA signaling during stomatal movement. Plant Physiol. 177 (4), 1666–1678. doi: 10.1104/pp.18.00377
Sorenson, R. S., Deshotel, M. J., Johnson, K., Adler, F. R., Sieburth, L. E. (2018). Arabidopsis mRNA decay landscape arises from specialized RNA decay substrates, decapping-mediated feedback, and redundancy. Proc. Natl. Acad. Sci. U.S.A. 13, E1485–E1494, 115(7). doi: 10.1073/pnas.1712312115
Sözen, C., Schenk, S. T., Boudsocq, M., Chardin, C., Almeida-Trapp, M., Krapp, A., et al. (2020). Wounding and insect feeding trigger two independent MAPK pathways with distinct regulation and kinetics. Plant Cell 32, 1988–2003. doi: 10.1105/tpc.19.00917
Sreekanta, S., Bethke, G., Hatsugai, N., Tsuda, K., Thao, A., Wang, L., et al. (2015). The receptor-like cytoplasmic kinase PCRK1 contributes to pattern-triggered immunity against Pseudomonas syringae in Arabidopsis thaliana. New Phyt. 207, 78–90. doi: 10.1111/nph.13345
Sridharamurthy, M., Kovach, A., Zhao, Y., Zhu, J. K., Xu, H. E., Swaminathan, K., et al. (2014). H2O2 inhibits ABA-signaling protein phosphatase HAB1. PloS One 9 (12), e113643. doi: 10.1371/journal.pone.0113643
Srivastava, P. K., Kumar, J., Prasad, S. M. (2021). Salt stress responses in plants: perception, signaling, omics and tolerance mechanisms (Nova Science Publishers Inc).
Sun, Y., Liu, Z., Guo, J., Zhu, Z., Zhou, Y., Guo, C., et al. (2020). WRKY33-PIF4 loop is required for the regulation of H2O2 homeostasis. Bioch. Biophys. Res. Commun. 527, 922–928. doi: 10.1016/j.bbrc.2020.05.041
Sun, T., Nitta, Y., Zhang, Q., Wu, D., Tian, H., Suk, J., et al. (2018). Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO rep 19. doi: 10.15252/embr.201745324
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Szymańska, K. P., Polkowska-Kowalczyk, L., Lichocka, M., Maszkowska, J., Dobrowolska, G. (2019). SNF1-related protein kinases SnRK2.4 and SnRK2.10 modulate ROS homeostasis in plant response to salt stress. Int. J. Mol. Sci. 20, 143. doi: 10.3390/ijms20010143
Tan, W.-J., Yang, Y.-C., Zhou, Y., Huang, L.-P., Xu, L., Chen, Q.-F., et al. (2018). DIACYLGLYCEROL ACYLTRANSFERASE and DIACYLGLYCEROL KINASE modulate triacylglycerol and phosphatidic acid production in the plant response to freezing stress. Plant Physiol. 177 (3), 1303–1318. doi: 10.1104/pp.18.00402
Tang, W., Luo, C. (2018). Overexpression of zinc finger transcription factor ZAT6 enhances salt tolerance. Open Life Sci. 13, 431–445. doi: 10.1515/biol-2018-0052
Tasseva, G., Richard, L., Zachowski, A. (2004). Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 566 (1-3), 115–120. doi: 10.1016/j.febslet.2004.04.015
Teige, M., Scheikl, E., Eulgem, T., Dóczi, R., Ichimura, K., Shinozaki, K. (2004). The MKK2 pathway mediates cold and salt stress signaling in arabidopsis. Mol. Cell 15 (1), 141–152. doi: 10.1016/j.molcel.2004.06.023
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., Gao, G. (2020). PlantRegMap: charting functional regulatory maps in plants. Nuc. Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz1020
Tripathi, D., Zhang, T., Koo, A. J., Stacey, G., Tanaka, K. (2018). Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiol. 176, 511–523. doi: 10.1104/pp.17.01477
Tsukagoshi, H., Busch, W., Benfey, P. N. (2010). Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616. doi: 10.1016/j.cell.2010.10.020
Ueoka-Nakanishi, H., Sazuka, T., Nakanishi, Y., Maeshima, M., Mori, H., Hisabori, T. (2013). Thioredoxin h regulates calcium dependent protein kinases in plasma membranes. FEBS J. 280, 3220–3231. doi: 10.1111/febs.12301
Ugalde, J. M., Fuchs, P., Nietzel, T., Cutolo, E. A., Homagk, M., Vothknecht, U. C., et al. (2021). Chloroplast-derived photo-oxidative stress causes changes in H2O2 and EGSH in other subcellular compartments. Plant Physiol. 186 (1), 125–141. doi: 10.1093/plphys/kiaa095
Van der Does, D., Boutrot, F., Engelsdorf, T., Rhodes, J., McKenna, J. F., Vernhettes, S., et al. (2017). The arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PloS Gen. 13 (6), e1006832. doi: 10.1371/journal.pgen.1006832
van Kleeff, P. J. M., Gao, J., Mol, S., Zwart, N., Zhang, H., Li, K. W., et al. (2018). The arabidopsis GORK k+-channel is phosphorylated by calcium-dependent protein kinase 21 (CPK21), which in turn is activated by 14-3-3 proteins. Plant Physiol. Biochem. 125, 219–231. doi: 10.1016/j.plaphy.2018.02.013
Verica, J. A., Chae, L., Tong, H., Ingmire, P., He, Z.-H. (2003). Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in arabidopsis. Plant Physiol. 133 (4), 1732–1746. doi: 10.1104/pp.103.028530
Wan, J., Tanaka, K., Zhang, X.-C., Son, G. H., Brechenmacher, L., Nguyen, T. H. N., et al. (2012). LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in arabidopsis. Plant Physiol. 160 (1), 396–406. doi: 10.1104/pp.112.201699
Wan, J., Zhang, X.-C., Neece, D., Ramonell, K. M., Clough, S., Kim, S.-Y., et al. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in arabidopsis. Plant Cell 20 (2), 471–481. doi: 10.1105/tpc.107.056754
Wang, B., Ding, H., Chen, Q., Ouyang, L., Li, S., Zhang, J. (2019). Enhanced tolerance to methyl viologen-mediated oxidative stress via AtGR2 expression from chloroplast genome. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01178
Wang, P., Du, Y., Li, Y., Ren, D., Song, C. P. (2010). Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in arabidopsis. Plant Cell 22 (9), 2981–2998. doi: 10.1105/tpc.109.072959
Wang, X., Hou, S., Wu, Q., Lin, M., Acharya, B. R., Wu, D., et al. (2017). IDL6-HAE/HSL2 impacts pectin degradation and resistance to Pseudomonas syringae pv tomato DC3000 in arabidopsis leaves. Plant J. 89, 250–263. doi: 10.1111/tpj.13380
Wang, T.-J., Huang, S., Zhang, A., Guo, P., Liu, Y., Xu, C., et al. (2021). JMJ17-WRKY40 and HY5-ABI5 modules regulate the expression of ABA-responsive genes in arabidopsis. New Phytol. 230 (2), 567–584. doi: 10.1111/nph.17177
Wang, G., Roux, B., Feng, F., Guy, E., Li, L., Li, N., et al. (2015). The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Micr. 18 (3), 285–295. doi: 10.1016/j.chom.2015.08.004
Wang, L., Wilkins, K. A., Davies, J. M. (2018). Arabidopsis DORN1 extracellular ATP receptor; activation of plasma membrane k+-and Ca2+-permeable conductances. New Phyt. 218 (4), 1301–1304. doi: 10.1111/nph.15111
Wani, S. H., Anand, S., Singh, B., Bohra, A., Joshi, R. (2021). WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085. doi: 10.1007/s00299-021-02691-8
Wu, Y., Deng, Z., Lai, J., Zhang, Y., Yang, C., Yin, B., et al. (2009). Dual function of arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19, 1279–1290. doi: 10.1038/cr.2009.108
Xiang, T., Zong, N., Zou, Y., Wu, Y., Zhang, J., Xing, W., et al. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18, 74–80. doi: 10.1016/j.cub.2007.12.020
Xie, C., Zhou, X., Deng, X., Guo, Y. J. (2010). PKS5, a SNF1-related kinase, interacts with and phosphorylates NPR1, and modulates expression of WRKY38 and WRKY62. Genet. Genomics 37, 359–369. doi: 10.1016/S1673-8527(09)60054-0
Xin, Z., Wang, A., Yang, G., Gao, P., Zheng, Z.-L. (2008). The arabidopsis A4 subfamily of lectin receptor kinases negatively regulates abscisic acid response in seed germination. Plant Physiol. 149 (1), 434–444. doi: 10.1104/pp.108.130583
Xu, X., Chen, C., Fan, B., Chen, Z. (2006). Physical and functional interactions between pathogen-induced arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18 (5), 1310–1326. doi: 10.1105/tpc.105.037523
Xu, J., Li, Y., Wang, Y., Liu, H., Lei, L., Yang, H., et al. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in arabidopsis. J. Biol. Chem. 283 (40), 26996–27006. doi: 10.1074/jbc.M801392200
Xue, D.-X., Li, C.-L., Xie, Z.-P., Staehelin, C. (2019). LYK4 is a component of a tripartite chitin receptor complex in Arabidopsis thaliana. J. Exp. Bot. 70 (19), 5507–5516. doi: 10.1093/jxb/erz313
Yamada, K., Yamaguchi, K., Shirakawa, T., Nakagami, H., Mine, A., Ishikawa, K., et al. (2016). The arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 35, 2468–2483. doi: 10.15252/embj.201694248
Yan, H., Zhao, Y., Shi, H., Li, J., Wang, Y., Tang, D. (2018). BRASSINOSTEROID-SIGNALING KINASE1 phosphorylates MAPKKK5 to regulate immunity in arabidopsis. Plant Physiol. 176 (4), 2991–3002. doi: 10.1104/pp.17.01757
Yang, Y., Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Y., Zhang, C., Tang, R.-J., Xu, H.-X., Lan, W.-Z., Zhao, F., et al. (2019). Calcineurin b-like proteins CBL4 and CBL10 mediate two independent salt tolerance pathways in arabidopsis. Int. J. Mol. Sci. 20, 2421. doi: 10.3390/ijms20102421
Yasuda, S., Okada, K., Saijo, Y. (2017). A look at plant immunity through the window of the multitasking coreceptor BAK1. Curr. Opin. Plant Biol. 38, 10–18. doi: 10.1016/j.pbi.2017.04.007
Ye, H., Liu, S., Tang, B., Chen, J., Xie, Z., Nolan, T. M., et al. (2017). RD26 mediates crosstalk between drought and brassinosteroid signaling pathways. Nat. Commun. 8, 14573. doi: 10.1038/ncomms14573
Yeh, Y.-H., Chang, Y.-H., Huang, P.-Y., Huang, J.-B., Zimmerli, L. (2015). Enhanced arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00322
Yeh, Y.-H., Panzeri, D., Kadota, Y., Huang, Y.-C., Huang, P.-Y., Tao, C.-N., et al. (2016). The arabidopsis malectin-Like/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 28 (7), 1701–1721. doi: 10.1105/tpc.16.00313
Yekondi, S., Liang, F.-C., Okuma, E., Radziejwoski, A., Mai, H.-W., Swain, S., et al. (2018). Nonredundant functions ofArabidopsisLecRK-V.2 and LecRK-VII.1 in controlling stomatal immunity and jasmonate-mediated stomatal closure. New Phytol. 218, 253–268. doi: 10.1111/nph.14953
Yu, Y., Xu, W., Wang, J., Wang, L., Yao, W., Yang, Y., et al. (2013). The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol. 200, 834–846. doi: 10.1111/nph.12418
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Zentgraf, U., Andrade-Galar, A. G., Bieker, S. (2022). Specificity of H2O2 signaling in leaf senescence: is the ratio of H2O2 contents in different cellular compartments sensed in arabidopsis plants? Cell Mol. Biol. Lett. 27, 4. doi: 10.1186/s11658-021-00300-w
Zhang, L., Chen, L., Yu, D. (2018). Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 176 (1), 790–803. doi: 10.1104/pp.17.00657
Zhang, J., Gao, J., Zhu, Z., Song, Y., Wang, X., Wang, X., et al. (2020). MKK4/MKK5 MPK1/MPK2 cascade mediates SA activated leaf senescence via phosphorylation of NPR1 in arabidopsis. Plant Mol. Biol. 102, 463–475. doi: 10.1007/s11103-019-00958-z
Zhang, Y., Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5, (1). doi: 10.1186/1471-2148-5-1
Zhang, H., Zhang, L., Ji, Y., Jing, Y., Li, L., Chen, Y., et al. (2022). Arabidpsis SIGMA FACTOR BINDING PROTEIN (SIB1) and SIB2 inhibit WRKY75 function in abscisic acid-mediated leaf senescence and seed germination. J. Exp. Bot. 73, 182–196. doi: 10.1093/jxb/erab391
Zhang, D., Zhu, Z., Gao, J., Zhou, X., Zhu, S., Wang, X., et al. (2021). The NPR1-WRKY46-WRKY6 signaling cascade mediates probenazole/salicylic acid-elicited leaf senescence in arabidopsis thaliana. J. Int. Plant Biol. 63 (5), 924–936. doi: 10.1111/jipb.13044
Zhao, R., Sun, H., Zhao, N., Jing, X., Shen, X., Chen, S. (2015). The arabidopsis Ca2+-dependent protein kinase CPK27 is required for plant response to salt-stress. Gene 563 (2), 203–214. doi: 10.1016/j.gene.2015.03.024
Zheng, Z., Qamar, S. A., Chen, Z., Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48 (4), 592–605. doi: 10.1111/j.1365-313X.2006.02901.x
Zhou, J., Wang, X., He, Y., Sang, T., Wang, P., Dai, S., et al. (2020). Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in arabidopsis. Plant Cell 32 (8), 2621–2638. doi: 10.1105/tpc.19.00971
Zhuang, Y., Wei, M., Ling, C., Liu, Y., Amin, A. K., Li, P., et al. (2021). EGY3 mediates chloroplastic ROS homeostasis and promotes retrograde signaling in response to salt stress in arabidopsis. Cell Rep. 36 (2), 109384. doi: 10.1016/j.celrep.2021.109384
Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J. D. G., Boller, T., et al. (2006). Perception of the bacterial PAMP EF-tu by the receptor EFR restricts agrobacterium-mediated transformation. Cell 125 (4), 749–760. doi: 10.1016/j.cell.2006.03.037
Keywords: SnRK2, stress signaling, salinity, oxidative stress, WRKY TFs
Citation: Rachowka J, Anielska-Mazur A, Bucholc M, Stephenson K and Kulik A (2023) SnRK2.10 kinase differentially modulates expression of hub WRKY transcription factors genes under salinity and oxidative stress in Arabidopsis thaliana. Front. Plant Sci. 14:1135240. doi: 10.3389/fpls.2023.1135240
Received: 31 December 2022; Accepted: 30 May 2023;
Published: 09 August 2023.
Edited by:
Andrzej Miroslaw Pacak, Adam Mickiewicz University, PolandReviewed by:
Stéphane Bourque, Université de Bourgogne, FranceAnja Thoe Fuglsang, University of Copenhagen, Denmark
Copyright © 2023 Rachowka, Anielska-Mazur, Bucholc, Stephenson and Kulik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Kulik, anja@ibb.waw.pl
 Julia Rachowka
Julia Rachowka Anna Anielska-Mazur
Anna Anielska-Mazur Maria Bucholc
Maria Bucholc Krystyna Stephenson
Krystyna Stephenson Anna Kulik
Anna Kulik