- 1Department of Genomic Research, State Research Center of Virology and Biotechnology “Vector”, Koltsovo, Russia
- 2Department of Microbiology and Immunology, Jacobs School of Medicine and Biomedical Sciences, University at Buffalo, Buffalo, NY, United States
- 3Department of Microbiology and Virology, Pacific State Medical University, Vladivostok, Russia
- 4Laboratory of Experimental Virology, Somov Institute of Epidemiology and Microbiology, Vladivostok, Russia
Hemorrhagic fever with renal syndrome (HFRS) is a public health problem in Vladivostok city, Russia. From 1997 to 2019, a study of hantaviruses in Norway rats (Rattus norvegicus), a natural reservoir of Seoul virus (SEOV), and in HFRS patients was conducted. We demonstrated the presence of SEOV in the local population of Norway rats and detected SEOV in 10, Amur virus (AMRV) in 4 and Hantaan virus (HTNV) in 1 out of 15 HFRS patients. Genetic analysis based on partial S, M and L segment sequences revealed that the Russian SEOV strains were related most closely to strains from Cambodia and Vietnam. We postulate that the SEOV strains found in the port city of Vladivostok have been spread from South-East Asia as a result of distribution of rats during standard shipping trade activities. Moreover, we suggest that city residents may have acquired AMRV and HTNV infection during visits to rural areas.
Introduction
Hemorrhagic fever with renal syndrome (HFRS) is endemic around the world. The disease is caused by viruses belonging to the genus Orthohantaviruses, that includes Hantaan virus (HTNV), Seoul virus (SEOV), Dobrava/Belgrade virus (DOBV), Puumala virus (PUUV) and Tula virus (TULV) (1, 2). Hantaviruses (family Hantaviridae) possess a negative-sense, single-stranded, tripartite RNA genome, comprising L, M, and S segments, which encode an RNA-dependent RNA polymerase (RdRp), envelope glycoproteins (Gn and Gc) and a nucleocapsid (N protein), respectively. HFRS-associated hantaviruses are transmitted by aerosolized excreta of their natural hosts, rodents belonging to the Muridae and Cricetidae families.
SEOV is the only hantavirus with a worldwide distribution: Asia (South Korea, Japan, China, Indonesia, Cambodia, Singapore, Vietnam), both Americas (USA, Brazil, Argentina), Africa (Egypt), Eurasia (Russia), and Europe (France, Belgium, United Kingdom, the Netherlands and Sweden) (3). SEOV is carried by wild Norway rats (Rattus norvegicus) and is classified as causing mild to moderate clinical forms of HFRS. SEOV has also been detected in other species of rats, e.g., Rattus rattus and Rattus losea (4). Phylogenetic analysis has identified at least seven distinct genetic lineages of SEOV (5). It has been suggested that, unlike other hantavirus species, which followed natural on-land migrations of their hosts, the main reservoir of SEOV, Norway rats, is omnipresent due to global trade and human migration (6, 7). For example, SEOV strains from France and Belgium do not show geographic clustering but are closely related to strains from Vietnam, Cambodia, Singapore, and Indonesia (5, 8, 9).
Russia included HFRS in the official reporting system of the Ministry of Public Health in 1978, and 131,590 cases of HFRS have been registered from 2000 to 2017 (10). In European Russia, reported cases are caused by PUUV and two types of DOBV, Kurkino virus and Sochi virus. HTNV, HTNV-related virus Amur (AMRV), and SEOV have been identified by us as causative agents of HFRS in Far Eastern Russia (11). SEOV has been detected in wild rats in the Asian part of Russia, in Primorsky, Amursk, and Sakhalin regions (12, 13). However, human cases of SEOV infection have only been seen in Vladivostok city (Primorsky region) since 1992 (11). The virus in Vladivostok was shown to cause generally mild to moderate HFRS, while 5.7% cases had severe clinical manifestations (12). Serologic differential diagnosis based on neutralization tests have demonstrated that the major cause of HFRS in Vladivostok city is SEOV (14). The genetic evidence for SEOV infection in both rodents and humans in Vladivostok city and the high percent of SEOV-associated HFRS cases (>60%) showed the importance of this hantavirus for public health in this part of Russia (15). Here, we investigate SEOV infection in wild rats and characterize etiologic agents of HFRS among the citizens of Vladivostok during a period of 23 years.
Materials and Methods
Norway rats were trapped throughout Vladivostok city according to well-established protocols (16). Hantavirus infection in rats was analyzed by detection of virus-specific antibodies (Ab) or/and hantavirus antigen (Ag). Sera were screened for IgG and/or IgM anti-hantavirus Ab by an indirect immunofluorescence assay (IFA) using Vero E6 cells infected with PUUV, HTNV and SEOV as antigens (17). Lung tissues were analyzed for whole hantavirus Ag by ELISA using “HANTAGNOST” kit (Russia).
Blood samples of suspected HFRS patients from Vladivostok city were collected between 1997 and 2019. Clinical diagnosis of HFRS was laboratory-confirmed by the presence of IgG and/or IgM anti-hantavirus Ab, detected by IFA using “HFRS diagnostic kit” (Federal State Unitary Enterprise on Manufacture of Bacterial and Viral Preparations of Chumakov Institute of Poliomyelitis & Viral Encephalitis). For each HFRS patient a 4-fold increase in antibody level over a 2-week interval was registered. For genetic analysis, moderate and severe clinical HFRS cases were selected. The clinical disease severity of HFRS patients was subdivided into mild, moderate, or severe following the standard Russian criteria (length of febrile phase, minimal blood pressure in the hypotonic phase, extent of hemorrhagic symptoms, minimal urine production, serum creatinine level, and extent of proteinuria) (18). Signed informed consent was obtained from each patient (article 20, Federal Law “Protection of Health Right of Citizens of Russian Federation” N323-FZ, 11.21.2011).
RNA was extracted from rat lung tissues and human blood clots using the RNeasy Mini Kit (Qiagen), and cDNA was synthesized using Revert Aid premium reverse transcriptase (Thermo Scientific). Samples were screened for hantaviruses by nested RT-PCR, targeting the conserved regions of the viral genome. Three sets of nested primers to recover partial S (nt 610-936), M (nt 2737-2980) or L (nt 175-511) segment sequences were used (Supplementary Table 1).
The distance-based neighbor-joining and maximum likelihood methods, supported by MEGA 5 (19), were used to construct phylogenetic trees.
Results
During the period 1997–2019, 750 human HFRS cases were laboratory-confirmed in Vladivostok city. 24.4% patients had a mild clinical course, 62.7% were moderate, and 12.9% had severe forms of disease. HFRS incident rate varied from 4.2 to 8.4 per 100,000 population (Table 1). At the same time, active circulation of hantavirus was detected in urban population of R. norvegicus, the primary reservoir for SEOV. A total of 1,903 Norway rats were captured in Vladivostok city in 1997 and between 2000 and 2014 (Table 1). Hantavirus-infected animals were found every year. The overall infection rate of R. norvegicus varied from 2.0% in 2011 to 31.4% in 2000. The highest incident rate in humans and a high infection rate among rats was registered during 1997–2007.
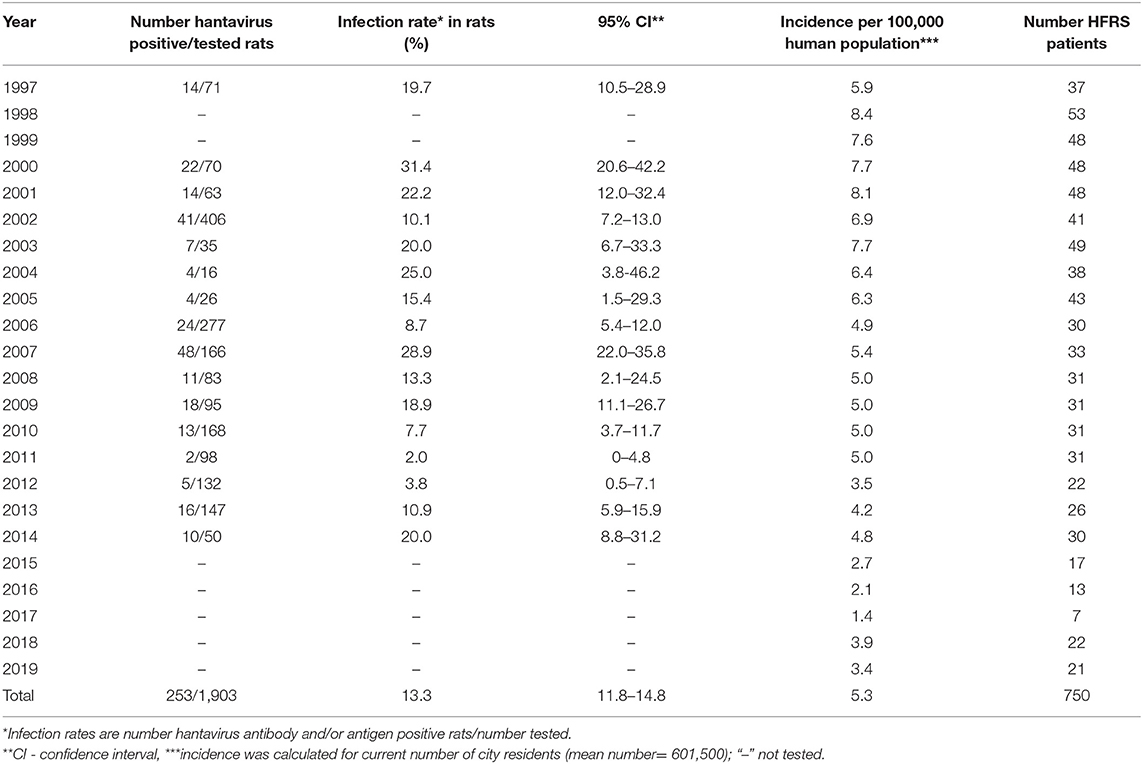
Table 1. Annual dynamic of Rattus norvegicus infection rate and HFRS morbidity in Vladivostok city, Russia (1997–2019).
For molecular typing of Russian SEOV strains, lung samples of Ag-positive rats, captured in 1997 and 2006, and human blood samples from Ab-positive HFRS patients, infected in 2000–2001, 2004, 2008, 2011, 2018–2019 were tested. Samples from patients with moderate and severe clinical forms of HFRS were selected for analysis. Hantaviral RNA (328, 244, and 337 base pairs for S, M, and L segments, respectively) was identified in 4/4 rat samples and in 15/28 human samples (Table 2). All recovered viral sequences were deposited in GenBank (accession numbers: MW073474-MW073498, MW088971-MW088982).
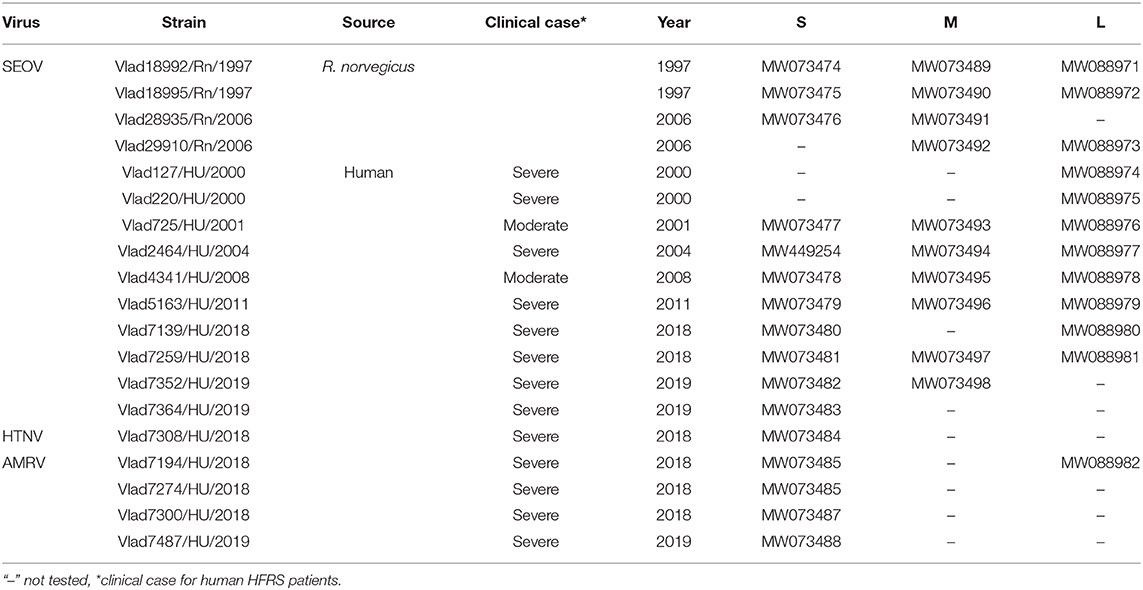
Table 2. Hantavirus strains identified in Rattus norvegicus and HFRS patients from Vladivostok city, Russia.
Phylogenetic analysis of partial S and M segment sequences recovered from R. norvegicus showed similarity to that of the SEOV strains previously identified in HFRS patients and rats from Vladivostok city and also in rats from south-eastern Asia (Cambodia, Vietnam, Indonesia, Singapore) (Figure 1). Phylogenetic analysis of sequences recovered from HFRS patients identified three different hantaviruses: SEOV in 10/15, AMRV in 4/15, and HTNV in 1/15 samples. The nucleotide sequence divergence among the newly identified SEOV strains were 0–1.4%, 0–0.9%, and 0–1.9% for the S, M, and L segments, respectively, and deduced amino acid sequences were identical. Compared with previously published SEOV S and M nucleotide sequences from south-eastern Asia, divergence did not exceed 1.6%, while the differences with the SEOV strains from other regions (China, Korea, Japan, USA) reached 5.7%. Partial S segment sequences from two rats, captured in 1997, and three HFRS patients infected in 2001, 2004, and 2008 were identical to those of strains 24D12 from Vietnam and strain Rn152 from Cambodia (20, 21). The partial amino acid sequences of the RdRp, Gc, and N proteins of the SEOV strains from Vladivostok were identical with that of SEOV strains from south-eastern Asia and strains found in France, USA and Belgium.
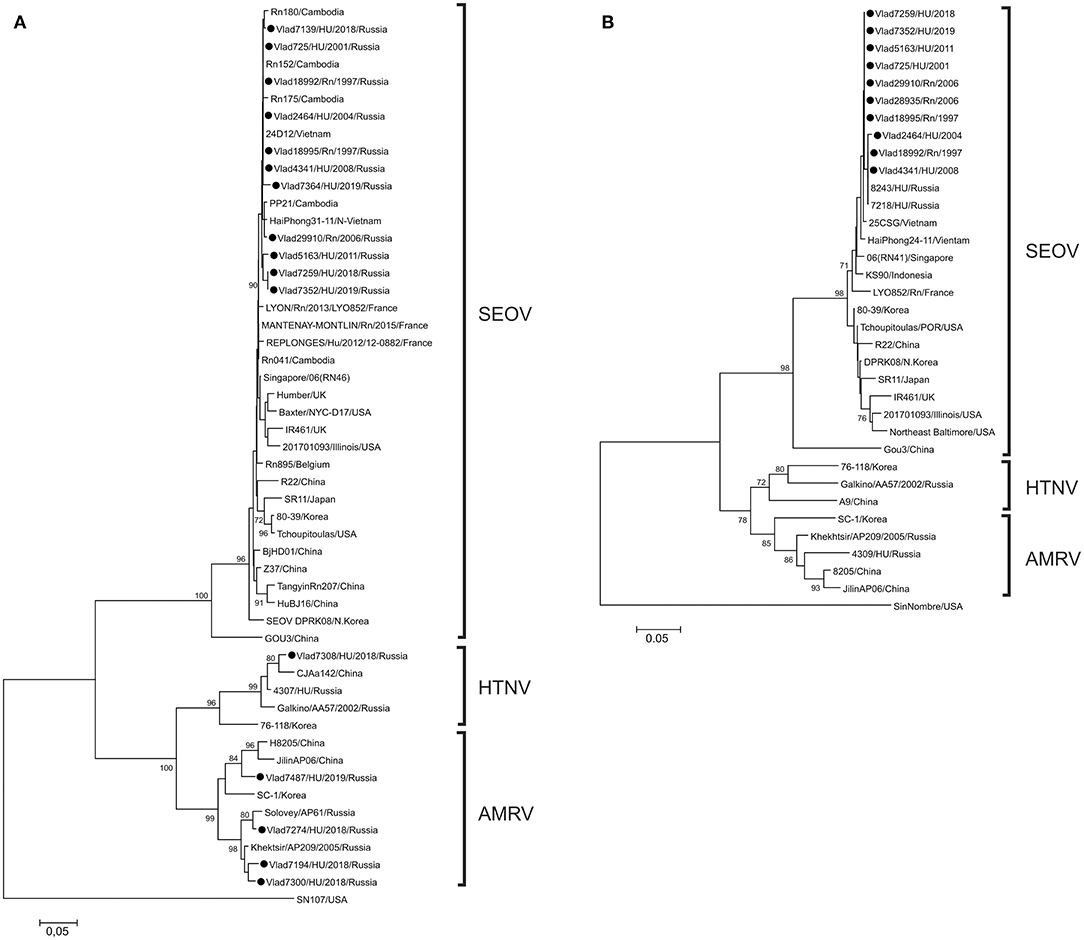
Figure 1. Phylogenetic trees of orthohantaviruses based on partial sequences of the (A) S (nt 610-936) and (B) M (nt 2737-2980) segment sequences. All analyses (neighbor-joining method) were performed using the MEGA software (19). Sin Nombre virus was used as the outgroup. Bootstrap values were calculated from 1,000 replicates; only values >70% are shown at the branch nodes. The trees constructed by using the maximum-likelihood method (not shown) had similar topology. Circles indicate sequences of strains detected in Norway rats and from the patients in this study.
Analysis of AMRV partial S segment sequences showed that patients from Vladivostok were infected by three genetic lineages of virus (Figure 1). Two of these lineages joined previously identified strains from far-eastern Russia and partial S segment divergence between sequences within each lineage did not exceed 5.2%. The third lineage was formed by a novel Russian strain and strains from the neighboring Jilin province of China.
As expected, the Vladivostok HTNV sequence was more closely related to HTNV strains from far-eastern Russia and the neighboring north-eastern area of China. The partial S sequence divergence between strain from Vladivostok and strains from north-eastern China (strains CJAa142, Fuyuan-Aa-26, Bao14, ShenyangAa13) was 2.7–4.3%. All AMRV- and HTNV-infected human patients had severe clinical manifestations of HFRS.
Discussion
The results of genetic analysis of SEOV strains from rats and HFRS patients over two decades demonstrate the high stability of SEOV strains circulating in Vladivostok city. Phylogenetic analysis of the short sequences of SEOV that we obtained does not allow further differentiation of the lineages. However, the complete identity of the partial S segment sequences from Vladivostok with those from Cambodia and Vietnam for five strains isolated during 10 years leads us to define the origin of the virus and its phylogenetic placement into lineage 7, joining strains from Cambodia, Vietnam, Indonesia, Singapore and imported strains in France and Belgium (5, 8). In the Primorsky region, SEOV was detected in Norway rats and HFRS patients only in Vladivostok city, while circulation of a closely related genetic variant of SEOV among R. norvegicus was registered in large areas of South-East Asia, including the southern and central highland area of Vietnam, and the southern area of Cambodia. So, we suggest that SEOV in Vladivostok port city originated as a result of movement of its natural host from South-East Asia during intensive shipping trade activities between Vietnam, Cambodia and Russia.
Among serologically confirmed HFRS cases associated with SEOV, 31.5% patients had a mild clinical course and 62.8% were moderate, while 5.7% had severe forms of HFRS (12). Our direct genetic evidence shows that the SEOV strains from Vladivostok can cause severe clinical manifestations of HFRS. Also, some patients in Vladivostok were infected with AMRV and HTNV (4/15 and 1/15, respectively) and these finding are consistent with serologic data from a previous report (14). Phylogenetic analysis demonstrated that AMRV cases belong to three genetic lineages of AMRV. So, most probably, these HFRS patients were infected in different parts of the Primorsky region, likely located in the suburbs of Vladivostok city. City residents may acquire AMRV and HTNV infection from Apodemus peninsulae (a natural host of AMRV) and A. agrarius (a natural host of HTNV) during visits to their vegetable gardens in neighborhoods of Vladivostok or on trips to forests for nut harvesting and hunting.
In our study, a limited number of strains were genetically typed and information regarding the location of exposure for some patients from Vladivostok was not available. However, our data are in accordance with previously published results. In the Primorsky region, SEOV circulation was registered only in Vladivostok city but not in rural areas (12, 22). As the same genetically stable variant of SEOV, distinct from strains in neighboring China, was detected both in rats (4 strains) and human patients (10 strains) in Vladivostok, we inferred that patients who acquired this SEOV strain did so in the city. Identification of genetically divergent AMRV (4 strains) and HTNV (1 strain) from HFRS patients supports our hypothesis that some city residents likely acquired AMRV and HTNV infection from areas outside Vladivostok, where these strains circulate. Also, the proportion of severe HFRS cases (12.9%) among residents of Vladivostok that we detected, exceed those (5.7%) among serologically confirmed SEOV-associated patients, as published previously (12).
In summary, our study indicates that the main causative agent of HFRS in Vladivostok is SEOV, originating from infected Norway rats traveling by ship from south-east Asian countries. The other HFRS pathogens, AMRV and HTNV, showed clear spatial association with local geography.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics committee of Somov Institute of Epidemiology and Microbiology, Vladivostok, Russia. The patients/participants provided their written informed consent to participate in this study. Ethical review and approval was not required for the animal study because Wild Norway rats were trapped according to well-established protocols (16).
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This study was supported in part by a grant from the Biotechnology Engagement Program, through the International Science and Technology Center (#0805.2), and state assignment of FBRI SRC VB VECTOR, Rospotrebnadzor.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2021.620279/full#supplementary-material
References
1. Jonsson CB, Figueiredo LTM, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. (2010) 23:412–41. doi: 10.1128/CMR.00062-09
2. Reynes JM, Carli D, Boukezia N, Debruyne M, Herti S. Tula hantavirus infection in a hospitalised patient, France, June 2015. Euro Surveill. (2015) 20:30095. doi: 10.2807/1560-7917.ES.2015.20.50.30095
3. Clement J, LeDuc JW, Lloyd G, Reynes J-M, McElhinney L, Ranst MV, et al. Wild rats, laboratory rats, pet rats: global Seoul hantavirus disease revisited. Viruses. (2019) 11:652. doi: 10.3390/v11070652
4. Wang H, Yoshimatsu K, Ebihara H, Ogino M, Araki K, Kariwa H, et al. Genetic diversity of hantaviruses isolated from Niviventer confucianus and Rattus rattus. Virology. (2000) 278:332–45. doi: 10.1006/viro.2000.0630
5. Plyusnina A, Heyman P, Baert K, Stuyck J, Cochez C, Plyusnin A. Genetic characterization of Seoul hantavirus originated from Norway rats (Rattus norvegicus) captured in Belgium. J Med Virol. (2012) 84:1298–303. doi: 10.1002/jmv.23321
6. Lin X-D, Guo W-P, Wang W, Zou Y, Hao Z-Y, Zhou D-J, et al. Migration of Norway rats resulted in the worldwide distribution of Seoul hantavirus. J Virol. (2012) 86:972–81. doi: 10.1128/JVI.00725-11
7. Plyusnin A, Morzunov SP. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr Top Microbiol Immunol. (2001) 256:47–75. doi: 10.1007/978-3-642-56753-7_4
8. Dupinay T, Pounder KC, Aural F, Laaberki M-H, Marston DA, Lacote S, et al. Detection and genetic characterization of Seoul virus from commensal brown rats in France. Virol J. (2014) 11:32. doi: 10.1186/1743-422X-11-32
9. Reynes JM, Carli D, Bour JB, Boudjeltia S, Dewilde A, Gerbier G, et al. Seoul virus infection in humans, France, 2014–2016. Emerg Infect Dis. (2017) 23:973–7. doi: 10.3201/eid2306.160927
10. Tkachenko EA, Ishmukhamedov AA, Dzagurova TK, Bernshtein AD, Morozov VG, et al. Hemorrhagic fever with renal syndrome, Russia. Emerg Infect Dis. (2019) 25:2325–7. doi: 10.3201/eid2512.181649
11. Yashina LN, Patrushev NA, Ivanov LI, Slonova RA, Mishin V, Kompanez GG, et al. Genetic diversity of hantaviruses associated with hemorrhagic fever with renal syndrome in the Far East of Russia. Virus Res. (2000) 70:31–44. doi: 10.1016/S0168-1702(00)00203-3
12. Slonova RA, Kompanets GG, Podogova LM, Astachova TI, Perminova LA, Khomenko TV, et al. Circulation of Seoul hantavirus in populations of synanthropic rodents and its significance in the incidence of hemorrhagic fever with renal syndrome in the Primorye territory (in Russian). Vopr Virusol. (1999) 5:213–7.
13. Marunich NA, Gavrilovskaya IN, Gorbachkova EA, Apekina NS, Figurnov VA, Yakunin KF, et al. Cases of hemorrhagic fever with the renal syndrome, caused by virus of serotype Rattus, in the Amur region (in Russian). J Microbiol Epidemiol Immunobiol. (1990) 3:48–52.
14. Kariwa H, Yoshikawa K, Tanikawa Y, Seto T, Sanada T, Saasa N, et al. Isolation and characterization of hantaviruses in Far East Russia and etiology of hemorrhagic fever with renal syndrome in the region. Am J Trop Med Hyg. (2012) 86:545–53. doi: 10.4269/ajtmh.2012.11-0297
15. Slonova RA, Yashina LN, Kompanets GG, Mishin VA. Antigenic and genetic characteristics of virus strains Seoul – a causative agent of hemorrhagic fever with renal syndrome (in Russian). Vopr Virusol. (2003) 3:10−4.
16. Mills JN, Childs JE, Ksiazek TG, Peters CJ. Methods for Trapping and Sampling Small Mammals for Virologic Testing. Atlanta: US Department of Health and Human Services, Center for Disease Control and Prevention (1995).
17. Dzagurova TK, Tkachenko EA, Petrov VF. Effectiveness of tissue culture antigens for serodiagnosis of HFRS by IFA test (in Russian). Vopr Virusol. (1988) 33:71–5.
18. Kruger DH, Tkachenko EA, Morozov VG, Yunicheva YV, Pilikova OM, et al. Life-threatening Sochi virus infections, Russia. Emerg Infect Dis. (2015) 21:2204–8. doi: 10.3201/eid2112.150891
19. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. (2011) 28:2731–9. doi: 10.1093/molbev/msr121
20. Luan VD, Yoshimatsu K, Endo R, Taruishi M, Huong VT, Dat DT, et al. Studies on hantaviral infection in small mammals captured in southern and central highland area of Vietnam. J Vet Med Sci. (2012) 74:1155–62. doi: 10.1292/jvms.11-0567
Keywords: hantavirus, Seoul virus, Rattus norvegicus, hemorrhagic fever with renal syndrome, Russia, Vladivostok
Citation: Yashina LN, Hay J, Smetannikova NA, Kushnareva TV, Iunikhina OV and Kompanets GG (2021) Hemorrhagic Fever With Renal Syndrome in Vladivostok City, Russia. Front. Public Health 9:620279. doi: 10.3389/fpubh.2021.620279
Received: 22 October 2020; Accepted: 11 January 2021;
Published: 05 February 2021.
Edited by:
Detlev H. Kruger, Charité – Universitätsmedizin Berlin, GermanyReviewed by:
Jean-Marc Reynes, Institut Pasteur, FranceJan Clement, KU Leuven, Belgium
Jin Won Song, Korea University, South Korea
Copyright © 2021 Yashina, Hay, Smetannikova, Kushnareva, Iunikhina and Kompanets. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liudmila N. Yashina, yashina@vector.nsc.ru; John Hay, jhay150@gmail.com
 Liudmila N. Yashina
Liudmila N. Yashina John Hay
John Hay Natalia A. Smetannikova
Natalia A. Smetannikova Tatiana V. Kushnareva
Tatiana V. Kushnareva Olga V. Iunikhina
Olga V. Iunikhina Galina G. Kompanets
Galina G. Kompanets