Antimicrobial peptides: Defending the mucosal epithelial barrier
- Department of Diagnostic and Biological Sciences, School of Dentistry, University of Minnesota, Minneapolis, MN, United States
The recent epidemic caused by aerosolized SARS-CoV-2 virus illustrates the importance and vulnerability of the mucosal epithelial barrier against infection. Antimicrobial proteins and peptides (AMPs) are key to the epithelial barrier, providing immunity against microbes. In primitive life forms, AMPs protect the integument and the gut against pathogenic microbes. AMPs have also evolved in humans and other mammals to enhance newer, complex innate and adaptive immunity to favor the persistence of commensals over pathogenic microbes. The canonical AMPs are helictical peptides that form lethal pores in microbial membranes. In higher life forms, this type of AMP is exemplified by the defensin family of AMPs. In epithelial tissues, defensins, and calprotectin (complex of S100A8 and S100A9) have evolved to work cooperatively. The mechanisms of action differ. Unlike defensins, calprotectin sequesters essential trace metals from microbes, which inhibits growth. This review focuses on defensins and calprotectin as AMPs that appear to work cooperatively to fortify the epithelial barrier against infection. The antimicrobial spectrum is broad with overlap between the two AMPs. In mice, experimental models highlight the contribution of both AMPs to candidiasis as a fungal infection and periodontitis resulting from bacterial dysbiosis. These AMPs appear to contribute to innate immunity in humans, protecting the commensal microflora and restricting the emergence of pathobionts and pathogens. A striking example in human innate immunity is that elevated serum calprotectin protects against neonatal sepsis. Calprotectin is also remarkable because of functional differences when localized in epithelial and neutrophil cytoplasm or released into the extracellular environment. In the cytoplasm, calprotectin appears to protect against invasive pathogens. Extracellularly, calprotectin can engage pathogen-recognition receptors to activate innate immune and proinflammatory mechanisms. In inflamed epithelial and other tissue spaces, calprotectin, DNA, and histones are released from degranulated neutrophils to form insoluble antimicrobial barriers termed neutrophil extracellular traps. Hence, calprotectin and other AMPs use several strategies to provide microbial control and stimulate innate immunity.
Introduction
Encounters with infectious agents or tissue injury cause inflammation, the initial response for tissue repair and innate immune defense [1]. The recent epidemic caused by SARS-CoV-2 resulted in millions of deaths worldwide in only 2 years [2–6]. Infections caused by this airborne virus [7, 8] reveal the vulnerability of our mucosal epithelial barriers against infections. Among the oldest known immune defense molecules, antimicrobial proteins/peptides (AMPs) are ubiquitous through evolution and across species. The first identified AMPs in Precambrian protozoans date to 1,000 million years ago [9]. Most prominent at the epithelial barrier where most infections occur, AMPs are crucial to host defense, and about 45 AMPs have been identified in human saliva and in the oral environment [10]. To understand the contribution to epithelial barrier defense, AMP function in higher animals must be distinguished from adaptive and other innate immune mechanisms. Some AMPs show broad-spectrum activity across phyla, whereas others are more specific. Even in the face of posited resistance mechanisms, therapeutic strategies are in development to fortify antimicrobial defense at the epithelial barrier. Indeed, translation of knowledge of the structure and function of AMPs may herald a post-antibiotic era.
The epithelial barrier in resistance to microbial infection
Barrier protection against infection by exogenous bacteria, fungi, and viruses is essential for survival. Formed by epithelia, the barrier physically partitions the underlying connective tissues from the external environment [11]. Barrier breaches are defended by host adaptive and innate immune mechanisms.
Innate responses are activated by the engagement of signaling pathways including Toll, Immune deficiency (IMD), and Janus Kinase and Signal Transducer and Activator of Transcription (JAK/STAT) [12]. These three pathways are found in virtually all life forms, both ancient and contemporary. For example, recognition and responses to pathogen-associated molecular patterns (PAMPs) in insects involve signaling pathways that are highly homologous to humans [12]. Crosstalk between the Toll and IMD signaling cascades induce the production of AMPs. The three pathways, however, partition antimicrobial responsibility. JAK/STAT is activated by infection or sepsis [13], producing the downstream effectors cytokines and stress response proteins [14]. The Toll family of receptors distinguish and signal in response to PAMPs originating from Gram-positive and Gram-negative bacteria, fungi, viruses [15, 16], and damage/danger-associated molecular patterns (DAMPs) that are released from injured tissues [16–18]. The IMD effects humoral immunity against Gram-negative bacteria and fungi [14], illustrating a redundancy in primitive immune function. Indeed, immunity in lower life forms reflects low specificity and broad, overlapping recognition and signaling responses to pathogens.
Functioning to protect epithelial barriers, the effectors of innate immune functions in the host span phylogeny and evolutionary time. Produced as part of the transcriptional response to engagement of PAMPs, many AMPs are highly conserved in invertebrates and serve as the primary humoral response [19]. In contrast, a minimally competent cell-mediated immune response is provided by hemocytes. In lower life forms, the AMP-mediated innate immune response is central to resistance against pathogenic microbes and survival.
To illustrate the scope of AMP production across species, Drosophila [19], the domesticated silk moth, Bombyx mori [20], and Rhynchophorus ferrugineus (red palm weevil) [21] use the Spätzle-mediated activation of the Toll pathway to upregulate genes for multiple AMPs. AMPs produced in response to activation of the Toll-Spz pathway also provide antimicrobial immunity in Manduca sexta [22], Antheraea pernyi [23], mosquito species [24], and shrimps [25]. Using genome wide analysis, Manduca potentially express 86 different AMPs [26]. Remarkably, Manduca also show intergenerational immunity, whereby offspring show increased AMP production after mothers were infected with Serratia marcescans [27]. The upregulated AMPs provide immunity against many common bacterial and fungal pathogens.
In the mealworm beetle, Tenebrio molitor, a frank breach in the cuticle epithelium accompanied by microbial challenge triggers local epithelial production of AMPs such as cecropins [28, 29] and melanin to thwart parasitic infection of plants and attenuate parasites and pathogens, including fungi [30]. Mealworms, then, illustrate that the cuticular epithelium can deploy different AMPs, eliciting somewhat specialized antimicrobial responses as an infection becomes more invasive.
More closely related to vertebrates, Tunicates including Ascidiae, inhabit marine environments, where they encounter infectious agents such as viruses, bacteria, and fungi in the pharynx [31]. The pharynx functions for breathing and food collection and is the primary immune organ [31]. Reflecting an innate response of greater sophistication, Ciona robusta utilize hemocytes in the hemolymph to initiate inflammation [32]. Unique to these invertebrates, C. robusta express innate immune receptors including secreted immunoglobulin Variable-region containing Chitin Binding Proteins (CrVCBPs) early in the response [32]. CrVCBPs behave as antibodies of restricted specificity enabling a response to lipopolysaccharides that are common to their colonizing microbes. AMPs produced by Tunicates act as the first line of epithelial defense against pathogens including bacteria, fungi, viruses, and parasites [33].
This discussion illustrates that the lowest life forms express AMPs, which enable organisms to resist human pathogens such as Staphylococcus aureus. In the skin of a frog, the antimicrobial products of different genes can work synergistically in the same tissue. The granular glands of the splendid leaf frog, Cruziohyla calcarifer, produce cruzioceptins which are 21–23 residue alpha helical cationic peptides with antimicrobial activity against E. coli, S. aureus, and C. albicans [34]. In the Australian tree frog, the three AMPs from the skin (e.g., aurein 1.2, maculatin 1.1, and caerin 1.1) function synergistically against Gram-negative Escherichia coli and Gram-positive Staphylococcus aureus [35]. Expressed by species through evolution, two or more unique AMPs work synergistically to provide overarching defense against infection by exogenous organisms and control or limit the growth of commensal species [9]. In lower animals, AMPs function typically in the absence of adaptive immunity. Virtually every life form employs AMPs to provide anti-bacterial and anti-fungal defense.
Since most human infections require that microbes breach the epithelial barrier, epithelial cells have evolved to contribute to resistance to infection by providing robust local immunity. During oral mucosal health, constitutive expression of AMPs likely serves to control the overgrowth of commensal species, the emergence of pathobionts, and dysbiosis. Antimicrobial proteins are expressed by epithelial cells from tissues ranging from the gingiva [36–38] to the cornea of the eye [39] and to the Paneth cells of the intestines [40]. These epithelial AMPs confer protection against infection by bacteria [41–43], protozoa [44], fungi [45, 46], and viruses [47, 48]. By studying interactions between AMPs and the transcriptional response of bacterial cells and applying artificial intelligence algorithms, AMPs could be engineered in the future to overcome microbial resistance mechanisms.
Microbes drive the development of innate oral immunity
The pressure of early colonizing commensal bacteria appears to drive epithelial development, maturation, and AMP expression. Using reconstituted human gingiva, incubation with the prominent salivary organisms Granulicatella, Veillonella, and Streptococcus promotes keratinocyte proliferation, thickening, and greater organization of the epithelial layer [49]. Epithelial maturation was accompanied by increased expression of several key regulated AMPs including elafin, hBD2, hBD3, adrenomedullin, and cathelicidin (LL-37), and secretion of antimicrobial AMPs (e.g., IL-6, CXCL8, CXCL1, CCL2). Except for hBD1, other AMPs were not studied. These in vitro data suggest that in the absence of leukocytes, the commensal microbiota drives the development of the epithelial barrier against infection.
The role of the oral microbiota in driving mucosal maturation including innate immunity is supported by in vivo studies in mice. In mouse neonates, microbes colonize the oral mucosa at high levels. The prenatal oral mucosa expresses the chemotactic cytokine, IL-17, which apparently recruited neutrophils since these phagocytes were not seen in Il-17−/− mice [50]. IL-17 and neutrophils virtually disappear by 4-week of age in the buccal mucosa, while persisting in the gingiva and the junctional epithelium into adulthood. The presence of IL-17 and neutrophils was directly associated with the presence of γδT cells. After weaning, the density of oral mucosal microorganisms reduces to adult levels, and the Streptococcaceae and other genuses outgrow the Pasteurellaceae in mice. The microbiota drive the maturation of the oral epithelium including loss of permeability, lower turnover rate, and increased expression of AMPs as marked by CRAMP, the murine analog of human LL-37, and β-defensins 4 and 14. Postnatally, the production of AMPs in saliva reduces the oral microbial load [50]. Microbial colonization is essential for normal mucosal maturation and the development of local innate immunity in this in vivo model.
In epithelia, the control of AMP expression involves pathogen-recognition receptors (PRRs) that are conserved in insects, worms, and in humans and other mammals [9]. By engaging a pathogen-associated molecular pattern (PAMP) representing virulence factors on microbes, PRRs activate the mTOR and NF-kB pathways to promote expression of many immune response genes including AMPs. The AMPs produced in higher life forms are generally conserved from lower life forms [9]. AMPs such as hCAP-18, which is cleaved to release the bioactive peptide LL-37, and calprotectin (e.g., complexed S100A8 and S100A9; S100A8/A9) are found in humans and other mammals and are inducible in epithelia [51–53]. The more evolutionarily modern epithelial AMPS such as calprotectin present new antimicrobial mechanisms and function by sequestering essential trace metals from microbes [54, 55], whereas more ancient AMPs form pores in microbial membranes [56]. Given the very different modes of action, the two mechanisms of AMP activity can function synergistically [57–59].
In humans and other mammals, AMPs in the extracellular milieu protect epithelia against tissue-invasive microbes. In this environment, insoluble neutrophil extracellular traps (NETs) form after dead and dying cells release AMPs, DNA, and histones [60–62]. These structures trap proximal microbes and use the bound AMPs to kill or prevent bacterial and fungal growth. Within viable resting cells, AMPs, including defensins and cathelicidins, typically localize in cytoplasmic granules [63–66] and calprotectin in the cytoplasm [67, 68].
Within cells, AMPs in granules and in the cytoplasm (e.g., calprotectin) contribute to antimicrobial defense. During microbial invasion or phagocytosis, AMP-containing granules fuse with phagosomes or endosomes where invasive microbes localize [69–72]. After release into the endosome or phagolysosome, AMPs cooperate with other antimicrobial mechanisms, including the production of reactive oxygen species to inactivate and kill intracellular pathogens.
AMPs found exclusively in the cytoplasm of viable cells include calprotectin and LL-37 and appear to protect cells from invasive pathogens that escape endosomes and phagosomes. Some invasive pathogens like Listeria monocytogenes use listeriolysin O (LLO) and lipases to digest the endosome and phagosome membranes and escape into the cytoplasm [73–75]. Cells have apparently co-evolved to protect against microbial invasion into the cytoplasm. Human α-defensins block LLO-dependent membrane pore formation, release of LLO from the bacteria, and intracellular multiplication [76]. Listeria counter by activating type I interferons, which suppress phagosome maturation and proteolysis of LLO and facilitate escape and spread of infection [77]. As Listeria exits the phagosome, however, calprotectin in the cytoplasm of epithelial cells and neutrophils limits intracellular bacterial growth [68]. The antimicrobial activity of calprotectin against Listeria is enhanced by ubiquitination of InlC, which stabilizes the S100A9 subunit of calprotectin and causes an increase in ROS production by neutrophils in mice [78]. At 2 h in the cytoplasm, growth of Listeria and Salmonella typhimurium are inhibited by calprotectin and LL-37 [68, 79, 80]. At the same time, up to 10–12% of epithelial cells show LLO-dependent calprotectin mobilization; mobilized calprotectin complexes with polymerized cytoplasmic microtubules [68]. When complexed with microtubules, anti-Listeria activity is subverted. Listeria also causes epithelial cells to upregulate IL-1a, which signals the IL-1 receptor in an autocrine loop to upregulate calprotectin [81–83]. Cytoplasmic AMPs, therefore, largely function to control microbes within cells or resist intracellular invasion.
Spectrum of AMP activity across microbial kingdoms
Many species of bacteria and fungi are cell invasive, and cytoplasmic AMPs limit the extent of intracellular invasion. Common fungi that can be identified in the oral epithelial tissues include Candida, Malassezia, Cryptococcus, and Trichoderma spp. [84]. Candida albicans and other fungal species persist as low abundance commensals which emerge prominently in the immunocompromised [85] (Figure 1). These fungi appear to be kept in check by neighboring mucosal microbiota and may overgrow because of interkingdom changes centering on the oral microbiome [87]. Antifungal proteins of the oral cavity include the histatins [88], α- and β-defensins [89], and calprotectin [90, 91]. In saliva, fungal growth is controlled by several known antimicrobial proteins including CCL28 [92], histatin 5 [93], the cystatins [94], and MUC7 [95]. Proximal to the mucosal surfaces, pathogenic Candida albicans, the most common pathogenic fungus in the oral cavity, secrete candidalysin from hyphae [96]. Candidalysin activates oral epithelial cells via EGFR to produce a robust antifungal response [97]. The epithelial response includes the release of anti-fungal hBD2 and hBD3, LL37, S100A8, and ATP, which appears to transactivate neighboring epithelial cells. Released S100 proteins and defensins from epithelial cells into the tissues serve to recruit neutrophils containing a potent redundant armamentarium of AMPs [97]. The antifungal responses of epithelial cells are made more robust by epithelial IL-17 signaling, which increases expression of β-defensin 3 and thwarts oropharyngeal candidiasis in mouse models [98]. Similarly, deficiency in mouse β-defensin-1 results in IL-17 deficiency and mucosal candidiasis upon infection with C. albicans [99] (Figure 1).
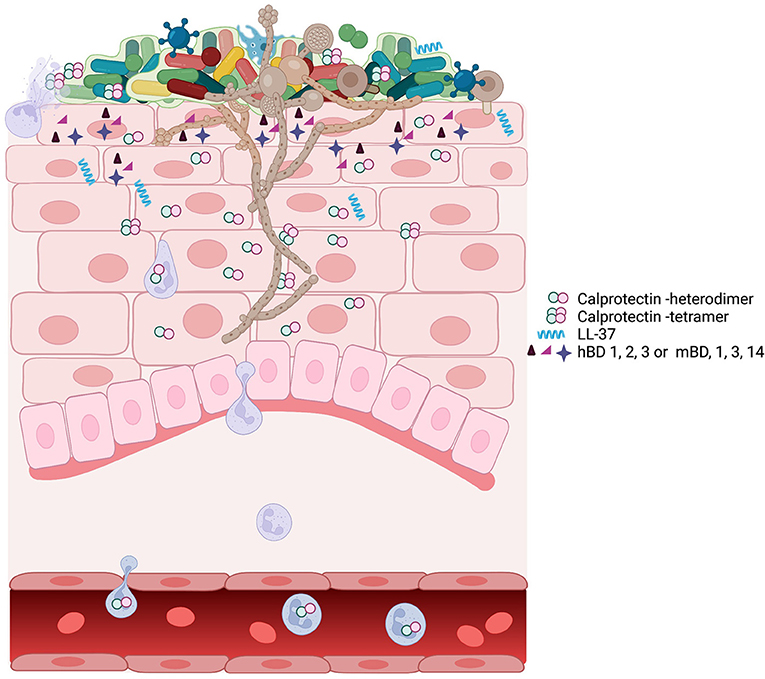
Figure 1. A schematic view of oropharyngeal candidiasis showing the complex participants in the infection. The oral microbiome associated with candidiasis is different from the oral microbiome in health [86]. The outgrowth of C. albicans hyphae may be facilitated by or drive the changes associated with the dysbiotic microbiome. C. albicans in a dysbiotic community leads to inflammation and upregulation of many AMPs including calprotectin, LL-37, and defensins. At the site of infection, the expression of AMPs, the dysbiotic microbiome, and the outgrowth of C. albicans can individually or collectively signal for infiltration by immune cells, including neutrophils, macrophage, and T cells. Neutrophils and macrophage phagocytose Candida. Neutrophil degranulation provides the components for neutrophil extracellular traps, which contain the spread of Candida amid high concentrations of AMP calprotectin [67].
Viruses that typically infect oral epithelial tissues include the herpesviruses: Herpes Simplex type 1 (HSV-1), Epstein-Barr virus (EBV), cytomegalovirus (CMV), and Kaposi's sarcoma-associated herpesvirus (KSHV) [100]. Antimicrobial peptides appear to mitigate infections by acting directly on herpes simplex type 1 [101–104], SARS-CoV-2 [105], and KSHV [48]. The AMP LL-37 appears to inhibit infection by KSHV [48], whereas α-defensin-derived peptide HD5 suppresses infection by CMV [106], and lactoferrin attenuates HSV-1 [107]. Generally acquired by sexual activity, human papilloma virus (HPV) is thwarted by α-defensin 5 [108] and theta-defensins [109], although clearance is multifactorial [110]. To produce a new generation of therapeutic agents, naturally occurring AMPs are being packaged [111] and re-engineered [112] to optimize direct antiviral activity.
Although not considered an oral virus, HIV-1 interactions with oral mucosal epithelium result in non-permissive infection and the virus does not replicate in the oral mucosa [113]. In the oral tissues, AMPs could contribute to local viral restrictions that curtail replication since the anatomically similar genital mucosa do become infected. Upon oral exposures, HIV-1 must translocate across mucosal epithelia to infect subepithelial CD4 T cells, which may be facilitated by the induction of epithelial-mesenchymal transition [114]. Translocation of HIV-1 may also be enabled by a co-infection with the periodontal pathogen, P. gingivalis [113, 115–117]. Rapid uptake by epithelial cells enables HIV-1 to escape inactivation by human saliva [118] without appearing to permit productive infection of keratinocytes [113]. Among the restrictions of HIV-1 replication in mucosal epithelial cells and vaginal fluid are cationic antimicrobial peptides including calprotectin, hBD-1 and 2, and the secretory leukocyte protease inhibitor (SLPI) [119]. Although LL-37 appears to inhibit HIV-1 replication in peripheral blood mononuclear cells [120, 121], this AMP enhances HIV-1 infection in intraepithelial Langerhans cells [122]. The contribution of AMPs to resist productive HIV-1 infection in the oral mucosa has not been definitively established.
Recently SARS-CoV-2 has been shown to infect salivary glands, which express the cognate ACE2 receptor [123]. In the salivary glands, SARS-CoV-2 harbors, replicates, and is subsequently released in saliva [124, 125]. Surprisingly, SARS-CoV-2 infections are mitigated by antimicrobial peptides (as is HSV-1) [105]. Binding of hBD-2 blocks ACE2-mediated entry of SARS-CoV-2 into target epithelial cells [126].
Tissue-specific expression of AMPs could explain tropisms that govern the mucosal sites of viral infections. Consider that influenza type A virus (IAV) infection is confined to the upper respiratory tract although it is contiguous with but does not infect the oral mucosa. The anatomic site sequestration is somewhat paradoxical. The major AMPs in the salivary film coating the oral mucous membranes include HNPs, hCAP18/LL-37, hBDs [127], and salivary mucins [128]. Studies in primates suggest that many more AMPs are expressed in the microbially responsive gingival epithelium [129]. Indeed, human saliva also contains the antifungal histatins [93, 130] and the antifungal/antibacterial AMP, calprotectin [131]. The known antiviral proteins/peptides are high molecular mass glycoproteins [132]. Similar to the oral content, the predominant AMPs produced at the mucosal surfaces of the upper airways are neutrophil α-defensins/HNPs, HBDs, LL-37, sPLA2-IIA [133], and calprotectin [134]. Perhaps critically, upon infection the airway secretions become acidified which mitigates the effectiveness of the AMPs. The saliva and oral mucosal surfaces contain mechanisms that limit acidification [135], with specialized microbial domes that appear to contain acid that demineralizes the tooth structure [136].
We speculate that acidification of the airway fluids during infection mitigates the effectiveness of the AMPs, facilitating common co- or secondary-infection with Streptococcus pneumoniae in the respiratory epithelium caused by influenza [137]. Proximal IAV infection would appear to be stimulated by the neuraminidase-expressing streptococci, which include certain oral mitis group streptococci. Resulting interferon signaling increases the production of mucins [138], contributing to the release and spread from epithelial cells [139, 140]. The streptococci would be expected to be sensitive to the local AMPs if the secretions were not acidified [133]. Furthermore, IAV replication in airway epithelial cells appears to be stimulated by uncharacterized products of S. oralis and S. mitis, among Mitis group streptococci [141]. Although Mitis group streptococci are the most frequently recovered bacteria from bronchoalveolar lavage during aspiration pneumonia [142, 143], the seeming resistance of oral tissues to IAV infection is surprising. During IAV infection, both type I interferon production [138] and TGF-β-mediated expression of host cell integrin receptors [144] promote bacterial co-infection. Signaling through the IL-17 receptor upregulates anti-fungal AMPs [98, 145]. Unlike pulmonary fluids, the buffering capacity of saliva may allow for sustained AMP activity and suppression of intruding S. pneumoniae and provide a mechanistic explanation for the resistance of the oral cavity to IAV infection.
Specific contribution of AMPs to antimicrobial defense
In higher animals, the efficacy of AMPs has been determined by isolating and purifying the respective proteins/peptides followed by in vitro testing for antimicrobial function. In the host, however, the contribution of AMPs to broad antimicrobial defense is less clear because of the complexity and seeming functional redundancies of the innate and adaptive immune responses. And indeed, the AMPs can signal and modify adaptive immune responses [146–149], thereby providing points of convergence with innate immunity.
To resolve the functions of AMPs with greater clarity, animal models are useful. Animal models can facilitate understanding of modes of action, including knowledge of how interactions with inflammatory responses and adaptive immunity affect the composition of the microbiome. The microbiome can profoundly affect host health. Shifts in the microbiome can lead to dysbiosis and the emergence of pathobionts or to the acquisition of pathogens that are related to many diseases, including metabolic and neurologic disorders. AMPs that are constitutively expressed or upregulated during inflammation can contribute to homeostasis of the microbiome.
Defensins function as pore-forming antimicrobials in humans and other mammals
Defensins are cationic peptides that bind and form pores in bacterial and fungal membranes causing increasing membrane permeability and cell death [150]. A query of the genome sequences of 29 vertebrate species revealed that humans have genes for 31 β-defensin peptides, mice have 38, rats have 41, and cattle have 42 [151]. Only a few of the gene protein products have been evaluated for antimicrobial activity and immune function. Since studies analyze one family member at a time, the seeming redundancy in defensins (and other AMPs) leaves open the question of whether AMPs synergize or compliment their activities. The effectiveness of the defensins may be limited by environmental factors including inactivation by high salt concentration and degradation by protease activity. Clearly, expression and AMP effectiveness can differ.
There are two main families of defensins found in saliva [152, 153]. The α-defensins (HNP1, HNP2, HNP3, HNP4) are made by neutrophils [154] and are released into the saliva during infections and inflammation. β-defensins (hBD1, hBD2, hBD3, hBD4) are synthesized by mucosal epithelial cells and can be found at the mucosal surface in gingival crevicular fluid and saliva [49]. Although regulated in response to viral agents [47, 155], human β-defensin 1 (HBD1) is generally constitutively expressed [156], whereas HBD2 and HBD3 are induced by microbial insults and pro-inflammatory cytokines in various mucosal epithelial tissues [49].
Oral candidiasis or thrush is common in denture wearers, in people with salivary gland defects, and in the immunocompromised. In a model of oral infection by Candida albicans, wild type mice (DEFB1+/+) showed an influx of neutrophils at the sites of intraepithelial infection and adjacent to the fungal hyphae [99] (Figure 1). Mice deficient in the constitutively expressed mouse β-defensin-1 (DEFB1−/−) showed more sparse neutrophil infiltrates and increased fungal burdens at days 3 and 7 after inoculation [99]. The increased susceptibility to infection by C. albicans in DEFB1−/− mice may also be explained by reduced induction of mucosal AMPs including cathelicidin antimicrobial peptides (CAMP), LL-37, mBD2, and mBD4. The risk of infection may also increase if levels of calprotectin, chemokine CXCL1, and cytokines Il-17A, IL-17F, Il-6, and IL-1β are also reduced. In the DEFB1−/− mice, which are also deficient in CAMP, mouse β-defensin-1 appears to contribute to persistent antifungal activity and regulate host inflammatory responses [99]. Neutrophils and macrophages from the DEFB1−/− and wild type mice were similar in in vitro neutrophil killing assays and IL-1β release. Mouse genotype-specific differences in infection by C. albicans could also be attributed to competition with co-colonizing microbiota and the responses of T cells and other immune response elements.
In DEFB1−/− mice, IL-17 cytokines are associated with susceptibility to oral Candida infection [98]. In wild type mice, IL-17 signaling recruits neutrophils to the oral epithelium which can clear infections [50]. The IL-17 receptor pathway and the expression of the IL-17 receptor on surface mucosal epithelial cells (keratin 13 expressing) are also crucial to upregulating the AMPs that aid in controlling the Candida infection [98]. IL-17R plays a part in regulating the oral epithelial cell response to infection by Candida. When challenged with Candida, the susceptibility to infection of Il17ra−/− and Il17r conditional deletion (Il17raΔK13) mice was similar and associated with induction of the AMP, murine β-defensin-3 [157]. DEFB1 expression was also similar in control mice and those with oral (local) or complete IL-17 receptor deficiency. The IL-17 receptor controls the expression of several antimicrobial peptides including S100A8, S100A9, lipocalin 2, and mBD3 [145]. DEFB3−/− mice had the same susceptibility to oral candidiasis as the Il17ra−/− mice, highlighting the importance of IL-17 receptor and murine β-defensin-3 in antifungal immunity by immunocompetent mice [98].
The different dependencies of mBD1 (DEFB1−/− mice) [99] and mBD3 (Il17ra−/− mice) [98] on mitigating oropharyngeal candidiasis is interesting. In both models, the expression of non-inducible mBD1 is expected, but mBD3 is upregulated, however, by IL-17 receptor signaling in association with lipocalin-2 and calprotectin [98]. IL-17 promotes a 10–40-fold increase in calprotectin expression in keratinocytes in vitro and in mouse oropharyngeal candidiasis [158]; inflammation caused by experimental periodontitis also upregulates calprotectin [159]. The diminished antifungal activity in the Il17ra−/− mice may reflect a loss of synergy between several different AMPs including mBD3, lipocalin-2, and calprotectin. Indeed, purified calprotectin with lactoferrin completely inhibits C. albicans in vitro. Calprotectin and lactoferrin can both be released from neutrophils suggesting that they could also work cooperatively at sites of inflammation to clear C. albicans [160].
In the oral cavity, β-defensins may also protect against bacterial infections. Using a mouse model, a P. gingivalis-soaked ligature was used to induce experimental periodontitis, and HBD3 was applied to the periodontal pocket [161]. After three applications of HBD3, ligated sites showed reduced osteoclast and alveolar bone loss, and markers of periodontitis: MMP-9, TNF-α and IL-6. Incubation of HBD3 with a macrophage cell line attenuated polarization into a proinflammatory M1 phenotype. While it is unclear whether the human BD3 affected the expression of mouse β-defensins, calprotectin, or other synergistic oral AMPs, application of HBD3 appears to have therapeutic potential.
Calprotectin as a location-dependent AMP
Extracellular calprotectin
Calprotectin, a heterodimer of S100A8 and S100A9 (S100A8/A9), is a multifunctional AMP expressed in the cytoplasm of mucosal epithelial cells and neutrophils. In neutrophils, calprotectin comprises about 45% of the total cytosolic protein. Functionally prominent in the oral cavity, calprotectin is also found in the saliva [162, 163] and gingival crevicular fluid [164–167]. During infection and inflammation, the function of calprotectin can change in response to the local conditions (Figure 2).
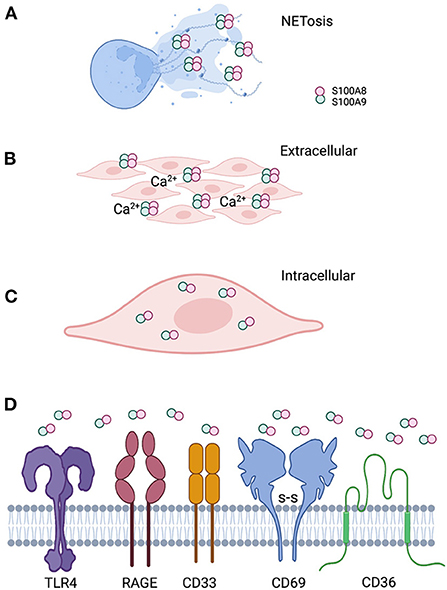
Figure 2. Localization of calprotectin at the epithelial barrier specifies function. (A) Calprotectin released from degranulated neutrophils and epithelial cells complex with DNA and histones to form antimicrobial neutrophil extracellular traps. (B) When calprotectin is released from cells in a high calcium concentration inflammatory environment, the soluble AMP forms heterotetramers, providing increased affinity for trace metal divalent cations. Successful sequestration of the trace metals from microbes results in reduced growth and “nutritional immunity.” (C) Calprotectin localized within the cytoplasm of epithelial cells or neutrophils appear to protect against invasive microbes that seek to reside intracellularly as part of their life cycle. Whether intracytoplasmic calprotectin with AMP activity exists as monomers, heterodimers. or heterotetramers, assuming increased cytoplasmic calcium upon release from intracellular stores, remains to be studied. (D) Released calprotectin can engage a range of receptors on cells in the inflammatory environment. Engagement with specific receptors link this AMP with a range of innate cellular immune responses.
Calprotectin has no signal sequence and is not secreted via the classical endoplasmic reticulum-Golgi pathway. At the mucosal surface, calprotectin enters the extracellular space when released by activated monocytes [168] or neutrophils [67]. Extracellular calprotectin can also undergo oxidation [169], thereby increasing its vulnerability to proteolysis and fragmentation into smaller peptides of yet unknown biological activity [170]. In the extracellular space, calprotectin is available to bind receptors (Table 1, Figure 2), assemble heterodimers and other oligomeric forms [189], and sequester metal ions [190]. Soluble calprotectin may be recognized as a damage-associated molecular pattern (DAMP) or alarmin to activate neutrophils or function as an antimicrobial protein to suppress microbial growth [191]. Antimicrobial activity is increased locally when calprotectin incorporates into neutrophil extracellular traps (NETs) [191].
NETs are an insoluble mesh formed primarily from the released granular and cytoplasmic contents of dead and dying neutrophils, which include DNA, histones, and calprotectin among other AMPs [192]. Calprotectin in NETs may be essential to clear infections in the tissues as observed in a mouse model of Candida albicans infection [67]. When complexed in NETs, calprotectin can activate neutrophils by upregulating CD11b and increasing cellular adhesion [191]. S100A9 released from the neutrophil can bind the neutrophil RAGE receptor and potentiate further release of NETs [178].
In the extracellular space, calprotectin sequesters nutritional metal ions which promotes antimicrobial “nutritional immunity” [190, 193, 194]. Each S100 subunit contains two Ca2+ binding sites [195, 196]. In the absence of Ca2+ ions, S100A8/A9 exists as a heterodimer, and upon Ca2+ binding, the two heterodimers self-associate to form a heterotetramer [189, 196]. Hetero-multimerization can also be promoted by oxidative cross-linking during inflammation [197]. In comparison to its human ortholog, murine calprotectin requires 10-fold more Ca2+ equivalents to form a heterotetramer [198]. Each heterodimer contains two distinct transition-metal-binding sites. When compared with the heterodimer, calcium-induced tetramers show enhanced binding affinity for Mn, Fe, Ni, Cu, and Zn and increased protease stability [189, 196]. By sequestering transition metals, calprotectin heterotetramers starve microbes that infect the surrounding inflammatory tissue. Calprotectin heterotetramers also lose DAMP activity because this multimer shows reduced access to the TLR4/MD2-binding site, which mitigates a broader inflammatory response [199]. By reducing the inflammatory response, off-target tissue damage is minimized during clearance of the infection [200–203].
Some bacterial pathogens have evolved to thwart nutritional immunity. To compete with calprotectin and commensal species for nutritional metal ions, pathogens utilize and can increase avidity of metal ion transporters [204, 205]. By sequestering Mn++, for example, Salmonella resists killing after phagocytosis by inhibiting neutrophil Mn++-dependent enzymes that detoxify reactive oxygen species [205].
Some Gram-positive species may benefit when calprotectin binds zinc. In Streptococcus pneumoniae, for example calprotectin reverses toxic sensitivity to zinc [190]. The zinc is bound by the solute-binding proteins of the ABC transport system. When calprotectin successfully chelates this divalent trace metal, however, the cells increase binding of Mn++, favoring growth and persistence [190]. Environment counts also, and the decreasing pH found in inflamed tissues reduces calprotectin heterotetramer formation and the chelation of trace metals [206]. By reducing the effectiveness of metal binding by calprotectin, solute binding proteins become more effective at capturing Mn++. Nutritional immunity mitigates against certain bacteria, including S. pneumoniae and S. aureus [190]. Indeed, many bacteria express solute-binding proteins as part of their ABC transporter systems, which bring nutritional solutes into the bacterial cell to aid cell growth. Many streptococci, including the oral species S. gordonii [207, 208], S. mutans [209], and other related species [210] have ABC transporter systems. In oral streptococci, calprotectin compromises the function of the ABC transporters and the utilization of trace metals, which negatively impacts growth [211]. Most microbes live in complex communities. More must be learned about how calprotectin affects commensal and pathogenic microbes existing in a complex community and whether sequestration of zinc or other trace metals might favor growth of commensal bacteria while limiting pathobionts and pathogens.
Intracellular calprotectin
Calprotectin localizes after synthesis in the cytoplasm of cells including neutrophils, monocytes, and mucosal squamous epithelial cells [212, 213]. In neutrophil cytoplasm, calprotectin is the most abundant protein on a molar basis. Cytosolic calprotectin appears to directly protect the interior of squamous epithelial cells against invasive microbes by antimicrobial activity [36, 68, 80, 195] and indirectly by activating NADPH oxidases leading to production of reactive oxygen species (ROS) [214, 215]. After phagocytosis, neutrophils effect indirect calprotectin-dependent antimicrobial mechanisms [216, 217]. In neutrophils, calprotectin appears to act as a calcium relay to activate antibacterial ROS production [218], but direct antimicrobial activity by calprotectin in the cytosol of neutrophils has not yet been shown definitively.
In epithelial cells, calprotectin expression increases in response to microbes including Fusobacterium nucleatum and P. gingivalis [219, 220] and PAMPs such as lipopolysaccharide (LPS) [221] and flagellin [222]. Indeed, when bacteria bind, epithelial cells upregulate IL1α, which is released and then engaged by the IL1 receptor [82, 83]. Engagement of the IL-1 receptor signals through the p38-MAP kinase pathway, increasing CCAAT/enhancer binding protein β (C/EBPβ) transcriptional activity, which upregulates expression of calprotectin.
To counter intracellular calprotectin, bacterial species have evolved countermeasures. For example, Listeria induces calprotectin co-localization with the actin cytoskeleton and reduction in antimicrobial activity [68]. Fungi such as Candida albicans survive by promoting AMP effectors and efflux pumps and regulating downstream signaling pathways [45]. Encounters with phagocytosed bacteria such as Porphyromonas gingivalis, a prominent periodontal pathobiont, can also trigger the release of calprotectin from neutrophils into the extracellular environment [223]. Both monomers are vulnerable to oxidation on methionine and cysteine residues, which mitigates antimicrobial activity [169]. Clearly, the role of calprotectin in antibacterial defense depends on the cell source, multimerization state, which optimizes nutritional immunity, and cooperative mechanisms such as activation of ROS production, the cellular or pericellular localization, and the ability of the microbe to evade or subvert calprotectin.
The antimicrobial effectiveness of calprotectin in vivo
In vivo studies can provide a deductive approach to understanding the contribution of calprotectin to innate immunity. In a study of experimental periodontitis in mice that compared wild type and S100A9−/− (calprotectinnull), expression of calprotectin minimized the emergence of a dysbiotic periodontal microflora, gingival inflammation, and periodontitis with loss of alveolar bone [159]. In wild type mice, the density of neutrophil infiltrate was lower and emergence of a dysbiotic microbial community was mitigated when compared to periodontitis in the calprotectinnull mouse. In the presence of calprotectin, the lower gingival inflammatory cell infiltrate could increase the alkaline pH environment, albeit with lower tissue Ca++ which is released from neutrophils. Inflammation is also generally accompanied by interstitial acidosis, which serves as another danger signal [224], and release of cellular calcium into the tissues [189]. In human periodontitis [164, 165], the neutrophil-rich inflammatory environment would also be expected to contain increased extracellular levels of calprotectin [191]. The elevation in calcium promotes tetramerization of calprotectin, increasing antimicrobial activity. In contrast, lower tissue pH would mitigate the ability of calprotectin to sequester Mn++ and tetramerize [206]. Tetramerization and stronger nutritional immunity in low tissue pH would, however, occur in the presence of higher Ca++ environments [190]. Given the complexity of inflammatory tissue environments, the effectiveness of antimicrobial calprotectin may be difficult to predict.
In wild type calprotectin-expressing mice, PMN-rich inflammation was dampened 2 days after ligature placement, whereas inflammation was robust in calprotectinnull mice [159]. At 5 days of ligature placement, inflammation was greater in the wild type mice. Hence, early inflammation was suppressed by calprotectin but over time inflammation was enhanced. The role of calprotectin appeared dichotomous over time. Eventually, extracellular soluble calprotectin heterodimer appears to serve as a proinflammatory alarmin by engaging TLR4 [171, 174, 176], RAGE [225–227], CD36 [184, 225], CD69 [186, 228], and perhaps other receptors. CD33, a highly conserved sialic-acid-binding immunoglobulin-like lectin (Siglec) expressed primarily on monocytes and myeloid progenitors [229], binds S100A9. Little is known, however, about engagement of other forms of calprotectin [18]. Signaling through these receptors activates proinflammatory signaling pathways (Table 1, Figure 2). Receptor engagement by calprotectin is cell type and tissue specific. Human oral epithelial cells and gingival fibroblasts express TLR4; human oral epithelial cells also express TLR2 and RAGE which, like TLR4, interact with the calprotectin heterodimer [177].
Calprotectin stimulates murine neutrophils to migrate and adhere to integrin substrates [183]. Loss of calprotectin in mouse models make neutrophils less sensitive to chemotactic agents including IL-8 [230]. The N1 neutrophil phenotype is more proinflammatory than the N2 phenotype. The N1 neutrophils respond to calprotectin (S100A9) by chemotaxis and elevated expression of NADPH-oxidase [231]. Hence, the response to calprotectin by neutrophils will depend on the prevalence of N1 and N2 phenotypes in vivo. In reported infection models to date, the predominance of N1 or N2 neutrophil phenotypes is generally not reported. For example, in a murine model of S. pneumoniae-induced lung infection, calprotectin is required for the appearance of a robust neutrophil infiltrate with the regulated production of G-CSF and a reduction of the infectious load in the lungs [232]. Whether calprotectin acts dichotomously as an alarmin or as an inflammation-dampening protein complex is strongly suggested to depend on the prevalence of a dominant N1 phenotype in the response to infection.
Calprotectin expression in human infants (and newborn mice) sustains the composition of the healthy developing gut microbiome [233]. Infants with high fecal concentrations of calprotectin had greater abundance of Actinobacteria and Bifidobacteriaceae and fewer Gammaproteobacteria including opportunistic Enterobacteriaceae. Low calprotectin expression was associated with sepsis and obesity by 2 years of age [233].
Stability of the human infant gut microbiome is supported by breast milk, which contains concentrations of calprotectin that are six-times higher than in the serum of newborns [234]. In infants, calprotectin is anti-inflammatory. After term vaginal delivery, calprotectin levels in breast milk are higher than after pre-term and Caesarian section deliveries. Two weeks after birth, breast milk calprotectin decreases to the levels in adult sera. At these levels, breast milk calprotectin significantly inhibited growth of Mn++-sensitive bacteria such as Staphylococcus aureus and group B streptococci (neonatal sepsis related bacteria) [234]. After term deliveries, higher levels of calcium in breast milk were associated with a reduction in the amount of calprotectin needed to inhibit pathogens, presumably due to heterotetramer formation. For comparison, in S100A9−/− mice, GI Enterobacteriaceae were more abundant when compared to wild-type mice. Simulating neonates with low levels of calprotectin, S100A9−/− mice also showed altered intestinal macrophage phenotypes and reduced numbers of T-regulatory cells [234]. These characteristics may predispose human and murine neonates to sepsis. Neonatal sepsis may be prevented or mitigated by the high concentration of calprotectin in breast milk and protect against lactational mastitis in the mother.
AMPs in the oral cavity
The oral cavity contains an estimated 700 microbial species that organize into communities reflecting the unique anatomic and nutritional features of the local habitats [235]. Hence, the communities residing on the tongue differ from those on the smooth enamel surfaces, which differ from those in the gingival crevice or within gingival epithelial cells. The microbes recovered in saliva cannot be viewed as a resident community since saliva in the mouth replenishes continually, with about 1 mL secreted and swallowed each minute by healthy adults. The salivary microbiome reflects the organisms shed from the oral surfaces on an ongoing basis [236] and can change due to underlying conditions such as inflammatory bowel disease [237].
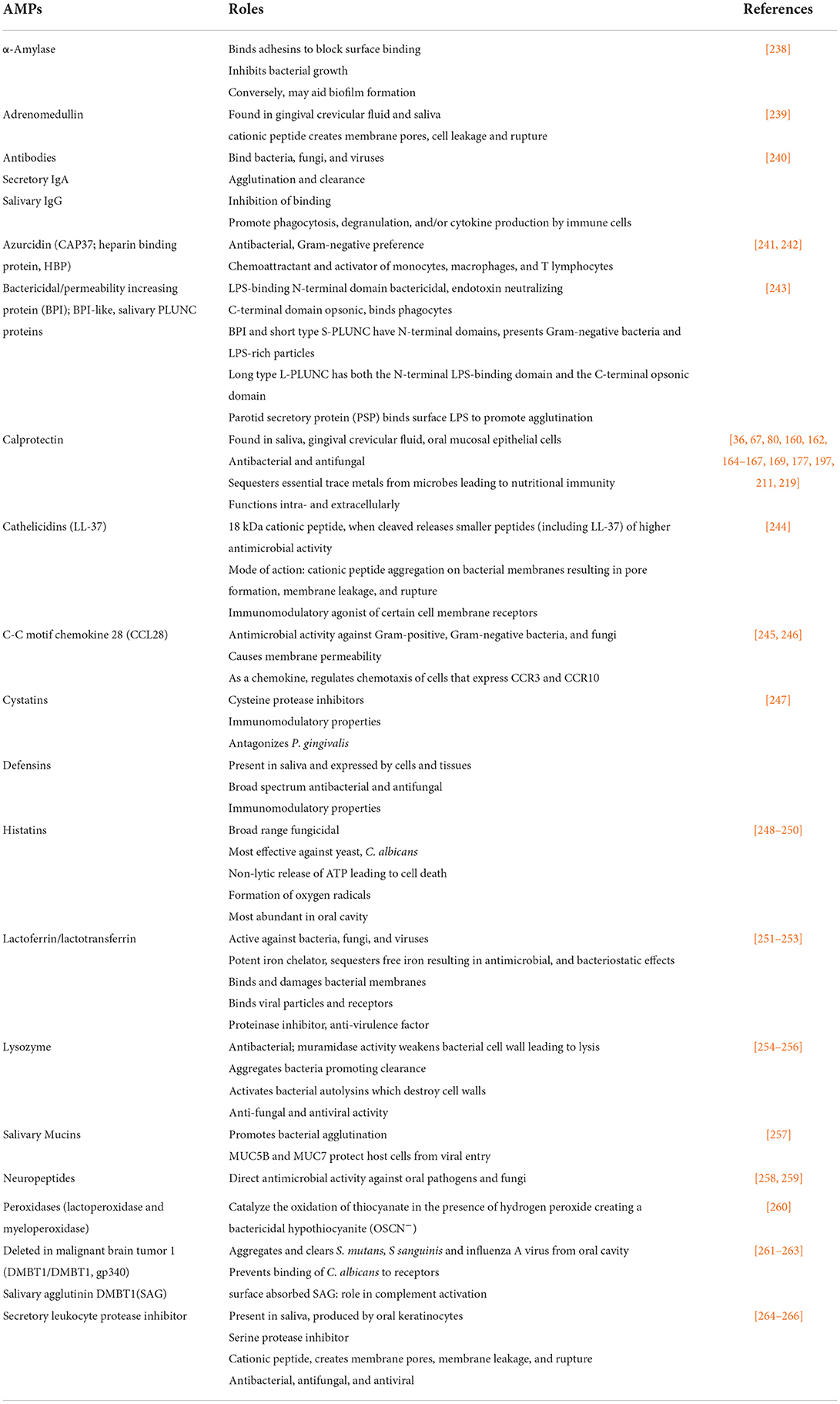
In the oral cavity, AMPs produced in different anatomic sites are among the factors that control the growth and complexity of resident microbes (Table 2). There are no fewer than 18 discrete molecular families that confer antimicrobial control in the oral cavity. The relative abundance of different AMPs in intraoral sites serves as an important ecological determinant that contributes to the abundance, organization, complexity, and stability of the resident microbial communities [99, 135, 159, 267, 268]. At the epithelial interfaces, each AMP and cooperation between them are poorly understood as influences of the composition and pathogenicity of the oral communities.
Use of mouse genetic models and the development of AMP therapeutics
Use of mouse models to identify candidate genes has proven invaluable in increasing our understanding of many diseases. As we illustrate, a gene of interest can be shown to contribute to a functional or disease phenotype. Typically, however, the results show only that the gene product is necessary but not necessarily sufficient or the sole protein of interest. In the case of studying AMPs and innate immunity, we see functional redundancy in the control of the microbiota. We can also be confounded by interactions between AMPs and immune cells that contribute to an observed phenotype. AMPs can contribute to the development of the murine immune system and sustain the development of a healthy microbiome. As investigators translate data about a protein/peptide into a treatment, results in the human clinical trials may not mirror the potency or effectiveness predicted by genetic data in mice. To identify the cells and tissues responsible for the AMP activity, the use of conditional knockout strains may be useful when expression is under control of cell and tissue-specific promoters. Knock-in studies that add or increase expression of a specific gene would make the experimental paradigm more robust. Bacteria and fungi are generally sensitive to AMPs including calprotectin and have co-evolved successfully, but resistance mechanisms in key pathogens have been characterized [269–273]. Engineered AMPs have been suggested as therapeutics to compliment or replace the use of antibiotics [274] where resistance is a major therapeutic problem [275–278]. To replace or serve as an adjunct to antibiotics, specific AMP therapeutics can be developed in animal models. One interesting strategy would target production by the native AMP-producing cells. Indeed, we may be able to develop AMP therapeutics that target the mucosal barrier epithelium where infections originate.
Conclusions
The contribution of AMPs to antimicrobial immunity is a crucial first defense of the epithelial barrier. The AMPs differ in structure and include the evolutionarily conserved families of pore-forming peptides and the essential trace metal binding proteins that create antimicrobial nutritional immunity. Functioning in affected tissues, AMPs serve cooperatively to thwart bacterial, fungal, and viral infections. In epithelial tissues spaces, the AMPs can present in microbe-trapping antimicrobial NETs to circumscribe and neutralize invading pathogens. Functioning intracellularly, AMPs protect cells against invasive microbes. Given their presence in different anatomic sites in the oral cavity, AMPs appear to confer site-specific microbial control. During the immune response to infection, parsing out the role of the AMPs even when mutated or deleted in genetic models can be challenging. Each AMP functions in the constellation of related proteins and peptides and adaptive immunity. Furthermore, crosstalk between systems can upregulate compensatory mechanisms. Nonetheless the evolution of AMPs in lower species indicates that innate immunity fortifies the epithelial barrier in the absence of the complex immunity common to humans and other mammals. Given that AMPs have broad-spectrum antimicrobial activity and do not readily promote microbial resistance, translational development of AMPs as therapies appears warranted in the face of growing microbial resistance to conventional antibiotics. Indeed, AMPs may be applied therapeutically to fortify the oral epithelial barrier against infection.
Author contributions
KJ and MH created the concept and drafted this manuscript. KJ created the figures using BioRender©. MH finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Investigations about the structure and function of AMPs in the authors' laboratory were supported by R21DE025711, R01DE021206, and RO1DE11831.
Acknowledgments
The authors thank Dr. Gay Herzberg for expert editorial assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Janeway CA Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. (2002) 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359
2. Elo IT, Luck A, Stokes AC, Hempstead K, Xie W, Preston SH. Evaluation of age patterns of COVID-19 mortality by race and ethnicity from March 2020 to October 2021 in the US. JAMA Netw Open. (2022) 5:e2212686. doi: 10.1001/jamanetworkopen.2022.12686
3. Batty GD, Gaye B, Gale CR, Hamer M, Lassale C. Explaining ethnic differentials in COVID-19 mortality: a cohort study. Am J Epidemiol. (2022) 191:275–81. doi: 10.1093/aje/kwab237
4. Auld SC, Harrington KRV, Adelman MW, Robichaux CJ, Overton EC, Caridi-Scheible M, et al. Trends in ICU mortality from coronavirus disease 2019: a tale of three surges. Crit Care Med. (2022) 50:245–55. doi: 10.1097/CCM.0000000000005185
5. Cuellar L, Torres I, Romero-Severson E, Mahesh R, Ortega N, Pungitore S, et al. Excess deaths reveal the true spatial, temporal and demographic impact of COVID-19 on mortality in ecuador. Int J Epidemiol. (2022) 51:54–62. doi: 10.1093/ije/dyab163
6. Kumar AKA, Mishra N. Mortality during the COVID-19 pandemic: the blind spots in statistics. Lancet Infect Dis. (2022) 22:428–9. doi: 10.1016/S1473-3099(21)00767-2
7. Horve PF, Dietz LG, Bowles G, MacCrone G, Olsen-Martinez A, Northcutt D, et al. Longitudinal analysis of built environment and aerosol contamination associated with isolated COVID-19 positive individuals. Sci Rep. (2022) 12:7395. doi: 10.1038/s41598-022-11303-8
8. Bozack A, Pierre S, DeFelice N, Colicino E, Jack D, Chillrud SN, et al. Long-Term air pollution exposure and COVID-19 mortality: a patient-level analysis from New York city. Am J Respir Crit Care Med. (2022) 205:651–62. doi: 10.1164/rccm.202104-0845OC
9. Christophers E, Schroder JM. Evolution of innate defense in human skin. Exp Dermatol. (2022) 31:304–11. doi: 10.1111/exd.14482
10. Gorr SU, Abdolhosseini M. Antimicrobial peptides and periodontal disease. J Clin Periodontol. (2011) 38 (Suppl. 11):126–41. doi: 10.1111/j.1600-051X.2010.01664.x
11. Lu Q, Stappenbeck TS. Local barriers configure systemic communications between the host and microbiota. Science. (2022) 376:950–5. doi: 10.1126/science.abo2366
12. Stokes BA, Yadav S, Shokal U, Smith LC, Eleftherianos I. Bacterial and fungal pattern recognition receptors in homologous innate signaling pathways of insects and mammals. Front Microbiol. (2015) 6:19. doi: 10.3389/fmicb.2015.00019
13. Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol Rev. (2004) 198:72–82. doi: 10.1111/j.0105-2896.2004.0133.x
14. Myllymaki H, Ramet M. JAK/STAT pathway in Drosophila immunity. Scand J Immunol. (2014) 79:377–85. doi: 10.1111/sji.12170
15. Lindsay SA, Wasserman SA. Conventional and non-conventional Drosophila toll signaling. Dev Comp Immunol. (2014) 42:16–24. doi: 10.1016/j.dci.2013.04.011
16. Le Noci V, Bernardo G, Bianchi F, Tagliabue E, Sommariva M, Sfondrini L. Toll like receptors as sensors of the tumor microbial dysbiosis: implications in cancer progression. Front Cell Dev Biol. (2021) 9:732192. doi: 10.3389/fcell.2021.732192
17. Behzadi P, Garcia-Perdomo HA, Karpinski TM. Toll-Like receptors: general molecular and structural biology. J Immunol Res. (2021) 2021:9914854. doi: 10.1155/2021/9914854
18. Garcia V, Perera YR, Chazin WJ. A structural perspective on calprotectin as a ligand of receptors mediating inflammation and potential drug target. Biomolecules. (2022) 12:519. doi: 10.3390/biom12040519
19. Jang HA, Patnaik BB, Ali Mohammadie Kojour M, Kim BB, Bae YM, Park KB, et al. TmSpz-like plays a fundamental role in response to E. coli but not S. aureus or C. albican infection in tenebrio molitor via regulation of antimicrobial peptide production. Int J Mol Sci. (2021) 22:10888. doi: 10.3390/ijms221910888
20. Yu B, Sang Q, Pan G, Li C, Zhou Z. A toll-spatzle pathway in the immune response of Bombyx mori. Insects. (2020) 11:586. doi: 10.3390/insects11090586
21. Muhammad A, Habineza P, Wang X, Xiao R, Ji T, Hou Y, et al. Spatzle homolog-mediated toll-like pathway regulates innate immune responses to maintain the homeostasis of gut microbiota in the red palm weevil, rhynchophorus ferrugineus olivier (Coleoptera: Dryophthoridae). Front Microbiol. (2020) 11:846. doi: 10.3389/fmicb.2020.00846
22. Wang Y, Yang F, Cao X, Zou Z, Lu Z, Kanost MR, et al. Hemolymph protease-5 links the melanization and toll immune pathways in the tobacco hornworm, Manduca sexta. Proc Natl Acad Sci U S A. (2020) 117:23581–7. doi: 10.1073/pnas.2004761117
23. Liu W, Wang Y, Zhou J, Zhang Y, Ma Y, Wang D, et al. Peptidoglycan recognition proteins regulate immune response of Antheraea pernyi in different ways. J Invertebr Pathol. (2019) 166:107204. doi: 10.1016/j.jip.2019.107204
24. Bevivino G, Arca B, Lombardo F. Effects of local and systemic immune challenges on the expression of selected salivary genes in the malaria mosquito Anopheles coluzzii. Pathogens. (2021) 10:1300. doi: 10.3390/pathogens10101300
25. Hong PP, Zhu XX, Yuan WJ, Niu GJ, Wang JX. Nitric oxide synthase regulates gut microbiota homeostasis by ERK-NF-kappaB pathway in shrimp. Front Immunol. (2021) 12:778098. doi: 10.3389/fimmu.2021.778098
26. He Y, Cao X, Li K, Hu Y, Chen YR, Blissard G, et al. A genome-wide analysis of antimicrobial effector genes and their transcription patterns in Manduca sexta. Insect Biochem Mol Biol. (2015) 62:23–37. doi: 10.1016/j.ibmb.2015.01.015
27. Roesel CL, Rosengaus RB, Smith W, Vollmer SV. Transcriptomics reveals specific molecular mechanisms underlying transgenerational immunity in Manduca sexta. Ecol Evol. (2020) 10:11251–61. doi: 10.1002/ece3.6764
28. Vigneron A, Jehan C, Rigaud T, Moret Y. Immune defenses of a beneficial pest: the mealworm beetle, Tenebrio molitor. Front Physiol. (2019) 10:138. doi: 10.3389/fphys.2019.00138
29. Brey PT, Lee WJ, Yamakawa M, Koizumi Y, Perrot S, Francois M, et al. Role of the integument in insect immunity: epicuticular abrasion and induction of cecropin synthesis in cuticular epithelial cells. Proc Natl Acad Sci USA. (1993) 90:6275–9. doi: 10.1073/pnas.90.13.6275
30. Kan H, Kim CH, Kwon HM, Park JW, Roh KB, Lee H, et al. Molecular control of phenoloxidase-induced melanin synthesis in an insect. J Biol Chem. (2008) 283:25316–23. doi: 10.1074/jbc.M804364200
31. Longo V, Parrinello D, Longo A, Parisi MG, Parrinello N, Colombo P, et al. The conservation and diversity of ascidian cells and molecules involved in the inflammatory reaction: the ciona robusta model. Fish Shellfish Immunol. (2021) 119:384–96. doi: 10.1016/j.fsi.2021.10.022
32. Dishaw LJ, Giacomelli S, Melillo D, Zucchetti I, Haire RN, Natale L, et al. A role for variable region-containing chitin-binding proteins (VCBPs) in host gut-bacteria interactions. Proc Natl Acad Sci USA. (2011) 108:16747–52. doi: 10.1073/pnas.1109687108
33. Tincu JA, Taylor SW. Antimicrobial peptides from marine invertebrates. Antimicrob Agents Chemother. (2004) 48:3645–54. doi: 10.1128/AAC.48.10.3645-3654.2004
34. Cuesta SA, Reinoso C, Morales F, Pilaquinga F, Moran-Marcillo G, Proano-Bolanos C, et al. Novel antimicrobial cruzioseptin peptides extracted from the splendid leaf frog, Cruziohyla calcarifer. Amino Acids. (2021) 53:853–68. doi: 10.1007/s00726-021-02986-w
35. Sani MA, Carne S, Overall SA, Poulhazan A, Separovic F. One pathogen two stones: are Australian tree frog antimicrobial peptides synergistic against human pathogens? Eur Biophys J. (2017) 46:639–46. doi: 10.1007/s00249-017-1215-9
36. Nisapakultorn K, Ross KF, Herzberg MC. Calprotectin expression in vitro by oral epithelial cells confers resistance to infection by Porphyromonas gingivalis. Infect Immun. (2001) 69:4242–7. doi: 10.1128/IAI.69.7.4242-4247.2001
37. Hans M, Madaan Hans V. Epithelial antimicrobial peptides: guardian of the oral cavity. Int J Pept. (2014) 2014:370297. doi: 10.1155/2014/370297
38. Li X, Duan D, Yang J, Wang P, Han B, Zhao L, et al. The expression of human beta-defensins (hBD-1, hBD-2, hBD-3, hBD-4) in gingival epithelia. Arch Oral Biol. (2016) 66:15–21. doi: 10.1016/j.archoralbio.2016.01.012
39. Mohammed I, Said DG, Dua HS. Human antimicrobial peptides in ocular surface defense. Prog Retin Eye Res. (2017) 61:1–22. doi: 10.1016/j.preteyeres.2017.03.004
40. Barreto EBL, Rattes IC, da Costa AV, Gama P. Paneth cells and their multiple functions. Cell Biol Int. (2022) 46:701–10. doi: 10.1002/cbin.11764
41. Mitchell G, Silvis MR, Talkington KC, Budzik JM, Dodd CE, Paluba JM, et al. Ceragenins and antimicrobial peptides kill bacteria through distinct mechanisms. mBio. (2022) 13:e0272621. doi: 10.1128/mbio.02726-21
42. Li C, Sutherland D, Hammond SA, Yang C, Taho F, Bergman L, et al. AMPlify: attentive deep learning model for discovery of novel antimicrobial peptides effective against WHO priority pathogens. BMC Genomics. (2022) 23:77. doi: 10.1186/s12864-022-08310-4
43. Lin B, Hung A, Li R, Barlow A, Singleton W, Matthyssen T, et al. Systematic comparison of activity and mechanism of antimicrobial peptides against nosocomial pathogens. Eur J Med Chem. (2022) 231:114135. doi: 10.1016/j.ejmech.2022.114135
44. Bao X, Wiehe R, Dommisch H, Schaefer AS. Entamoeba gingivalis causes oral inflammation and tissue destruction. J Dent Res. (2020) 99:561–7. doi: 10.1177/0022034520901738
45. Swidergall M, Ernst JF. Interplay between Candida albicans and the antimicrobial peptide armory. Eukaryot Cell. (2014) 13:950–7. doi: 10.1128/EC.00093-14
46. Buda De Cesare G, Cristy SA, Garsin DA, Lorenz MC. Antimicrobial peptides: a new frontier in antifungal therapy. mBio. (2020) 11:e02123–20. doi: 10.1128/mBio.02123-20
47. Ryan LK, Dai J, Yin Z, Megjugorac N, Uhlhorn V, Yim S, et al. Modulation of human beta-defensin-1 (hBD-1) in plasmacytoid dendritic cells (PDC), monocytes, and epithelial cells by influenza virus, herpes simplex virus, and sendai virus and its possible role in innate immunity. J Leukoc Biol. (2011) 90:343–56. doi: 10.1189/jlb.0209079
48. Brice DC, Toth Z, Diamond G. LL-37 disrupts the Kaposi's sarcoma-associated herpesvirus envelope and inhibits infection in oral epithelial cells. Antiviral Res. (2018) 158:25–33. doi: 10.1016/j.antiviral.2018.07.025
49. Shang L, Deng D, Buskermolen JK, Janus MM, Krom BP, Roffel S, et al. Multi-species oral biofilm promotes reconstructed human gingiva epithelial barrier function. Sci Rep. (2018) 8:16061. doi: 10.1038/s41598-018-34390-y
50. Koren N, Zubeidat K, Saba Y, Horev Y, Barel O, Wilharm A, et al. Maturation of the neonatal oral mucosa involves unique epithelium-microbiota interactions. Cell Host Microbe. (2021) 29:197–209.e5. doi: 10.1016/j.chom.2020.12.006
51. Frohm M, Agerberth B, Ahangari G, Stahle-Backdahl M, Liden S, Wigzell H, et al. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J Biol Chem. (1997) 272:15258–63. doi: 10.1074/jbc.272.24.15258
52. Harder J, Schroder JM. Antimicrobial peptides in human skin. Chem Immunol Allergy. (2005) 86:22–41. doi: 10.1159/000086650
53. Boehncke WH. The alpha-defensins HNP-1 and HNP-2 are dominant self-peptides presented by HLA class-II molecules in lesional psoriatic skin. Eur J Dermatol. (2004) 14:142–5.
54. Harman JL, Loes AN, Warren GD, Heaphy MC, Lampi KJ, Harms MJ. Evolution of multifunctionality through a pleiotropic substitution in the innate immune protein S100A9. Elife. (2020) 9:e54100. doi: 10.7554/eLife.54100
55. Antelo GT, Vila AJ, Giedroc DP, Capdevila DA. Molecular evolution of transition metal bioavailability at the host-pathogen interface. Trends Microbiol. (2021) 29:441–57. doi: 10.1016/j.tim.2020.08.001
56. Last NB, Miranker AD. Common mechanism unites membrane poration by amyloid and antimicrobial peptides. Proc Natl Acad Sci USA. (2013) 110:6382–7. doi: 10.1073/pnas.1219059110
57. Duong L, Gross SP, Siryaporn A. A novel antibacterial strategy: histone and antimicrobial peptide synergy. Microb Cell. (2020) 7:309–11. doi: 10.15698/mic2020.11.736
58. Doolin T, Amir HM, Duong L, Rosenzweig R, Urban LA, Bosch M, et al. Mammalian histones facilitate antimicrobial synergy by disrupting the bacterial proton gradient and chromosome organization. Nat Commun. (2020) 11:3888. doi: 10.1038/s41467-020-17699-z
59. Duong L, Gross SP, Siryaporn A. Developing antimicrobial synergy with AMPs. Front Med Technol. (2021) 3:640981. doi: 10.3389/fmedt.2021.640981
60. Cooper PR, Palmer LJ, Chapple IL. Neutrophil extracellular traps as a new paradigm in innate immunity: friend or foe? Periodontol. (2013) 63:165–97. doi: 10.1111/prd.12025
61. Dominguez-Diaz C, Varela-Trinidad GU, Munoz-Sanchez G, Solorzano-Castanedo K, Avila-Arrezola KE, Iniguez-Gutierrez L, et al. To trap a pathogen: neutrophil extracellular traps and their role in mucosal epithelial and skin diseases. Cells. (2021) 10:1469. doi: 10.3390/cells10061469
62. Drury B, Hardisty G, Gray RD, Ho GT. Neutrophil extracellular traps in inflammatory bowel disease: pathogenic mechanisms and clinical translation. Cell Mol Gastroenterol Hepatol. (2021) 12:321–33. doi: 10.1016/j.jcmgh.2021.03.002
63. Lueschow SR, McElroy SJ. The paneth cell: the curator and defender of the immature small intestine. Front Immunol. (2020) 11:587. doi: 10.3389/fimmu.2020.00587
64. Minns D, Smith KJ, Alessandrini V, Hardisty G, Melrose L, Jackson-Jones L, et al. The neutrophil antimicrobial peptide cathelicidin promotes Th17 differentiation. Nat Commun. (2021) 12:1285. doi: 10.1038/s41467-021-21533-5
65. Zhang QY, Yan ZB, Meng YM, Hong XY, Shao G, Ma JJ, et al. Antimicrobial peptides: mechanism of action, activity and clinical potential. Mil Med Res. (2021) 8:48. doi: 10.1186/s40779-021-00343-2
66. Elieh Ali Komi D, Kuebler WM. Significance of mast cell formed extracellular traps in microbial defense. Clin Rev Allergy Immunol. (2022) 62:160–79. doi: 10.1007/s12016-021-08861-6
67. Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, et al. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. (2009) 5:e1000639. doi: 10.1371/journal.ppat.1000639
68. Zaia AA, Sappington KJ, Nisapakultorn K, Chazin WJ, Dietrich EA, Ross KF, et al. Subversion of antimicrobial calprotectin (S100A8/S100A9 complex) in the cytoplasm of TR146 epithelial cells after invasion by listeria monocytogenes. Mucosal Immunol. (2009) 2:43–53. doi: 10.1038/mi.2008.63
69. Salvatori O, Pathirana RU, Kay JG, Edgerton M. Candida albicans Ras1 inactivation increases resistance to phagosomal killing by human neutrophils. Infect Immun. (2018) 86:e00685–18. doi: 10.1128/IAI.00685-18
70. Hu W, Zhang L, Li MX, Shen J, Liu XD, Xiao ZG, et al. Vitamin D3 activates the autolysosomal degradation function against Helicobacter pylori through the PDIA3 receptor in gastric epithelial cells. Autophagy. (2019) 15:707–25. doi: 10.1080/15548627.2018.1557835
71. Dang AT, Teles RM, Liu PT, Choi A, Legaspi A, Sarno EN, et al. Autophagy links antimicrobial activity with antigen presentation in langerhans cells. JCI Insight. (2019) 4:e126955. doi: 10.1172/jci.insight.126955
72. Pidwill GR, Gibson JF, Cole J, Renshaw SA, Foster SJ. The role of macrophages in Staphylococcus aureus infection. Front Immunol. (2020) 11:620339. doi: 10.3389/fimmu.2020.620339
73. Czuczman MA, Fattouh R, van Rijn JM, Canadien V, Osborne S, Muise AM, et al. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature. (2014) 509:230–4. doi: 10.1038/nature13168
74. Nguyen BN, Peterson BN, Portnoy DA. Listeriolysin O: a phagosome-specific cytolysin revisited. Cell Microbiol. (2019) 21:e12988. doi: 10.1111/cmi.12988
75. Gluschko A, Farid A, Herb M, Grumme D, Kronke M, Schramm M. Macrophages target listeria monocytogenes by two discrete non-canonical autophagy pathways. Autophagy. (2021) 18:1090–107. doi: 10.1080/15548627.2021.1969765
76. Arnett E, Lehrer RI, Pratikhya P, Lu W, Seveau S. Defensins enable macrophages to inhibit the intracellular proliferation of listeria monocytogenes. Cell Microbiol. (2011) 13:635–51. doi: 10.1111/j.1462-5822.2010.01563.x
77. Tan JMJ, Garner ME, Regeimbal JM, Greene CJ, Marquez JDR, Ammendolia DA, et al. Listeria exploits IFITM3 to suppress antibacterial activity in phagocytes. Nat Commun. (2021) 12:4999. doi: 10.1038/s41467-021-24982-0
78. Gouin E, Balestrino D, Rasid O, Nahori MA, Villiers V, Impens F, et al. Ubiquitination of listeria virulence factor inlc contributes to the host response to infection. mBio. (2019) 10:e02778–19. doi: 10.1128/mBio.02778-19
79. Leonard BC, Chu H, Johns JL, Gallo RL, Moore PF, Marks SL, et al. Expression and activity of a novel cathelicidin from domestic cats. PLoS ONE. (2011) 6:e18756. doi: 10.1371/journal.pone.0018756
80. Zou X, Sorenson BS, Ross KF, Herzberg MC. Augmentation of epithelial resistance to invading bacteria by using mRNA transfections. Infect Immun. (2013) 81:3975–83. doi: 10.1128/IAI.00539-13
81. Hiroshima Y, Bando M, Kataoka M, Inagaki Y, Herzberg MC, Ross KF, et al. Regulation of antimicrobial peptide expression in human gingival keratinocytes by interleukin-1alpha. Arch Oral Biol. (2011) 56:761–7. doi: 10.1016/j.archoralbio.2011.01.004
82. Sorenson BS, Khammanivong A, Guenther BD, Ross KF, Herzberg MC. IL-1 receptor regulates S100A8/A9-dependent keratinocyte resistance to bacterial invasion. Mucosal Immunol. (2012) 5:66–75. doi: 10.1038/mi.2011.48
83. Bando M, Zou X, Hiroshima Y, Kataoka M, Ross KF, Shinohara Y, et al. Mechanism of interleukin-1alpha transcriptional regulation of S100A9 in a human epidermal keratinocyte cell line. Biochim Biophys Acta. (2013) 1829:954–62. doi: 10.1016/j.bbagrm.2013.03.010
84. Baraniya D, Chen T, Nahar A, Alakwaa F, Hill J, Tellez M, et al. Supragingival mycobiome and inter-kingdom interactions in dental caries. J Oral Microbiol. (2020) 12:1729305. doi: 10.1080/20002297.2020.1729305
85. Diaz PI, Dongari-Bagtzoglou A. Critically appraising the significance of the oral mycobiome. J Dent Res. (2021) 100:133–40. doi: 10.1177/0022034520956975
86. Lyu X, Zheng H, Wang X, Zhang H, Gao L, Xun Z, et al. Oral microbiota composition and function changes during chronic erythematous candidiasis. Front Cell Infect Microbiol. (2021) 11:691092. doi: 10.3389/fcimb.2021.691092
87. Bertolini M, Vazquez Munoz R, Archambault L, Shah S, Souza JGS, Costa RC, et al. Mucosal bacteria modulate Candida albicans Virulence in oropharyngeal candidiasis. mBio. (2021) 12:e0193721. doi: 10.1128/mBio.01937-21
88. Edgerton M, Koshlukova SE, Lo TE, Chrzan BG, Straubinger RM, Raj PA. Candidacidal activity of salivary histatins. Identification of a histatin 5-binding protein on Candida albicans. J Biol Chem. (1998) 273:20438–47. doi: 10.1074/jbc.273.32.20438
89. Sawaki K, Mizukawa N, Yamaai T, Fukunaga J, Sugahara T. Immunohistochemical study on expression of alpha-defensin and beta-defensin-2 in human buccal epithelia with candidiasis. Oral Dis. (2002) 8:37–41. doi: 10.1034/j.1601-0825.2002.1o770.x
90. Brandtzaeg P, Jones DB, Flavell DJ, Fagerhol MK. Mac 387 antibody and detection of formalin resistant myelomonocytic L1 antigen. J Clin Pathol. (1988) 41:963–70. doi: 10.1136/jcp.41.9.963
91. Steinbakk M, Naess-Andresen CF, Lingaas E, Dale I, Brandtzaeg P, Fagerhol MK. Antimicrobial actions of calcium binding leucocyte L1 protein, calprotectin. Lancet. (1990) 336:763–5. doi: 10.1016/0140-6736(90)93237-J
92. He J, Thomas MA, de Anda J, Lee MW, Van Why E, Simpson P, et al. Chemokine CCL28 is a potent therapeutic agent for oropharyngeal candidiasis. Antimicrob Agents Chemother. (2020) 64:e00210–20. doi: 10.1128/AAC.00210-20
93. Zolin GVS, Fonseca FHD, Zambom CR, Garrido SS. Histatin 5 metallopeptides and their potential against candida albicans pathogenicity and drug resistance. Biomolecules. (2021) 11:1209. doi: 10.3390/biom11081209
94. Lindh E, Brannstrom J, Jones P, Wermeling F, Hassler S, Betterle C, et al. Autoimmunity and cystatin SA1 deficiency behind chronic mucocutaneous candidiasis in autoimmune polyendocrine syndrome type 1. J Autoimmun. (2013) 42:1–6. doi: 10.1016/j.jaut.2012.10.001
95. Xu D, Pavlidis P, Thamadilok S, Redwood E, Fox S, Blekhman R, et al. Recent evolution of the salivary mucin MUC7. Sci Rep. (2016) 6:31791. doi: 10.1038/srep31791
96. Naglik JR, Gaffen SL, Hube B. Candidalysin: discovery and function in Candida albicans infections. Curr Opin Microbiol. (2019) 52:100–9. doi: 10.1016/j.mib.2019.06.002
97. Ho J, Wickramasinghe DN, Nikou SA, Hube B, Richardson JP, Naglik JR. Candidalysin is a potent trigger of alarmin and antimicrobial peptide release in epithelial cells. Cells. (2020) 9:699. doi: 10.3390/cells9030699
98. Conti HR, Bruno VM, Childs EE, Daugherty S, Hunter JP, Mengesha BG, et al. IL-17 receptor signaling in oral epithelial cells is critical for protection against oropharyngeal candidiasis. Cell Host Microbe. (2016) 20:606–17. doi: 10.1016/j.chom.2016.10.001
99. Tomalka J, Azodi E, Narra HP, Patel K, O'Neill S, Cardwell C, et al. beta-Defensin 1 plays a role in acute mucosal defense against Candida albicans. J Immunol. (2015) 194:1788–95. doi: 10.4049/jimmunol.1203239
100. Thakkar P, Banks JM, Rahat R, Brandini DA, Naqvi AR. Viruses of the oral cavity: prevalence, pathobiology and association with oral diseases. Rev Med Virol. (2021) 32:e2311. doi: 10.1002/rmv.2311
101. Yang X, Xiang Z, Sun Z, Ji F, Ren K, Pan D. Host MOV10 is induced to restrict herpes simplex virus 1 lytic infection by promoting type I interferon response. PLoS Pathog. (2022) 18:e1010301. doi: 10.1371/journal.ppat.1010301
102. Lloyd MG, Liu D, Legendre M, Markovitz DM, Moffat JF. H84T BanLec has broad spectrum antiviral activity against human herpesviruses in cells, skin, and mice. Sci Rep. (2022) 12:1641. doi: 10.1038/s41598-022-05580-6
103. Ma Y, Li J, Dong H, Yang Z, Zhou L, Xu P. PML body component Sp100A restricts wild-type herpes simplex virus 1 infection. J Virol. (2022) 96:e0027922. doi: 10.1128/jvi.00279-22
104. Vilas Boas LC, de Lima LM, Migliolo L, Mendes GD, de Jesus MG, Franco OL, et al. Linear antimicrobial peptides with activity against herpes simplex virus 1 and aichi virus. Biopolymers. (2017) 108:1–6. doi: 10.1002/bip.22871
105. Drozdzik A, Drozdzik M. Oral pathology in COVID-19 and SARS-CoV-2 infection-molecular aspects. Int J Mol Sci. (2022) 23:1431. doi: 10.3390/ijms23031431
106. Boffert R, Businger R, Preiss H, Ehmann D, Truffault V, Simon C, et al. The human alpha-defensin-derived peptide HD5(1-9) inhibits cellular attachment and entry of human cytomegalovirus. Antiviral Res. (2020) 177:104779. doi: 10.1016/j.antiviral.2020.104779
107. Valimaa H, Tenovuo J, Waris M, Hukkanen V. Human lactoferrin but not lysozyme neutralizes HSV-1 and inhibits HSV-1 replication and cell-to-cell spread. Virol J. (2009) 6:53. doi: 10.1186/1743-422X-6-53
108. Wiens ME, Smith JG. Alpha-defensin HD5 inhibits human papillomavirus 16 infection via capsid stabilization and redirection to the lysosome. mBio. (2017) 8:e02304–16. doi: 10.1128/mBio.02304-16
109. Skeate JG, Segerink WH, Garcia MD, Fernandez DJ, Prins R, Luhen KP, et al. Theta-Defensins inhibit high-risk human papillomavirus infection through charge-driven capsid clustering. Front Immunol. (2020) 11:561843. doi: 10.3389/fimmu.2020.561843
110. Antonsson A, de Souza M, Wood ZC, Carroll A, Van K, Paterson L, et al. Natural history of oral HPV infection: longitudinal analyses in prospective cohorts from Australia. Int J Cancer. (2021) 148:1964–72. doi: 10.1002/ijc.33442
111. Ron-Doitch S, Sawodny B, Kuhbacher A, David MMN, Samanta A, Phopase J, et al. Reduced cytotoxicity and enhanced bioactivity of cationic antimicrobial peptides liposomes in cell cultures and 3D epidermis model against HSV. J Control Release. (2016) 229:163–71. doi: 10.1016/j.jconrel.2016.03.025
112. Shartouny JR, Jacob J. Mining the tree of life: host defense peptides as antiviral therapeutics. Semin Cell Dev Biol. (2019) 88:147–55. doi: 10.1016/j.semcdb.2018.03.001
113. Vacharaksa A, Asrani AC, Gebhard KH, Fasching CE, Giacaman RA, Janoff EN, et al. Oral keratinocytes support non-replicative infection and transfer of harbored HIV-1 to permissive cells. Retrovirology. (2008) 5:66. doi: 10.1186/1742-4690-5-66
114. Lien K, Mayer W, Herrera R, Rosbe K, Tugizov SM. HIV-1 proteins gp120 and tat induce the epithelial-mesenchymal transition in oral and genital mucosal epithelial cells. PLoS ONE. (2019) 14:e0226343. doi: 10.1371/journal.pone.0226343
115. Giacaman RA, Nobbs AH, Ross KF, Herzberg MC. Porphyromonas gingivalis selectively up-regulates the HIV-1 coreceptor CCR5 in oral keratinocytes. J Immunol. (2007) 179:2542–50. doi: 10.4049/jimmunol.179.4.2542
116. Giacaman RA, Asrani AC, Gebhard KH, Dietrich EA, Vacharaksa A, Ross KF, et al. Porphyromonas gingivalis induces CCR5-dependent transfer of infectious HIV-1 from oral keratinocytes to permissive cells. Retrovirology. (2008) 5:29. doi: 10.1186/1742-4690-5-29
117. Dong XH, Ho MH, Liu B, Hildreth J, Dash C, Goodwin JS, et al. Role of Porphyromonas gingivalis outer membrane vesicles in oral mucosal transmission of HIV. Sci Rep. (2018) 8:8812. doi: 10.1038/s41598-018-27284-6
118. Dietrich EA, Gebhard KH, Fasching CE, Giacaman RA, Kappes JC, Ross KF, et al. Short communication: HIV type 1 escapes inactivation by saliva via rapid escape into oral epithelial cells. AIDS Res Hum Retroviruses. (2012) 28:1574–8. doi: 10.1089/aid.2011.0069
119. Venkataraman N, Cole AL, Svoboda P, Pohl J, Cole AM. Cationic polypeptides are required for anti-HIV-1 activity of human vaginal fluid. J Immunol. (2005) 175:7560–7. doi: 10.4049/jimmunol.175.11.7560
120. Bergman P, Walter-Jallow L, Broliden K, Agerberth B, Soderlund J. The antimicrobial peptide LL-37 inhibits HIV-1 replication. Curr HIV Res. (2007) 5:410–5. doi: 10.2174/157016207781023947
121. Wang G, Watson KM, Buckheit RW Jr. Anti-human immunodeficiency virus type 1 activities of antimicrobial peptides derived from human and bovine cathelicidins. Antimicrob Agents Chemother. (2008) 52:3438–40. doi: 10.1128/AAC.00452-08
122. Ogawa Y, Kawamura T, Matsuzawa T, Aoki R, Gee P, Yamashita A, et al. Antimicrobial peptide LL-37 produced by HSV-2-infected keratinocytes enhances HIV infection of langerhans cells. Cell Host Microbe. (2013) 13:77–86. doi: 10.1016/j.chom.2012.12.002
123. Huang N, Perez P, Kato T, Mikami Y, Okuda K, Gilmore RC, et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat Med. (2021) 27:892–903. doi: 10.1038/s41591-021-01296-8
124. Connor MC, Copeland M, Curran T. Investigation of saliva, tongue swabs and buccal swabs as alternative specimen types to nasopharyngeal swabs for SARS-CoV-2 testing. J Clin Virol. (2022) 146:105053. doi: 10.1016/j.jcv.2021.105053
125. Carrouel F, Gadea E, Esparcieux A, Dimet J, Langlois ME, Perrier H, et al. Saliva quantification of SARS-CoV-2 in real-time PCR from asymptomatic or mild COVID-19 adults. Front Microbiol. (2021) 12:786042. doi: 10.3389/fmicb.2021.786042
126. Zhang L, Ghosh SK, Basavarajappa SC, Chen Y, Shrestha P, Penfield J, et al. HBD-2 binds SARS-CoV-2 RBD and blocks viral entry: strategy to combat COVID-19. iScience. (2022) 25:103856. doi: 10.1016/j.isci.2022.103856
127. Guncu GN, Yilmaz D, Kononen E, Gursoy UK. salivary antimicrobial peptides in early detection of periodontitis. Front Cell Infect Microbiol. (2015) 5:99. doi: 10.3389/fcimb.2015.00099
128. McQuaid IK, Dorfman JR, Mall AS. The comparative inhibitory potency of salivary mucins against human immunodeficiency virus type 1. Virology. (2021) 553:1–8. doi: 10.1016/j.virol.2020.10.004
129. Ebersole JL, Kirakodu S, Nguyen L, Gonzalez OA. Gingival transcriptome of innate antimicrobial factors and the oral microbiome with aging and periodontitis. Front Oral Health. (2022) 3:817249. doi: 10.3389/froh.2022.817249
130. Puri S, Edgerton M. How does it kill?: understanding the candidacidal mechanism of salivary histatin 5. Eukaryot Cell. (2014) 13:958–64. doi: 10.1128/EC.00095-14
131. Challacombe SJ, Sweet SP. Salivary and mucosal immune responses to HIV and its co-pathogens. Oral Dis. (1997) 3 (Suppl. 1):S79–84. doi: 10.1111/j.1601-0825.1997.tb00381.x
132. Malamud D, Abrams WR, Barber CA, Weissman D, Rehtanz M, Golub E. Antiviral activities in human saliva. Adv Dent Res. (2011) 23:34–7. doi: 10.1177/0022034511399282
133. Geitani R, Moubareck CA, Xu Z, Karam Sarkis D, Touqui L. Expression and roles of antimicrobial peptides in innate defense of airway mucosa: potential implication in cystic fibrosis. Front Immunol. (2020) 11:1198. doi: 10.3389/fimmu.2020.01198
134. Kotsiou OS, Papagiannis D, Papadopoulou R, Gourgoulianis KI. Calprotectin in lung diseases. Int J Mol Sci. (2021) 22:1706. doi: 10.3390/ijms22041706
135. Rosier BT, Marsh PD, Mira A. Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J Dent Res. (2018) 97:371–80. doi: 10.1177/0022034517742139
136. Kim D, Barraza JP, Arthur RA, Hara A, Lewis K, Liu Y, et al. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc Natl Acad Sci USA. (2020) 117:12375–86. doi: 10.1073/pnas.1919099117
137. Okahashi N, Sumitomo T, Nakata M, Kawabata S. Secondary streptococcal infection following influenza. Microbiol Immunol. (2022) doi: 10.1111/1348-0421.12965
138. Zangari T, Ortigoza MB, Lokken-Toyli KL, Weiser JN. Type I interferon signaling is a common factor driving streptococcus pneumoniae and influenza A virus shedding and transmission. mBio. (2021) 12:e03589–20. doi: 10.1128/mBio.03589-20
139. Nishikawa T, Shimizu K, Tanaka T, Kuroda K, Takayama T, Yamamoto T, et al. Bacterial neuraminidase rescues influenza virus replication from inhibition by a neuraminidase inhibitor. PLoS ONE. (2012) 7:e45371. doi: 10.1371/journal.pone.0045371
140. Kamio N, Imai K, Shimizu K, Cueno ME, Tamura M, Saito Y, et al. Neuraminidase-producing oral mitis group streptococci potentially contribute to influenza viral infection and reduction in antiviral efficacy of zanamivir. Cell Mol Life Sci. (2015) 72:357–66. doi: 10.1007/s00018-014-1669-1
141. Nishioka K, Kyo M, Nakaya T, Shime N. Proteins produced by streptococcus species in the lower respiratory tract can modify antiviral responses against influenza virus in respiratory epithelial cells. Microbes Infect. (2021) 23:104764. doi: 10.1016/j.micinf.2020.09.010
142. Akata K, Yatera K, Yamasaki K, Kawanami T, Naito K, Noguchi S, et al. The significance of oral streptococci in patients with pneumonia with risk factors for aspiration: the bacterial floral analysis of 16S ribosomal RNA gene using bronchoalveolar lavage fluid. BMC Pulm Med. (2016) 16:79. doi: 10.1186/s12890-016-0235-z
143. Noguchi S, Mukae H, Kawanami T, Yamasaki K, Fukuda K, Akata K, et al. Bacteriological assessment of healthcare-associated pneumonia using a clone library analysis. PLoS ONE. (2015) 10:e0124697. doi: 10.1371/journal.pone.0124697
144. Li N, Ren A, Wang X, Fan X, Zhao Y, Gao GF, et al. Influenza viral neuraminidase primes bacterial coinfection through TGF-beta-mediated expression of host cell receptors. Proc Natl Acad Sci USA. (2015) 112:238–43. doi: 10.1073/pnas.1414422112
145. Kolls JK, McCray PB Jr, Chan YR. Cytokine-mediated regulation of antimicrobial proteins. Nat Rev Immunol. (2008) 8:829–35. doi: 10.1038/nri2433
146. Magrone T, Russo MA, Jirillo E. Antimicrobial peptides: phylogenic sources and biological activities. First of two parts. Curr Pharm Des. (2018) 24:1043–53. doi: 10.2174/1381612824666180403123736
147. Ngo VL, Abo H, Kuczma M, Szurek E, Moore N, Medina-Contreras O, et al. IL-36R signaling integrates innate and adaptive immune-mediated protection against enteropathogenic bacteria. Proc Natl Acad Sci USA. (2020) 117:27540–8. doi: 10.1073/pnas.2004484117
148. Alecu M, Coman G, Musetescu A, Coman OA. Antimicrobial peptides as an argument for the involvement of innate immunity in psoriasis (review). Exp Ther Med. (2020) 20:192. doi: 10.3892/etm.2020.9322
149. Guryanova SV, Ovchinnikova TV. Immunomodulatory and allergenic properties of antimicrobial peptides. Int J Mol Sci. (2022) 23:2499. doi: 10.3390/ijms23052499
150. Rani L, Arora A, Majhi S, Mishra A, Mallajosyula SS. Experimental and simulation studies reveal mechanism of action of human defensin derivatives. Biochim Biophys Acta Biomembr. (2022) 1864:183824. doi: 10.1016/j.bbamem.2021.183824
151. Tu J, Li D, Li Q, Zhang L, Zhu Q, Gaur U, et al. Molecular evolutionary analysis of beta-defensin peptides in vertebrates. Evol Bioinform Online. (2015) 11:105–14. doi: 10.4137/EBO.S25580
152. De Smet K, Contreras R. Human antimicrobial peptides: defensins, cathelicidins and histatins. Biotechnol Lett. (2005) 27:1337–47. doi: 10.1007/s10529-005-0936-5
153. Wiesner J, Vilcinskas A. Antimicrobial peptides: the ancient arm of the human immune system. Virulence. (2010) 1:440–64. doi: 10.4161/viru.1.5.12983
154. Wattanarat O, Nirunsittirat A, Piwat S, Manmontri C, Teanpaisan R, Pahumunto N, et al. Significant elevation of salivary human neutrophil peptides 1-3 levels by probiotic milk in preschool children with severe early childhood caries: a randomized controlled trial. Clin Oral Investig. (2021) 25:2891–903. doi: 10.1007/s00784-020-03606-9
155. Ryan LK, Diamond G. Modulation of human beta-defensin-1 production by viruses. Viruses. (2017) 9:153. doi: 10.3390/v9060153
156. Mathews M, Jia HP, Guthmiller JM, Losh G, Graham S, Johnson GK, et al. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. (1999) 67:2740–5. doi: 10.1128/IAI.67.6.2740-2745.1999
157. Bals R, Wang X, Meegalla RL, Wattler S, Weiner DJ, Nehls MC, et al. Mouse beta-defensin 3 is an inducible antimicrobial peptide expressed in the epithelia of multiple organs. Infect Immun. (1999) 67:3542–7. doi: 10.1128/IAI.67.7.3542-3547.1999
158. Christmann C, Zenker S, Martens L, Hubner J, Loser K, Vogl T, et al. Interleukin 17 promotes expression of alarmins S100A8 and S100A9 during the inflammatory response of keratinocytes. Front Immunol. (2020) 11:599947. doi: 10.3389/fimmu.2020.599947
159. Johnstone KF, Wei Y, Bittner-Eddy PD, Vreeman GW, Stone IA, Clayton JB, et al. Calprotectin (S100A8/A9) is an innate immune effector in experimental periodontitis. Infect Immun. (2021) 89:e0012221. doi: 10.1128/IAI.00122-21
160. Okutomi T, Tanaka T, Yui S, Mikami M, Yamazaki M, Abe S, et al. Anti-Candida activity of calprotectin in combination with neutrophils or lactoferrin. Microbiol Immunol. (1998) 42:789–93. doi: 10.1111/j.1348-0421.1998.tb02353.x
161. Cui D, Lyu J, Li H, Lei L, Bian T, Li L, et al. Human beta-defensin 3 inhibits periodontitis development by suppressing inflammatory responses in macrophages. Mol Immunol. (2017) 91:65–74. doi: 10.1016/j.molimm.2017.08.012
162. Cuida M, Brun JG, Tynning T, Jonsson R. Calprotectin levels in oral fluids: the importance of collection site. Eur J Oral Sci. (1995) 103:8–10. doi: 10.1111/j.1600-0722.1995.tb00003.x
163. Kim HD, Karna S, Shin Y, Vu H, Cho HJ, Kim S. S100A8 and S100A9 in saliva, blood and gingival crevicular fluid for screening established periodontitis: a cross-sectional study. BMC Oral Health. (2021) 21:388. doi: 10.1186/s12903-021-01749-z
164. Kido J, Nakamura T, Kido R, Ohishi K, Yamauchi N, Kataoka M, et al. Calprotectin, a leukocyte protein related to inflammation, in gingival crevicular fluid. J Periodontal Res. (1998) 33:434–7. doi: 10.1111/j.1600-0765.1998.tb02340.x
165. Kaner D, Bernimoulin JP, Dietrich T, Kleber BM, Friedmann A. Calprotectin levels in gingival crevicular fluid predict disease activity in patients treated for generalized aggressive periodontitis. J Periodontal Res. (2011) 46:417–26. doi: 10.1111/j.1600-0765.2011.01355.x
166. Farina R, Guarnelli ME, Figuero E, Herrera D, Sanz M, Trombelli L. Microbiological profile and calprotectin expression in naturally occurring and experimentally induced gingivitis. Clin Oral Investig. (2012) 16:1475–84. doi: 10.1007/s00784-011-0643-7
167. Gao H, Xu J, He L, Meng H, Hou J. Calprotectin levels in gingival crevicular fluid and serum of patients with chronic periodontitis and type 2 diabetes mellitus before and after initial periodontal therapy. J Periodontal Res. (2021) 56:121–30. doi: 10.1111/jre.12800
168. Rammes A, Roth J, Goebeler M, Klempt M, Hartmann M, Sorg C. Myeloid-related protein (MRP) 8 and MRP14, calcium-binding proteins of the S100 family, are secreted by activated monocytes via a novel, tubulin-dependent pathway. J Biol Chem. (1997) 272:9496–502. doi: 10.1074/jbc.272.14.9496
169. Sroussi HY, Kohler GA, Agabian N, Villines D, Palefsky JM. Substitution of methionine 63 or 83 in S100A9 and cysteine 42 in S100A8 abrogate the antifungal activities of S100A8/A9: potential role for oxidative regulation. FEMS Immunol Med Microbiol. (2009) 55:55–61. doi: 10.1111/j.1574-695X.2008.00498.x
170. Edwards TS, Dickerhof N, Magon NJ, Paton LN, Sly PD, Kettle AJ. Formation of calprotectin-derived peptides in the airways of children with cystic fibrosis. J Immunol. (2022) 208:979–90. doi: 10.4049/jimmunol.2001017
171. Ma L, Sun P, Zhang JC, Zhang Q, Yao SL. Proinflammatory effects of S100A8/A9 via TLR4 and RAGE signaling pathways in BV-2 microglial cells. Int J Mol Med. (2017) 40:31–8. doi: 10.3892/ijmm.2017.2987
172. Cheng P, Corzo CA, Luetteke N, Yu B, Nagaraj S, Bui MM, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. (2008) 205:2235–49. doi: 10.1084/jem.20080132
173. Huang M, Wu R, Chen L, Peng Q, Li S, Zhang Y, et al. S100A9 regulates MDSCs-mediated immune suppression via the RAGE and TLR4 signaling pathways in colorectal carcinoma. Front Immunol. (2019) 10:2243. doi: 10.3389/fimmu.2019.02243
174. Vogl T, Tenbrock K, Ludwig S, Leukert N, Ehrhardt C, van Zoelen MA, et al. Mrp8 and Mrp14 are endogenous activators of toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. (2007) 13:1042–9. doi: 10.1038/nm1638
175. Tsai SY, Segovia JA, Chang TH, Morris IR, Berton MT, Tessier PA, et al. DAMP molecule S100A9 acts as a molecular pattern to enhance inflammation during influenza A virus infection: role of DDX21-TRIF-TLR4-MyD88 pathway. PLoS Pathog. (2014) 10:e1003848. doi: 10.1371/journal.ppat.1003848
176. Loser K, Vogl T, Voskort M, Lueken A, Kupas V, Nacken W, et al. The toll-like receptor 4 ligands Mrp8 and Mrp14 are crucial in the development of autoreactive CD8+ T cells. Nat Med. (2010) 16:713–7. doi: 10.1038/nm.2150
177. Nishikawa Y, Kajiura Y, Lew JH, Kido JI, Nagata T, Naruishi K. Calprotectin induces IL-6 and MCP-1 production via toll-like receptor 4 signaling in human gingival fibroblasts. J Cell Physiol. (2017) 232:1862–71. doi: 10.1002/jcp.25724
178. Tatsiy O, de Carvalho Oliveira V, Mosha HT, McDonald PP. Early and late processes driving NET formation, and the autocrine/paracrine role of endogenous RAGE ligands. Front Immunol. (2021) 12:675315. doi: 10.3389/fimmu.2021.675315
179. Wang Y, Gao H, Kessinger CW, Schmaier A, Jaffer FA, Simon DI. Myeloid-related protein-14 regulates deep vein thrombosis. JCI Insight. (2017) 2:e91356. doi: 10.1172/jci.insight.91356
180. Boyd JH, Kan B, Roberts H, Wang Y, Walley KR. S100A8 and S100A9 mediate endotoxin-induced cardiomyocyte dysfunction via the receptor for advanced glycation end products. Circ Res. (2008) 102:1239–46. doi: 10.1161/CIRCRESAHA.107.167544
181. Gebhardt C, Riehl A, Durchdewald M, Nemeth J, Furstenberger G, Muller-Decker K, et al. RAGE signaling sustains inflammation and promotes tumor development. J Exp Med. (2008) 205:275–85. doi: 10.1084/jem.20070679
182. Narumi K, Miyakawa R, Ueda R, Hashimoto H, Yamamoto Y, Yoshida T, et al. Proinflammatory proteins S100A8/S100A9 activate NK cells via interaction with RAGE. J Immunol. (2015) 194:5539–48. doi: 10.4049/jimmunol.1402301
183. Ryckman C, Vandal K, Rouleau P, Talbot M, Tessier PA. Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J Immunol. (2003) 170:3233–42. doi: 10.4049/jimmunol.170.6.3233
184. Kerkhoff C, Sorg C, Tandon NN, Nacken W. Interaction of S100A8/S100A9-arachidonic acid complexes with the scavenger receptor CD36 may facilitate fatty acid uptake by endothelial cells. Biochemistry. (2001) 40:241–8. doi: 10.1021/bi001791k
185. Wang Y, Fang C, Gao H, Bilodeau ML, Zhang Z, Croce K, et al. Platelet-derived S100 family member myeloid-related protein-14 regulates thrombosis. J Clin Invest. (2014) 124:2160–71. doi: 10.1172/JCI70966
186. Lin CR, Wei TY, Tsai HY, Wu YT, Wu PY, Chen ST. Glycosylation-dependent interaction between CD69 and S100A8/S100A9 complex is required for regulatory T-cell differentiation. FASEB J. (2015) 29:5006–17. doi: 10.1096/fj.15-273987
187. Tsai SY, Segovia JA, Chang TH, Shil NK, Pokharel SM, Kannan TR, et al. Regulation of TLR3 activation by S100A9. J Immunol. (2015) 195:4426–37. doi: 10.4049/jimmunol.1500378
188. Moles A, Murphy L, Wilson CL, Chakraborty JB, Fox C, Park EJ, et al. A TLR2/S100A9/CXCL-2 signaling network is necessary for neutrophil recruitment in acute and chronic liver injury in the mouse. J Hepatol. (2014) 60:782–91. doi: 10.1016/j.jhep.2013.12.005
189. Silvers R, Stephan JR, Griffin RG, Nolan EM. Molecular basis of Ca(II)-induced tetramerization and transition-metal sequestration in human calprotectin. J Am Chem Soc. (2021) 143:18073–90. doi: 10.1021/jacs.1c06402
190. Rosen T, Wang KA, Nolan EM. Metal sequestration by S100 proteins in chemically diverse environments. Trends Microbiol. (2022) 30:654–64. doi: 10.1016/j.tim.2021.12.006
191. Sprenkeler EGG, Zandstra J, van Kleef ND, Goetschalckx I, Verstegen B, Aarts CEM, et al. S100A8/A9 is a marker for the release of neutrophil extracellular traps and induces neutrophil activation. Cells. (2022) 11:236. doi: 10.3390/cells11020236
192. von Kockritz-Blickwede M, Winstel V. Molecular prerequisites for neutrophil extracellular trap formation and evasion mechanisms of Staphylococcus aureus. Front Immunol. (2022) 13:836278. doi: 10.3389/fimmu.2022.836278
193. Zackular JP, Chazin WJ, Skaar EP. Nutritional immunity: S100 proteins at the host-pathogen interface. J Biol Chem. (2015) 290:18991–8. doi: 10.1074/jbc.R115.645085
194. Nelson CE, Huang W, Zygiel EM, Nolan EM, Kane MA, Oglesby AG. The human innate immune protein calprotectin elicits a multimetal starvation response in Pseudomonas aeruginosa. Microbiol Spectr. (2021) 9:e0051921. doi: 10.1128/Spectrum.00519-21
195. Champaiboon C, Sappington KJ, Guenther BD, Ross KF, Herzberg MC. Calprotectin S100A9 calcium-binding loops I and II are essential for keratinocyte resistance to bacterial invasion. J Biol Chem. (2009) 284:7078–90. doi: 10.1074/jbc.M806605200
196. Adhikari J, Stephan JR, Rempel DL, Nolan EM, Gross ML. Calcium binding to the innate immune protein human calprotectin revealed by integrated mass spectrometry. J Am Chem Soc. (2020) 142:13372–83. doi: 10.1021/jacs.9b11950
197. Hoskin TS, Crowther JM, Cheung J, Epton MJ, Sly PD, Elder PA, et al. Oxidative cross-linking of calprotectin occurs in vivo, altering its structure and susceptibility to proteolysis. Redox Biol. (2019) 24:101202. doi: 10.1016/j.redox.2019.101202
198. Hadley RC, Gagnon DM, Ozarowski A, Britt RD, Nolan EM. Murine calprotectin coordinates Mn(II) at a hexahistidine site with Ca(II)-dependent affinity. Inorg Chem. (2019) 58:13578–90. doi: 10.1021/acs.inorgchem.9b00763
199. Vogl T, Stratis A, Wixler V, Voller T, Thurainayagam S, Jorch SK, et al. Autoinhibitory regulation of S100A8/S100A9 alarmin activity locally restricts sterile inflammation. J Clin Invest. (2018) 128:1852–66. doi: 10.1172/JCI89867
200. Vitkov L, Minnich B, Knopf J, Schauer C, Hannig M, Herrmann M. NETs are double-edged swords with the potential to aggravate or resolve periodontal inflammation. Cells. (2020) 9:2614. doi: 10.3390/cells9122614
201. Estrada Brull A, Panetti C, Joller N. Moving to the outskirts: interplay between regulatory T cells and peripheral tissues. Front Immunol. (2022) 13:864628. doi: 10.3389/fimmu.2022.864628
202. Gaboriaud C, Lorvellec M, Rossi V, Dumestre-Perard C, Thielens NM. Complement system and alarmin HMGB1 crosstalk: for better or worse. Front Immunol. (2022) 13:869720. doi: 10.3389/fimmu.2022.869720
203. Silberberg E, Filep JG, Ariel A. Weathering the storm: harnessing the resolution of inflammation to limit COVID-19 pathogenesis. Front Immunol. (2022) 13:863449. doi: 10.3389/fimmu.2022.863449
204. Kehl-Fie TE, Zhang Y, Moore JL, Farrand AJ, Hood MI, Rathi S, et al. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun. (2013) 81:3395–405. doi: 10.1128/IAI.00420-13
205. Diaz-Ochoa VE, Lam D, Lee CS, Klaus S, Behnsen J, Liu JZ, et al. Salmonella mitigates oxidative stress and thrives in the inflamed gut by evading calprotectin-mediated manganese sequestration. Cell Host Microbe. (2016) 19:814–25. doi: 10.1016/j.chom.2016.05.005
206. Rosen T, Nolan EM. Metal sequestration and antimicrobial activity of human calprotectin are pH-dependent. Biochemistry. (2020) 59:2468–78. doi: 10.1021/acs.biochem.0c00359
207. Kolenbrander PE, Andersen RN, Baker RA, Jenkinson HF. The adhesion-associated sca operon in Streptococcus gordonii encodes an inducible high-affinity ABC transporter for Mn2+ uptake. J Bacteriol. (1998) 180:290–5. doi: 10.1128/JB.180.2.290-295.1998
208. Jakubovics NS, Smith AW, Jenkinson HF. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol Microbiol. (2000) 38:140–53. doi: 10.1046/j.1365-2958.2000.02122.x
209. Paik S, Brown A, Munro CL, Cornelissen CN, Kitten T. The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J Bacteriol. (2003) 185:5967–75. doi: 10.1128/JB.185.20.5967-5975.2003
210. Biswas S, Biswas I. Role of VltAB, an ABC transporter complex, in viologen tolerance in Streptococcus mutans. Antimicrob Agents Chemother. (2011) 55:1460–9. doi: 10.1128/AAC.01094-10
211. Kajfasz JK, Katrak C, Ganguly T, Vargas J, Wright L, Peters ZT, et al. Manganese uptake, mediated by SloABC and MntH, is essential for the fitness of Streptococcus mutans. mSphere. (2020) 5:e00764–19. doi: 10.1128/mSphere.00764-19
212. Dale I, Brandtzaeg P, Fagerhol MK, Scott H. Distribution of a new myelomonocytic antigen (L1) in human peripheral blood leukocytes. Immunofluorescence and immunoperoxidase staining features in comparison with lysozyme and lactoferrin. Am J Clin Pathol. (1985) 84:24–34. doi: 10.1093/ajcp/84.1.24
213. Eversole LR, Miyasaki KT, Christensen RE. The distribution of the antimicrobial protein, calprotectin, in normal oral keratinocytes. Arch Oral Biol. (1992) 37:963–8. doi: 10.1016/0003-9969(92)90068-J
214. Benedyk M, Sopalla C, Nacken W, Bode G, Melkonyan H, Banfi B, et al. HaCaT keratinocytes overexpressing the S100 proteins S100A8 and S100A9 show increased NADPH oxidase and NF-kappaB activities. J Invest Dermatol. (2007) 127:2001–11. doi: 10.1038/sj.jid.5700820
215. Voss A, Bode G, Sopalla C, Benedyk M, Varga G, Bohm M, et al. Expression of S100A8/A9 in HaCaT keratinocytes alters the rate of cell proliferation and differentiation. FEBS Lett. (2011) 585:440–6. doi: 10.1016/j.febslet.2010.12.037
216. Doussiere J, Bouzidi F, Vignais PV. The S100A8/A9 protein as a partner for the cytosolic factors of NADPH oxidase activation in neutrophils. Eur J Biochem. (2002) 269:3246–55. doi: 10.1046/j.1432-1033.2002.03002.x
217. Berthier S, Nguyen MV, Baillet A, Hograindleur MA, Paclet MH, Polack B, et al. Molecular interface of S100A8 with cytochrome b558 and NADPH oxidase activation. PLoS ONE. (2012) 7:e40277. doi: 10.1371/journal.pone.0040277
218. Steinckwich N, Schenten V, Melchior C, Brechard S, Tschirhart EJ. An essential role of STIM1, orai1, and S100A8-A9 proteins for Ca2+ signaling and FcgammaR-mediated phagosomal oxidative activity. J Immunol. (2011) 186:2182–91. doi: 10.4049/jimmunol.1001338
219. Milward MR, Chapple IL, Wright HJ, Millard JL, Matthews JB, Cooper PR. Differential activation of NF-kappaB and gene expression in oral epithelial cells by periodontal pathogens. Clin Exp Immunol. (2007) 148:307–24. doi: 10.1111/j.1365-2249.2007.03342.x
220. Hiroshima Y, Sakamoto E, Yoshida K, Abe K, Naruishi K, Yamamoto T, et al. Advanced glycation end-products and Porphyromonas gingivalis lipopolysaccharide increase calprotectin expression in human gingival epithelial cells. J Cell Biochem. (2018) 119:1591–603. doi: 10.1002/jcb.26319
221. Henke MO, Renner A, Rubin BK, Gyves JI, Lorenz E, Koo JS. Up-regulation of S100A8 and S100A9 protein in bronchial epithelial cells by lipopolysaccharide. Exp Lung Res. (2006) 32:331–47. doi: 10.1080/01902140600959580
222. Gao N, Sang Yoon G, Liu X, Mi X, Chen W, Standiford TJ, et al. Genome-wide transcriptional analysis of differentially expressed genes in flagellin-pretreated mouse corneal epithelial cells in response to Pseudomonas aeruginosa: involvement of S100A8/A9. Mucosal Immunol. (2013) 6:993–1005. doi: 10.1038/mi.2012.137
223. Kido J, Kido R, Suryono, Kataoka M, Fagerhol MK, Nagata T. Calprotectin release from human neutrophils is induced by Porphyromonas gingivalis lipopolysaccharide via the CD-14-Toll-like receptor-nuclear factor kappaB pathway. J Periodontal Res. (2003) 38:557–63. doi: 10.1034/j.1600-0765.2003.00691.x
224. Rajamaki K, Nordstrom T, Nurmi K, Akerman KE, Kovanen PT, Oorni K, et al. Extracellular acidosis is a novel danger signal alerting innate immunity via the NLRP3 inflammasome. J Biol Chem. (2013) 288:13410–9. doi: 10.1074/jbc.M112.426254
225. Robinson MJ, Tessier P, Poulsom R, Hogg N. The S100 family heterodimer, MRP-8/14, binds with high affinity to heparin and heparan sulfate glycosaminoglycans on endothelial cells. J Biol Chem. (2002) 277:3658–65. doi: 10.1074/jbc.M102950200
226. Argyris PP, Slama ZM, Ross KF, Khammanivong A, Herzberg MC. Calprotectin and the initiation and progression of head and neck cancer. J Dent Res. (2018) 97:674–82. doi: 10.1177/0022034518756330
227. Page TH, Chiappo D, Brunini F, Garnica J, Blackburn J, Dudhiya F, et al. Danger-associated molecular pattern molecules and the receptor for advanced glycation end products enhance ANCA-induced responses. Rheumatology. (2022) 61:834–45. doi: 10.1093/rheumatology/keab413
228. Shimada S, Nakamura M, Tanaka Y, Tsutsumi K, Katano M, Masuko K, et al. Crosslinking of the CD69 molecule enhances S100A9 production in activated neutrophils. Microbiol Immunol. (2007) 51:87–98. doi: 10.1111/j.1348-0421.2007.tb03878.x
229. Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. (2007) 7:255–66. doi: 10.1038/nri2056
230. Manitz MP, Horst B, Seeliger S, Strey A, Skryabin BV, Gunzer M, et al. Loss of S100A9 (MRP14) results in reduced interleukin-8-induced CD11b surface expression, a polarized microfilament system, and diminished responsiveness to chemoattractants in vitro. Mol Cell Biol. (2003) 23:1034–43. doi: 10.1128/MCB.23.3.1034-1043.2003
231. Mihaila AC, Ciortan L, Macarie RD, Vadana M, Cecoltan S, Preda MB, et al. Transcriptional profiling and functional analysis of N1/N2 neutrophils reveal an immunomodulatory effect of S100A9-blockade on the pro-inflammatory N1 subpopulation. Front Immunol. (2021) 12:708770. doi: 10.3389/fimmu.2021.708770
232. De Filippo K, Neill DR, Mathies M, Bangert M, McNeill E, Kadioglu A, et al. A new protective role for S100A9 in regulation of neutrophil recruitment during invasive pneumococcal pneumonia. FASEB J. (2014) 28:3600–8. doi: 10.1096/fj.13-247460
233. Willers M, Ulas T, Vollger L, Vogl T, Heinemann AS, Pirr S, et al. S100A8 and S100A9 are important for postnatal development of gut microbiota and immune system in mice and infants. Gastroenterology. (2020) 159:2130–45.e5. doi: 10.1053/j.gastro.2020.08.019
234. Pirr S, Richter M, Fehlhaber B, Pagel J, Hartel C, Roth J, et al. High amounts of S100-alarmins confer antimicrobial activity on human breast milk targeting pathogens relevant in neonatal sepsis. Front Immunol. (2017) 8:1822. doi: 10.3389/fimmu.2017.01822
235. Zhang M, Whiteley M, Lewin GR. Polymicrobial interactions of oral microbiota: a historical review and current perspective. mBio. (2022) 13:e0023522. doi: 10.1128/mbio.00235-22
236. Relvas M, Regueira-Iglesias A, Balsa-Castro C, Salazar F, Pacheco JJ, Cabral C, et al. Relationship between dental and periodontal health status and the salivary microbiome: bacterial diversity, co-occurrence networks and predictive models. Sci Rep. (2021) 11:929. doi: 10.1038/s41598-020-79875-x
237. Qi Y, Wu HM, Yang Z, Zhou YF, Jin L, Yang MF, et al. New insights into the role of oral microbiota dysbiosis in the pathogenesis of inflammatory bowel disease. Dig Dis Sci. (2022) 67:42–55. doi: 10.1007/s10620-021-06837-2
238. Lahiri D, Nag M, Banerjee R, Mukherjee D, Garai S, Sarkar T, et al. Amylases: biofilm inducer or biofilm inhibitor? Front Cell Infect Microbiol. (2021) 11:660048. doi: 10.3389/fcimb.2021.660048
239. Allaker RP, Grosvenor PW, McAnerney DC, Sheehan BE, Srikanta BH, Pell K, et al. Mechanisms of adrenomedullin antimicrobial action. Peptides. (2006) 27:661–6. doi: 10.1016/j.peptides.2005.09.003
240. Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. (2014) 162 (2 Pt. A):22–38. doi: 10.1016/j.imlet.2014.08.017
241. Laputkova G, Schwartzova V, Banovcin J, Alexovic M, Sabo J. Salivary protein roles in oral health and as predictors of caries risk. Open Life Sci. (2018) 13:174–200. doi: 10.1515/biol-2018-0023
242. Chertov O, Michiel DF, Xu L, Wang JM, Tani K, Murphy WJ, et al. Identification of defensin-1, defensin-2, and CAP37/azurocidin as T-cell chemoattractant proteins released from interleukin-8-stimulated neutrophils. J Biol Chem. (1996) 271:2935–40. doi: 10.1074/jbc.271.6.2935
243. Fabian TK, Hermann P, Beck A, Fejerdy P, Fabian G. Salivary defense proteins: their network and role in innate and acquired oral immunity. Int J Mol Sci. (2012) 13:4295–320. doi: 10.3390/ijms13044295
244. Tokajuk J, Deptula P, Piktel E, Daniluk T, Chmielewska S, Wollny T, et al. Cathelicidin LL-37 in health and diseases of the oral cavity. Biomedicines. (2022) 10:1086. doi: 10.3390/biomedicines10051086
245. Hieshima K, Ohtani H, Shibano M, Izawa D, Nakayama T, Kawasaki Y, et al. CCL28 has dual roles in mucosal immunity as a chemokine with broad-spectrum antimicrobial activity. J Immunol. (2003) 170:1452–61. doi: 10.4049/jimmunol.170.3.1452
246. Mohan T, Deng L, Wang BZ. CCL28 chemokine: an anchoring point bridging innate and adaptive immunity. Int Immunopharmacol. (2017) 51:165–70. doi: 10.1016/j.intimp.2017.08.012
247. Amerongen AV, Veerman EC. Saliva–the defender of the oral cavity. Oral Dis. (2002) 8:12–22. doi: 10.1034/j.1601-0825.2002.1o816.x
248. Koshlukova SE, Lloyd TL, Araujo MW, Edgerton M. Salivary histatin 5 induces non-lytic release of ATP from Candida albicans leading to cell death. J Biol Chem. (1999) 274:18872–9. doi: 10.1074/jbc.274.27.18872
249. Helmerhorst EJ, Troxler RF, Oppenheim FG. The human salivary peptide histatin 5 exerts its antifungal activity through the formation of reactive oxygen species. Proc Natl Acad Sci USA. (2001) 98:14637–42. doi: 10.1073/pnas.141366998
250. Sharma P, Chaudhary M, Khanna G, Rishi P, Kaur IP. Envisaging antifungal potential of histatin 5: a physiological salivary peptide. J Fungi. (2021) 7:1070. doi: 10.3390/jof7121070
251. Zarzosa-Moreno D, Avalos-Gomez C, Ramirez-Texcalco LS, Torres-Lopez E, Ramirez-Mondragon R, Hernandez-Ramirez JO, et al. Lactoferrin and its derived peptides: an alternative for combating virulence mechanisms developed by pathogens. Molecules. (2020) 25:5763. doi: 10.3390/molecules25245763
252. Dashper SG, Pan Y, Veith PD, Chen YY, Toh EC, Liu SW, et al. Lactoferrin inhibits Porphyromonas gingivalis proteinases and has sustained biofilm inhibitory activity. Antimicrob Agents Chemother. (2012) 56:1548–56. doi: 10.1128/AAC.05100-11
253. Velliyagounder K, Bahdila D, Pawar S, Fine DH. Role of lactoferrin and lactoferrin-derived peptides in oral and maxillofacial diseases. Oral Dis. (2019) 25:652–69. doi: 10.1111/odi.12868
254. Laible NJ, Germaine GR. Bactericidal activity of human lysozyme, muramidase-inactive lysozyme, and cationic polypeptides against Streptococcus sanguis and Streptococcus faecalis: inhibition by chitin oligosaccharides. Infect Immun. (1985) 48:720–8. doi: 10.1128/iai.48.3.720-728.1985
255. Samaranayake YH, Samaranayake LP, Wu PC, So M. The antifungal effect of lactoferrin and lysozyme on Candida krusei and Candida albicans. APMIS. (1997) 105:875–83. doi: 10.1111/j.1699-0463.1997.tb05097.x
256. Ibrahim HR, Thomas U, Pellegrini A. A helix-loop-helix peptide at the upper lip of the active site cleft of lysozyme confers potent antimicrobial activity with membrane permeabilization action. J Biol Chem. (2001) 276:43767–74. doi: 10.1074/jbc.M106317200
257. Peacocke J, Lotz Z, de Beer C, Roux P, Mall AS. The role of crude saliva and purified salivary mucins in the inhibition of the human immunodeficiency virus type 1. Virol J. (2012) 9:177. doi: 10.1186/1743-422X-9-177
258. El Karim IA, Linden GJ, Orr DF, Lundy FT. Antimicrobial activity of neuropeptides against a range of micro-organisms from skin, oral, respiratory and gastrointestinal tract sites. J Neuroimmunol. (2008) 200:11–6. doi: 10.1016/j.jneuroim.2008.05.014
259. Lundy FT, El Karim IA, Linden GJ. Neuropeptide Y (NPY) and NPY Y1 receptor in periodontal health and disease. Arch Oral Biol. (2009) 54:258–62. doi: 10.1016/j.archoralbio.2008.10.002
260. Ihalin R, Loimaranta V, Tenovuo J. Origin, structure, and biological activities of peroxidases in human saliva. Arch Biochem Biophys. (2006) 445:261–8. doi: 10.1016/j.abb.2005.07.004
261. Madsen J, Mollenhauer J, Holmskov U. Review: Gp-340/DMBT1 in mucosal innate immunity. Innate Immun. (2010) 16:160–7. doi: 10.1177/1753425910368447
262. Boks MA, Gunput ST, Kosten I, Gibbs S, van Vliet SJ, Ligtenberg AJ, et al. The human glycoprotein salivary agglutinin inhibits the interaction of DC-SIGN and langerin with oral micro-organisms. J Innate Immun. (2016) 8:350–61. doi: 10.1159/000443016
263. Leito JT, Ligtenberg AJ, van Houdt M, van den Berg TK, Wouters D. The bacteria binding glycoprotein salivary agglutinin (SAG/gp340) activates complement via the lectin pathway. Mol Immunol. (2011) 49:185–90. doi: 10.1016/j.molimm.2011.08.010
264. McNeely TB, Dealy M, Dripps DJ, Orenstein JM, Eisenberg SP, Wahl SM. Secretory leukocyte protease inhibitor: a human saliva protein exhibiting anti-human immunodeficiency virus 1 activity in vitro. J Clin Invest. (1995) 96:456–64. doi: 10.1172/JCI118056
265. Williams SE, Brown TI, Roghanian A, Sallenave JM. SLPI and elafin: one glove, many fingers. Clin Sci. (2006) 110:21–35. doi: 10.1042/CS20050115
266. Jana NK, Gray LR, Shugars DC. Human immunodeficiency virus type 1 stimulates the expression and production of secretory leukocyte protease inhibitor (SLPI) in oral epithelial cells: a role for SLPI in innate mucosal immunity. J Virol. (2005) 79:6432–40. doi: 10.1128/JVI.79.10.6432-6440.2005
267. Grant M, Kilsgard O, Akerman S, Klinge B, Demmer RT, Malmstrom J, et al. The human salivary antimicrobial peptide profile according to the oral microbiota in health, periodontitis and smoking. J Innate Immun. (2019) 11:432–44. doi: 10.1159/000494146
268. Wu L, Li F, Ran L, Gao Y, Xie P, Yang J, et al. Insight into the effects of nisin and cecropin on the oral microbial community of rats by high-throughput sequencing. Front Microbiol. (2020) 11:1082. doi: 10.3389/fmicb.2020.01082
269. Gaddy JA, Radin JN, Cullen TW, Chazin WJ, Skaar EP, Trent MS, et al. Helicobacter pylori resists the antimicrobial activity of calprotectin via lipid a modification and associated biofilm formation. mBio. (2015) 6:e01349–15. doi: 10.1128/mBio.01349-15
270. Jean S, Juneau RA, Criss AK, Cornelissen CN. Neisseria gonorrhoeae evades calprotectin-mediated nutritional immunity and survives neutrophil extracellular traps by production of TdfH. Infect Immun. (2016) 84:2982–94. doi: 10.1128/IAI.00319-16
271. Lonergan ZR, Nairn BL, Wang J, Hsu YP, Hesse LE, Beavers WN, et al. An acinetobacter baumannii, zinc-regulated peptidase maintains cell wall integrity during immune-mediated nutrient sequestration. Cell Rep. (2019) 26:2009–18.e6. doi: 10.1016/j.celrep.2019.01.089
272. Hesse LE, Lonergan ZR, Beavers WN, Skaar EP. The acinetobacter baumannii znu system overcomes host-imposed nutrient zinc limitation. Infect Immun. (2019) 87:e00746–19. doi: 10.1128/IAI.00746-19
273. Zackular JP, Knippel RJ, Lopez CA, Beavers WN, Maxwell CN, Chazin WJ, et al. ZupT facilitates clostridioides difficile resistance to host-mediated nutritional immunity. mSphere. (2020) 5:e00061–20. doi: 10.1128/mSphere.00061-20
274. Mortensen BL, Rathi S, Chazin WJ, Skaar EP. Acinetobacter baumannii response to host-mediated zinc limitation requires the transcriptional regulator zur. J Bacteriol. (2014) 196:2616–26. doi: 10.1128/JB.01650-14
275. Galarde-Lopez M, Velazquez-Meza ME, Bobadilla-Del-Valle M, Cornejo-Juarez P, Carrillo-Quiroz BA, Ponce-de-Leon A, et al. Antimicrobial resistance patterns and clonal distribution of E. coli, Enterobacter spp. and Acinetobacter spp. Strains isolated from two hospital wastewater plants. Antibiotics. (2022) 11:601. doi: 10.3390/antibiotics11050601
276. Marciano DC, Wang C, Hsu TK, Bourquard T, Atri B, Nehring RB, et al. Evolutionary action of mutations reveals antimicrobial resistance genes in Escherichia coli. Nat Commun. (2022) 13:3189. doi: 10.1038/s41467-022-30889-1
277. Wozniak TM, Cuningham W, Ledingham K, McCulloch K. Contribution of socio-economic factors in the spread of antimicrobial resistant infections in Australian primary healthcare clinics. J Glob Antimicrob Resist. [Preprint]. (2022). doi: 10.1016/j.jgar.2022.06.005.
Keywords: epithelium, antimicrobial peptides/proteins, calprotectin, defensins, LL-37, health, disease
Citation: Johnstone KF and Herzberg MC (2022) Antimicrobial peptides: Defending the mucosal epithelial barrier. Front. Oral. Health 3:958480. doi: 10.3389/froh.2022.958480
Received: 31 May 2022; Accepted: 30 June 2022;
Published: 01 August 2022.
Edited by:
Octavio Alberto Gonzalez, University of Kentucky, United StatesReviewed by:
Ronaldo Lira-Junior, Karolinska Institutet (KI), SwedenGill Diamond, University of Louisville, United States
Robert Allaker, Queen Mary University of London, United Kingdom
Copyright © 2022 Johnstone and Herzberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark C. Herzberg, mcherzb@umn.edu
 Karen F. Johnstone
Karen F. Johnstone  Mark C. Herzberg
Mark C. Herzberg