Assigning the Origin of Microbial Natural Products by Chemical Space Map and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. NPAtlas Dataset
2.2. MAP4 Fingerprint
2.3. TMAP Layout
2.4. Properties Calculation
2.5. TMAP Color Gradients
2.6. Support Vector Machine (SVM) and k-Nearest Neighbor (k-NN) Classifiers
2.7. Classifiers Evaluation Metrics
3. Results and Discussion
3.1. The TMAP of NPAtlas
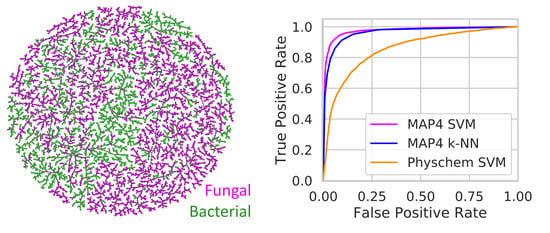
3.2. Distinguishing Between Bacterial and Fungal NPs
3.3. Predicting the Origin of Newly Discovered NPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A Review of the Microbial Production of Bioactive Natural Products and Biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; de Bruyn Kops, C.; Kirchmair, J. Data Resources for the Computer-Guided Discovery of Bioactive Natural Products. J. Chem. Inf. Model. 2017, 57, 2099–2111. [Google Scholar] [CrossRef] [PubMed]
- Osada, H.; Nogawa, T. Systematic isolation of microbial metabolites for natural products depository (NPDepo). Pure Appl. Chem. 2011, 84, 1407–1420. [Google Scholar] [CrossRef]
- Grabowski, K.; Baringhaus, K.-H.; Schneider, G. Scaffold diversity of natural products: Inspiration for combinatorial library design. Nat. Prod. Rep. 2008, 25, 892–904. [Google Scholar] [CrossRef] [PubMed]
- Grisoni, F.; Merk, D.; Consonni, V.; Hiss, J.A.; Tagliabue, S.G.; Todeschini, R.; Schneider, G. Scaffold hopping from natural products to synthetic mimetics by holistic molecular similarity. Commun. Chem. 2018, 1, 1–9. [Google Scholar] [CrossRef]
- Fraser, L.-A.; Mulholland, D.A.; Fraser, D.D. Classification of limonoids and protolimonoids using neural networks. Phytochem. Anal. 1997, 8, 301–311. [Google Scholar] [CrossRef]
- Martínez-Treviño, S.H.; Uc-Cetina, V.; Fernández-Herrera, M.A.; Merino, G. Prediction of Natural Product Classes Using Machine Learning and 13C NMR Spectroscopic Data. J. Chem. Inf. Model. 2020, 7, 3376–3386. [Google Scholar] [CrossRef]
- Rupp, M.; Bauer, M.R.; Wilcken, R.; Lange, A.; Reutlinger, M.; Boeckler, F.M.; Schneider, G. Machine Learning Estimates of Natural Product Conformational Energies. PLoS Comput. Biol. 2014, 10, e1003400. [Google Scholar] [CrossRef]
- Chen, Y.; Stork, C.; Hirte, S.; Kirchmair, J. NP-Scout: Machine Learning Approach for the Quantification and Visualization of the Natural Product-Likeness of Small Molecules. Biomolecules 2019, 9, 43. [Google Scholar] [CrossRef] [Green Version]
- Rupp, M.; Schroeter, T.; Steri, R.; Zettl, H.; Proschak, E.; Hansen, K.; Rau, O.; Schwarz, O.; Müller-Kuhrt, L.; Schubert-Zsilavecz, M.; et al. From Machine Learning to Natural Product Derivatives that Selectively Activate Transcription Factor PPARγ. Chem. Med. Chem. 2010, 5, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Awale, M.; Sirockin, F.; Stiefl, N.; Reymond, J.-L. Drug Analogs from Fragment-Based Long Short-Term Memory Generative Neural Networks. J. Chem. Inf. Model. 2019, 59, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jafari, M.; Tang, Y.; Tang, J. Predicting Meridian in Chinese traditional medicine using machine learning approaches. PLoS Comput. Biol. 2019, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Li, X.; Zhang, X.; Qin, H.; Xiao, W. Machine learning approaches for elucidating the biological effects of natural products. Nat. Prod. Rep. 2020. [Google Scholar] [CrossRef] [PubMed]
- Van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dice, L.R. Measures of the Amount of Ecologic Association between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Rogers, D.; Hahn, M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. [Google Scholar] [CrossRef]
- Capecchi, A.; Probst, D.; Reymond, J.-L. One molecular fingerprint to rule them all: Drugs, biomolecules, and the metabolome. J. Cheminform. 2020, 12, 43. [Google Scholar] [CrossRef]
- Carhart, R.E.; Smith, D.H.; Venkataraghavan, R. Atom pairs as molecular features in structure-activity studies: Definition and applications. J. Chem. Inf. Comput. Sci. 1985, 25, 64–73. [Google Scholar] [CrossRef]
- Jin, X.; Awale, M.; Zasso, M.; Kostro, D.; Patiny, L.; Reymond, J.L. PDB-Explorer: A web-based interactive map of the protein data bank in shape space. BMC Bioinform. 2015, 16, 339. [Google Scholar] [CrossRef] [Green Version]
- Di Bonaventura, I.; Jin, X.; Visini, R.; Probst, D.; Javor, S.; Gan, B.H.; Michaud, G.; Natalello, A.; Doglia, S.M.; Kohler, T.; et al. Chemical space guided discovery of antimicrobial bridged bicyclic peptides against Pseudomonas aeruginosa and its biofilms. Chem. Sci. 2017, 8, 6784–6798. [Google Scholar] [CrossRef] [Green Version]
- Capecchi, A.; Awale, M.; Probst, D.; Reymond, J.-L. PubChem and ChEMBL beyond Lipinski. Mol. Inform. 2019. [Google Scholar] [CrossRef] [PubMed]
- Capecchi, A.; Zhang, A.; Reymond, J.-L. Populating Chemical Space with Peptides Using a Genetic Algorithm. J. Chem. Inf. Model. 2020, 60, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Probst, D.; Reymond, J.-L. A probabilistic molecular fingerprint for big data settings. J. Cheminform. 2018, 10, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Probst, D.; Reymond, J.-L. Visualization of very large high-dimensional data sets as minimum spanning trees. J. Cheminform. 2020, 12, 12. [Google Scholar] [CrossRef] [Green Version]
- Van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Schneider, N.; Sayle, R.A.; Landrum, G.A. Get Your Atoms in Order—An Open-Source Implementation of a Novel and Robust Molecular Canonicalization Algorithm. J. Chem. Inf. Model. 2015, 55, 2111–2120. [Google Scholar] [CrossRef]
- RDKit. Available online: https://www.rdkit.org/ (accessed on 25 September 2018).
- Dang, Q.H. Secure Hash Standard; National Institute of Standards and Technology: Gaithersburg, MD, USA, 2015. [Google Scholar]
- Broder, A.Z.; Charikar, M.; Frieze, A.M.; Mitzenmacher, M. Min-wise Independent Permutations. J. Comput. Syst. Sci. 1998, 60, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Bawa, M.; Condie, T.; Ganesan, P. LSH forest: Self-tuning indexes for similarity search. In Proceedings of the 14th international conference on World Wide Web, Chiba, Japan, 10–14 May 2005; Association for Computing Machinery: New York, NY, USA, 2005; pp. 651–660. [Google Scholar]
- Kruskal, J.B. On the shortest spanning subtree of a graph and the traveling salesman problem. Proc. Am. Math. Soc. 1956, 7, 48–50. [Google Scholar] [CrossRef]
- Probst, D.; Reymond, J.-L.; Wren, J. FUn: A framework for interactive visualizations of large, high-dimensional datasets on the web. Bioinformatics 2018, 34, 1433–1435. [Google Scholar] [CrossRef] [Green Version]
- Wildman, S.A.; Crippen, G.M. Prediction of Physicochemical Parameters by Atomic Contributions. J. Chem. Inf. Comput. Sci. 1999, 39, 868–873. [Google Scholar] [CrossRef]
- Shi, C.; Borchardt, T.B. JRgui: A Python Program of Joback and Reid Method. ACS Omega 2017, 2, 8682–8688. [Google Scholar] [CrossRef] [PubMed]
- Joback, K.G.; Reid, R.C. Estimation of Pure-Component Properties from Group-Contributions. Chem. Eng. Commun. 1987, 57, 233–243. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Daylight. Available online: https://www.daylight.com/ (accessed on 17 July 2020).
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [Green Version]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Platt, J.C. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. In Advances in Large Margin Classifiers; MIT Press: Cambridge, MA, USA, 1999; pp. 61–74. [Google Scholar]
- Vert, J.P.; Tsuda, K.; Schölkopf, B. A Primer on Kernel Methods: In Kernel Methods in Computational Biology; Biologische Kybernetik: Cambridge, MA, USA, 2004; pp. 35–70. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Gallegos, D.A.; Saurí, J.; Cohen, R.D.; Wan, X.; Videau, P.; Vallota-Eastman, A.O.; Shaala, L.A.; Youssef, D.T.A.; Williamson, R.T.; Martin, G.E.; et al. Jizanpeptins, Cyanobacterial Protease Inhibitors from a Symploca sp. Cyanobacterium Collected in the Red Sea. J. Nat. Prod. 2018, 81, 1417–1425. [Google Scholar] [CrossRef]
- Mao, X.-M.; Xu, W.; Li, D.; Yin, W.-B.; Chooi, Y.-H.; Li, Y.-Q.; Tang, Y.; Hu, Y. Epigenetic Genome Mining of an Endophytic Fungus Leads to the Pleiotropic Biosynthesis of Natural Products. Angew. Chem. Int. Ed. 2015, 54, 7592–7596. [Google Scholar] [CrossRef] [Green Version]
- Dion, H.W.; Woo, P.W.K.; Willmer, N.E.; Kern, D.L.; Onaga, J.; Fusari, S.A. Butirosin, a New Aminoglycosidic Antibiotic Complex: Isolation and Characterization. Antimicrob. Agents Chemother. 1972, 2, 84–88. [Google Scholar] [CrossRef] [Green Version]
- Tatsuda, D.; Momose, I.; Someno, T.; Sawa, R.; Kubota, Y.; Iijima, M.; Kunisada, T.; Watanabe, T.; Shibasaki, M.; Nomoto, A. Quinofuracins A–E, Produced by the Fungus Staphylotrichum boninense PF1444, Show p53-Dependent Growth Suppression. J. Nat. Prod. 2015, 78, 188–195. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, S.; Liu, H.; Liu, X.; Che, Y. Cycloaspeptides F and G, Cyclic Pentapeptides from a Cordyceps-Colonizing Isolate of Isaria farinosa. J. Nat. Prod. 2009, 72, 1364–1367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji, N.; Kobayashi, M.; Kamigauchi, T.; Yoshimura, Y.; Terui, Y. New glycopeptide antibiotics. I. The structures of orienticins. J. Antibiot. 1988, 41, 819–822. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.C.; Hwang, E.; Kim, T.; Ham, J.; Kim, S.Y.; Kwon, H.C. Nocatriones A and B, Photoprotective Tetracenediones from a Marine-Derived Nocardiopsis sp. J. Nat. Prod. 2014, 77, 2326–2330. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-B.; Zhou, Y.-H.; Zhu, R.-X.; Chang, W.-Q.; Yuan, H.-Q.; Gao, W.; Zhang, L.-L.; Zhao, Z.-T.; Lou, H.-X. Identification and Biological Evaluation of Secondary Metabolites from the Endolichenic Fungus Aspergillus versicolor. Chem. Biodivers. 2015, 12, 575–592. [Google Scholar] [CrossRef] [PubMed]
- Spyere, A.; Rowley, D.C.; Jensen, P.R.; Fenical, W. New Neoverrucosane Diterpenoids Produced by the Marine Gliding Bacterium Saprospira grandis. J. Nat. Prod. 2003, 66, 818–822. [Google Scholar] [CrossRef]
- Yamamoto, T.; Izumi, N.; Ui, H.; Sueki, A.; Masuma, R.; Nonaka, K.; Hirose, T.; Sunazuka, T.; Nagai, T.; Yamada, H.; et al. Wickerols A and B: Novel anti-influenza virus diterpenes produced by Trichoderma atroviride FKI-3849. Tetrahedron 2012, 68, 9267–9271. [Google Scholar] [CrossRef]
- Mitchell, J.B.O. Machine learning methods in chemoinformatics. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2014, 4, 468–481. [Google Scholar] [CrossRef] [Green Version]
- Lanzoni, O.; Sabaneyeva, E.; Modeo, L.; Castelli, M.; Lebedeva, N.; Verni, F.; Schrallhammer, M.; Potekhin, A.; Petroni, G. Diversity and environmental distribution of the cosmopolitan endosymbiont “Candidatus Megaira”. Sci. Rep. 2019, 9, 1179. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Hou, C.; Yuan, W.; Wang, Z.; Zhang, J.; Jiang, L.; Karthik, L.; Li, B.; Ren, B.; Lv, K.; et al. Molecular networking assisted discovery and biosynthesis elucidation of the antimicrobial spiroketals epicospirocins. Chem. Commun. 2020. [Google Scholar] [CrossRef]
- Cheng, X.; Liang, X.; Zheng, Z.-H.; Zhang, X.-X.; Lu, X.-H.; Yao, F.-H.; Qi, S.-H. Penicimeroterpenoids A–C, Meroterpenoids with Rearrangement Skeletons from the Marine-Derived Fungus Penicillium sp. SCSIO 41512. Org. Lett. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.; Shin, J.; Nam, K.; An, J.S.; Yang, S.-H.; Hong, S.-H.; Bae, M.; Moon, K.; Cho, Y.; Woo, J.; et al. Rhizolutin, a novel 7/10/6-tricyclic dilactone, dissociates misfolded protein aggregates and reduces apoptosis/inflammation associated with Alzheimer’s disease. Angew. Chem. Int. Ed. 2020. [Google Scholar] [CrossRef]
- Xu, Z.F.; Bo, S.T.; Wang, M.J.; Shi, J.; Jiao, R.H.; Sun, Y.; Xu, Q.; Tan, R.; Ge, H.M. Discovery and biosynthesis of bosamycin from Streptomyces sp. 120454. Chem. Sci. 2020. [Google Scholar] [CrossRef]
- Luyen, N.D.; Huong, L.M.; Thi Hong Ha, T.; Cuong, L.H.; Thi Hai Yen, D.; Nhiem, N.X.; Tai, B.H.; Gardes, A.; Kopprio, G.; Van Kiem, P. Aspermicrones A-C, novel dibenzospiroketals from the seaweed-derived endophytic fungus Aspergillus micronesiensis. J. Antibiot. 2019, 72, 843–847. [Google Scholar] [CrossRef]
- Kosemura, S. Meroterpenoids from Penicillium citreo-viride B. IFO 4692 and 6200 hybrid. Tetrahedron 2003, 59, 5055–5072. [Google Scholar] [CrossRef]
- Endo, A. Monacolin K, a new hypocholesterolemic agent that specifically inhibits 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Antibiot. 1980, 33, 334–336. [Google Scholar] [CrossRef] [Green Version]
- Ji, G.; Beavis, R.; Novick, R.P. Bacterial Interference Caused by Autoinducing Peptide Variants. Science 1997, 276, 2027–2030. [Google Scholar] [CrossRef]
- Wu, Y.; Liao, H.; Liu, L.-Y.; Sun, F.; Chen, H.-F.; Jiao, W.-H.; Zhu, H.-R.; Yang, F.; Huang, G.; Zeng, D.-Q.; et al. Phakefustatins A–C: Kynurenine-Bearing Cycloheptapeptides as RXRα Modulators from the Marine Sponge Phakellia fusca. Org. Lett. 2020. [Google Scholar] [CrossRef]
- Naman, C.B.; Rattan, R.; Nikoulina, S.E.; Lee, J.; Miller, B.W.; Moss, N.A.; Armstrong, L.; Boudreau, P.D.; Debonsi, H.M.; Valeriote, F.A.; et al. Integrating Molecular Networking and Biological Assays To Target the Isolation of a Cytotoxic Cyclic Octapeptide, Samoamide A, from an American Samoan Marine Cyanobacterium. J. Nat. Prod. 2017, 80, 625–633. [Google Scholar] [CrossRef]
- Brinkmann, C.M.; Marker, A.; Kurtböke, D.İ. An Overview on Marine Sponge-Symbiotic Bacteria as Unexhausted Sources for Natural Product Discovery. Diversity 2017, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Han, M.; Liu, F.; Zhang, F.; Li, Z.; Lin, H. Bacterial and archaeal symbionts in the South China Sea sponge Phakellia fusca: Community structure, relative abundance, and ammonia-oxidizing populations. Mar. Biotechnol. 2012, 14, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, M.; Steinbeck, C. Review on natural products databases: Where to find data in 2020. J. Cheminform. 2020, 12, 20. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Kirchmair, J. Cheminformatics in Natural Product-Based Drug Discovery. Mol. Inform. 2020. [Google Scholar] [CrossRef] [PubMed]



| Property | Min. Value | Max. Value | 25% Quantile | 50% Quantile | 75% Quantile |
|---|---|---|---|---|---|
| Molecular weight A | 70.1 | 2901.3 (1000 F) | 292 | 408.9 | 562.6 |
| Sp3 C fraction A | 0.0 | 1.0 | 0.4 | 0.6 | 0.7 |
| HBA count A,B | 0 | 68 (20 F) | 4 | 6 | 9 |
| HBD count A,C | 0 | 47 (10 F) | 3 | 2 | 4 |
| AlogP A,D | −28.9 (−2 G) | 33.8 (8 F) | 1.2 | 2.5 | 4.1 |
| TPSA A,E | 0.0 | 1135.81 (500 F) | 69.64 | 99.66 | 152.8 |
| Boiling point A,H | 311.5 | 7806.5 (2000 F) | 890.8 | 1141.6 | 1518.5 |
| Is Lipinski | Categorical: yes/no | ||||
| Substructures I | Categorical: contains dipeptide moiety/contains glycoside moiety/contains dipeptide and glycoside moieties | ||||
| Origin | Categorical: Bacterial/Fungal | ||||
| MAP4 SVM J prediction | Categorical: Bacterial/Fungal | ||||
| MAP4 SVM J performances | Categorical: correct/wrong | ||||
| Fungal A | Bacterial A | |
|---|---|---|
| NPAtlas entries (≥1000 Da) | 15,759 (347) | 9764 (1392) |
| Unique publications B | 6110 (145) | 4653 (711) |
| Peptides (≥1000 Da) C | 722 (311) | 2144 (901) |
| Glycosides (≥1000 Da) D | 814 (12) | 1616 (421) |
| Glycopeptides (≥1000 Da) E | 1 (0) | 112 (89) |
| Aromatic NPs (≥1000 Da) F | 1322 (0) | 800 (31) |
| Aliphatic NPs (≥1000 Da) G | 2184 (59) | 1366 (220) |
| Classifier | ROC AUC A | F1 Score A | Balanced Accuracy A | MCC A |
|---|---|---|---|---|
| MAP4 SVM B | 0.97 | 0.91 | 0.93 | 0.86 |
| MAP4 k-NN C | 0.96 | 0.88 | 0.90 | 0.81 |
| Physchem SVM D | 0.86 | 0.73 | 0.78 | 0.56 |
| Natural Product | MAP4 SVM A Fungal, Bacterial | Training Set Nearest Neighbor (NN) | JD from NN B |
|---|---|---|---|
| Epicospirocin 1 | 0.99, 0.01 | Aspermicrone A (NPA024935) | 0.66 |
| Penicimeroterpenoid A | 1.0, 0.0 | Isocitreohybridone H (NPA016454) | 0.63 |
| Rhizolutin | 0.83, 0.17 | Monacolin K (NPA009354) | 0.80 |
| Bosamycin A | 0.04, 0.96 | AIP I (NPA010987) | 0.77 |
| Phakefustatin A | 0.12, 0.88 | Samoamide A (NPA022212) | 0.68 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capecchi, A.; Reymond, J.-L. Assigning the Origin of Microbial Natural Products by Chemical Space Map and Machine Learning. Biomolecules 2020, 10, 1385. https://doi.org/10.3390/biom10101385
Capecchi A, Reymond J-L. Assigning the Origin of Microbial Natural Products by Chemical Space Map and Machine Learning. Biomolecules. 2020; 10(10):1385. https://doi.org/10.3390/biom10101385
Chicago/Turabian StyleCapecchi, Alice, and Jean-Louis Reymond. 2020. "Assigning the Origin of Microbial Natural Products by Chemical Space Map and Machine Learning" Biomolecules 10, no. 10: 1385. https://doi.org/10.3390/biom10101385