Current Perspectives in Cancer Immunotherapy
Abstract
:1. Introduction
2. Immunotherapeutic Approaches against Cancer
2.1. Cancer Immunotherapy with Immune Checkpoint Blockade
2.1.1. Monoclonal Antibodies against CTLA-4 and the PD-1/PD-L1 Axis
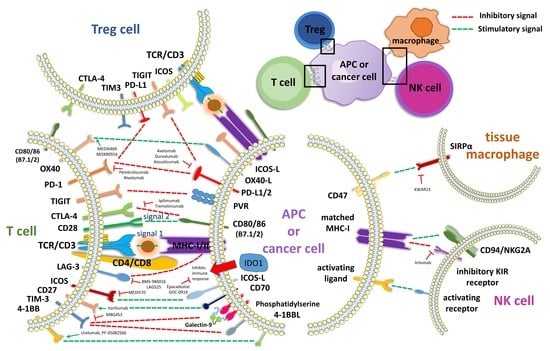
2.1.2. Further Co-Inhibitory and Co-Stimulatory Markers
2.1.3. Adverse Effects Associated with Checkpoint Blockade
2.2. Cancer Immunotherapy with Cytokine Therapy
2.3. Cancer Immunotherapy with T-Cells Redirected against Tumor Antigens
2.3.1. TCR Engineered-T Cell Immunotherapy
2.3.2. CAR Engineered T-Cell Immunotherapy versus T-Cell Activating Polyspecific Antibodies
3. Therapeutic Vaccines
4. Mutational and Epigenetic Landscape in Tumor Progression and Response to Therapy
5. Modulation of Gut Microbiota Composition and Response to Immunotherapy
6. Evolution of the Landscape on Cancer Neoepitopes during Immunotherapy
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Falzone, L.; Salomone, S.; Libra, M. Evolution of Cancer Pharmacological Treatments at the Turn of the Third Millennium. Front. Pharmacol. 2018, 9, 1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamid, O.; Robert, C.; Daud, A.; Hodi, F.S.; Hwu, W.J.; Kefford, R.; Wolchok, J.D.; Hersey, P.; Joseph, R.W.; Weber, J.S.; et al. Safety and Tumor Responses with Lambrolizumab (Anti–PD-1) in Melanoma. N. Engl. J. Med. 2013, 369, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Soria, J.-C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef] [Green Version]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.M.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Tykodi, S.S. PD-1 as an emerging therapeutic target in renal cell carcinoma: Current evidence. Onco Targets Ther. 2014, 7, 1349–1359. [Google Scholar] [CrossRef]
- Kato, Y.; Tabata, K.; Kimura, T.; Yachie-Kinoshita, A.; Ozawa, Y.; Yamada, K.; Ito, J.; Tachino, S.; Hori, Y.; Matsuki, M.; et al. Lenvatinib plus anti-PD-1 antibody combination treatment activates CD8+ T cells through reduction of tumor-associated macrophage and activation of the interferon pathway. PLoS ONE 2019, 14, e0212513. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Saibil, S.D.; Sotov, V.; Le, M.X.; Khoja, L.; Ghazarian, D.; Bonilla, L.; Majeed, H.; Hogg, D.; Joshua, A.M.; et al. Phase II clinical trial of adoptive cell therapy for patients with metastatic melanoma with autologous tumor-infiltrating lymphocytes and low-dose interleukin-2. Cancer Immunol. Immunother. 2019, 68, 773–785. [Google Scholar] [CrossRef] [Green Version]
- Fountain, E.; Bassett, R.L.; Cain, S.; Posada, L.; Gombos, D.S.; Hwu, P.; Bedikian, A.; Patel, S.P. Adjuvant Ipilimumab in High-Risk Uveal Melanoma. Cancers 2019, 11, 152. [Google Scholar] [CrossRef]
- Inozume, T.; Yaguchi, T.; Ariyasu, R.; Togashi, Y.; Ohnuma, T.; Honobe, A.; Nishikawa, H.; Kawakami, Y.K.T. Analysis of the Tumor Reactivity of Tumor-Infiltrating Lymphocytes in a Metastatic Melanoma Lesion that Lost Major Histocompatibility Complex Class I Expression after Anti-PD-1 Therapy. J. Investig. Dermatol. 2019, 139, 1490–1496. [Google Scholar] [CrossRef]
- Sebastian, M.; Schröder, A.; Scheel, B.; Hong, H.S.; Muth, A.; von Boehmer, L.; Zippelius, A.; Mayer, F.; Reck, M.; Atanackovic, D.; et al. A phase I/IIa study of the mRNA-based cancer immunotherapy CV9201 in patients with stage IIIB/IV non-small cell lung cancer. Cancer Immunol. Immunother. 2019, 68, 799–812. [Google Scholar] [CrossRef] [PubMed]
- Pignon, J.C.; Jegede, O.; Shukla, S.A.; Braun, D.A.; Horak, C.E.; Wind-Rotolo, M.; Ishii, Y.; Catalano, P.J.; Grosha, J.; Flaifel, A.; et al. irRECIST for the Evaluation of Candidate Biomarkers of Response to Nivolumab in Metastatic Clear Cell Renal Cell Carcinoma: Analysis of a Phase II Prospective Clinical Trial. Clin. Cancer Res. 2019, 25, 2174–2184. [Google Scholar] [CrossRef] [PubMed]
- Duffy, A.G.; Ulahannan, S.V.; Makorova-Rusher, O.; Rahma, O.; Wedemeyer, H.; Pratt, D.; Davis, J.L.; Hughes, M.S.; Heller, T.; ElGindi, M.; et al. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J. Hepatol. 2017, 66, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; He, Q.; Yu, X.; Khan, K.; Weng, X.; Guan, M. Complete response associated with immune checkpoint inhibitors in advanced non-small-cell lung cancer: A meta-analysis of nine randomized controlled trials. Cancer Manag. Res. 2019, 11, 1623–1629. [Google Scholar] [CrossRef]
- Ning, Y.; Suzman, D.; Maher, V.E.; Zhang, L.; Tang, S.; Ricks, T.; Palmby, T.; Fu, W.; Liu, Q.; Goldberg, K.B.; et al. FDA Approval Summary: Atezolizumab for the Treatment of Patients with Progressive Advanced Urothelial Carcinoma after Platinum-Containing Chemotherapy. Oncologist 2017, 22, 743–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heery, C.R.; O’Sullivan-Coyne, G.; Madan, R.A.; Cordes, L.; Rajan, A.; Rauckhorst, M.; Lamping, E.; Oyelakin, I.; Marté, J.L.; Lepone, L.M.; et al. Avelumab for metastatic or locally advanced previously treated solid tumours (JAVELIN Solid Tumor): A phase 1a, multicohort, dose-escalation trial. Lancet Oncol. 2017, 18, 587–598. [Google Scholar] [CrossRef]
- Syn, N.L.; Teng, M.W.L.; Mok, T.S.K.; Soo, R.A. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017, 18, e731–e741. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, Activity and Immune Correlates of Anti–PD-1 Antibody in Cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef]
- Powles, T.; O’Donnell, P.H.; Massard, C.; Arkenau, H.-T.; Friedlander, T.W.; Hoimes, C.; Lee, J.; Ong, M.; Sridhar, S.S.; Vogelzang, N.J.; et al. Updated efficacy and tolerability of durvalumab in locally advanced or metastatic urothelial carcinoma. J. Clin. Oncol. 2017, 35, 286. [Google Scholar] [CrossRef]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Kurata, T.; Chiappori, A.; Lee, K.H.; De Wit, M.; et al. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N. Engl. J. Med. 2018, 379, 2342–2350. [Google Scholar] [CrossRef]
- Kjeldsen, J.W.; Iversen, T.Z.; Engell-Noerregaard, L.; Mellemgaard, A.; Andersen, M.H.; Svane, I.M. Durable clinical responses and long-term follow-up of stage III-IV non-small-cell lung cancer (NSCLC) patients treated with IDO peptide vaccine in a phase i study-a brief research report. Front. Immunol. 2018, 9, 2145. [Google Scholar] [CrossRef] [PubMed]
- Uyttenhove, C.; Pilotte, L.; Théate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van Den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhou, W.; Zhang, X.; Ding, Y.; Du, Q.; Hu, R. 1-L-MT, an IDO inhibitor, prevented colitis-associated cancer by inducing CDC20 inhibition-mediated mitotic death of colon cancer cells. Int. J. Cancer 2018, 143, 1516–1529. [Google Scholar] [CrossRef]
- Zakharia, Y.; Rixe, O.; Ward, J.H.; Drabick, J.J.; Shaheen, M.F.; Milhem, M.M.; Munn, D.; Kennedy, E.P.; Vahanian, N.N.; Link, C.J.; et al. Phase 2 trial of the IDO pathway inhibitor indoximod plus checkpoint inhibition for the treatment of patients with advanced melanoma. J. Clin. Oncol. 2018, 36, 9512. [Google Scholar] [CrossRef]
- Jha, G.G.; Gupta, S.; Tagawa, S.T.; Koopmeiners, J.S.; Vivek, S.; Dudek, A.Z.; Cooley, S.A.; Blazar, B.R.; Miller, J.S. A phase II randomized, double-blind study of sipuleucel-T followed by IDO pathway inhibitor, indoximod or placebo in the treatment of patients with metastatic castration resistant prostate cancer (mCRPC). J. Clin. Oncol. 2017, 35 (Suppl. S15), 3066. [Google Scholar] [CrossRef]
- Bravo-Alegria, J.; Maegawa, S.; Callegari, K.; Kennis, B.; Ma, W.; Brugmann, W.; Rao, A.; Lee, D.A.; Khatua, S.; Zaky, W.; et al. IMMU-22. Natural Killer Cell Immunotherapy For Diffuse Intrinsic Pontine Glioma. Neuro Oncol. 2018, 20, 103. [Google Scholar] [CrossRef]
- Emadi, A.; Holtzman, N.G.; Imran, M.; El Chaer, F.; Koka, M.; Singh, Z.; Shahlaee, A.; Sausville, E.A.; Law, J.; Lee, S.T.; et al. Indoximod in combination with idarubicin and cytarabine for upfront treatment of patients with newly diagnosed acute myeloid leukemia (AML): Phase 1 report. Haematologica 2017, 102, 375. [Google Scholar]
- Pilotte, L.; Larrieu, P.; Stroobant, V.; Colau, D.; Dolušić, E.; Frédérick, R.; De Plaen, E.; Uyttenhove, C.; Wouters, J.; Masereel, B.; et al. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc. Natl. Acad. Sci. USA 2012, 109, 2497–2502. [Google Scholar] [CrossRef] [Green Version]
- Nayak-Kapoor, A.; Hao, Z.; Sadek, R.; Dobbins, R.; Marshall, L.; Vahanian, N.N.; Jay Ramsey, W.; Kennedy, E.; Mautino, M.R.; Link, C.J.; et al. Phase Ia study of the indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor navoximod (GDC-0919) in patients with recurrent advanced solid tumors. J. Immunother. Cancer 2018, 6, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, K.H.; LoRusso, P.M.; Burris, H.A.; Gordon, M.S.; Bang, Y.-J.; Hellmann, M.D.; Cervantes, A.; de Olza, M.O.; Marabelle, A.; Hodi, F.S.; et al. Phase I Study of the Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Navoximod (GDC-0919) Administered with PD-L1 Inhibitor (Atezolizumab) in Advanced Solid Tumors. Clin. Cancer Res. 2019, 25, 3220–3228. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, T.C.; Hamid, O.; Smith, D.C.; Bauer, T.M.; Wasser, J.S.; Olszanski, A.J.; Luke, J.J.; Balmanoukian, A.S.; Schmidt, E.V.; Zhao, Y.; et al. Epacadostat Plus Pembrolizumab in Patients With Advanced Solid Tumors: Phase I Results From a Multicenter, Open-Label Phase I/II Trial (ECHO-202/KEYNOTE-037). J. Clin. Oncol. 2018, 36, 3223. [Google Scholar] [CrossRef] [PubMed]
- Rexin, P.; Tauchert, A.; Hänze, J.; Heers, H.; Schmidt, A.; Hofmann, R.; Hegele, A. The Immune Checkpoint Molecule CD200 Is Associated with Tumor Grading and Metastasis in Bladder Cancer. Anticancer Res. 2018, 38, 2749–2754. [Google Scholar] [PubMed]
- Burris, H.A.; Infante, J.R.; Ansell, S.M.; Nemunaitis, J.J.; Weiss, G.R.; Villalobos, V.M.; Sikic, B.I.; Taylor, M.H.; Northfelt, D.W.; Carson, W.E.; et al. Safety and activity of varlilumab, a novel and first-in-class agonist anti-CD27 antibody, in patients with advanced solid tumors. J. Clin. Oncol. 2017, 35, 2028–2036. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, V.; Sundarapandiyan, K.; Zhao, B.; Bylesjo, M.; Marsh, H.C.; Keler, T. Characterization of the human T cell response to in vitro CD27 costimulation with varlilumab. J. Immunother. Cancer 2015, 3, 37. [Google Scholar] [CrossRef] [Green Version]
- Yanagita, T.; Murata, Y.; Tanaka, D.; Motegi, S.; Arai, E.; Daniwijaya, E.W.; Hazama, D.; Washio, K.; Saito, Y.; Kotani, T.; et al. Anti-SIRPα antibodies as a potential new tool for cancer immunotherapy. JCI Insight 2017, 2, e89140. [Google Scholar] [CrossRef] [PubMed]
- Ring, N.G.; Herndler-Brandstetter, D.; Weiskopf, K.; Shan, L.; Volkmer, J.-P.; George, B.M.; Lietzenmayer, M.; McKenna, K.M.; Naik, T.J.; McCarty, A.; et al. Anti-SIRPα antibody immunotherapy enhances neutrophil and macrophage antitumor activity. Proc. Natl. Acad. Sci. USA 2017, 114, E10578–E10585. [Google Scholar] [CrossRef]
- Sanmamed, M.F.; Rodriguez, I.; Schalper, K.A.; Oñate, C.; Azpilikueta, A.; Rodriguez-Ruiz, M.E.; Morales-Kastresana, A.; Labiano, S.; Perez-Gracia, J.L.; Martin-Algarra, S.; et al. Nivolumab and urelumab enhance antitumor activity of human T lymphocytes engrafted in Rag2-/-IL2Rγnull immunodeficient mice. Cancer Res. 2015, 75, 3466–3478. [Google Scholar] [CrossRef]
- Segal, N.H.; Logan, T.F.; Hodi, F.S.; McDermott, D.; Melero, I.; Hamid, O.; Schmidt, H.; Robert, C.; Chiarion-Sileni, V.; Ascierto, P.A.; et al. Results from an integrated safety analysis of urelumab, an agonist anti-CD137 monoclonal antibody. Clin. Cancer Res. 2017, 23, 1929–1936. [Google Scholar] [CrossRef]
- Faden, D.L.; Concha-Benavente, F.; Chakka, A.B.; McMichael, E.L.; Chandran, U.; Ferris, R.L. Immunogenomic correlates of response to cetuximab monotherapy in head and neck squamous cell carcinoma. Head Neck 2019, 41, 2591–2601. [Google Scholar] [CrossRef]
- Kohrt, H.E.; Thielens, A.; Marabelle, A.; Sagiv-Barfi, I.; Sola, C.; Chanuc, F.; Fuseri, N.; Bonnafous, C.; Czerwinski, D.; Rajapaksa, A.; et al. Anti-KIR antibody enhancement of anti-lymphoma activity of natural killer cells as monotherapy and in combination with anti-CD20 antibodies. Blood 2014, 123, 678–686. [Google Scholar] [CrossRef] [Green Version]
- Yalniz, F.F.; Daver, N.; Rezvani, K.; Kornblau, S.; Ohanian, M.; Borthakur, G.; DiNardo, C.D.; Konopleva, M.; Burger, J.; Gasior, Y.; et al. A Pilot Trial of Lirilumab With or Without Azacitidine for Patients With Myelodysplastic Syndrome. Clin. Lymphoma Myeloma Leuk. 2018, 18, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Masarova, L.; Patel, K.P.; Newberry, K.J.; Cortes, J.; Borthakur, G.; Konopleva, M.; Estrov, Z.; Kantarjian, H.; Verstovsek, S. Pegylated interferon alfa-2a in patients with essential thrombocythaemia or polycythaemia vera: A post-hoc, median 83 month follow-up of an open-label, phase 2 trial. Lancet Haematol. 2017, 4, e165–e175. [Google Scholar] [CrossRef]
- Atkins, M.B.; Kunkel, L.; Sznol, M.; Rosenberg, S.A. High-dose recombinant interleukin-2 therapy in patients with metastatic melanoma: Long-term survival update. Cancer J. Sci. Am. 2000, 6 (Suppl. S1), S11–S14. [Google Scholar] [PubMed]
- Rosenberg, S.A.; Yang, J.C.; Topalian, S.L.; Schwartzentruber, D.J.; Weber, J.S.; Parkinson, D.R.; Seipp, C.A.; Einhorn, J.H.; White, D.E. Treatment of 283 Consecutive Patients With Metastatic Melanoma or Renal Cell Cancer Using High-Dose Bolus Interleukin 2. JAMA J. Am. Med. Assoc. 1994, 271, 907–913. [Google Scholar] [CrossRef]
- Mocellin, S.; Pasquali, S.; Rossi, C.R.; Nitti, D. Interferon alpha adjuvant therapy in patients with high-risk melanoma: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2010, 102, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Maude, S.L.; Laetsch, T.W.; Buechner, J.; Rives, S.; Boyer, M.; Bittencourt, H.; Bader, P.; Verneris, M.R.; Stefanski, H.E.; Myers, G.D.; et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018, 378, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.J.; Bishop, M.R.; Tam, C.S.; Waller, E.K.; Borchmann, P.; McGuirk, J.P.; Jager, U.; Jaglowski, S.; Andreadis, C.; Westin, J.R.; et al. Tisagenlecleucel in Adult Relapsed or Refractory DLBCL. N. Engl. J. Med. 2019, 380, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Neelapu, S.S.; Locke, F.L.; Bartlett, N.L.; Lekakis, L.J.; Miklos, D.B.; Jacobson, C.A.; Braunschweig, I.; Oluwole, O.O.; Siddiqi, T.; Lin, Y.; et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-Cell lymphoma. N. Engl. J. Med. 2017, 377, 2531–2544. [Google Scholar] [CrossRef] [PubMed]
- Locke, F.L.; Ghobadi, A.; Jacobson, C.A.; Miklos, D.B.; Lekakis, L.J.; Oluwole, O.O.; Lin, Y.; Braunschweig, I.; Hill, B.T.; Timmerman, J.M.; et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): A single-arm, multicentre, phase 1–2 trial. Lancet Oncol. 2019, 20, 31–42. [Google Scholar] [CrossRef]
- You, F.; Jiang, L.; Zhang, B.; Lu, Q.; Zhou, Q.; Liao, X.; Wu, H.; Du, K.; Zhu, Y.; Meng, H.; et al. Phase 1 clinical trial demonstrated that MUC1 positive metastatic seminal vesicle cancer can be effectively eradicated by modified Anti-MUC1 chimeric antigen receptor transduced T cells. Sci. China Life Sci. 2016, 59, 386–397. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.Y.; Yu, K.R.; Kenderian, S.S.; Ruella, M.; Chen, S.; Shin, T.H.; Aljanahi, A.A.; Schreeder, D.; Klichinsky, M.; Shestova, O.; et al. Genetic Inactivation of CD33 in Hematopoietic Stem Cells to Enable CAR T Cell Immunotherapy for Acute Myeloid Leukemia. Cell 2018, 173, 1439–1453. [Google Scholar] [CrossRef]
- Kurokawa, T.; Oelke, M.; Mackensen, A. Induction and clonal expansion of tumor-specific cytotoxic T lymphocytes from renal cell carcinoma patients after stimulation with autologous dendritic cells loaded with tumor cells. Int. J. Cancer 2001, 91, 749–756. [Google Scholar] [CrossRef]
- Carreno, B.M.; Magrini, V.; Becker-Hapak, M.; Kaabinejadian, S.; Hundal, J.; Petti, A.A.; Ly, A.; Lie, W.R.; Hildebrand, W.H.; Mardis, E.R.; et al. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015, 348, 803–808. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Parsonage, G.; Zhuang, X.; Machado, L.R.; James, C.H.; Salman, A.; Searle, P.F.; Hui, E.P.; Chan, A.T.; Lee, S.P. Human leukocyte antigen (HLA) A∗1101-restricted epstein-barr virus-specific t-cell receptorgenetransfer to target nasopharyngeal carcinoma. Cancer Immunol. Res. 2015, 3, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Chemin, I. Evaluation of a hepatitis B vaccination program in Taiwan: Impact on hepatocellular carcinoma development. Future Oncol. 2010, 6, 21–23. [Google Scholar] [CrossRef] [PubMed]
- Mammas, I.N.; Sourvinos, G.; Zaravinos, A.; Spandidos, D.A. Vaccination against human papilloma virus (HPV): Epidemiological evidence of HPV in non-genital cancers. Pathol. Oncol. Res. 2011, 17, 103–119. [Google Scholar] [CrossRef]
- Kreimer, A.R. Prospects for prevention of HPV-driven oropharynx cancer. Oral Oncol. 2014, 50, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Paavonen, J.; Naud, P.; Salmerón, J.; Wheeler, C.M.; Chow, S.N.; Apter, D.; Kitchener, H.; Castellsague, X.; Teixeira, J.C.; Skinner, S.R.; et al. Efficacy of human papillomavirus (HPV)-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by oncogenic HPV types (PATRICIA): Final analysis of a double-blind, randomised study in young women. Lancet 2009, 374, 301–314. [Google Scholar] [CrossRef]
- Kantoff, P.W.; Higano, C.S.; Shore, N.D.; Berger, E.R.; Small, E.J.; Penson, D.F.; Redfern, C.H.; Ferrari, A.C.; Dreicer, R.; Sims, R.B.; et al. Sipuleucel-T Immunotherapy for Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2010, 363, 411–422. [Google Scholar] [CrossRef] [Green Version]
- Schwartzentruber, D.J.; Lawson, D.H.; Richards, J.M.; Conry, R.M.; Miller, D.M.; Treisman, J.; Gailani, F.; Riley, L.; Conlon, K.; Pockaj, B.; et al. gp100 Peptide Vaccine and Interleukin-2 in Patients with Advanced Melanoma. N. Engl. J. Med. 2011, 364, 2119–2127. [Google Scholar] [CrossRef] [Green Version]
- Fu, J.; Kanne, D.B.; Leong, M.; Glickman, L.H.; McWhirter, S.M.; Lemmens, E.; Mechette, K.; Leong, J.J.; Lauer, P.; Liu, W.; et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med. 2015, 7, 283. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Wang-Gillam, A.; Picozzi, V.; Greten, T.F.; Crocenzi, T.; Springett, G.; Morse, M.; Zeh, H.; Cohen, D.; Fine, R.L.; et al. Safety and survival with GVAX pancreas prime and Listeria monocytogenes-expressing mesothelin (CRS-207) boost vaccines for metastatic pancreatic cancer. J. Clin. Oncol. 2015, 33, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Gubin, M.M.; Zhang, X.; Schuster, H.; Caron, E.; Ward, J.P.; Noguchi, T.; Ivanova, Y.; Hundal, J.; Arthur, C.D.; Krebber, W.J.; et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014, 515, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Chen, L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat. Rev. Immunol. 2004, 4, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Hamanishi, J.; Mandai, M.; Matsumura, N.; Abiko, K.; Baba, T.; Konishi, I. PD-1/PD-L1 blockade in cancer treatment: Perspectives and issues. Int. J. Clin. Oncol. 2016, 21, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Chen, L. Inhibitory B7-family molecules in the tumour microenvironment. Nat. Rev. Immunol. 2008, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Redman, J.M.; Gibney, G.T.; Atkins, M.B. Advances in immunotherapy for melanoma. BMC Med. 2016, 14, 20. [Google Scholar] [CrossRef] [PubMed]
- Rini, B.I.; Plimack, E.R.; Stus, V.; Gafanov, R.; Hawkins, R.; Nosov, D.; Pouliot, F.; Alekseev, B.; Soulières, D.; Melichar, B.; et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2019, 380, 1116–1127. [Google Scholar] [CrossRef]
- Donskov, F.; McDermott, D.F.; Lee, J.L.; Szczylik, C.; Malik, J.; Alekseev, B.Y.; Larkin, J.M.G.; Matveev, V.B.; Airatovich Gafanov, R.; Tomczak, P.; et al. 871PKEYNOTE-427 cohort A: Pembrolizumab monotherapy as first-line therapy in advanced clear cell renal cell carcinoma (ccRCC). Ann. Oncol. 2018, 29 (Suppl. S8), viii303–viii331. [Google Scholar] [CrossRef]
- Economopoulou, P.; Agelaki, S.; Perisanidis, C.; Giotakis, E.I.; Psyrri, A. The promise of immunotherapy in head and neck squamous cell carcinoma. Ann. Oncol. 2016, 27, 1675–1685. [Google Scholar] [CrossRef] [PubMed]
- Zaravinos, A. An updated overview of HPV-associated head and neck carcinomas. Oncotarget 2014, 5, 3956–3969. [Google Scholar] [CrossRef]
- Schoppy, D.W.; Sunwoo, J.B. Immunotherapy for Head and Neck Squamous Cell Carcinoma. Hematol. Oncol. Clin. N. Am. 2015, 29, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Sznol, M.; McDermott, D.F.; Kluger, H.M.; Carvajal, R.D.; Sharfman, W.H.; Brahmer, J.R.; Lawrence, D.P.; Atkins, M.B.; Powderly, J.D.; et al. Survival, durable tumor remission and long-term safety in patients with advanced melanoma receiving nivolumab. J. Clin. Oncol. 2014, 32, 1020–1030. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.M.; Hwu, W.-J.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K.; et al. Safety and Activity of Anti–PD-L1 Antibody in Patients with Advanced Cancer. N. Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Hirano, F.; Kaneko, K.; Tamura, H.; Dong, H.; Wang, S.; Ichikawa, M.; Rietz, C.; Flies, D.B.; Lau, J.S.; Zhu, G.; et al. Blockade of B7-H1 and PD-1 by monoclonal antibodies potentiates cancer therapeutic immunity. Cancer Res. 2005, 65, 1089–1096. [Google Scholar]
- Armand, P.; Nagler, A.; Weller, E.A.; Devine, S.M.; Avigan, D.E.; Chen, Y.B.; Kaminski, M.S.; Holland, H.K.; Winter, J.N.; Mason, J.R.; et al. Disabling immune tolerance by programmed death-1 blockade with pidilizumab after autologous hematopoietic stem-cell transplantation for diffuse large b-cell lymphoma: Results of an international phase II trial. J. Clin. Oncol. 2013, 31, 4199–4206. [Google Scholar] [CrossRef]
- Bryan, L.J.; Gordon, L.I. Pidilizumab in the treatment of diffuse large B-cell lymphoma. Expert Opin. Biol. Ther. 2014, 14, 1361–1368. [Google Scholar] [CrossRef]
- Hamilou, Z.; Lavaud, P.; Loriot, Y. Atezolizumab in urothelial bladder carcinoma. Future Oncol. 2018, 14, 331–341. [Google Scholar] [CrossRef]
- Gkolfinopoulos, S.; Mountzios, G. Recent clinical trials of immunotherapy in non-small-cell lung cancer. Immunotherapy 2019, 11, 461–466. [Google Scholar] [CrossRef]
- Passiglia, F.; Galvano, A.; Rizzo, S.; Incorvaia, L.; Listì, A.; Bazan, V.; Russo, A. Looking for the best immune-checkpoint inhibitor in pre-treated NSCLC patients: An indirect comparison between nivolumab, pembrolizumab and atezolizumab. Int. J. Cancer 2018, 142, 1277–1284. [Google Scholar] [CrossRef]
- Maio, M.; Scherpereel, A.; Calabrò, L.; Aerts, J.; Perez, S.C.; Bearz, A.; Nackaerts, K.; Fennell, D.A.; Kowalski, D.; Tsao, A.S.; et al. Tremelimumab as second-line or third-line treatment in relapsed malignant mesothelioma (DETERMINE): A multicentre, international, randomised, double-blind, placebo-controlled phase 2b trial. Lancet Oncol. 2017, 18, 1261–1273. [Google Scholar] [CrossRef]
- Thomas, A.; Hassan, R. CTLA-4 blockade in mesothelioma: Ineffective or reason for optimism? Lancet Oncol. 2017, 18, 1150–1151. [Google Scholar] [CrossRef]
- Li, J.; Ni, L.; Dong, C. Immune checkpoint receptors in cancer: Redundant by design? Curr. Opin. Immunol. 2017, 45, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; Hamid, O.; Daud, A.; Hodi, F.S.; Wolchok, J.D.; Kefford, R.; Joshua, A.M.; Patnaik, A.; Hwu, W.-J.; Weber, J.S.; et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA 2016, 19, 1600–1609. [Google Scholar] [CrossRef] [PubMed]
- Valsecchi, M.E. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 1270. [Google Scholar] [PubMed]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef]
- Schwartz, R.H. Costimulation of T lymphocytes: The role of CD28, CTLA-4 and B7/BB1 in interleukin-2 production and immunotherapy. Cell 1992, 71, 1065–1068. [Google Scholar] [CrossRef]
- DeBenedette, M.A.; Randall Chu, N.; Pollok, K.E.; Hurtado, J.; Wade, W.F.; Kwon, B.S.; Watts, T.H. Role of 4-1BB ligand in costimulation of T lymphocyte growth and its upregulation on M12 B lymphomas by camp. J. Exp. Med. 1995, 181, 985–992. [Google Scholar] [CrossRef]
- Gramaglia, I.; Weinberg, A.D.; Lemon, M.; Croft, M. Ox-40 ligand: A potent costimulatory molecule for sustaining primary CD4 T cell responses. J. Immunol. 1998, 161, 6510–6517. [Google Scholar]
- Tesselaar, K.; Arens, R.; Van Schijndel, G.M.W.; Baars, P.A.; Van Der Valk, M.A.; Borst, J.; Van Oers, M.H.J.; Van Lier, R.A.W. Lethal T cell immunodeficiency induced by chronic costimulation via CD27-CD70 interactions. Nat. Immunol. 2003, 4, 49–54. [Google Scholar] [CrossRef]
- Hutloff, A.; Dittrich, A.M.; Beier, K.C.; Eljaschewitsch, B.; Kraft, R.; Anagnostopoulos, I.; Kroczek, R.A. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature 1999, 397, 263–266. [Google Scholar] [CrossRef]
- Nocentini, G.; Giunchi, L.; Ronchetti, S.; Krausz, L.T.; Bartoli, A.; Moraca, R.; Migliorati, G.; Riccardi, C. A new member of the tumor necrosis factor/nerve growth factor receptor family inhibits T cell receptor-induced apoptosis. Proc. Natl. Acad. Sci. USA 1997, 94, 6216–6221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janakiram, M.; Chinai, J.M.; Zhao, A.; Sparano, J.A.; Zang, X. HHLA2 and TMIGD2: New immunotherapeutic targets of the B7 and CD28 families. Oncoimmunology 2015, 4, e1026534. [Google Scholar] [CrossRef] [PubMed]
- Tamada, K.; Shimozaki, K.; Chapoval, A.I.; Zhai, Y.; Su, J.; Chen, S.F.; Hsieh, S.L.; Nagata, S.; Ni, J.; Chen, L. LIGHT, a TNF-Like Molecule, Costimulates T Cell Proliferation and Is Required for Dendritic Cell-Mediated Allogeneic T Cell Response. J. Immunol. 2000, 164, 4105–4110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoenberger, S.P.; Toes, R.E.M.; Van Dervoort, E.I.H.; Offringa, R.; Melief, C.J.M. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD4OL interactions. Nature 1998, 393, 480–483. [Google Scholar] [CrossRef]
- Tahara-Hanaoka, S.; Miyamoto, A.; Hara, A.; Honda, S.I.; Shibuya, K.; Shibuya, A. Identification and characterization of murine DNAM-1 (CD226) and its poliovirus receptor family ligands. Biochem. Biophys. Res. Commun. 2005, 329, 996–1000. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.-M.; Bhawan, S.; Majima, T.; Wei, H.; Nishimura, M.I.; Yagita, H.; Kumar, V. Cutting Edge: The NK Cell Receptor 2B4 Augments Antigen-Specific T Cell Cytotoxicity Through CD48 Ligation on Neighboring T Cells. J. Immunol. 2003, 170, 4881–4885. [Google Scholar] [CrossRef]
- Seiffert, M.; Brossart, P.; Cant, C.; Cella, M.; Colonna, M.; Brugger, W.; Ullrich, A.; Bühring, H.J. Signal-regulatory protein α (SIRPα) but not SIRPβ is involved in T-cell activation, binds to CD47 with high affinity and is expressed on immature CD34+CD38- hematopoietic cells. Blood 2001, 97, 2741–2749. [Google Scholar] [CrossRef]
- Fernandez, N.C.; Treiner, E.; Vance, R.E.; Jamieson, A.M.; Lemieux, S.; Raulet, D.H. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood 2005, 105, 4416–4423. [Google Scholar] [CrossRef] [Green Version]
- Hodge, J.W.; Greiner, J.W.; Tsang, K.Y.; Sabzevari, H.; Kudo-Saito, C.; Grosenbach, D.W.; Gulley, J.L.; Arlen, P.M.; Marshall, J.L.; Panicali, D.; et al. Costimulatory molecules as adjuvants for immunotherapy. Front. Biosci. 2006, 11, 788–803. [Google Scholar] [CrossRef]
- Van Seventer, G.A.; Shimizu, Y.; Horgan, K.J.; Shaw, S. The LFA-1 ligand ICAM-1 provides an important costimulatory signal for T cell receptor-mediated activation of resting T cells. J. Immunol. 1990, 144, 4579–4586. [Google Scholar]
- Cohen, C.J.; Zhao, Y.; Zheng, Z.; Rosenberg, S.A.; Morgan, R.A. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 2006, 66, 8878–8886. [Google Scholar] [CrossRef] [PubMed]
- Kessels, H.W.H.G.; Wolkers, M.C.; Van Den Boom, M.D.; Van Den Valk, M.A.; Schumacher, T.N.M. Immunotherapy through TCR gene transfer. Nat. Immunol. 2001, 2, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Wing, K.; Onishi, Y.; Prieto-Martin, P.; Yamaguchi, T.; Miyara, M.; Fehervari, Z.; Nomura, T.; Sakaguchi, S. CTLA-4 control over Foxp3+ regulatory T cell function. Science 2008, 322, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Keir, M.E.; Liang, S.C.; Guleria, I.; Latchman, Y.E.; Qipo, A.; Albacker, L.A.; Koulmanda, M.; Freeman, G.J.; Sayegh, M.H.; Sharpe, A.H. Tissue expression of PD-L1 mediates peripheral T cell tolerance. J. Exp. Med. 2006, 203, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Butte, M.J.; Keir, M.E.; Phamduy, T.B.; Sharpe, A.H.; Freeman, G.J. Programmed Death-1 Ligand 1 Interacts Specifically with the B7-1 Costimulatory Molecule to Inhibit T Cell Responses. Immunity 2007, 27, 111–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opitz, C.A.; Litzenburger, U.M.; Sahm, F.; Ott, M.; Tritschler, I.; Trump, S.; Schumacher, T.; Jestaedt, L.; Schrenk, D.; Weller, M.; et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011, 478, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, M.L.; Lucas, C.L.; Buhler, L.; Rayat, G.; Rodriguez-Barbosa, J.I. HVEM/LIGHT/BTLA/CD160 cosignaling pathways as targets for immune regulation. J. Leukoc. Biol. 2010, 87, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Monney, L.; Sabatos, C.A.; Gaglia, J.L.; Ryu, A.; Waldner, H.; Chernova, T.; Manning, S.; Greenfield, E.A.; Coyle, A.J.; Sobel, R.A.; et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 2002, 415, 536–541. [Google Scholar] [CrossRef]
- Sakuishi, K.; Apetoh, L.; Sullivan, J.M.; Blazar, B.R.; Kuchroo, V.K.; Anderson, A.C. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J. Exp. Med. 2011, 208, 1331. [Google Scholar] [CrossRef]
- Yu, X.; Harden, K.; Gonzalez, L.C.; Francesco, M.; Chiang, E.; Irving, B.; Tom, I.; Ivelja, S.; Refino, C.J.; Clark, H.; et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009, 10, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.J.; Andrews, D.M.; McLaughlin, N.M.; Yagita, H.; Gilfillan, S.; Colonna, M.; Smyth, M.J. DNAM-1/CD155 Interactions Promote Cytokine and NK Cell-Mediated Suppression of Poorly Immunogenic Melanoma Metastases. J. Immunol. 2010, 184, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Paniccia, A.; Schulick, A.C.; Chen, W.; Koenig, M.R.; Byers, J.T.; Yao, S.; Bevers, S.; Edil, B.H. Identification of CD112R as a novel checkpoint for human T cells. J. Exp. Med. 2016, 213, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Stanietsky, N.; Simic, H.; Arapovic, J.; Toporik, A.; Levy, O.; Novik, A.; Levine, Z.; Beiman, M.; Dassa, L.; Achdout, H.; et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 17858–17863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohta, A.; Gorelik, E.; Prasad, S.J.; Ronchese, F.; Lukashev, D.; Wong, M.K.K.; Huang, X.; Caldwell, S.; Liu, K.; Smith, P.; et al. A2A adenosine receptor protects tumors from antitumor T cells. Proc. Natl. Acad. Sci. USA 2006, 103, 13132–13137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kretz-Rommel, A.; Qin, F.; Dakappagari, N.; Ravey, E.P.; McWhirter, J.; Oltean, D.; Frederickson, S.; Maruyama, T.; Wild, M.A.; Nolan, M.J.; et al. CD200 Expression on Tumor Cells Suppresses Antitumor Immunity: New Approaches to Cancer Immunotherapy. J. Immunol. 2007, 178, 5595–5605. [Google Scholar] [CrossRef] [Green Version]
- Suh, W.K.; Gajewska, B.U.; Okada, H.; Gronski, M.A.; Bertram, E.M.; Dawicki, W.; Duncan, G.S.; Bukczynski, J.; Plyte, S.; Elia, A.; et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat. Immunol. 2003, 4, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Husain, B.; Ramani, S.R.; Chiang, E.; Lehoux, I.; Paduchuri, S.; Arena, T.A.; Patel, A.; Wilson, B.; Chan, P.; Franke, Y.; et al. A platform for extracellular interactome discovery identifies novel functional binding partners for the immune receptors B7-H3/CD276 and PVR/CD155. Mol. Cell. Proteom. 2019. [Google Scholar] [CrossRef]
- Sica, G.L.; Choi, I.H.; Zhu, G.; Tamada, K.; Wang, S.D.; Tamura, H.; Chapoval, A.I.; Flies, D.B.; Bajorath, J.; Chen, L. B7-H4, a Molecule of the B7 Family, Negatively Regulates T Cell Immunity. Immunity 2003, 18, 849–861. [Google Scholar] [CrossRef] [Green Version]
- Lines, J.L.; Sempere, L.F.; Broughton, T.; Wang, L.; Noelle, R. VISTA Is a Novel Broad-Spectrum Negative Checkpoint Regulator for Cancer Immunotherapy. Cancer Immunol. Res. 2014, 2, 510–517. [Google Scholar] [CrossRef]
- Wang, J.; Wu, G.; Manick, B.; Hernandez, V.; Renelt, M.; Erickson, C.; Guan, J.; Singh, R.; Rollins, S.; Solorz, A.; et al. VSIG-3 as a ligand of VISTA inhibits human T-cell function. Immunology 2019, 156, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Chinai, J.M.; Buhl, S.; Scandiuzzi, L.; Ray, A.; Jeon, H.; Ohaegbulam, K.C.; Ghosh, K.; Zhao, A.; Scharff, M.D.; et al. HHLA2 is a member of the B7 family and inhibits human CD4 and CD8 T-cell function. Proc. Natl. Acad. Sci. USA 2013, 110, 9879–9884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruggeri, L.; Capanni, M.; Urbani, E.; Perruccio, K.; Shlomchik, W.D.; Tosti, A.; Posati, S.; Rogaia, D.; Frassoni, F.; Aversa, F.; et al. Effectiveness of donor natural killer cell aloreactivity in mismatched hematopoietic transplants. Science 2002, 295, 2097–2100. [Google Scholar] [CrossRef] [PubMed]
- Kisielow, M.; Kisielow, J.; Capoferri-Sollami, G.; Karjalainen, K. Expression of lymphocyte activation gene 3 (LAG-3) on B cells is induced by T cells. Eur. J. Immunol. 2005, 35, 2081–2088. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sanmamed, M.F.; Datar, I.; Su, T.T.; Ji, L.; Sun, J.; Chen, L.; Chen, Y.; Zhu, G.; Yin, W.; et al. Fibrinogen-like Protein 1 Is a Major Immune Inhibitory Ligand of LAG-3. Cell 2019, 176, 334–347. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.R.; Turnis, M.E.; Goldberg, M.V.; Bankoti, J.; Selby, M.; Nirschl, C.J.; Bettini, M.L.; Gravano, D.M.; Vogel, P.; Liu, C.L.; et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 2012, 72, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Sprengers, D.; Boor, P.P.C.; Doukas, M.; Schutz, H.; Mancham, S.; Pedroza-Gonzalez, A.; Polak, W.G.; de Jonge, J.; Gaspersz, M.; et al. Antibodies Against Immune Checkpoint Molecules Restore Functions of Tumor-Infiltrating T Cells in Hepatocellular Carcinomas. Gastroenterology 2017, 153, 1107–1119. [Google Scholar] [CrossRef]
- Huard, B.; Gaulard, P.; Faure, F.; Hercend, T.; Triebel, F. Cellular expression and tissue distribution of the human LAG-3-encoded protein, an MHC class II ligand. Immunogenetics 1994, 39, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Triebel, F.; Jitsukawa, S.; Baixeras, E.; Roman-Roman, S.; Genevee, C.; Viegas-Pequignot, E.; Hercend, T. LAG-3, a novel lymphocyte activation gene closely related to CD4. J. Exp. Med. 1990, 171, 1393–1405. [Google Scholar] [CrossRef]
- Workman, C.J.; Wang, Y.; El Kasmi, K.C.; Pardoll, D.M.; Murray, P.J.; Drake, C.G.; Vignali, D.A.A. LAG-3 Regulates Plasmacytoid Dendritic Cell Homeostasis. J. Immunol. 2009, 182, 1885–1891. [Google Scholar] [CrossRef] [Green Version]
- Huard, B.; Prigent, P.; Tournier, M.; Bruniquel, D.; Triebel, F. CD4/major histocompatibility complex class II interaction analyzed with CD4- and lymphocyte activation gene-3 (LAG-3)-Ig fusion proteins. Eur. J. Immunol. 1995, 25, 2718–2721. [Google Scholar] [CrossRef] [PubMed]
- Huard, B.; Tournier, M.; Hercend, T.; Triebel, F.; Faure, F. Lymphocyte-activation gene 3/major histocompatibility complex class II interaction modulates the antigenic response of CD4+ T lymphocytes. Eur. J. Immunol. 1994, 24, 3216–3221. [Google Scholar] [CrossRef] [PubMed]
- Grosso, J.F.; Goldberg, M.V.; Getnet, D.; Bruno, T.C.; Yen, H.-R.; Pyle, K.J.; Hipkiss, E.; Vignali, D.A.A.; Pardoll, D.M.; Drake, C.G. Functionally Distinct LAG-3 and PD-1 Subsets on Activated and Chronically Stimulated CD8 T Cells. J. Immunol. 2009, 182, 6659–6669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg, M.V.; Drake, C.G. LAG-3 in cancer immunotherapy. Curr. Top. Microbiol. Immunol. 2010, 344, 269–278. [Google Scholar]
- Camisaschi, C.; Casati, C.; Rini, F.; Perego, M.; De Filippo, A.; Triebel, F.; Parmiani, G.; Belli, F.; Rivoltini, L.; Castelli, C. LAG-3 Expression Defines a Subset of CD4 + CD25highFoxp3+ Regulatory T Cells That Are Expanded at Tumor Sites. J. Immunol. 2010, 184, 6545–6551. [Google Scholar] [CrossRef] [PubMed]
- Loo, D.; Alderson, R.F.; Chen, F.Z.; Huang, L.; Zhang, W.; Gorlatov, S.; Burke, S.; Ciccarone, V.; Li, H.; Yang, Y.; et al. Development of an Fc-enhanced anti-B7-H3 monoclonal antibody with potent antitumor activity. Clin. Cancer Res. 2012, 18, 3834–3845. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Liu, Z.; Zhang, Y.; Quan, Q.; Huang, L.; Xu, Y.; Cao, L.; Zhang, X. Association of increased B7 protein expression by infiltrating immune cells with progression of gastric carcinogenesis. Medicine (Baltimore) 2019, 98, e14663. [Google Scholar] [CrossRef]
- Iizuka, A.; Nonomura, C.; Ashizawa, T.; Kondou, R.; Ohshima, K.; Sugino, T.; Mitsuya, K.; Hayashi, N.; Nakasu, Y.; Maruyama, K.; et al. A T-cell-engaging B7-H4/CD3 bispecific Fab-scFv antibody targets human breast cancer. Clin. Cancer Res. 2019, 25, 2925–2934. [Google Scholar] [CrossRef]
- Altan, M.; Kidwell, K.M.; Pelekanou, V.; Carvajal-Hausdorf, D.E.; Schalper, K.A.; Toki, M.I.; Thomas, D.G.; Sabel, M.S.; Hayes, D.F.; Rimm, D.L. Association of B7-H4, PD-L1 and tumor infiltrating lymphocytes with outcomes in breast cancer. NPJ Breast Cancer 2018, 4, 40. [Google Scholar] [CrossRef]
- Azuma, T.; Sato, Y.; Ohno, T.; Azuma, M.; Kume, H. Serum soluble B7-H4 is a prognostic marker for patients with non-metastatic clear cell renal cell carcinoma. PLoS ONE 2018, 13, e0199719. [Google Scholar] [CrossRef]
- Yao, Y.; Luo, F.; Tang, C.; Chen, D.; Qin, Z.; Hua, W.; Xu, M.; Zhong, P.; Yu, S.; Chen, D.; et al. Molecular subgroups and B7-H4 expression levels predict responses to dendritic cell vaccines in glioblastoma: An exploratory randomized phase II clinical trial. Cancer Immunol. Immunother. 2018, 67, 1777–1788. [Google Scholar] [CrossRef] [PubMed]
- Radichev, I.A.; Maneva-Radicheva, L.V.; Amatya, C.; Salehi, M.; Parker, C.; Ellefson, J.; Burn, P.; Savinov, A.Y. Loss of Peripheral Protection in Pancreatic Islets by Proteolysis-Driven Impairment of VTCN1 (B7-H4) Presentation Is Associated with the Development of Autoimmune Diabetes. J. Immunol. 2016, 196, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Lines, J.L.; Pantazi, E.; Mak, J.; Sempere, L.F.; Wang, L.; O’Connell, S.; Ceeraz, S.; Suriawinata, A.A.; Yan, S.; Ernstoff, M.S.; et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014, 74, 1924–1932. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Ward, J.F.; Pettaway, C.A.; Shi, L.Z.; Subudhi, S.K.; Vence, L.M.; Zhao, H.; Chen, J.; Chen, H.; Efstathiou, E.; et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat. Med. 2017, 23, 551–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villarroel-Espindola, F.; Yu, X.; Datar, I.; Mani, N.; Sanmamed, M.; Velcheti, V.; Syrigos, K.; Toki, M.; Zhao, H.; Chen, L.; et al. Spatially resolved and quantitative analysis of vista/pd-1h as a novel immunotherapy target in human non–small cell lung cancer. Clin. Cancer Res. 2018, 24, 1562–1573. [Google Scholar] [CrossRef]
- Dong, C.; Juedes, A.E.; Temann, U.A.; Shresta, S.; Allison, J.P.; Ruddle, N.H.; Flavell, R.A. ICOS co-stimulatory receptor is essential for T-cell activation and function. Nature 2001, 409, 97–101. [Google Scholar] [CrossRef]
- McAdam, A.J.; Greenwald, R.J.; Levin, M.A.; Chernova, T.; Malenkovich, N.; Ling, V.; Freeman, G.J.; Sharpe, A.H. Icos is critical for CD40-mediated antibody class switching. Nature 2001, 409, 102–105. [Google Scholar] [CrossRef]
- Fan, X.; Quezada, S.A.; Sepulveda, M.A.; Sharma, P.; Allison, J.P. Engagement of the ICOS pathway markedly enhances efficacy of CTLA-4 blockade in cancer immunotherapy. J. Exp. Med. 2014, 211, 715–725. [Google Scholar] [CrossRef] [Green Version]
- Amatore, F.; Gorvel, L.; Olive, D. Inducible co-stimulator (ICOS) as a potential therapeutic target for anti-cancer therapy. Expert Opin. Ther. Targets 2018, 22, 343–351. [Google Scholar] [CrossRef]
- Wright, G.J.; Puklavec, M.J.; Willis, A.C.; Hoek, R.M.; Sedgwick, J.D.; Brown, M.H.; Barclay, A.N. Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function. Immunity 2000, 13, 233–242. [Google Scholar] [CrossRef]
- Caserta, S.; Nausch, N.; Sawtell, A.; Drummond, R.; Barr, T.; MacDonald, A.S.; Mutapi, F.; Zamoyska, R. Chronic infection drives expression of the inhibitory receptor CD200R and its ligand CD200, by mouse and human CD4 T cells. PLoS ONE 2012, 7, e35466. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Pan, Y.; Zhang, Q.; Sun, W. Role of CD200/CD200R Signaling Pathway in Regulation of CD4+T Cell Subsets During Thermal Ablation of Hepatocellular Carcinoma. Med. Sci. Monit. 2019, 25, 1718–1728. [Google Scholar] [CrossRef] [PubMed]
- Rijkers, E.S.K.; de Ruiter, T.; Baridi, A.; Veninga, H.; Hoek, R.M.; Meyaard, L. The inhibitory CD200R is differentially expressed on human and mouse T and B lymphocytes. Mol. Immunol. 2008, 45, 1126–1135. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, K.W.; Kwon, Y.R.; Kim, B.M.; Kim, Y.J. Forced expression of CD200 improves the differentiation capability and immunoregulatory functions of mesenchymal stromal cells. Biotechnol. Lett. 2018, 40, 1425–1433. [Google Scholar] [CrossRef] [PubMed]
- Rygiel, T.P.; Karnam, G.; Goverse, G.; Van Der Marel, A.P.J.; Greuter, M.J.; Van Schaarenburg, R.A.; Visser, W.F.; Brenkman, A.B.; Molenaar, R.; Hoek, R.M.; et al. CD200-CD200R signaling suppresses anti-tumor responses independently of CD200 expression on the tumor. Oncogene 2012, 31, 2979–2988. [Google Scholar] [CrossRef] [PubMed]
- Johnston, R.J.; Comps-Agrar, L.; Hackney, J.; Yu, X.; Huseni, M.; Yang, Y.; Park, S.; Javinal, V.; Chiu, H.; Irving, B.; et al. The Immunoreceptor TIGIT Regulates Antitumor and Antiviral CD8 + T Cell Effector Function. Cancer Cell 2014, 26, 923–937. [Google Scholar] [CrossRef] [Green Version]
- Chauvin, J.M.; Pagliano, O.; Fourcade, J.; Sun, Z.; Wang, H.; Sander, C.; Kirkwood, J.M.; Chen, T.T.; Maurer, M.; Korman, A.J.; et al. TIGIT and PD-1 impair tumor antigen-specific CD8 + T cells in melanoma patients. J. Clin. Investig. 2015, 125, 2046–2058. [Google Scholar] [CrossRef]
- Kurtulus, S.; Sakuishi, K.; Ngiow, S.F.; Joller, N.; Tan, D.J.; Teng, M.W.L.; Smyth, M.J.; Kuchroo, V.K.; Anderson, A.C. TIGIT predominantly regulates the immune response via regulatory T cells. J. Clin. Investig. 2015, 125, 4053–4062. [Google Scholar] [CrossRef] [Green Version]
- Lens, S.M.A.; Tesselaar, K.; Van Oers, M.H.J.; Van Lier, R.A.W. Control of lymphocyte function through CD27-CD70 interactions. Semin. Immunol. 1998, 10, 491–499. [Google Scholar] [CrossRef]
- Kobata, T.; Jacquot, S.; Kozlowski, S.; Agematsu, K.; Schlossman, S.F.; Morimoto, C. CD27-CD70 interactions regulate B-cell activation by T cells. Proc. Natl. Acad. Sci. USA 2006, 92, 11249–11253. [Google Scholar] [CrossRef]
- Borst, J.; Hendriks, J.; Xiao, Y. CD27 and CD70 in T cell and B cell activation. Curr. Opin. Immunol. 2005, 17, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, J.; Yamazaki, S.; Takahashi, T.; Ishida, Y.; Sakaguchi, S. Stimulation of CD25 + CD4 + regulatory T cells through GITR breaks immunological self-tolerance. Nat. Immunol. 2002, 3, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, R.; Maruhashi, T.; Sugiura, D.; Shimizu, K.; Watada, M.; Okazaki, I.; Okazaki, T. PD-1 efficiently inhibits T cell activation even in the presence of co-stimulation through CD27 and GITR. Biochem. Biophys. Res. Commun. 2019, 511, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Van Beek, A.A.; Zhou, G.; Doukas, M.; Boor, P.P.C.; Noordam, L.; Mancham, S.; Campos Carrascosa, L.; Van Der Heide-Mulder, M.; Polak, W.G.; Ijzermans, J.N.M.; et al. GITR ligation enhances functionality of tumor-infiltrating T cells in hepatocellular carcinoma. Int. J. Cancer 2019, 145, 1111–1124. [Google Scholar] [CrossRef] [PubMed]
- Tsai, R.K.; Discher, D.E. Inhibition of “self” engulfment through deactivation of myosin-II at the phagocytic synapse between human cells. J. Cell. Biol. 2008, 180, 989–1003. [Google Scholar] [CrossRef]
- Barclay, A.N. Signal regulatory protein alpha (SIRPα)/CD47 interaction and function. Curr. Opin. Immunol. 2009, 21, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Stefanidakis, M.; Newton, G.; Lee, W.Y.; Parkos, C.A.; Luscinskas, F.W. Endothelial CD47 interaction with SIRP{gamma} is required for human T-cell transendothelial migration under shear flow conditions in vitro. Blood 2008, 112, 1280–1289. [Google Scholar] [CrossRef]
- Weiskopf, K.; Ring, A.M.; Ho, C.C.M.; Volkmer, J.P.; Levin, A.M.; Volkmer, A.K.; Özkan, E.; Fernhoff, N.B.; Van De Rijn, M.; Weissman, I.L.; et al. Engineered SIRPα variants as immunotherapeutic adjuvants to anticancer antibodies. Science 2013, 341, 88–91. [Google Scholar] [CrossRef]
- Ganong, A.H.; Cotman, C.W. Kynurenic Acid and Quinolinic Acid Act at N-Methyl-D-Aspartate Receptors in the Rat Hippocampus. J. Pharmacol. Exp. Ther. 1986, 236, 293–299. [Google Scholar]
- Hilmas, C.; Pereira, E.F.; Alkondon, M.; Rassoulpour, A.; Schwarcz, R.; Albuquerque, E.X. The brain metabolite kynurenic acid inhibits alpha7 nicotinic receptor activity and increases non-alpha7 nicotinic receptor expression: Physiopathological implications. J. Neurosci. 2001, 21, 7463–7473. [Google Scholar] [CrossRef]
- Prendergast, G.C.; Smith, C.; Thomas, S.; Mandik-Nayak, L.; Laury-Kleintop, L.; Metz, R.; Muller, A.J. Indoleamine 2,3-dioxygenase pathways of pathogenic inflammation and immune escape in cancer. Cancer Immunol. Immunother. 2014, 63, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Munn, D.H.; Mellor, A.L. IDO in the Tumor Microenvironment: Inflammation, Counter-Regulation and Tolerance. Trends Immunol. 2016, 37, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.W.; Hajjar, J.; Hwu, P.; Naing, A. Targeting the indoleamine 2,3-dioxygenase pathway in cancer. J. Immunother. Cancer 2015, 3, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munn, D.H.; Mellor, A.L. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013, 34, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Marti, L.C.; Pavon, L.; Severino, P.; Sibov, T.; Guilhen, D.; Moreira-Filho, C.A. Vascular endothelial growth factor-A enhances indoleamine 2,3-dioxygenase expression by dendritic cells and subsequently impacts lymphocyte proliferation. Mem. Inst. Oswaldo Cruz 2014, 109, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Xiao, C.; Evans, K.S.; Theivanthiran, T.; DeVito, N.; Holtzhausen, A.; Liu, J.; Liu, X.; Boczkowski, D.; Nair, S.; et al. Paracrine Wnt5a-β-Catenin Signaling Triggers a Metabolic Program that Drives Dendritic Cell Tolerization. Immunity 2018, 48, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Argentiero, A.; De Summa, S.; Di Fonte, R.; Iacobazzi, R.M.; Porcelli, L.; Da Vià, M.C.; Brunetti, O.; Azzariti, A.; Silvestris, N.; Solimando, A.G. Gene expression comparison between the lymph node-positive and -negative reveals a peculiar immune microenvironment signature and a theranostic role for WNT targeting in pancreatic ductal adenocarcinoma: A pilot study. Cancers 2019, 11, 942. [Google Scholar] [CrossRef]
- Jiang, T.; Sun, Y.; Yin, Z.; Feng, S.; Sun, L.; Li, Z. Research progress of indoleamine 2,3-dioxygenase inhibitors. Future Med. Chem. 2015, 7, 185–201. [Google Scholar] [CrossRef]
- Fox, E.; Oliver, T.; Rowe, M.; Thomas, S.; Zakharia, Y.; Gilman, P.B.; Muller, A.J.; Prendergast, G.C. Indoximod: An Immunometabolic Adjuvant That Empowers T Cell Activity in Cancer. Front. Oncol. 2018, 8, 370. [Google Scholar] [CrossRef] [Green Version]
- Borrego, F.; Masilamani, M.; Marusina, A.I.; Tang, X.; Coligan, J.E. The CD94/NKG2 family of receptors: From molecules and cells to clinical relevance. Immunol. Res. 2006, 35, 263–277. [Google Scholar] [CrossRef]
- Colonna, M.; Moretta, A.; Vély, F.; Vivier, E. A high-resolution view of NK-cell receptors: Structure and function. Immunol. Today 2000, 21, 428–431. [Google Scholar] [CrossRef]
- Yokoyama, W.M.; Plougastel, B.F.M. Immune functions encoded by the natural killer gene complex. Nat. Rev. Immunol. 2003, 3, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L. NK cell recognition. Annu. Rev. Immunol. 2005, 23, 225–274. [Google Scholar] [CrossRef] [PubMed]
- Yawata, M.; Yawata, N.; Abi-Rached, L.; Parham, P. Variation Within the Human Killer Cell Immunoglobulin-Like Receptor (KIR) Gene Family. Crit. Rev. Immunol. 2012, 22, 20. [Google Scholar] [CrossRef]
- Bashirova, A.A.; Martin, M.P.; McVicar, D.W.; Carrington, M. The Killer Immunoglobulin-Like Receptor Gene Cluster: Tuning the Genome for Defense. Annu. Rev. Genom. Hum. Genet. 2006, 7, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Raulet, D.H.; Vance, R.E.; McMahon, C.W. Regulation of the natural killer cell receptor repertoire. Annu. Rev. Immunol. 2001, 19, 291–330. [Google Scholar] [CrossRef] [PubMed]
- Vilches, C.; Parham, P. KIR: Diverse, Rapidly Evolving Receptors of Innate and Adaptive Immunity. Annu. Rev. Immunol. 2002, 20, 217–251. [Google Scholar] [CrossRef]
- Vey, N.; Karlin, L.; Sadot-Lebouvier, S.; Broussais, F.; Berton-Rigaud, D.; Rey, J.; Charbonnier, A.; Marie, D.; André, P.; Paturel, C.; et al. A phase 1 study of lirilumab (antibody against killer immunoglobulin-like receptor antibody KIR2D; IPH2102) in patients with solid tumors and hematologic malignancies. Oncotarget 2018, 9, 17675–17688. [Google Scholar] [CrossRef] [Green Version]
- Vey, N.; Dumas, P.; Recher, C.; Gastaud, L.; Lioure, B.; Bulabois, C.; Pautas, C.; Marolleau, J.; Leprêtre, S.; Raffoux, E.; et al. Randomized Phase 2 Trial of Lirilumab (anti-KIR monoclonal antibody, mAb) As Maintenance Treatment in Elderly Patients (pts) with Acute Myeloid Leukemia (AML): Results of the Effikir Trial. Blood 2017, 130 (Suppl. S1), 889. [Google Scholar]
- Seidel, J.A.; Otsuka, A.; Kabashima, K. Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy and Limitations. Front. Oncol. 2018, 8, 86. [Google Scholar] [CrossRef]
- Draghi, A.; Chamberlain, C.A.; Furness, A.; Donia, M. Acquired resistance to cancer immunotherapy. Semin. Immunopathol. 2019, 41, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Myers, G. Immune-related adverse events of immune checkpoint inhibitors: A brief review. Curr. Oncol. 2018, 25, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, D.J.; Carlino, M.S. Immune Checkpoint Inhibitor Toxicity. Curr. Oncol. Rep. 2018, 20, 72. [Google Scholar] [CrossRef] [PubMed]
- Kroschinsky, F.; Stölzel, F.; von Bonin, S.; Beutel, G.; Kochanek, M.; Kiehl, M.; Schellongowski, P. New drugs, new toxicities: Severe side effects of modern targeted and immunotherapy of cancer and their management. Crit. Care 2017, 21, 89. [Google Scholar] [CrossRef] [PubMed]
- Baldo, B.A. Adverse events to monoclonal antibodies used for cancer therapy focus on hypersensitivity responses. Oncoimmunology 2013, 2, e26333. [Google Scholar] [CrossRef]
- Ntali, G.; Kassi, E.; Alevizaki, M. Endocrine sequelae of immune checkpoint inhibitors. Hormones 2017, 16, 341–350. [Google Scholar] [PubMed]
- Mearns, E.S.; Bell, J.A.; Galaznik, A.; Puglielli, S.M.; Cichewicz, A.B.; Boulanger, T.; Garcia-Ribas, I. Gastrointestinal adverse events with combination of checkpoint inhibitors in advanced melanoma: A systematic review. Melanoma Manag. 2018, 5, MMT01. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Valkenburg, K.C.; De Groot, A.E.; Pienta, K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018, 15, 366–381. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Albini, A.; Bruno, A.; Noonan, D.M.; Mortara, L. Contribution to tumor angiogenesis from innate immune cells within the tumor microenvironment: Implications for immunotherapy. Front. Immunol. 2018, 9, 527. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramamonjisoa, N.; Ackerstaff, E. Characterization of the tumor microenvironment and tumor-stroma interaction by non-invasive preclinical imaging. Front. Oncol. 2017, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Fowler, J.A.; Edwards, C.M.; Croucher, P.I. Tumor-host cell interactions in the bone disease of myeloma. Bone 2011, 48, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Kolstad, P. Follow-up study of 232 patients with stage Ia1 and 411 patients with stage Ia2 squamous cell carcinoma of the cervix (microinvasive carcinoma). Gynecol. Oncol. 1989, 33, 265–272. [Google Scholar] [CrossRef]
- Solimando, A.G.; Da Vià, M.C.; Cicco, S.; Leone, P.; Di Lernia, G.; Giannico, D.; Desantis, V.; Frassanito, M.A.; Morizio, A.; Delgado, T.J.; et al. High-Risk Multiple Myeloma: Integrated Clinical and Omics Approach Dissects the Neoplastic Clone and the Tumor Microenvironment. J. Clin. Med. 2019, 8, 997. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Witzig, T.E.; Timm, M.; Haug, J.; Wellik, L.; Kimlinger, T.K.; Greipp, P.R.; Rajkumar, S.V. Bone marrow angiogenic ability and expression of angiogenic cytokines in myeloma: Evidence favoring loss of marrow angiogenesis inhibitory activity with disease progression. Blood 2004, 104, 1159–1165. [Google Scholar] [CrossRef]
- Hose, D.; Moreaux, J.; Meissner, T.; Seckinger, A.; Goldschmidt, H.; Benner, A.; Mahtouk, K.; Hillengass, J.; Rème, T.; De Vos, J.; et al. Induction of angiogenesis by normal and malignant plasma cells. Blood 2009, 114, 128–143. [Google Scholar] [CrossRef] [PubMed]
- Leone, P.; Di Lernia, G.; Solimando, A.G.; Cicco, S.; Saltarella, I.; Lamanuzzi, A.; Ria, R.; Frassanito, M.A.; Ponzoni, M.; Ditonno, P.; et al. Bone marrow endothelial cells sustain a tumor-specific CD8 + T cell subset with suppressive function in myeloma patients. Oncoimmunology 2019, 8, e1486949. [Google Scholar] [CrossRef] [PubMed]
- Croucher, D.C.; Chesi, M.; Li, Z.; Garbitt, V.M.; Sharik, M.E.; Waller, D.; Bergsagel, P.L.; Pugh, T.J.; Trudel, S. A single-cell transcriptional analysis of tumour cells and the immune microenvironment during disease evolution in a transgenic mouse model of myeloma. Blood 2018, 132 (Suppl. S1), 56. [Google Scholar]
- Seymour, F.; Cavenagh, J.; Gribben, J.G. Characterising the Immunological Microenvironment in Newly Diagnosed Multiple Myeloma Bone Marrow By Time of Flight Cytometry Reveals Abnormalities in Antigen Presenting and Effector Lymphocyte Populations with Prognostic Significance. Blood 2018, 132 (Suppl. S1), 58. [Google Scholar]
- Ribatti, D.; Nico, B.; Vacca, A. Multiple myeloma as a model for the role of bone marrow niches in the control of angiogenesis. Int. Rev. Cell. Mol. Biol. 2015, 314, 259–282. [Google Scholar] [PubMed]
- Servais, C.; Erez, N. From sentinel cells to inflammatory culprits: Cancer-associated fibroblasts in tumour-related inflammation. J. Pathol. 2013, 229, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.; Shani, O.; Raz, Y.; Sharon, Y.; Hoffman, D.; Abramovitz, L.; Erez, N. Fibroblasts drive an immunosuppressive and growth-promoting microenvironment in breast cancer via secretion of Chitinase 3-like 1. Oncogene 2017, 36, 4457–4468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteran, L.; Erez, N. The Dark Side of Fibroblasts: Cancer-Associated Fibroblasts as Mediators of Immunosuppression in the Tumor Microenvironment. Front. Immunol. 2019, 10, 1835. [Google Scholar] [CrossRef] [Green Version]
- Hasselbalch, H.C.; Bjørn, M.E. MPNs as Inflammatory Diseases: The Evidence, Consequences and Perspectives. Mediat. Inflamm. 2015, 2015, 1–16. [Google Scholar] [CrossRef]
- Hasselbalch, H.C.; Holmström, M.O. Perspectives on interferon-alpha in the treatment of polycythemia vera and related myeloproliferative neoplasms: Minimal residual disease and cure? Semin. Immunopathol. 2019, 41, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Kiladjian, J.J.; Cassinat, B.; Turlure, P.; Cambier, N.; Roussel, M.; Bellucci, S.; Menot, M.L.; Massonnet, G.; Dutel, J.L.; Ghomari, K.; et al. High molecular response rate of polycythemia vera patients treated with pegylated interferon α-2a. Blood 2006, 108, 2037–2040. [Google Scholar] [CrossRef]
- Masarova, L.; Yin, C.C.; Cortes, J.E.; Konopleva, M.; Borthakur, G.; Newberry, K.J.; Kantarjian, H.M.; Bueso-Ramos, C.E.; Verstovsek, S. Histomorphological responses after therapy with pegylated interferon α-2a in patients with essential thrombocythemia (ET) and polycythemia vera (PV). Exp. Hematol. Oncol. 2017, 6, 30. [Google Scholar]
- Crisà, E.; Cerrano, M.; Beggiato, E.; Benevolo, G.; Lanzarone, G.; Manzini, P.M.; Borchiellini, A.; Riera, L.; Boccadoro, M.; Ferrero, D. Can pegylated interferon improve the outcome of polycythemia vera patients? J. Hematol. Oncol. 2017, 10, 15. [Google Scholar] [CrossRef] [Green Version]
- Pestka, S.; Krause, C.D.; Walter, M.R. Interferons, interferon-like cytokines and their receptors. Immunol. Rev. 2004, 202, 8–32. [Google Scholar] [CrossRef]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Pestka, S. The interferons: 50 Years after their discovery, there is much more to learn. J. Biol. Chem. 2007, 282, 20047–20051. [Google Scholar] [CrossRef] [PubMed]
- Billiau, A. Interferon: The pathways of discovery I. Molecular and cellular aspects. Cytokine Growth Factor Rev. 2006, 17, 381–409. [Google Scholar] [CrossRef] [PubMed]
- Bracci, L.; Proietti, E.; Belardelli, F. IFN-α and novel strategies of combination therapy for cancer. Ann. N. Y. Acad. Sci. 2007, 1112, 256–268. [Google Scholar] [CrossRef]
- Xu, D.; Erickson, S.; Szeps, M.; Gruber, A.; Sangfelt, O.; Einhorn, S.; Pisa, P.; Grandér, D. Interferon alpha down-regulates telomerase reverse transcriptase and telomerase activity in human malignant and nonmalignant hematopoietic cells. Blood 2000, 96, 4313–4318. [Google Scholar] [PubMed]
- Hammill, J.A.; VanSeggelen, H.; Helsen, C.W.; Denisova, G.F.; Evelegh, C.; Tantalo, D.G.M.; Bassett, J.D.; Bramson, J.L. Designed ankyrin repeat proteins are effective targeting elements for chimeric antigen receptors. J. Immunother. Cancer 2015, 3, 55. [Google Scholar] [CrossRef] [Green Version]
- Rao, L.; De Veirman, K.; Giannico, D.; Saltarella, I.; Desantis, V.; Frassanito, M.A.; Solimando, A.G.; Ribatti, D.; Prete, M.; Harstrick, A.; et al. Targeting angiogenesis in multiple myeloma by the VEGF and HGF blocking DARPin® protein MP0250: A preclinical study. Oncotarget 2018, 9, 13366–13381. [Google Scholar] [CrossRef]
- Morgan, D.A.; Ruscetti, F.W.; Gallo, R. Selective in vitro growth of T lymphocytes from normal human bone marrows. Science 1976, 193, 1007–1008. [Google Scholar] [CrossRef]
- Rosenberg, S.A. Raising the bar: The curative potential of human cancer immunotherapy. Sci. Transl. Med. 2012, 4, 127. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Yang, J.C.; Sherry, R.M.; Kammula, U.S.; Hughes, M.S.; Phan, G.Q.; Citrin, D.E.; Restifo, N.P.; Robbins, P.F.; Wunderlich, J.R.; et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 2011, 17, 4550–4557. [Google Scholar] [CrossRef] [PubMed]
- Besser, M.J.; Shapira-Frommer, R.; Itzhaki, O.; Treves, A.J.; Zippel, D.B.; Levy, D.; Kubi, A.; Shoshani, N.; Zikich, D.; Ohayon, Y.; et al. Adoptive transfer of tumor-infiltrating lymphocytes in patients with metastatic melanoma: Intent-to-treat analysis and efficacy after failure to prior immunotherapies. Clin. Cancer Res. 2013, 19, 4792–4800. [Google Scholar] [CrossRef] [PubMed]
- Sukari, A.; Abdallah, N.; Nagasaka, M. Unleash the power of the mighty T cells-basis of adoptive cellular therapy. Crit. Rev. Oncol. Hematol. 2019, 136, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Debets, R.; Donnadieu, E.; Chouaib, S.; Coukos, G. TCR-engineered T cells to treat tumors: Seeing but not touching? Semin. Immunol. 2016, 28, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Spear, T.T.; Nagato, K.; Nishimura, M.I. Strategies to genetically engineer T cells for cancer immunotherapy. Cancer Immunol. Immunother. 2016, 65, 631–649. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, L. The Emerging World of TCR-T Cell Trials Against Cancer: A Systematic Review. Technol. Cancer Res. Treat. 2019, 18, 153303381983106. [Google Scholar] [CrossRef]
- Leung, W.; Heslop, H.E. Adoptive Immunotherapy with Antigen-Specific T Cells Expressing a Native TCR. Cancer Immunol. Res. 2019, 7, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Wolf, B.; Zimmermann, S.; Arber, C.; Irving, M.; Trueb, L.; Coukos, G. Safety and Tolerability of Adoptive Cell Therapy in Cancer. Drug Saf. 2019, 42, 315–334. [Google Scholar] [CrossRef] [Green Version]
- Sharpe, M.E. T-cell Immunotherapies and the Role of Nonclinical Assessment: The Balance between Efficacy and Pathology. Toxicol. Pathol. 2018, 46, 131–146. [Google Scholar] [CrossRef] [Green Version]
- Chuntova, P.; Downey, K.M.; Hegde, B.; Almeida, N.D.; Okada, H. Genetically Engineered T-Cells for Malignant Glioma: Overcoming the Barriers to Effective Immunotherapy. Front. Immunol. 2019, 9, 3062. [Google Scholar] [CrossRef] [Green Version]
- Tendeiro Rego, R.; Morris, E.C.; Lowdell, M.W. T-cell receptor gene-modified cells: Past promises, present methodologies and future challenges. Cytotherapy 2019, 21, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Xia, A.-L.; Wang, X.-C.; Lu, Y.-J.; Lu, X.-J.; Sun, B. Chimeric-antigen receptor T (CAR-T) cell therapy for solid tumors: Challenges and opportunities. Oncotarget 2017, 8, 90521–90531. [Google Scholar] [CrossRef] [PubMed]
- Zabel, M.; Tauber, P.A.; Pickl, W.F. The making and function of CAR cells. Immunol. Lett. 2019, 212, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Kohn, D.B.; Dotti, G.; Brentjens, R.; Savoldo, B.; Jensen, M.; Cooper, L.J.N.; June, C.H.; Rosenberg, S.; Sadelain, M.; Heslop, H.E. CARs on track in the clinic. Mol. Ther. 2011, 19, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Kochenderfer, J.N.; Wilson, W.H.; Janik, J.E.; Dudley, M.E.; Stetler-Stevenson, M.; Feldman, S.A.; Maric, I.; Raffeld, M.; Nathan, D.A.; Lanier, B.J.; et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 2010, 116, 4099–4102. [Google Scholar] [CrossRef]
- Büning, H.; Uckert, W.; Cichutek, K.; Hawkins, R.E.; Abken, H. Do CARs need a driver’s license? Adoptive cell therapy with chimeric antigen receptor-redirected T cells has caused serious adverse events. Hum. Gene. Ther. 2010, 21, 1039–1042. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Weiner, G.J.; Pardoll, D.M. Cancer immunotherapy comes of age. J. Clin. Oncol. 2011, 29, 4828–4836. [Google Scholar] [CrossRef]
- Xu, D.; Jin, G.; Chai, D.; Zhou, X.; Gu, W.; Chong, Y.; Song, J.; Zheng, J. The development of CAR design for tumor CAR-T cell therapy. Oncotarget 2018, 9, 13991–14004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinkove, R.; George, P.; Dasyam, N.; McLellan, A.D. Selecting costimulatory domains for chimeric antigen receptors: Functional and clinical considerations. Clin. Transl. Immunol. 2019, 8, e1049. [Google Scholar] [CrossRef] [PubMed]
- Van Schandevyl, S.; Kerre, T. Chimeric antigen receptor T-cell therapy: Design improvements and therapeutic strategies in cancer treatment. Acta Clin. Belg. 2018, 1–7. [Google Scholar] [CrossRef]
- Ormhøj, M.; Bedoya, F.; Frigault, M.J.; Maus, M.V. CARs in the Lead Against Multiple Myeloma. Curr. Hematol. Malig. Rep. 2017, 12, 119–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadelain, M.; Brentjens, R.; Rivière, I. The Basic Principles of Chimeric Antigen Receptor Design. Cancer Discov. 2013, 3, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Chmielewski, M.; Hombach, A.A.; Abken, H. Of CARs and TRUCKs: Chimeric antigen receptor (CAR) T cells engineered with an inducible cytokine to modulate the tumor stroma. Immunol. Rev. 2014, 257, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, Y.; Han, W.-D. Chimeric antigen receptor modified T-cells for cancer treatment. Chronic Dis. Transl. Med. 2018, 4, 225–243. [Google Scholar] [CrossRef]
- Kalos, M.; Levine, B.L.; Porter, D.L.; Katz, S.; Grupp, S.A.; Bagg, A.; June, C.H. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med. 2011, 3, 95ra73. [Google Scholar] [CrossRef] [PubMed]
- Grupp, S.A.; Kalos, M.; Barrett, D.; Aplenc, R.; Porter, D.L.; Rheingold, S.R.; Teachey, D.T.; Chew, A.; Hauck, B.; Wright, J.F.; et al. Chimeric Antigen Receptor–Modified T Cells for Acute Lymphoid Leukemia. N. Engl. J. Med. 2013, 368, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Brentjens, R.J.; Davila, M.L.; Riviere, I.; Park, J.; Wang, X.; Cowell, L.G.; Bartido, S.; Stefanski, J.; Taylor, C.; Olszewska, M.; et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med. 2013, 5, 177ra38. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.-L.; Hua, Z.-C. Chimeric Antigen Receptor T-cell (CAR T) Therapy for Hematologic and Solid Malignancies: Efficacy and Safety—A Systematic Review with Meta-Analysis. Cancers 2019, 11, 47. [Google Scholar] [CrossRef]
- Holzinger, A.; Barden, M.; Abken, H. The growing world of CAR T cell trials: A systematic review. Cancer Immunol. Immunother. 2016, 65, 1433–1450. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Shi, J.; Liu, H. Chimeric antigen receptor T cell targeting B cell maturation antigen immunotherapy is promising for multiple myeloma. Ann. Hematol. 2019, 98, 813–822. [Google Scholar] [CrossRef] [Green Version]
- Heyman, B.; Yang, Y. Chimeric Antigen Receptor T Cell Therapy for Solid Tumors: Current Status, Obstacles and Future Strategies. Cancers 2019, 11, 191. [Google Scholar] [CrossRef] [PubMed]
- Knochelmann, H.M.; Smith, A.S.; Dwyer, C.J.; Wyatt, M.M.; Mehrotra, S.; Paulos, C.M. CAR T Cells in Solid Tumors: Blueprints for Building Effective Therapies. Front. Immunol. 2018, 9, 1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, W.; Huang, K.; Zhang, Y.; Kupfer, G.; Zhao, Q. Chimeric antigen receptor T cell (CAR-T) immunotherapy for solid tumors: Lessons learned and strategies for moving forward. J. Hematol. Oncol. 2018, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Brudno, J.N.; Kochenderfer, J.N. Recent advances in CAR T-cell toxicity: Mechanisms, manifestations and management. Blood Rev. 2019, 34, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Morello, A.; Tano, Z.; Adusumilli, P.S. CAR T-cell intrinsic PD-1 checkpoint blockade: A two-in-one approach for solid tumor immunotherapy. Oncoimmunology 2017, 6, e1273302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yakoub-Agha, I.; Moreau, A.-S.; Ahmad, I.; Borel, C.; Hadhoum, N.; Masouridi-Levrat, S.; Naudin, J.; Nicolas-Virelizier, E.; Ouachée-Chardin, M.; Platon, L.; et al. Prise en charge pratique du syndrome de relargage des cytokines (CRS) post-CAR-T cells chez l’adulte et l’enfant: Recommandation de la Société francophone de greffe de moelle et de thérapie cellulaire (SFGM-TC). Bull. Cancer 2019, 106, S102–S109. [Google Scholar] [CrossRef] [PubMed]
- Solimando, A.G.; Ribatti, D.; Vacca, A.; Einsele, H. Targeting B-cell non Hodgkin lymphoma: New and old tricks. Leuk. Res. 2016, 42, 93–104. [Google Scholar] [CrossRef]
- Kobold, S.; Pantelyushin, S.; Rataj, F.; Vom Berg, J. Rationale for Combining Bispecific T Cell Activating Antibodies With Checkpoint Blockade for Cancer Therapy. Front. Oncol. 2018, 8, 285. [Google Scholar] [CrossRef]
- Runcie, K.; Budman, D.R.; John, V.; Seetharamu, N. Bi-specific and tri-specific antibodies- the next big thing in solid tumor therapeutics. Mol. Med. 2018, 24, 50. [Google Scholar] [CrossRef]
- Stieglmaier, J.; Benjamin, J.; Nagorsen, D. Utilizing the BiTE (bispecific T-cell engager) platform for immunotherapy of cancer. Expert Opin. Biol. Ther. 2015, 15, 1093–1099. [Google Scholar] [CrossRef]
- Klein, J.S.; Gnanapragasam, P.N.P.; Galimidi, R.P.; Foglesong, C.P.; West, A.P.; Bjorkman, P.J. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc. Natl. Acad. Sci. USA 2009, 106, 7385–7390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkmann, U.; Kontermann, R.E. The making of bispecific antibodies. MAbs 2017, 9, 182–212. [Google Scholar] [CrossRef] [PubMed]
- Goebeler, M.E.; Knop, S.; Viardot, A.; Kufer, P.; Topp, M.S.; Einsele, H.; Noppeney, R.; Hess, G.; Kallert, S.; Mackensen, A.; et al. Bispecific T-cell engager (BiTE) antibody construct Blinatumomab for the treatment of Patients with relapsed/refractory non-Hodgkin lymphoma: Final results from a phase I study. J. Clin. Oncol. 2016, 34, 1104–1111. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.L.; Sherman, M.; McElroy, P.L.; Lofgren, J.A.; Moody, G.; Baeuerle, P.A.; Coxon, A.; Arvedson, T. Bispecific T cell engager (BiTE®) antibody constructs can mediate bystander tumor cell killing. PLoS ONE 2017, 12, e0183390. [Google Scholar]
- Velasquez, M.P.; Torres, D.; Iwahori, K.; Kakarla, S.; Arber, C.; Rodriguez-Cruz, T.; Szoor, A.; Bonifant, C.L.; Gerken, C.; Cooper, L.J.; et al. T cells expressing CD19-specific Engager Molecules for the Immunotherapy of CD19-positive Malignancies. Sci. Rep. 2016, 6, 27130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Yang, Y.; Fan, D.; Xiong, D. The development of bispecific antibodies and their applications in tumor immune escape. Exp. Hematol. Oncol. 2017, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Wang, Z.; Hao, M.; Li, J. Bispecific antibodies and their applications. J. Hematol. Oncol. 2015, 8, 130. [Google Scholar] [CrossRef] [PubMed]
- Felices, M.; Lenvik, T.R.; Davis, Z.B.; Miller, J.S.; Vallera, D.A. Generation of BiKEs and TriKEs to improve NK cell-mediated targeting of tumor cells. Methods Mol. Biol. 2016, 1441, 333–346. [Google Scholar]
- Gleason, M.K.; Verneris, M.R.; Todhunter, D.A.; Zhang, B.; McCullar, V.; Zhou, S.X.; Panoskaltsis-Mortari, A.; Weiner, L.M.; Vallera, D.A.; Miller, J.S. Bispecific and trispecific killer cell engagers directly activate human NK cells through CD16 signaling and induce cytotoxicity and cytokine production. Mol. Cancer Ther. 2012, 11, 2674–2684. [Google Scholar] [CrossRef] [PubMed]
- Tay, S.S.; Carol, H.; Biro, M. TriKEs and BiKEs join CARs on the cancer immunotherapy highway. Hum. Vaccin. Immunother. 2016, 12, 2790–2796. [Google Scholar] [CrossRef] [PubMed]
- Mammas, I.N.; Theodoridou, M.; Kramvis, A.; Thiagarajan, P.; Gardner, S.; Papaioannou, G.; Melidou, A.; Koutsaki, M.; Kostagianni, G.; Achtsidis, V.; et al. Paediatric virology: A rapidly increasing educational challenge (Review). Exp. Ther. Med. 2017, 13, 364–377. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.H. Immune Regulation by Self-Recognition: Novel Possibilities for Anticancer Immunotherapy. J. Natl. Cancer Inst. 2015, 107, 154. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.H. The T-win® technology: Immune-modulating vaccines. Semin. Immunopathol. 2019, 41, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.H. Anti-cancer immunotherapy: Breakthroughs and future strategies. Semin. Immunopathol. 2019, 41, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, C.; Vogelstein, B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015, 347, 78–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helleday, T.; Eshtad, S.; Nik-Zainal, S. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet. 2014, 15, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. The Path to Cancer—Three Strikes and You’re Out. N. Engl. J. Med. 2015, 373, 1895–1898. [Google Scholar] [CrossRef] [PubMed]
- Sottoriva, A.; Kang, H.; Ma, Z.; Graham, T.A.; Salomon, M.P.; Zhao, J.; Marjoram, P.; Siegmund, K.; Press, M.F.; Shibata, D.; et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 2015, 47, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Nowell, P. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.J.; Werner, B.; Barnes, C.P.; Graham, T.A.; Sottoriva, A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 2016, 48, 238–244. [Google Scholar] [CrossRef] [Green Version]
- Baca, S.C.; Prandi, D.; Lawrence, M.S.; Mosquera, J.M.; Romanel, A.; Drier, Y.; Park, K.; Kitabayashi, N.; MacDonald, T.Y.; Ghandi, M.; et al. Punctuated Evolution of Prostate Cancer Genomes. Cell 2013, 153, 666–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martincorena, I.; Roshan, A.; Gerstung, M.; Ellis, P.; Van Loo, P.; McLaren, S.; Wedge, D.C.; Fullam, A.; Alexandrov, L.B.; Tubio, J.M.; et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015, 348, 880–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martincorena, I.; Raine, K.M.; Gerstung, M.; Dawson, K.J.; Haase, K.; Van Loo, P.; Davies, H.; Stratton, M.R.; Campbell, P.J. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 2017, 171, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Miñana, B.; Valcárcel, J.; Gabaldón, T.; Lehner, B. Synonymous Mutations Frequently Act as Driver Mutations in Human Cancers. Cell 2014, 156, 1324–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warneford, S.G.; Witton, L.J.; Townsend, M.L.; Rowe, P.B.; Reddel, R.R.; Dalla-Pozza, L.; Symonds, G. Germ-line splicing mutation of the p53 gene in a cancer-prone family. Cell Growth Differ. 1992, 3, 839–846. [Google Scholar] [PubMed]
- Soudon, J.; Caron de Fromentel, C.; Bernard, O.; Larsen, C.J. Inactivation of the p53 gene expression by a splice donor site mutation in a human T-cell leukemia cell line. Leukemia 1991, 5, 917–920. [Google Scholar]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instabilities in human cancers. Nature 1998, 396, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Preston, B.D.; Albertson, T.M.; Herr, A.J. DNA replication fidelity and cancer. Semin. Cancer Biol. 2010, 20, 281–293. [Google Scholar] [CrossRef] [Green Version]
- Maple, J.T.; Smyrk, T.C.; Boardman, L.A.; Johnson, R.A.; Thibodeau, S.N.; Chari, S.T.; Real, F.X. Defective DNA mismatch repair in long-term (≥3 years) survivors with pancreatic cancer. Pancreatology 2005, 5, 220–228. [Google Scholar] [CrossRef]
- Plaschke, J.; Engel, C.; Krüger, S.; Holinski-Feder, E.; Pagenstecher, C.; Mangold, E.; Moeslein, G.; Schulmann, K.; Gebert, J.; von Knebel Doeberitz, M.; et al. Lower Incidence of Colorectal Cancer and Later Age of Disease Onset in 27 Families With Pathogenic MSH6 Germline Mutations Compared With Families With MLH1 or MSH2 Mutations: The German Hereditary Nonpolyposis Colorectal Cancer Consortium. J. Clin. Oncol. 2004, 22, 4486–4494. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.F.; Johansen, J.; Bjørnevoll, I.; Sylvander, A.E.; Steinsbekk, K.S.; Sætrom, P.; Sandvik, A.K.; Drabløs, F.; Sjursen, W. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Fam. Cancer 2015, 14, 437–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palles, C.; Cazier, J.-B.; Howarth, K.M.; Domingo, E.; Jones, A.M.; Broderick, P.; Kemp, Z.; Spain, S.L.; Guarino, E.; Salguero, I.; et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013, 45, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Poynter, J.N.; Siegmund, K.D.; Weisenberger, D.J.; Long, T.I.; Thibodeau, S.N.; Lindor, N.; Young, J.; Jenkins, M.A.; Hopper, J.L.; Baron, J.A.; et al. Molecular Characterization of MSI-H Colorectal Cancer by MLHI Promoter Methylation, Immunohistochemistry and Mismatch Repair Germline Mutation Screening. Cancer Epidemiol. Biomark. Prev. 2008, 17, 3208–3215. [Google Scholar] [CrossRef] [PubMed]
- Salemi, R.; Falzone, L.; Madonna, G.; Polesel, J.; Cinà, D.; Mallardo, D.; Ascierto, P.A.; Libra, M.; Candido, S. MMP-9 as a Candidate Marker of Response to BRAF Inhibitors in Melanoma Patients With BRAFV600E Mutation Detected in Circulating-Free DNA. Front. Pharmacol. 2018, 9, 856. [Google Scholar] [CrossRef]
- DuPage, M.; Mazumdar, C.; Schmidt, L.M.; Cheung, A.F.; Jacks, T. Expression of tumour-specific antigens underlies cancer immunoediting. Nature 2012, 482, 405–409. [Google Scholar] [CrossRef] [Green Version]
- Matsushita, H.; Vesely, M.D.; Koboldt, D.C.; Rickert, C.G.; Uppaluri, R.; Magrini, V.J.; Arthur, C.D.; White, J.M.; Chen, Y.-S.; Shea, L.K.; et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012, 482, 400–404. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Furness, A.J.S.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T.; et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar] [CrossRef] [Green Version]
- Verdegaal, E.M.E.; de Miranda, N.F.C.C.; Visser, M.; Harryvan, T.; Van Buuren, M.M.; Andersen, R.S.; Hadrup, S.R.; Van Der Minne, C.E.; Schotte, R.; Spits, H.; et al. Neoantigen landscape dynamics during human melanoma–T cell interactions. Nature 2016, 536, 91–95. [Google Scholar] [CrossRef]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017, 355, eaaf8399. [Google Scholar] [CrossRef]
- Parcesepe, P.; Giordano, G.; Laudanna, C.; Febbraro, A.; Pancione, M. Cancer-Associated Immune Resistance and Evasion of Immune Surveillance in Colorectal Cancer. Gastroenterol. Res. Pract. 2016, 2016, 1–8. [Google Scholar] [CrossRef]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S.; et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Angelova, M.; Charoentong, P.; Hackl, H.; Fischer, M.L.; Snajder, R.; Krogsdam, A.M.; Waldner, M.J.; Bindea, G.; Mlecnik, B.; Galon, J.; et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015, 16, 64. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, V.P.; Luksza, M.; Zhao, J.N.; Makarov, V.; Moral, J.A.; Remark, R.; Herbst, B.; Askan, G.; Bhanot, U.; Senbabaoglu, Y.; et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017, 551, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Miao, D.; Margolis, C.A.; Vokes, N.I.; Liu, D.; Taylor-Weiner, A.; Wankowicz, S.M.; Adeegbe, D.; Keliher, D.; Schilling, B.; Tracy, A.; et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet. 2018, 50, 1271–1281. [Google Scholar] [CrossRef]
- Bin Lim, S.; Chua, M.L.K.; Yeong, J.P.S.; Tan, S.J.; Lim, W.-T.; Lim, C.T. Pan-cancer analysis connects tumor matrisome to immune response. NPJ Precis Oncol. 2019, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Tapia Rico, G.; Price, T.J. Atezolizumab for the treatment of colorectal cancer: The latest evidence and clinical potential. Expert Opin. Biol. Ther. 2018, 18, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Nakamura, J.; Noshiro, H. Promising immunotherapies for esophageal cancer. Expert Opin. Biol. Ther. 2017, 17, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Efremova, M.; Finotello, F.; Rieder, D.; Trajanoski, Z. Neoantigens Generated by Individual Mutations and Their Role in Cancer Immunity and Immunotherapy. Front. Immunol. 2017, 8, 1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, S.D.; Warren, R.L.; Gibb, E.A.; Martin, S.D.; Spinelli, J.J.; Nelson, B.H.; Holt, R.A. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014, 24, 743–750. [Google Scholar] [CrossRef] [Green Version]
- Boissière-Michot, F.; Lazennec, G.; Frugier, H.; Jarlier, M.; Roca, L.; Duffour, J.; Du Paty, E.; Laune, D.; Blanchard, F.; Le Pessot, F.; et al. Characterization of an adaptive immune response in microsatellite-instable colorectal cancer. Oncoimmunology 2014, 3, e29256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehnert, J.M.; Panda, A.; Zhong, H.; Hirshfield, K.; Damare, S.; Lane, K.; Sokol, L.; Stein, M.N.; Rodriguez-Rodriquez, L.; Kaufman, H.L.; et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J. Clin. Investig. 2016, 126, 2334–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef] [Green Version]
- Howitt, B.E.; Shukla, S.A.; Sholl, L.M.; Ritterhouse, L.L.; Watkins, J.C.; Rodig, S.; Stover, E.; Strickland, K.C.; D’Andrea, A.D.; Wu, C.J.; et al. Association of Polymerase e–Mutated and Microsatellite-Instable Endometrial Cancers With Neoantigen Load, Number of Tumor-Infiltrating Lymphocytes and Expression of PD-1 and PD-L1. JAMA Oncol. 2015, 1, 1319. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Bindea, G.; Angell, H.K.; Maby, P.; Angelova, M.; Tougeron, D.; Church, S.E.; Lafontaine, L.; Fischer, M.; Fredriksen, T.; et al. Integrative Analyses of Colorectal Cancer Show Immunoscore is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016, 44, 698–711. [Google Scholar] [CrossRef]
- Capietto, A.-H.; Jhunjhunwala, S.; Delamarre, L. Characterizing neoantigens for personalized cancer immunotherapy. Curr. Opin. Immunol. 2017, 46, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Vitale, I.; Sistigu, A.; Manic, G.; Rudqvist, N.-P.; Trajanoski, Z.; Galluzzi, L. Mutational and Antigenic Landscape in Tumor Progression and Cancer Immunotherapy. Trends Cell Biol. 2019, 29, 396–416. [Google Scholar] [CrossRef]
- Pan, R.-Y.; Chung, W.-H.; Chu, M.-T.; Chen, S.-J.; Chen, H.-C.; Zheng, L.; Hung, S.I. Recent Development and Clinical Application of Cancer Vaccine: Targeting Neoantigens. J. Immunol. Res. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Barrero, M. Epigenetic Strategies to Boost Cancer Immunotherapies. Int. J. Mol. Sci. 2017, 18, 1108. [Google Scholar] [CrossRef]
- Dunn, J.; Rao, S. Epigenetics and immunotherapy: The current state of play. Mol. Immunol. 2017, 87, 227–239. [Google Scholar] [CrossRef]
- Falzone, L.; Salemi, R.; Travali, S.; Scalisi, A.; McCubrey, J.; Candido, S.; Libra, M. MMP-9 overexpression is associated with intragenic hypermethylation of MMP9 gene in melanoma. Aging (Albany NY) 2017, 8, 933–944. [Google Scholar] [CrossRef]
- Goltz, D.; Gevensleben, H.; Grünen, S.; Dietrich, J.; Kristiansen, G.; Landsberg, J.; Dietrich, D. PD-L1 (CD274) promoter methylation predicts survival in patients with acute myeloid leukemia. Leukemia 2017, 31, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Kryczek, I.; Nagarsheth, N.; Zhao, L.; Wei, S.; Wang, W.; Sun, Y.; Zhao, E.; Vatan, L.; Szeliga, W.; et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature 2015, 527, 249–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juergens, R.A.; Wrangle, J.; Vendetti, F.P.; Murphy, S.C.; Zhao, M.; Coleman, B.; Sebree, R.; Rodgers, K.; Hooker, C.M.; Franco, N.; et al. Combination Epigenetic Therapy Has Efficacy in Patients with Refractory Advanced Non-Small Cell Lung Cancer. Cancer Discov. 2011, 1, 598–607. [Google Scholar] [CrossRef] [PubMed]
- Falzone, L.; Lupo, G.; La Rosa, G.R.M.; Crimi, S.; Anfuso, C.D.; Salemi, R.; Rapisarda, E.; Libra, M.; Candido, S. Identification of Novel MicroRNAs and Their Diagnostic and Prognostic Significance in Oral Cancer. Cancers 2019, 11, 610. [Google Scholar] [CrossRef]
- Falzone, L.; Romano, G.L.; Salemi, R.; Bucolo, C.; Tomasello, B.; Lupo, G.; Anfuso, C.D.; Spandidos, D.A.; Libra, M.; Candido, S. Prognostic significance of deregulated microRNAs in uveal melanomas. Mol. Med. Rep. 2019, 19, 2599–2610. [Google Scholar] [CrossRef] [Green Version]
- Battaglia, R.; Palini, S.; Vento, M.E.; La Ferlita, A.; Lo Faro, M.J.; Caroppo, E.; Borzì, P.; Falzone, L.; Barbagallo, D.; Ragusa, M.; et al. Identification of extracellular vesicles and characterization of miRNA expression profiles in human blastocoel fluid. Sci. Rep. 2019, 9, 84. [Google Scholar] [CrossRef]
- Falzone, L.; Scola, L.; Zanghì, A.; Biondi, A.; Di Cataldo, A.; Libra, M.; Candido, S. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging (Albany NY) 2018, 10, 1000–1014. [Google Scholar] [CrossRef]
- Polo, A.; Crispo, A.; Cerino, P.; Falzone, L.; Candido, S.; Giudice, A.; De Petro, G.; Ciliberto, G.; Montella, M.; Budillon, A.; et al. Environment and bladder cancer: Molecular analysis by interaction networks. Oncotarget 2017, 8, 65240–65252. [Google Scholar] [CrossRef]
- Falzone, L.; Candido, S.; Salemi, R.; Basile, M.S.; Scalisi, A.; McCubrey, J.A.; Torino, F.; Signorelli, S.S.; Montella, M.; Libra, M. Computational identification of microRNAs associated to both epithelial to mesenchymal transition and NGAL/MMP-9 pathways in bladder cancer. Oncotarget 2016, 7, 72758–72766. [Google Scholar] [CrossRef] [Green Version]
- Hafsi, S.; Candido, S.; Maestro, R.; Falzone, L.; Soua, Z.; Bonavida, B.; Spandidos, D.A.; Libra, M. Correlation between the overexpression of Yin Yang 1 and the expression levels of miRNAs in Burkitt’s lymphoma: A computational study. Oncol. Lett. 2016, 11, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Boeri, M.; Milione, M.; Proto, C.; Signorelli, D.; Lo Russo, G.; Galeone, C.; Verri, C.; Mensah, M.; Centonze, G.; Martinetti, A.; et al. Circulating miRNAs and PD-L1 Tumor Expression Are Associated with Survival in Advanced NSCLC Patients Treated with Immunotherapy: A Prospective Study. Clin. Cancer Res. 2019, 25, 2166–2173. [Google Scholar] [CrossRef]
- Romano, G.; Kwong, L.N. Diagnostic and therapeutic applications of miRNA-based strategies to cancer immunotherapy. Cancer Metastasis Rev. 2018, 37, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut Microbiota in Health and Disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vivarelli, S.; Falzone, L.; Basile, M.S.; Nicolosi, D.; Genovese, C.; Libra, M.; Salmeri, M. Benefits of using probiotics as adjuvants in anticancer therapy (Review). World Acad. Sci. J. 2019, 1, 125–135. [Google Scholar] [CrossRef]
- Vivarelli, S.; Salemi, R.; Candido, S.; Falzone, L.; Santagati, M.; Stefani, S.; Torino, F.; Banna, G.L.; Tonini, G.; Libra, M. Gut Microbiota and Cancer: From Pathogenesis to Therapy. Cancers 2019, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Banna, G.L.; Torino, F.; Marletta, F.; Santagati, M.; Salemi, R.; Cannarozzo, E.; Falzone, L.; Ferraù, F.; Libra, M. Lactobacillus rhamnosus GG: An Overview to Explore the Rationale of Its Use in Cancer. Front. Pharmacol. 2017, 8, 603. [Google Scholar] [CrossRef] [Green Version]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Vetizou, M.; Pitt, J.M.; Daillere, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.M.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [Green Version]
- Derosa, L.; Hellmann, M.D.; Spaziano, M.; Halpenny, D.; Fidelle, M.; Rizvi, H.; Long, N.; Plodkowski, A.J.; Arbour, K.C.; Chaft, J.E.; et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 2018, 29, 1437–1444. [Google Scholar] [CrossRef]
- Zitvogel, L.; Ma, Y.; Raoult, D.; Kroemer, G.; Gajewski, T.F. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018, 359, 1366–1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, B.B.; Light, N.; Fabrizio, D.; Zatzman, M.; Fuligni, F.; De Borja, R.; Davidson, S.; Edwards, M.; Elvin, J.A.; Hodel, K.P.; et al. Comprehensive Analysis of Hypermutation in Human Cancer. Cell 2017, 171, 1042–1056. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.A.; Gordenin, D.A. Hypermutation in human cancer genomes: Footprints and mechanisms. Nat. Rev. Cancer 2014, 14, 786–800. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, A.; Zanetti, M. Neoantigen prediction and the need for validation. Nat. Biotechnol. 2017, 35, 815–817. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.; Jhunjhunwala, S.; Phung, Q.T.; Lupardus, P.; Tanguay, J.; Bumbaca, S.; Franci, C.; Cheung, T.K.; Fritsche, J.; Weinschenk, T.; et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014, 515, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.W.; McPherson, A.; Milne, K.; Kroeger, D.R.; Hamilton, P.T.; Miranda, A.; Funnell, T.; Little, N.; de Souza, C.P.E.; Laan, S.; et al. Interfaces of Malignant and Immunologic Clonal Dynamics in Ovarian Cancer. Cell 2018, 173, 1755–1769. [Google Scholar] [CrossRef]
- Jiménez-Sánchez, A.; Memon, D.; Pourpe, S.; Veeraraghavan, H.; Li, Y.; Vargas, H.A.; Gill, M.B.; Park, K.J.; Zivanovic, O.; Konner, J.; et al. Heterogeneous Tumor-Immune Microenvironments among Differentially Growing Metastases in an Ovarian Cancer Patient. Cell 2017, 170, 927–938. [Google Scholar] [CrossRef]
- Angelova, M.; Mlecnik, B.; Vasaturo, A.; Bindea, G.; Fredriksen, T.; Lafontaine, L.; Buttard, B.; Morgand, E.; Bruni, D.; Jouret-Mourin, A.; et al. Evolution of Metastases in Space and Time under Immune Selection. Cell 2018, 175, 751–765. [Google Scholar] [CrossRef]
- Matsushita, H.; Hasegawa, K.; Oda, K.; Yamamoto, S.; Nishijima, A.; Imai, Y.; Asada, K.; Ikeda, Y.; Karasaki, T.; Fujiwara, K.; et al. The frequency of neoantigens per somatic mutation rather than overall mutational load or number of predicted neoantigens per se is a prognostic factor in ovarian clear cell carcinoma. Oncoimmunology 2017, 6, e1338996. [Google Scholar] [CrossRef]
- Milo, I.; Bedora-Faure, M.; Garcia, Z.; Thibaut, R.; Périé, L.; Shakhar, G.; Deriano, L.; Bousso, P. The immune system profoundly restricts intratumor genetic heterogeneity. Sci. Immunol. 2018, 3, eaat1435. [Google Scholar] [CrossRef]
- Andor, N.; Graham, T.A.; Jansen, M.; Xia, L.C.; Aktipis, C.A.; Petritsch, C.; Ji, H.P.; Maley, C.C. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 2016, 22, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Anagnostou, V.; Smith, K.N.; Forde, P.M.; Niknafs, N.; Bhattacharya, R.; White, J.; Zhang, T.; Adleff, V.; Phallen, J.; Wali, N.; et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non–Small Cell Lung Cancer. Cancer Discov. 2017, 7, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Efremova, M.; Rieder, D.; Klepsch, V.; Charoentong, P.; Finotello, F.; Hackl, H.; Hermann-Kleiter, N.; Löwer, M.; Baier, G.; Krogsdam, A.; et al. Targeting immune checkpoints potentiates immunoediting and changes the dynamics of tumor evolution. Nat. Commun. 2018, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Riaz, N.; Havel, J.J.; Makarov, V.; Desrichard, A.; Urba, W.J.; Sims, J.S.; Hodi, F.S.; Martín-Algarra, S.; Mandal, R.; Sharfman, W.H.; et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949. [Google Scholar] [CrossRef] [PubMed]
- Robert, L.; Tsoi, J.; Wang, X.; Emerson, R.; Homet, B.; Chodon, T.; Mok, S.; Huang, R.R.; Cochran, A.J.; Comin-Anduix, B.; et al. CTLA4 Blockade Broadens the Peripheral T-Cell Receptor Repertoire. Clin. Cancer Res. 2014, 20, 2424–2432. [Google Scholar] [CrossRef] [PubMed]
- Kvistborg, P.; Philips, D.; Kelderman, S.; Hageman, L.; Ottensmeier, C.; Joseph-Pietras, D.; Welters, M.J.P.; Van Der Burg, S.; Kapiteijn, E.; Michielin, O.; et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8 + T cell response. Sci. Transl. Med. 2014, 6, 254ra128. [Google Scholar] [CrossRef]
- Kansy, B.A.; Shayan, G.; Jie, H.-B.; Gibson, S.P.; Lei, Y.L.; Brandau, S.; Lang, S.; Schmitt, N.C.; Ding, F.; Lin, Y.; et al. T cell receptor richness in peripheral blood increases after cetuximab therapy and correlates with therapeutic response. Oncoimmunology 2018, 7, e1494112. [Google Scholar] [CrossRef] [Green Version]
- Aoki, H.; Ueha, S.; Shichino, S.; Ogiwara, H.; Hashimoto, S.; Kakimi, K.; Ito, S.; Matsushima, K. TCR Repertoire Analysis Reveals Mobilization of Novel CD8 + T Cell Clones Into the Cancer-Immunity Cycle Following Anti-CD4 Antibody Administration. Front. Immunol. 2019, 9, 3185. [Google Scholar] [CrossRef]
- Kvistborg, P.; Shu, C.J.; Heemskerk, B.; Fankhauser, M.; Thrue, C.A.; Toebes, M.; Van Rooij, N.; Linnemann, C.; Van Buuren, M.M.; Urbanus, J.H.M.; et al. TIL therapy broadens the tumor-reactive CD8 + T cell compartment in melanoma patients. Oncoimmunology 2012, 1, 409–418. [Google Scholar] [CrossRef]
- Chen, P.-L.; Roh, W.; Reuben, A.; Cooper, Z.A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Bassett, R.L.; Gopalakrishnan, V.; Wani, K.; et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov. 2016, 6, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Hu-Lieskovan, S.; Berent-Maoz, B.; Pang, J.; Chmielowski, B.; Cherry, G.; et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Immune Checkpoint Blockade | Cancer | Organism | References |
|---|---|---|---|
| Ipilimumab (anti–CTLA-4) | Melanoma | Humans | [3] |
| Tremelimumab (anti–CTLA-4) | Hepatocellular carcinoma (HCC) | Humans | [13] |
| Atezolizumab (anti-PD-L1) | Bladder, NSCLC | Humans | [14,15] |
| Avelumab (anti-PD-L1) | Merkel cell carcinoma | Humans | [16] |
| Pembrolizumab (anti-PD-1) | NSCLC, Melanoma, cHL, RCC, HNSCC, dMMR or MSI+ tumors | Humans | [17] |
| Nivolumab (anti-PD-1) | Humans | [18] | |
| Durvalumab (anti-PD-L1) | NSCLC, Urothelial carcinoma | Humans | [19,20] |
| IDO5 (IDO inhibitor) | NSCLC | Humans | [21] |
| 1-L-MT (IDO1 inhibitor) | Mastocytoma, CRC | Mice, cell lines | [22,23] |
| Indoximod (IDO1 inhibitor) | Melanoma, Prostate, Brain, AML | Humans | [24,25,26,27] |
| 680C91 & LM10 (TDO inhibitors) | Various cancer types | Mice, cell lines | [28] |
| Navoximod (IDO1 inhibitor) | Advanced solid tumors | Humans | [29,30] |
| Epacadostat (IDO1 inhibitor) | Multiple advanced solid tumors | Humans | [31] |
| Samalizumab (anti-CD200) | Bladder carcinoma | Humans | [32] |
| Varlilumab (anti-CD27) | Advanced refractory solid tumors | Humans | [33,34] |
| KWAR23 (anti-SIRPa) | Burkitt’s lymphoma, RCC, melanoma | Mice, cell lines | [35,36] |
| Urelumab (anti-CD137) | CRC, Gastric, Lymphoma | Mice, Humans | [37,38] |
| Lirilumab (anti-KIR2D mAb) | HNSCC, Lymphoma, myeloid malignancies | Humans, Mice, cell lines | [39,40,41] |
| Cytokine Therapy | |||
| Interleukin-2 (IL-2) | Melanoma, kidney, polycythemia vera | Humans | [42,43,44] |
| Interferon alpha (IFN-α) | Melanoma | Humans | [45] |
| Cellular Therapy | |||
| Tisagenlecleucel (anti-CD19) | NHL, ALL, DLBCL | Humans | [46,47] |
| Axicabtagene ciloleucel (anti-CD19) | Large B-Cell Lymphoma, NHL | Humans | [48,49] |
| anti-MUC1 CAR-T cells | Seminal vesicle carcinoma | Humans | [50] |
| CD33 knockout hematopoietic stem and progenitor cells (HSPCs) | Acute myeloid leukemia (AML) | Macaques, mice | [51] |
| Tumor antigen-loaded dendritic cells | Renal cell carcinoma (RCC) | Cell lines | [52] |
| IL-12p70-producing DCs | Melanoma | Humans | [53] |
| coTCRcys-transduced T cells | Nasopharyngeal | Cell lines | [54] |
| Therapeutic Vaccines | |||
| Hepatitis B virus (HBV) | HCC | Humans | [55] |
| Human papilloma virus (HPV) | Cervical, HNSCC, Oropharyngeal | Humans | [56,57,58] |
| Sipuleucel-T | Prostate | Humans | [59] |
| anti-gp100 | Melanoma | Humans | [60] |
| STINGVAX & anti–PD-1(G4) | Melanoma, Pancreatic, Colon, Tongue | Mice | [61] |
| GVAX & CRS-207 | Pancreatic | Humans | [62] |
| Immune Receptors | Cancer Cell or APC | T, Treg, M, NK Cells | References |
|---|---|---|---|
| co-stimulatory | CD80/86 (B7.1/2) | CD28 | [87] |
| 4-1BBL | CD137 (4-1BB) | [88] | |
| OX-40L | OX40 | [89] | |
| CD70 | CD27 | [90] | |
| ICOSL (B7RP1) | ICOS | [91] | |
| GITRL | GITR | [92] | |
| B7-H7 (HHLA2) | TMIGD2 (CD28H) | [93] | |
| LIGHT | HVEM (CD270) | [94] | |
| CD40 | CD40L | [95] | |
| PVR (CD155) | DNAM-1 (CD226) | [96] | |
| CD48 | 2B4 (CD244) | [97] | |
| CD47 | SIRPa | [98] | |
| MHC-I | CD94/NKG2 | [99] | |
| LFA3 (CD58) | CD2 | [100] | |
| ICAM | LFA1 | [101] | |
| MHC-I or II | TCR/CD3 | [102,103] | |
| co-inhibitory | CD80/86 (B7.1/2) | CTLA-4 (CD152) | [104] |
| PD-L1/2 | PD-1 | [105] | |
| PD-L1 | B7-1 (CD80) | [106] | |
| IDO1/2, TDO | Tryptophan | [22,107] | |
| HVEM (CD270) | BTLA, CD160 | [108] | |
| GAL9, PtdSer, HMGB1, Ceacam-1 | TIM3 | [109,110] | |
| PVR (CD155) | CD96, TIGIT, DNAM-1 | [111,112] | |
| PVRL2 (CD112) | CD112R, TIGIT | [113,114] | |
| Adenosine | A2aR | [115] | |
| CD200 | CD200R | [116] | |
| B7-H3 (CD276) | ?, IL20RA | [117,118] | |
| B7-H4 (B7S1, VTCN1) | ? | [119] | |
| B7-H5 (VISTA) | ?, VSIG-3 | [120,121] | |
| B7-H7 (HHLA2) | ? | [122] | |
| MHC-I | KIR | [123] | |
| MHC-I or II, FGL1 | LAG-3 | [124,125] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christofi, T.; Baritaki, S.; Falzone, L.; Libra, M.; Zaravinos, A. Current Perspectives in Cancer Immunotherapy. Cancers 2019, 11, 1472. https://doi.org/10.3390/cancers11101472
Christofi T, Baritaki S, Falzone L, Libra M, Zaravinos A. Current Perspectives in Cancer Immunotherapy. Cancers. 2019; 11(10):1472. https://doi.org/10.3390/cancers11101472
Chicago/Turabian StyleChristofi, Theodoulakis, Stavroula Baritaki, Luca Falzone, Massimo Libra, and Apostolos Zaravinos. 2019. "Current Perspectives in Cancer Immunotherapy" Cancers 11, no. 10: 1472. https://doi.org/10.3390/cancers11101472