In Vitro 3D Cultures to Model the Tumor Microenvironment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Models of 3D Cell Culture
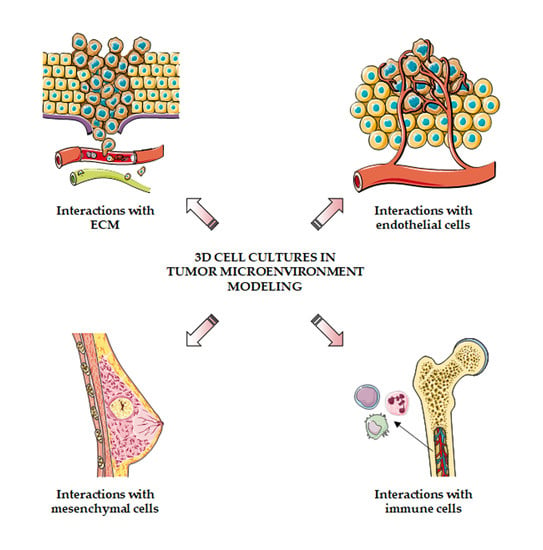
3. Dissecting Tumor–Stroma Interactions in 3D Models
3.1. Interactions with Extracellular Matrix
3.2. Interactions with Endothelial Cells
3.3. Interactions with Mesenchymal Cells
3.4. Interactions with Immune Cells
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tlsty, T.D.; Coussens, L.M. Tumor stroma and regulation of cancer development. Annu. Rev. Pathol. Mech. Dis. 2006, 1, 119–150. [Google Scholar] [CrossRef]
- Pietras, K.; Östman, A. Hallmarks of cancer: Interactions with the tumor stroma. Exp. Cell Res. 2010, 316, 1324–1331. [Google Scholar] [CrossRef]
- Witz, I.P. Tumor–Microenvironment Interactions: Dangerous Liaisons. Adv. Cancer Res. 2008, 100, 203–229. [Google Scholar] [PubMed]
- Najafi, M.; Goradel, N.H.; Farhood, B.; Salehi, E.; Solhjoo, S.; Toolee, H.; Kharazinejad, E.; Mortezaee, K. Tumor microenvironment: Interactions and therapy. J. Cell. Physiol. 2019, 234, 5700–5721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X. Growth factors in tumor microenvironment. Front. Biosci. 2010, 15, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landskron, G.; De la Fuente, M.; Thuwajit, P.; Thuwajit, C.; Hermoso, M.A. Chronic Inflammation and Cytokines in the Tumor Microenvironment. J. Immunol. Res. 2014, 2014, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Tao, S.-C.; Guo, S.-C. Role of extracellular vesicles in tumour microenvironment. Cell Commun. Signal. 2020, 18, 163. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Tian, Y.; Niu, G.; Cao, C. Role of microRNAs in remodeling the tumor microenvironment (Review). Int. J. Oncol. 2019, 56, 407–416. [Google Scholar] [CrossRef] [Green Version]
- McMillin, D.W.; Negri, J.M.; Mitsiades, C.S. The role of tumour–stromal interactions in modifying drug response: Challenges and opportunities. Nat. Rev. Drug Discov. 2013, 12, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Valkenburg, K.C.; de Groot, A.E.; Pienta, K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018, 15, 366–381. [Google Scholar] [CrossRef]
- Fontana, F.; Carollo, E.; Melling, G.E.; Carter, D.R.F. Extracellular Vesicles: Emerging Modulators of Cancer Drug Resistance. Cancers 2021, 13, 749. [Google Scholar] [CrossRef]
- Nishida-Aoki, N.; Gujral, T.S. Emerging approaches to study cell-cell interactions in tumor microenvironment. Oncotarget 2019, 10, 785–797. [Google Scholar] [CrossRef] [Green Version]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Asghar, W.; El Assal, R.; Shafiee, H.; Pitteri, S.; Paulmurugan, R.; Demirci, U. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today 2015, 18, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and Animal Models: Are 3D Cultures the Ideal Tool to Study Cancer-Microenvironment Interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, F.; Raimondi, M.; Marzagalli, M.; Sommariva, M.; Gagliano, N.; Limonta, P. Three-Dimensional Cell Cultures as an In Vitro Tool for Prostate Cancer Modeling and Drug Discovery. Int. J. Mol. Sci. 2020, 21, 6806. [Google Scholar] [CrossRef] [PubMed]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2016, 14, 910–919. [Google Scholar] [CrossRef]
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery (Review). Oncol. Lett. 2017, 14, 6999–7010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meijer, T.G.; Naipal, K.A.; Jager, A.; van Gent, D.C. Ex vivo tumor culture systems for functional drug testing and therapy response prediction. Future Sci. OA 2017, 3, FSO190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, M.P.; Chavali, M.S. Recent advances in biomaterials for 3D scaffolds: A review. Bioact. Mater. 2019, 4, 271–292. [Google Scholar] [CrossRef]
- Park, Y.; Huh, K.M.; Kang, S.-W. Applications of Biomaterials in 3D Cell Culture and Contributions of 3D Cell Culture to Drug Development and Basic Biomedical Research. Int. J. Mol. Sci. 2021, 22, 2491. [Google Scholar] [CrossRef] [PubMed]
- Ryu, N.-E.; Lee, S.-H.; Park, H. Spheroid Culture System Methods and Applications for Mesenchymal Stem Cells. Cells 2019, 8, 1620. [Google Scholar] [CrossRef] [Green Version]
- Bordanaba-Florit, G.; Madarieta, I.; Olalde, B.; Falcón-Pérez, J.M.; Royo, F. 3D Cell Cultures as Prospective Models to Study Extracellular Vesicles in Cancer. Cancers 2021, 13, 307. [Google Scholar] [CrossRef] [PubMed]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef]
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D In Vitro Model (R)evolution: Unveiling Tumor–Stroma Interactions. Trends Cancer 2021, 7, 249–264. [Google Scholar] [CrossRef]
- Fontana, F.; Raimondi, M.; Marzagalli, M.; Sommariva, M.; Limonta, P.; Gagliano, N. Epithelial-To-Mesenchymal Transition Markers and CD44 Isoforms Are Differently Expressed in 2D and 3D Cell Cultures of Prostate Cancer Cells. Cells 2019, 8, 143. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Luo, Z.; Lee, J.; Kim, H.-J.; Lee, K.; Tebon, P.; Feng, Y.; Dokmeci, M.R.; Sengupta, S.; Khademhosseini, A. Organ-on-a-Chip for Cancer and Immune Organs Modeling. Adv. Healthc. Mater. 2019, 8, 1801363. [Google Scholar] [CrossRef]
- Trujillo-de Santiago, G.; Flores-Garza, B.G.; Tavares-Negrete, J.A.; Lara-Mayorga, I.M.; González-Gamboa, I.; Zhang, Y.S.; Rojas-Martínez, A.; Ortiz-López, R.; Álvarez, M.M. The Tumor-on-Chip: Recent Advances in the Development of Microfluidic Systems to Recapitulate the Physiology of Solid Tumors. Materials 2019, 12, 2945. [Google Scholar] [CrossRef] [Green Version]
- Sapudom, J.; Rubner, S.; Martin, S.; Kurth, T.; Riedel, S.; Mierke, C.T.; Pompe, T. The phenotype of cancer cell invasion controlled by fibril diameter and pore size of 3D collagen networks. Biomaterials 2015, 52, 367–375. [Google Scholar] [CrossRef] [Green Version]
- Seo, B.R.; Chen, X.; Ling, L.; Song, Y.H.; Shimpi, A.A.; Choi, S.; Gonzalez, J.; Sapudom, J.; Wang, K.; Eguiluz, R.C.A.; et al. Collagen microarchitecture mechanically controls myofibroblast differentiation. Proc. Natl. Acad. Sci. USA 2020, 117, 11387–11398. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.R.; Bhardwaj, P.; Choi, S.; Gonzalez, J.; Eguiluz, R.C.A.; Wang, K.; Mohanan, S.; Morris, P.G.; Du, B.; Zhou, X.K.; et al. Obesity-dependent changes in interstitial ECM mechanics promote breast tumorigenesis. Sci. Transl. Med. 2015, 7, ra130–ra301. [Google Scholar] [CrossRef] [Green Version]
- Keely, P.J. Mechanisms by Which the Extracellular Matrix and Integrin Signaling Act to Regulate the Switch Between Tumor Suppression and Tumor Promotion. J. Mammary Gland Biol. Neoplasia 2011, 16, 205–219. [Google Scholar] [CrossRef]
- Bordeleau, F.; Tang, L.N.; Reinhart-King, C.A. Topographical guidance of 3D tumor cell migration at an interface of collagen densities. Phys. Biol. 2013, 10, 065004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sapudom, J.; Rubner, S.; Martin, S.; Pompe, T. Mimicking Tissue Boundaries by Sharp Multiparameter Matrix Interfaces. Adv. Healthc. Mater. 2016, 5, 1861–1867. [Google Scholar] [CrossRef] [PubMed]
- Morris, B.A.; Burkel, B.; Ponik, S.M.; Fan, J.; Condeelis, J.S.; Aguirre-Ghiso, J.A.; Castracane, J.; Denu, J.M.; Keely, P.J. Collagen Matrix Density Drives the Metabolic Shift in Breast Cancer Cells. EBioMedicine 2016, 13, 146–156. [Google Scholar] [CrossRef] [Green Version]
- Sapudom, J.; Mohamed, W.K.E.; Garcia-Sabaté, A.; Alatoom, A.; Karaman, S.; Mahtani, N.; Teo, J.C.M. Collagen Fibril Density Modulates Macrophage Activation and Cellular Functions during Tissue Repair. Bioengineering 2020, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Friedemann, M.; Kalbitzer, L.; Franz, S.; Moeller, S.; Schnabelrauch, M.; Simon, J.-C.; Pompe, T.; Franke, K. Instructing Human Macrophage Polarization by Stiffness and Glycosaminoglycan Functionalization in 3D Collagen Networks. Adv. Healthc. Mater. 2017, 6, 1600967. [Google Scholar] [CrossRef]
- Kuczek, D.E.; Larsen, A.M.H.; Thorseth, M.-L.; Carretta, M.; Kalvisa, A.; Siersbæk, M.S.; Simões, A.M.C.; Roslind, A.; Engelholm, L.H.; Noessner, E.; et al. Collagen density regulates the activity of tumor-infiltrating T cells. J. Immunother. Cancer 2019, 7, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lokeshwar, V.B.; Mirza, S.; Jordan, A. Targeting Hyaluronic Acid Family for Cancer Chemoprevention and Therapy. Adv. Cancer Res. 2014, 123, 35–65. [Google Scholar] [PubMed] [Green Version]
- Sapudom, J.; Ullm, F.; Martin, S.; Kalbitzer, L.; Naab, J.; Möller, S.; Schnabelrauch, M.; Anderegg, U.; Schmidt, S.; Pompe, T. Molecular weight specific impact of soluble and immobilized hyaluronan on CD44 expressing melanoma cells in 3D collagen matrices. Acta Biomater. 2017, 50, 259–270. [Google Scholar] [CrossRef] [Green Version]
- Tavianatou, A.-G.; Piperigkou, Z.; Barbera, C.; Beninatto, R.; Masola, V.; Caon, I.; Onisto, M.; Franchi, M.; Galesso, D.; Karamanos, N.K. Molecular size-dependent specificity of hyaluronan on functional properties, morphology and matrix composition of mammary cancer cells. Matrix Biol. Plus 2019, 3, 100008. [Google Scholar] [CrossRef] [PubMed]
- Sapudom, J.; Müller, C.D.; Nguyen, K.-T.; Martin, S.; Anderegg, U.; Pompe, T. Matrix Remodeling and Hyaluronan Production by Myofibroblasts and Cancer-Associated Fibroblasts in 3D Collagen Matrices. Gels 2020, 6, 33. [Google Scholar] [CrossRef]
- Cabral-Pacheco, G.A.; Garza-Veloz, I.; La Rosa, C.C.-D.; Ramirez-Acuña, J.M.; Perez-Romero, B.A.; Guerrero-Rodriguez, J.F.; Martinez-Avila, N.; Martinez-Fierro, M.L. The Roles of Matrix Metalloproteinases and Their Inhibitors in Human Diseases. Int. J. Mol. Sci. 2020, 21, 9739. [Google Scholar] [CrossRef]
- Levin, M.; Udi, Y.; Solomonov, I.; Sagi, I. Next generation matrix metalloproteinase inhibitors—Novel strategies bring new prospects. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1927–1939. [Google Scholar] [CrossRef]
- Liu, W.; Li, L.; Wang, X.; Ren, L.; Wang, X.; Wang, J.; Tu, Q.; Huang, X.; Wang, J. An integrated microfluidic system for studying cell-microenvironmental interactions versatilely and dynamically. Lab Chip 2010, 10, 1717. [Google Scholar] [CrossRef]
- Sung, K.E.; Yang, N.; Pehlke, C.; Keely, P.J.; Eliceiri, K.W.; Friedl, A.; Beebe, D.J. Transition to invasion in breast cancer: A microfluidic in vitro model enables examination of spatial and temporal effects. Integr. Biol. 2011, 3, 439–450. [Google Scholar] [CrossRef] [Green Version]
- Koch, T.M.; Münster, S.; Bonakdar, N.; Butler, J.P.; Fabry, B. 3D Traction Forces in Cancer Cell Invasion. PLoS ONE 2012, 7, e33476. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.-I.; Abaci, H.E.; Krupski, Y.; Weng, L.-C.; Burdick, J.A.; Gerecht, S. Hyaluronic acid hydrogel stiffness and oxygen tension affect cancer cell fate and endothelial sprouting. Biomater. Sci. 2014, 2, 655. [Google Scholar] [CrossRef] [Green Version]
- Mosadegh, B.; Lockett, M.R.; Minn, K.T.; Simon, K.A.; Gilbert, K.; Hillier, S.; Newsome, D.; Li, H.; Hall, A.B.; Boucher, D.M.; et al. A paper-based invasion assay: Assessing chemotaxis of cancer cells in gradients of oxygen. Biomaterials 2015, 52, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.M.; Sixt, M. Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 2019, 20, 738–752. [Google Scholar] [CrossRef]
- Doyle, A.D.; Yamada, K.M. Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp. Cell Res. 2016, 343, 60–66. [Google Scholar] [CrossRef] [Green Version]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-Dimensional Cell Culture: A Breakthrough in Vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Tamaki, M.; McDonald, W.; Amberger, V.R.; Moore, E.; Del Maestro, R.F. Implantation of C6 astrocytoma spheroid into collagen type I gels: Invasive, proliferative, and enzymatic characterizations. J. Neurosurg. 1997, 87, 602–609. [Google Scholar] [CrossRef] [Green Version]
- Lakka, S.S.; Gondi, C.S.; Yanamandra, N.; Olivero, W.C.; Dinh, D.H.; Gujrati, M.; Rao, J.S. Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene 2004, 23, 4681–4689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilina, O.; Bakker, G.-J.; Vasaturo, A.; Hofmann, R.M.; Friedl, P. Two-photon laser-generated microtracks in 3D collagen lattices: Principles of MMP-dependent and -independent collective cancer cell invasion. Phys. Biol. 2011, 8, 015010. [Google Scholar] [CrossRef] [PubMed]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell–ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef] [Green Version]
- Tu, T.-Y.; Wang, Z.; Bai, J.; Sun, W.; Peng, W.K.; Huang, R.Y.-J.; Thiery, J.-P.; Kamm, R.D. Rapid Prototyping of Concave Microwells for the Formation of 3D Multicellular Cancer Aggregates for Drug Screening. Adv. Healthc. Mater. 2014, 3, 609–616. [Google Scholar] [CrossRef] [Green Version]
- Attieh, Y.; Clark, A.G.; Grass, C.; Richon, S.; Pocard, M.; Mariani, P.; Elkhatib, N.; Betz, T.; Gurchenkov, B.; Vignjevic, D.M. Cancer-associated fibroblasts lead tumor invasion through integrin-β3–dependent fibronectin assembly. J. Cell Biol. 2017, 216, 3509–3520. [Google Scholar] [CrossRef] [Green Version]
- Erdogan, B.; Ao, M.; White, L.M.; Means, A.L.; Brewer, B.M.; Yang, L.; Washington, M.K.; Shi, C.; Franco, O.E.; Weaver, A.M.; et al. Cancer-associated fibroblasts promote directional cancer cell migration by aligning fibronectin. J. Cell Biol. 2017, 216, 3799–3816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazaki, K.; Oyanagi, J.; Hoshino, D.; Togo, S.; Kumagai, H.; Miyagi, Y. Cancer cell migration on elongate protrusions of fibroblasts in collagen matrix. Sci. Rep. 2019, 9, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Jiang, B.; Lee, M.H. A Novel 3D Model for Visualization and Tracking of Fibroblast-Guided Directional Cancer Cell Migration. Biology 2020, 9, 328. [Google Scholar] [CrossRef] [PubMed]
- Labernadie, A.; Kato, T.; Brugués, A.; Serra-Picamal, X.; Derzsi, S.; Arwert, E.; Weston, A.; González-Tarragó, V.; Elosegui-Artola, A.; Albertazzi, L.; et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat. Cell Biol. 2017, 19, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-K.; Jang, S.D.; Kim, H.; Chung, S.; Park, J.K.; Kuh, H.-J. Phenotypic Heterogeneity and Plasticity of Cancer Cell Migration in a Pancreatic Tumor Three-Dimensional Culture Model. Cancers 2020, 12, 1305. [Google Scholar] [CrossRef]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef] [Green Version]
- Brassard-Jollive, N.; Monnot, C.; Muller, L.; Germain, S. In vitro 3D Systems to Model Tumor Angiogenesis and Interactions With Stromal Cells. Front. Cell Dev. Biol. 2020, 8, 594903. [Google Scholar] [CrossRef] [PubMed]
- Ghiabi, P.; Jiang, J.; Pasquier, J.; Maleki, M.; Abu-Kaoud, N.; Rafii, S.; Rafii, A. Endothelial Cells Provide a Notch-Dependent Pro-Tumoral Niche for Enhancing Breast Cancer Survival, Stemness and Pro-Metastatic Properties. PLoS ONE 2014, 9, e112424. [Google Scholar] [CrossRef]
- Ghiabi, P.; Jiang, J.; Pasquier, J.; Maleki, M.; Abu-Kaoud, N.; Halabi, N.; Guerrouahen, B.S.; Rafii, S.; Rafii, A. Breast cancer cells promote a notch-dependent mesenchymal phenotype in endothelial cells participating to a pro-tumoral niche. J. Transl. Med. 2015, 13, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiew, G.G.Y.; Wei, N.; Sultania, S.; Lim, S.; Luo, K.Q. Bioengineered three-dimensional co-culture of cancer cells and endothelial cells: A model system for dual analysis of tumor growth and angiogenesis. Biotechnol. Bioeng. 2017, 114, 1865–1877. [Google Scholar] [CrossRef] [PubMed]
- Guerrouahen, B.S.; Pasquier, J.; Kaoud, N.A.; Maleki, M.; Beauchamp, M.-C.; Yasmeen, A.; Ghiabi, P.; Lis, R.; Vidal, F.; Saleh, A.; et al. Akt-Activated Endothelium Constitutes the Niche for Residual Disease and Resistance to Bevacizumab in Ovarian Cancer. Mol. Cancer Ther. 2014, 13, 3123–3136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef]
- Davis, F.M.; Stewart, T.A.; Thompson, E.W.; Monteith, G.R. Targeting EMT in cancer: Opportunities for pharmacological intervention. Trends Pharmacol. Sci. 2014, 35, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Aref, A.R.; Huang, R.Y.-J.; Yu, W.; Chua, K.-N.; Sun, W.; Tu, T.-Y.; Bai, J.; Sim, W.-J.; Zervantonakis, I.K.; Thiery, J.P.; et al. Screening therapeutic EMT blocking agents in a three-dimensional microenvironment. Integr. Biol. 2013, 5, 381–389. [Google Scholar] [CrossRef] [Green Version]
- Bai, J.; Tu, T.-Y.; Kim, C.; Thiery, J.P.; Kamm, R.D. Identification of drugs as single agents or in combination to prevent carcinoma dissemination in a microfluidic 3D environment. Oncotarget 2015, 6, 36603–36614. [Google Scholar] [CrossRef] [Green Version]
- Ehsan, S.M.; Welch-Reardon, K.M.; Waterman, M.L.; Hughes, C.C.W.; George, S.C. A three-dimensional in vitro model of tumor cell intravasation. Integr. Biol. 2014, 6, 603. [Google Scholar] [CrossRef]
- Bray, L.J.; Binner, M.; Holzheu, A.; Friedrichs, J.; Freudenberg, U.; Hutmacher, D.W.; Werner, C. Multi-parametric hydrogels support 3D in vitro bioengineered microenvironment models of tumour angiogenesis. Biomaterials 2015, 53, 609–620. [Google Scholar] [CrossRef] [Green Version]
- Nishiguchi, A.; Matsusaki, M.; Kano, M.R.; Nishihara, H.; Okano, D.; Asano, Y.; Shimoda, H.; Kishimoto, S.; Iwai, S.; Akashi, M. In vitro 3D blood/lymph-vascularized human stromal tissues for preclinical assays of cancer metastasis. Biomaterials 2018, 179, 144–155. [Google Scholar] [CrossRef]
- Bourland, J.; Fradette, J.; Auger, F.A. Tissue-engineered 3D melanoma model with blood and lymphatic capillaries for drug development. Sci. Rep. 2018, 8, 13191. [Google Scholar] [CrossRef] [Green Version]
- Shiga, K.; Hara, M.; Nagasaki, T.; Sato, T.; Takahashi, H.; Takeyama, H. Cancer-Associated Fibroblasts: Their Characteristics and Their Roles in Tumor Growth. Cancers 2015, 7, 2443–2458. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, E.; Makowski, L.; DiGiovanni, J.; Kolonin, M.G. Cancer as a Matter of Fat: The Crosstalk between Adipose Tissue and Tumors. Trends Cancer 2018, 4, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Cuiffo, B.G.; Karnoub, A.E. Mesenchymal stem cells in tumor development. Cell Adh. Migr. 2012, 6, 220–230. [Google Scholar] [CrossRef]
- Eder, T.; Weber, A.; Neuwirt, H.; Grünbacher, G.; Ploner, C.; Klocker, H.; Sampson, N.; Eder, I. Cancer-Associated Fibroblasts Modify the Response of Prostate Cancer Cells to Androgen and Anti-Androgens in Three-Dimensional Spheroid Culture. Int. J. Mol. Sci. 2016, 17, 1458. [Google Scholar] [CrossRef] [Green Version]
- Brancato, V.; Comunanza, V.; Imparato, G.; Corà, D.; Urciuolo, F.; Noghero, A.; Bussolino, F.; Netti, P.A. Bioengineered tumoral microtissues recapitulate desmoplastic reaction of pancreatic cancer. Acta Biomater. 2017, 49, 152–166. [Google Scholar] [CrossRef] [PubMed]
- Santo, V.E.; Estrada, M.F.; Rebelo, S.P.; Abreu, S.; Silva, I.; Pinto, C.; Veloso, S.C.; Serra, A.T.; Boghaert, E.; Alves, P.M.; et al. Adaptable stirred-tank culture strategies for large scale production of multicellular spheroid-based tumor cell models. J. Biotechnol. 2016, 221, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Devarasetty, M.; Dominijanni, A.; Herberg, S.; Shelkey, E.; Skardal, A.; Soker, S. Simulating the human colorectal cancer microenvironment in 3D tumor-stroma co-cultures in vitro and in vivo. Sci. Rep. 2020, 10, 9832. [Google Scholar] [CrossRef]
- Majety, M.; Pradel, L.P.; Gies, M.; Ries, C.H. Fibroblasts Influence Survival and Therapeutic Response in a 3D Co-Culture Model. PLoS ONE 2015, 10, e0127948. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Zhang, Z.; Fouladdel, S.; Deol, Y.; Ingram, P.N.; McDermott, S.P.; Azizi, E.; Wicha, M.S.; Yoon, E. Single cell dual adherent-suspension co-culture micro-environment for studying tumor–stromal interactions with functionally selected cancer stem-like cells. Lab Chip 2016, 16, 2935–2945. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, H.; Sugano, M.; Miyashita, T.; Hashimoto, H.; Ochiai, A.; Suzuki, K.; Tsuboi, M.; Ishii, G. Organoid culture containing cancer cells and stromal cells reveals that podoplanin-positive cancer-associated fibroblasts enhance proliferation of lung cancer cells. Lung Cancer 2019, 134, 100–107. [Google Scholar] [CrossRef]
- Pape, J.; Magdeldin, T.; Stamati, K.; Nyga, A.; Loizidou, M.; Emberton, M.; Cheema, U. Cancer-associated fibroblasts mediate cancer progression and remodel the tumouroid stroma. Br. J. Cancer 2020, 123, 1178–1190. [Google Scholar] [CrossRef]
- Lugo-Cintrón, K.M.; Gong, M.M.; Ayuso, J.M.; Tomko, L.A.; Beebe, D.J.; Virumbrales-Muñoz, M.; Ponik, S.M. Breast Fibroblasts and ECM Components Modulate Breast Cancer Cell Migration through the Secretion of MMPs in a 3D Microfluidic Co-Culture Model. Cancers 2020, 12, 1173. [Google Scholar] [CrossRef]
- Zhao, H.; Jiang, E.; Shang, Z. 3D Co-culture of Cancer-Associated Fibroblast with Oral Cancer Organoids. J. Dent. Res. 2021, 100, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Ishii, K.; Nakagawa, Y.; Matsuda, C.; Katoh, D.; Ichishi, M.; Shirai, T.; Hirokawa, Y.; Fujiwara, M.; Sugimura, Y.; Watanabe, M. Heterogeneous induction of an invasive phenotype in prostate cancer cells by coculturing with patient-derived fibroblasts. J. Cell. Biochem. 2021, 122, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Giannakou, A.; Wyman, S.; Gruzas, J.; Golas, J.; Zhong, W.; Loreth, C.; Sridharan, L.; Yamin, T.-T.; Damelin, M.; et al. Cancer-associated fibroblasts suppress SOX2-induced dysplasia in a lung squamous cancer coculture. Proc. Natl. Acad. Sci. USA 2018, 115, E11671–E11680. [Google Scholar] [CrossRef] [Green Version]
- Hume, R.D.; Berry, L.; Reichelt, S.; D’Angelo, M.; Gomm, J.; Cameron, R.E.; Watson, C.J. An Engineered Human Adipose/Collagen Model for In Vitro Breast Cancer Cell Migration Studies. Tissue Eng. Part A 2018, 24, 1309–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buschhaus, J.M.; Humphries, B.A.; Eckley, S.S.; Robison, T.H.; Cutter, A.C.; Rajendran, S.; Haley, H.R.; Bevoor, A.S.; Luker, K.E.; Luker, G.D. Targeting disseminated estrogen-receptor-positive breast cancer cells in bone marrow. Oncogene 2020, 39, 5649–5662. [Google Scholar] [CrossRef]
- Lewis, K.J.R.; Hall, J.K.; Kiyotake, E.A.; Christensen, T.; Balasubramaniam, V.; Anseth, K.S. Epithelial-mesenchymal crosstalk influences cellular behavior in a 3D alveolus-fibroblast model system. Biomaterials 2018, 155, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Saini, H.; Rahmani Eliato, K.; Veldhuizen, J.; Zare, A.; Allam, M.; Silva, C.; Kratz, A.; Truong, D.; Mouneimne, G.; LaBaer, J.; et al. The role of tumor-stroma interactions on desmoplasia and tumorigenicity within a microengineered 3D platform. Biomaterials 2020, 247, 119975. [Google Scholar] [CrossRef] [PubMed]
- Norberg, K.J.; Liu, X.; Moro, C.F.; Strell, C.; Nania, S.; Blümel, M.; Balboni, A.; Bozóky, B.; Heuchel, R.L.; Löhr, J.M. A novel pancreatic tumour and stellate cell 3D co-culture spheroid model. BMC Cancer 2020, 20, 475. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, S.-K.; Khawar, I.A.; Jeong, S.-Y.; Chung, S.; Kuh, H.-J. Microfluidic co-culture of pancreatic tumor spheroids with stellate cells as a novel 3D model for investigation of stroma-mediated cell motility and drug resistance. J. Exp. Clin. Cancer Res. 2018, 37, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, C.-W.; Han, H.-W.; Tien, Y.-W.; Hsu, S. Biomaterial substrate-derived compact cellular spheroids mimicking the behavior of pancreatic cancer and microenvironment. Biomaterials 2019, 213, 119202. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.J.; Oh, M.-S.; Lee, D.W.; Kuh, H.-J. Multiplex quantitative analysis of stroma-mediated cancer cell invasion, matrix remodeling, and drug response in a 3D co-culture model of pancreatic tumor spheroids and stellate cells. J. Exp. Clin. Cancer Res. 2019, 38, 258. [Google Scholar] [CrossRef] [Green Version]
- Bersini, S.; Jeon, J.S.; Dubini, G.; Arrigoni, C.; Chung, S.; Charest, J.L.; Moretti, M.; Kamm, R.D. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials 2014, 35, 2454–2461. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, L.; Maffini, M.V.; Soto, A.; Sonnenschein, C.; Kaplan, D.L. A complex 3D human tissue culture system based on mammary stromal cells and silk scaffolds for modeling breast morphogenesis and function. Biomaterials 2010, 31, 3920–3929. [Google Scholar] [CrossRef] [Green Version]
- Morales, D.; Lombart, F.; Truchot, A.; Maire, P.; Hussein, M.; Hamitou, W.; Vigneron, P.; Galmiche, A.; Lok, C.; Vayssade, M. 3D Coculture Models Underline Metastatic Melanoma Cell Sensitivity to Vemurafenib. Tissue Eng. Part A 2019, 25, 1116–1126. [Google Scholar] [CrossRef] [PubMed]
- Lamberti, M.J.; Morales Vasconsuelo, A.B.; Ferrara, M.G.; Rumie Vittar, N.B. Recapitulation of Hypoxic Tumor–stroma Microenvironment to Study Photodynamic Therapy Implications. Photochem. Photobiol. 2020, 96, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Lamberti, M.J.; Rettel, M.; Krijgsveld, J.; Rivarola, V.A.; Vittar, N.B.R. Secretome profiling of heterotypic spheroids suggests a role of fibroblasts in HIF-1 pathway modulation and colorectal cancer photodynamic resistance. Cell. Oncol. 2019, 42, 173–196. [Google Scholar] [CrossRef]
- Kock, A.; Bergqvist, F.; Steinmetz, J.; Elfman, L.H.M.; Korotkova, M.; Johnsen, J.I.; Jakobsson, P.; Kogner, P.; Larsson, K. Establishment of an in vitro 3D model for neuroblastoma enables preclinical investigation of combined tumor-stroma drug targeting. FASEB J. 2020, 34, 11101–11114. [Google Scholar] [CrossRef]
- Houshmand, M.; Soleimani, M.; Atashi, A.; Saglio, G.; Abdollahi, M.; Zarif, M.N. Mimicking the Acute Myeloid Leukemia Niche for Molecular Study and Drug Screening. Tissue Eng. Part C Methods 2017, 23, 72–85. [Google Scholar] [CrossRef]
- Dhimolea, E.; Simoes, R.D.M.; Kansara, D.; Weng, X.; Sharma, S.; Awate, P.; Liu, Z.; Gao, D.; Mitsiades, N.; Schwab, J.H.; et al. Pleiotropic Mechanisms Drive Endocrine Resistance in the Three-Dimensional Bone Microenvironment. Cancer Res. 2021, 81, 371–383. [Google Scholar] [CrossRef]
- Toh, Y.-C.; Lim, T.C.; Tai, D.; Xiao, G.; van Noort, D.; Yu, H. A microfluidic 3D hepatocyte chip for drug toxicity testing. Lab Chip 2009, 9, 2026. [Google Scholar] [CrossRef]
- Lee, J.Y.; Chaudhuri, O. Modeling the tumor immune microenvironment for drug discovery using 3D culture. APL Bioeng. 2021, 5, 010903. [Google Scholar] [CrossRef]
- Marzagalli, M.; Ebelt, N.D.; Manuel, E.R. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin. Cancer Biol. 2019, 59, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Shelton, S.E.; Nguyen, H.T.; Barbie, D.A.; Kamm, R.D. Engineering approaches for studying immune-tumor cell interactions and immunotherapy. iScience 2021, 24, 101985. [Google Scholar] [CrossRef]
- Park, N.; Pandey, K.; Chang, S.K.; Kwon, A.-Y.; Cho, Y.B.; Hur, J.; Katwal, N.B.; Kim, S.K.; Lee, S.A.; Son, G.W.; et al. Preclinical platform for long-term evaluation of immuno-oncology drugs using hCD34+ humanized mouse model. J. Immunother. Cancer 2020, 8, e001513. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wang, X.-J.; Chen, D.-X.; Liu, X.-N.; Wang, X.-J. Humanized mouse model: A review on preclinical applications for cancer immunotherapy. Am. J. Cancer Res. 2020, 10, 4568–4584. [Google Scholar] [PubMed]
- Tian, H.; Lyu, Y.; Yang, Y.-G.; Hu, Z. Humanized Rodent Models for Cancer Research. Front. Oncol. 2020, 10, 1696. [Google Scholar] [CrossRef]
- Boucherit, N.; Gorvel, L.; Olive, D. 3D Tumor Models and Their Use for the Testing of Immunotherapies. Front. Immunol. 2020, 11, 603640. [Google Scholar] [CrossRef] [PubMed]
- Rocha, S.; Basto, A.P.; Ijsselsteijn, M.E.; Teles, S.P.; Azevedo, M.M.; Gonçalves, G.; Gullo, I.; Almeida, G.M.; Maqueda, J.J.; Oliveira, M.I.; et al. Immunophenotype of Gastric Tumors Unveils a Pleiotropic Role of Regulatory T Cells in Tumor Development. Cancers 2021, 13, 421. [Google Scholar] [CrossRef]
- Majedi, F.S.; Hasani-Sadrabadi, M.M.; Thauland, T.J.; Li, S.; Bouchard, L.-S.; Butte, M.J. T-cell activation is modulated by the 3D mechanical microenvironment. Biomaterials 2020, 252, 120058. [Google Scholar] [CrossRef]
- Courau, T.; Bonnereau, J.; Chicoteau, J.; Bottois, H.; Remark, R.; Assante Miranda, L.; Toubert, A.; Blery, M.; Aparicio, T.; Allez, M.; et al. Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment. J. Immunother. Cancer 2019, 7, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surendran, V.; Rutledge, D.; Colmon, R.; Chandrasekaran, A. A novel tumor-immune microenvironment (TIME)-on-Chip mimics three dimensional neutrophil-tumor dynamics and neutrophil extracellular traps (NETs)-mediated collective tumor invasion. Biofabrication 2021, 13, 035029. [Google Scholar] [CrossRef]
- Tang, M.; Xie, Q.; Gimple, R.C.; Zhong, Z.; Tam, T.; Tian, J.; Kidwell, R.L.; Wu, Q.; Prager, B.C.; Qiu, Z.; et al. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res. 2020, 30, 833–853. [Google Scholar] [CrossRef] [PubMed]
- Marrella, A.; Dondero, A.; Aiello, M.; Casu, B.; Olive, D.; Regis, S.; Bottino, C.; Pende, D.; Meazza, R.; Caluori, G.; et al. Cell-Laden Hydrogel as a Clinical-Relevant 3D Model for Analyzing Neuroblastoma Growth, Immunophenotype, and Susceptibility to Therapies. Front. Immunol. 2019, 10, 1876. [Google Scholar] [CrossRef] [Green Version]
- Appleton, K.M.; Elrod, A.K.; Lassahn, K.A.; Shuford, S.; Holmes, L.M.; DesRochers, T.M. PD-1/PD-L1 checkpoint inhibitors in combination with olaparib display antitumor activity in ovarian cancer patient-derived three-dimensional spheroid cultures. Cancer Immunol. Immunother. 2021, 70, 843–856. [Google Scholar] [CrossRef] [PubMed]


| Technique | Pros | Cons | Ref. |
|---|---|---|---|
| Scaffolds | Accurate tissue recapitulation | Expensive, variability in polymer composition | [14,17,18] |
| Agitation-based methods | Easy to perform, inexpensive, appropriate for multicellular spheroid generation | Variability in spheroid size, extracellular matrix not addable, inappropriate for migration assays | [19,20,21,22] |
| Suspension cell cultures | Easy to perform, inexpensive, appropriate for multicellular spheroid generation | Variability in spheroid size, extracellular matrix not addable, inappropriate for migration assays | [19,20,21,22] |
| Hanging drop | Inexpensive, spheroid size uniformity | Difficult to perform, extracellular matrix not addable, inappropriate for migration assays | [19,20,21,22] |
| Organ-on-a-chip | Rapid spheroid formation, spheroid size uniformity, constant perfusion and distribution of oxygen and nutrients | Expensive, difficult to perform, specialized equipment and expertise | [23,24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fontana, F.; Marzagalli, M.; Sommariva, M.; Gagliano, N.; Limonta, P. In Vitro 3D Cultures to Model the Tumor Microenvironment. Cancers 2021, 13, 2970. https://doi.org/10.3390/cancers13122970
Fontana F, Marzagalli M, Sommariva M, Gagliano N, Limonta P. In Vitro 3D Cultures to Model the Tumor Microenvironment. Cancers. 2021; 13(12):2970. https://doi.org/10.3390/cancers13122970
Chicago/Turabian StyleFontana, Fabrizio, Monica Marzagalli, Michele Sommariva, Nicoletta Gagliano, and Patrizia Limonta. 2021. "In Vitro 3D Cultures to Model the Tumor Microenvironment" Cancers 13, no. 12: 2970. https://doi.org/10.3390/cancers13122970