Experimental Models of Hepatocellular Carcinoma—A Preclinical Perspective
Abstract
:Simple Summary
Abstract
1. Introduction
2. General Aspects of HCC
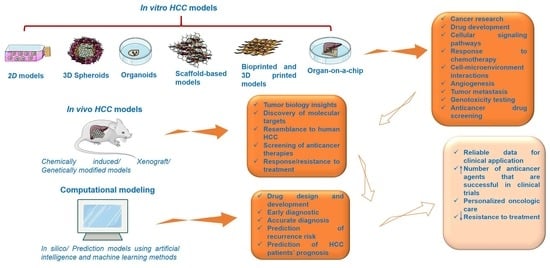
3. Preclinical Experimental Models for HCC
3.1. In Vitro HCC Models
3.1.1. 2D HCC Models
3.1.2. 3D HCC Models
HCC Co-Cultures
HCC Spheroids
HCC Organoids
Scaffold-Based HCC Models
Bioprinted and 3D-Printed HCC Models
HCC-on-a-Chip
3.2. In Vivo Experimental Models for HCC
3.2.1. Mouse HCC Models
Chemically Induced HCC Mouse Models
Xenograft HCC Models
Genetically Engineered HCC Mouse Models
Humanized HCC Mouse Models
3.2.2. Non-Mouse HCC Models
Rat HCC Model
Woodchuck HCC Model
Zebrafish HCC Model
3.3. Computational Modeling of HCC
3.3.1. In Silico Models
3.3.2. Prediction Models of HCC Using Artificial Intelligence and Machine Learning Methods
4. Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Rebouissou, S.; Nault, J.C. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J. Hepatol. 2020, 72, 215–229. [Google Scholar] [CrossRef] [Green Version]
- Molina-Sánchez, P.; Lujambio, A. Experimental Models for Preclinical Research in Hepatocellular Carcinoma. In Hepatocellular Carcinoma: Translational Precision Medicine Approaches; Hoshida, Y., Ed.; Humana Press: Cham, Switzerland, 2019; pp. 333–358. [Google Scholar]
- Hirschfield, H.; Bian, C.B.; Higashi, T.; Nakagawa, S.; Zeleke, T.Z.; Nair, V.D.; Fuchs, B.C.; Hoshida, Y. In vitro modeling of hepatocellular carcinoma molecular subtypes for anti-cancer drug assessment. Exp. Mol. Med. 2018, 50, e419. [Google Scholar] [CrossRef]
- Villanueva, A. Hepato-cellular carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [Green Version]
- Newell, P.; Villanueva, A.; Friedman, S.L.; Koike, K.; Llovet, J.M. Experimental models of hepatocellular carcinoma. J. Hepatol. 2008, 48, 858–879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connor, F.; Rayner, T.F.; Aitken, S.J.; Feig, C.; Lukk, M.; Santoyo-Lopez, J.; Odom, D.T. Mutational landscape of a chemically-induced mouse model of liver cancer. J. Hepatol. 2018, 69, 840–850. [Google Scholar] [CrossRef]
- Ghavimi, S.; Apfel, T.; Azimi, H.; Persaud, A.; Pyrsopoulos, N.T. Management and treatment of hepatocellular carcinoma with immunotherapy: A review of current and future options. J. Clin. Transl. Hepatol. 2020, 8, 168–176. [Google Scholar] [CrossRef]
- Neureiter, D.; Stintzing, S.; Kiesslich, T.; Ocker, M. Hepatocellular carcinoma: Therapeutic advances in signaling, epigenetic and immune targets. World J. Gastroenterol. 2019, 25, 3136–3150. [Google Scholar] [CrossRef]
- Piñero, F.; Silva, M.; Iavarone, M. Sequencing of systemic treatment for hepatocellular carcinoma: Second line competitors. World J. Gastroenterol. 2020, 26, 1888–1900. [Google Scholar] [CrossRef]
- Khawar, I.A.; Park, J.K.; Jung, E.S.; Lee, M.A.; Chang, S.; Kuh, H.J. Three Dimensional Mixed-Cell Spheroids Mimic Stroma-Mediated Chemoresistance and Invasive Migration in hepatocellular carcinoma. Neoplasia 2018, 20, 800–812. [Google Scholar] [CrossRef]
- Fan, H.; Demirci, U.; Chen, P. Emerging organoid models: Leaping forward in cancer research. J. Hematol. Oncol. 2019, 12, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Yim, S.Y.; Shim, J.J.; Sohn, B.H.; Lee, J.S. Experimental Models of Liver Cancer: Genomic Assessment of Experimental Models. In The Liver: Biology and Pathobiology, 6th ed.; Irwin, M.A., Harvey, J.A., James, L.B., David, E.C., David, A.S., Snorri, S.T., Allan, W., Eds.; John Wiley & Sons Ltd.: Oxford, UK, 2020; pp. 747–757. [Google Scholar]
- Grandhi, M.S.; Kim, A.K.; Ronnekleiv-Kelly, S.M.; Kamel, I.R.; Ghasebeh, M.A.; Pawlik, T.M. Hepatocellular carcinoma: From diagnosis to treatment. Surg. Oncol. 2016, 25, 74–85. [Google Scholar] [CrossRef]
- Ghouri, Y.A.; Mian, I.; Rowe, J.H. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J. Carcinog. 2017, 16, 1. [Google Scholar]
- Geh, D.; Anstee, Q.M.; Reeves, H.L. NAFLD-Associated HCC: Progress and Opportunities. J. Hepatocell. Carcinoma 2021, 8, 223–239. [Google Scholar] [CrossRef]
- Farci, P.; Niro, G.A.; Zamboni, F.; Diaz, G. Hepatitis D virus and hepatocellular carcinoma. Viruses 2021, 13, 830. [Google Scholar] [CrossRef]
- Clark, T.; Maximin, S.; Meier, J.; Pokharel, S.; Bhargava, P. Hepatocellular Carcinoma: Review of Epidemiology, Screening, Imaging Diagnosis, Response Assessment, and Treatment. Curr. Probl. Diagn. Radiol. 2015, 44, 479–486. [Google Scholar] [CrossRef]
- Malik, A.; Thanekar, U.; Amarachintha, S.; Mourya, R.; Nalluri, S.; Bondoc, A.; Shivakumar, P. “Complimenting the Complement”: Mechanistic Insights and Opportunities for Therapeutics in Hepatocellular Carcinoma. Front. Oncol. 2021, 10, 1–30. [Google Scholar] [CrossRef]
- Bozward, A.G.; Warricker, F.; Oo, Y.H.; Khakoo, S.I. Natural Killer Cells and Regulatory T Cells Cross Talk in Hepatocellular Carcinoma: Exploring Therapeutic Options for the Next Decade. Front. Immunol. 2021, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Vij, M.; Calderaro, J. Pathologic and molecular features of hepatocellular carcinoma: An update. World J. Hepatol. 2021, 13, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Schulze, K.; Nault, J.C.; Villanueva, A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J. Hepatol. 2016, 65, 1031–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Z.S.; Niu, X.J.; Wang, W.H. Genetic alterations in hepatocellular carcinoma: An update. World J. Gastroenterol. 2016, 22, 9069–9095. [Google Scholar] [CrossRef] [PubMed]
- Nault, J.C.; Zucman-Rossi, J. TERT promoter mutations in primary liver tumors. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Inokawa, Y.; Inaoka, K.; Sonohara, F.; Hayashi, M.; Kanda, M.; Nomoto, S. Molecular alterations in the carcinogenesis and progression of hepatocellular carcinoma: Tumor factors and background liver factors (Review). Oncol. Lett. 2016, 12, 3662–3668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalaf, A.M.; Fuentes, D.; Morshid, A.I.; Burke, M.R.; Kaseb, A.O.; Hassan, M.; Hazle, J.D.; Elsayes, K.M. Role of Wnt/β-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J. Hepatocell. Carcinoma 2018, 5, 61–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galicia-Moreno, M.; Silva-Gomez, J.A.; Lucano-Landeros, S.; Santos, A.; Monroy-Ramirez, H.C.; Armendariz-Borunda, J. Liver Cancer: Therapeutic Challenges and the Importance of Experimental Models. Can. J. Gastroenterol. Hepatol. 2021, 2021, 8837811. [Google Scholar] [CrossRef]
- Wang, H.; Yang, J.; Zhang, K.; Liu, J.; Li, Y.; Su, W.; Song, N. Advances of Fibroblast Growth Factor/Receptor Signaling Pathway in Hepatocellular Carcinoma and its Pharmacotherapeutic Targets. Front. Pharmacol. 2021, 12, 1–13. [Google Scholar]
- Tenen, D.G.; Chai, L.; Tan, J.L. Metabolic alterations and vulnerabilities in hepatocellular carcinoma. Gastroenterol. Rep. 2021, 9, 1–13. [Google Scholar] [CrossRef]
- van Tienderen, G.S.; Koerkamp, B.G.; Ijzermans, J.N.M.; van der Laan, L.J.W.; Verstegen, M.M.A. Recreating tumour complexity in a dish: Organoid models to study liver cancer cells and their extracellular environment. Cancers 2019, 11, 1706. [Google Scholar] [CrossRef] [Green Version]
- Sevic, I.; Spinelli, F.M.; Cantero, M.J.; Reszegi, A.; Kovalszky, I.; García, M.G.; Alaniz, L. The Role of the Tumor Microenvironment in the Development and Progression of Hepatocellular Carcinoma. In Hepatocellular Carcinoma; Tirnitz-Parker, J., Ed.; Codon Publications: Brisbane, Austrilia, 2019. [Google Scholar]
- Chedid, M.F.; Kruel, C.R.P.; Pinto, M.A.; Grezzana-Filho, T.J.M.; Leipnitz, I.; Kruel, C.D.P.; Scaffaro, L.A.; Chedid, A.D. Hepatocellular Carcinoma: Diagnosis and Operative Management. ABCD Arq. Bras. Cir. Dig. 2017, 30, 272–278. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Y.N.; Wang, K.T.; Chen, L. Lenvatinib for hepatocellular carcinoma: From preclinical mechanisms to anti-cancer therapy. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188391. [Google Scholar] [CrossRef]
- Costa, E.; Ferreira-Gonçalves, T.; Chasqueira, G.; Cabrita, A.S.; Figueiredo, I.V.; Reis, C.P. Experimental models as refined translational tools for breast cancer research. Sci. Pharm. 2020, 88, 32. [Google Scholar] [CrossRef]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In vitro tumor models: Advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef]
- Mirabelli, P.; Coppola, L.; Salvatore, M. Cancer cell lines are useful model systems for medical research. Cancers 2019, 11, 1098. [Google Scholar] [CrossRef] [Green Version]
- Saydé, T.; Hamoui, O.E.; Alies, B.; Gaudin, K.; Lespes, G.; Battu, S. Biomaterials for three-dimensional cell culture: From applications in oncology to nanotechnology. Nanomaterials 2021, 11, 481. [Google Scholar] [CrossRef]
- Kimlin, L.C.; Casagrande, G.; Virador, V.M. In vitro three-dimensional (3D) models in cancer research: An update. Mol. Carcinog. 2013, 52, 167–182. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Leung, M. Chitosan-Alginate Scaffold Culture System for Hepatocellular Carcinoma Increases Malignancy and Drug Resistance. Bone 2011, 23, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Chiew, G.G.Y.; Wei, N.; Sultania, S.; Lim, S.; Luo, K.Q. Bioengineered three-dimensional co-culture of cancer cells and endothelial cells: A model system for dual analysis of tumor growth and angiogenesis. Biotechnol. Bioeng. 2017, 114, 1865–1877. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, R.; Everett, W.; Lim, S.; Natasha, G.; Loizidou, M.; Jell, G.; Tan, A.; Seifalian, A.M. Personalized in vitro cancer modeling—Fantasy or reality? Transl. Oncol. 2014, 7, 657–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koledova, Z. 3D Cell Culture: An Introduction. Methods Mol. Biol. 2017, 1612, 325–344. [Google Scholar]
- Wang, C.; Tang, Z.; Zhao, Y.; Yao, R.; Li, L.; Sun, W. Three-dimensional in vitro cancer models: A short review. Biofabrication 2014, 6, 022001. [Google Scholar] [CrossRef]
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Paul Solomon, F.D. 3D cell culture systems: Advantages and applications. J. Cell. Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef]
- Collins, S.D.; Yuen, G.; Tu, T.; Budzinska, M.A.; Spring, K.; Bryant, K.; Shackel, N.A. In Vitro Models of the Liver: Disease Modeling, Drug Discovery and Clinical Applications. In Hepatocellular Carcinoma; Tirnitz-Parker, J., Ed.; Codon Publications: Brisbane, Austrilia, 2019. [Google Scholar]
- Qiu, Z.; Zou, K.; Zhuang, L.; Qin, J.; Li, H.; Li, C.; Zhang, Z.; Chen, X.; Cen, J.; Meng, Z.; et al. Hepatocellular carcinoma cell lines retain the genomic and transcriptomic landscapes of primary human cancers. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Fukuyama, K.; Asagiri, M.; Sugimoto, M.; Tsushima, H.; Seo, S.; Taura, K.; Uemoto, S.; Iwaisako, K. Gene expression profiles of liver cancer cell lines reveal two hepatocyte-like and fibroblast-like clusters. PLoS ONE 2021, 16, e0245939. [Google Scholar] [CrossRef]
- Qiu, G.H.; Xie, X.; Xu, F.; Shi, X.; Wang, Y.; Deng, L. Distinctive pharmacological differences between liver cancer cell lines HepG2 and Hep3B. Cytotechnology 2015, 67, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, J.; Wei, W.; Chen, Z.; Lin, B.; Zhao, W.; Luo, Y.; Zhang, X. Engineered liver-on-a-chip platform to mimic liver functions and its biomedical applications: A review. Micromachines 2019, 10, 676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinken, M.; Rogiers, V. Culture and Functional Characterization of Human Hepatoma HepG2 Cells María. Protoc. Vitr. Hepatocyte Res. 2015, 1250, 1–390. [Google Scholar]
- Mavri-Damelin, D.; Damelin, L.H.; Eaton, S.; Rees, M.; Selden, C.; Hodgson, H.J. Cells for bioartificial liver devices: The human hepatoma-derived cell line C3A produces urea but does not detoxify ammonia. Biotechnol. Bioeng. 2008, 99, 644–651. [Google Scholar] [CrossRef]
- van Wenum, M.; Adam, A.A.A.; Hakvoort, T.B.M.; Hendriks, E.J.; Shevchenko, V.; van Gulik, T.M.; Chamuleau, R.A.F.M.; Hoekstra, R. Selecting cells for bioartificial liver devices and the importance of a 3D culture environment: A functional comparison between the hepaRG and C3A cell lines. Int. J. Biol. Sci. 2016, 12, 964–978. [Google Scholar] [CrossRef]
- Goyak, K.M.O.; Laurenzana, E.M.; Omiecinski, C.J. Hepatocyte Differentiation. Methods Mol. Biol. 2010, 640, 115–138. [Google Scholar]
- Lacoste, B.; Raymond, V.A.; Cassim, S.; Lapierre, P.; Bilodeau, M. Highly tumorigenic hepatocellular carcinoma cell line with cancer stem cell-like properties. PLoS ONE 2017, 12, e0171215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ATCC:The Global Bioresource Center. Available online: https://www.atcc.org/ (accessed on 10 June 2021).
- Wei, J.C.; Meng, F.D.; Qu, K.; Wang, Z.X.; Wu, Q.F.; Zhang, L.Q.; Pang, Q.; Liu, C. Sorafenib inhibits proliferation and invasion of human hepatocellular carcinoma cells via up-regulation of p53 and suppressing FoxM1. Acta Pharmacol. Sin. 2015, 36, 241–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Liu, Y.; Meng, L.; Ji, B.; Yang, D. Synergistic antitumor effect of sorafenib in combination with ATM inhibitor in hepatocellular carcinoma cells. Int. J. Med. Sci. 2017, 14, 523–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Qin, S. Apatinib targets both tumor and endothelial cells in hepatocellular carcinoma. Cancer Med. 2018, 7, 4570–4583. [Google Scholar] [CrossRef] [Green Version]
- Hoshi, T.; Watanabe Miyano, S.; Watanabe, H.; Sonobe, R.M.K.; Seki, Y.; Ohta, E.; Nomoto, K.; Matsui, J.; Funahashi, Y. Lenvatinib induces death of human hepatocellular carcinoma cells harboring an activated FGF signaling pathway through inhibition of FGFR–MAPK cascades. Biochem. Biophys. Res. Commun. 2019, 513, 1–7. [Google Scholar] [CrossRef]
- Shi, T.; Fujita, K.; Gong, J.; Nakahara, M.; Iwama, H.; Liu, S.; Yoneyama, H.; Morishita, A.; Nomura, T.; Tani, J.; et al. Aspirin inhibits hepatocellular carcinoma cell proliferation in vitro and in vivo via inducing cell cycle arrest and apoptosis. Oncol. Rep. 2020, 44, 457–468. [Google Scholar] [CrossRef]
- Lin, J.; Schyschka, L.; Mühl-Benninghaus, R.; Neumann, J.; Hao, L.; Nussler, N.; Dooley, S.; Liu, L.; Stöckle, U.; Nussler, A.K.; et al. Comparative analysis of phase I and II enzyme activities in 5 hepatic cell lines identifies Huh-7 and HCC-T cells with the highest potential to study drug metabolism. Arch. Toxicol. 2012, 86, 87–95. [Google Scholar] [CrossRef]
- Zhai, J.M.; Yin, X.Y.; Lai, Y.R.; Hou, X.; Cai, J.P.; Hao, X.Y.; Liang, L.J.; Zhang, L.J. Sorafenib enhances the chemotherapeutic efficacy of S-1 against hepatocellular carcinoma through downregulation of transcription factor E2F-1. Cancer Chemother. Pharmacol. 2013, 71, 1255–1264. [Google Scholar] [CrossRef]
- Hoekstra, R.; Nibourg, G.A.A.; Van Der Hoeven, T.V.; Ackermans, M.T.; Hakvoort, T.B.M.; Van Gulik, T.M.; Lamers, W.H.; Elferink, R.P.O.; Chamuleau, R.A.F.M. The HepaRG cell line is suitable for bioartificial liver application. Int. J. Biochem. Cell Biol. 2011, 43, 1483–1489. [Google Scholar] [CrossRef]
- Andersson, T.B.; Kanebratt, K.P.; Kenna, J.G. The HepaRG cell line: A unique in vitro tool for understanding drug metabolism and toxicology in human. Expert Opin. Drug Metab. Toxicol. 2012, 8, 909–920. [Google Scholar] [CrossRef]
- Xie, F.; Sun, L.; Pang, Y.; Xu, G.; Jin, B.; Xu, H.; Lu, X.; Xu, Y.; Du, S.; Wang, Y.; et al. Three-dimensional bio-printing of primary human hepatocellular carcinoma for personalized medicine. Biomaterials 2021, 265, 120416. [Google Scholar] [CrossRef]
- Sonntag, R.; Gassler, N.; Bangen, J.M.; Trautwein, C.; Liedtke, C. Pro-apoptotic Sorafenib signaling in murine hepatocytes depends on malignancy and is associated with PUMA expression in vitro and in vivo. Cell Death Dis. 2014, 5, e1030-12. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.R.; Park, S.C.; Choi, S.J.; Lee, J.C.; Kim, Y.C.; Han, C.J.; Kim, J.; Yang, K.Y.; Kim, Y.J.; Noh, G.Y.; et al. Combined treatment with silibinin and either sorafenib or gefitinib enhances their growth-inhibiting effects in hepatocellular carcinoma cells. Clin. Mol. Hepatol. 2015, 21, 49–59. [Google Scholar] [CrossRef]
- Mountcastle, S.E.; Cox, S.C.; Sammons, R.L.; Jabbari, S.; Shelton, R.M.; Kuehne, S.A. A review of co-culture models to study the oral microenvironment and disease. J. Oral Microbiol. 2020, 12, 1773122. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Ono, K.; Hata, S.; Suzuki, T.; Kumamoto, H.; Sasano, H. The advantages of co-culture over mono cell culture in simulating in vivo environment. J. Steroid Biochem. Mol. Biol. 2012, 131, 68–75. [Google Scholar] [CrossRef]
- Shuichi, I.; Shimada, M.; Morine, Y.; Imura, S.; Ikemoto, T.; Saito, Y.; Yamada, S.; Rui, F. The effect of hepatic stellate cells on hepatocellular carcinoma progression. J. Clin. Oncol. 2019, 37, 265. [Google Scholar]
- Chen, W.; Wu, J.; Shi, H.; Wang, Z.; Zhang, G.; Cao, Y.; Jiang, C.; Ding, Y. Hepatic stellate cell coculture enables sorafenib resistance in Huh7 cells through HGF/c-Met/Akt and Jak2/Stat3 pathways. Biomed. Res. Int. 2014, 2014, 764981. [Google Scholar] [CrossRef] [PubMed]
- Fasolino, I.; Guarino, V.; Marrese, M.; Cirillo, V.; Vallifuoco, M.; Tamma, M.L.; Vassallo, V.; Bracco, A.; Calise, F.; Ambrosio, L. HepG2 and human healthy hepatocyte in vitro culture and co-culture in PCL electrospun platforms. Biomed. Mater. 2018, 13, 015017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.W.; Kook, Y.M.; Lee, H.J.; Park, H.; Koh, W.G. A three-dimensional co-culture of HepG2 spheroids and fibroblasts using double-layered fibrous scaffolds incorporated with hydrogel micropatterns. RSC Adv. 2014, 4, 61005–61011. [Google Scholar] [CrossRef]
- Coulouarn, C.; Corlu, A.; Glaise, D.; Guénon, I.; Thorgeirsson, S.S.; Clément, B. Hepatocyte-stellate cell cross-talk in the liver engenders a permissive inflammatory microenvironment that drives progression in hepatocellular carcinoma. Cancer Res. 2012, 72, 2533–2542. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.R.; Kang, H.M.; Ryu, J.W.; Kim, D.S.; Noh, K.H.; Kim, E.S.; Lee, H.J.; Chung, K.S.; Cho, H.S.; Kim, N.S.; et al. Cell Spheroids with Enhanced Aggressiveness to Mimic Human Liver Cancer in Vitro and in Vivo. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Štampar, M.; Tomc, J.; Filipič, M.; Žegura, B. Development of in vitro 3D cell model from hepatocellular carcinoma (HepG2) cell line and its application for genotoxicity testing. Arch. Toxicol. 2019, 93, 3321–3333. [Google Scholar] [CrossRef]
- Takai, A.; Fako, V.; Dang, H.; Forgues, M.; Yu, Z.; Budhu, A.; Wang, X.W. Three-dimensional Organotypic Culture Models of Human Hepatocellular Carcinoma. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Takeishi, K.; Li, Z.; Cervantes-Alvarez, E.; de L’Hortet, L.C.; Guzman-Lepe, J.; Cui, X.; Zhu, J. Microenvironment of a tumor-organoid system enhances hepatocellular carcinoma malignancyrelated hallmarks. Organogenesis 2017, 13, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Taymour, R.; Kilian, D.; Ahlfeld, T.; Gelinsky, M.; Lode, A. 3D bioprinting of hepatocytes: Core–shell structured co-cultures with fibroblasts for enhanced functionality. Sci. Rep. 2021, 11, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, H.; Wang, Y.; Zhang, X.; Jin, B.; Xie, F.; Jin, Y.; Pang, Y.; Zhao, H.; Lu, X.; et al. Application of a 3D Bioprinted Hepatocellular Carcinoma Cell Model in Antitumor Drug Research. Front. Oncol. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Yu, C.; Wang, P.; Xu, W.; Wan, X.; Lai, C.S.E.; Liu, J.; Koroleva-Maharajh, A.; Chen, S. Rapid 3D bioprinting of decellularized extracellular matrix with regionally varied mechanical properties and biomimetic microarchitecture. Biomaterials 2018, 185, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Velliou, E.; Gupta, P.; Ricci, C.; Danti, S. Biomaterial-based in vitro models for pancreatic cancer. In Materials Today, Biomaterials for 3D Tumor Modeling; Subhas, C.K., Rui, L.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 235–249. [Google Scholar]
- Khanna, S.; Bhatt, A.N.; Dwarakanath, B.S. Multicellular Spheroid: 3-D Tissue Culture Model for Cancer Research. In Animal Biotechnology; Academic Press: New York, NY, USA, 2014; pp. 195–210. [Google Scholar]
- Fiorini, E.; Veghini, L.; Corbo, V. Modeling Cell Communication in Cancer With Organoids: Making the Complex Simple. Front. Cell Dev. Biol. 2020, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Rolver, M.G.; Elingaard-Larsen, L.O.; Pedersen, S.F. Assessing cell viability and death in 3d spheroid cultures of cancer cells. J. Vis. Exp. 2019, 2019, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Kim, J.S.; Kim, S.H.; Park, Y.K.; Yu, E.; Kim, K.H.; Seo, E.J.; Oh, H.B.; Lee, H.C.; Kim, K.M.; et al. Patient-derived multicellular tumor spheroids towards optimized treatment for patients with hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 1–13. [Google Scholar] [CrossRef]
- Rijal, G.; Li, W. A versatile 3D tissue matrix scaffold system for tumor modeling and drug screening. Sci. Adv. 2017, 3, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2016, 14, 910–919. [Google Scholar] [CrossRef] [PubMed]
- Porter, R.J.; Murray, G.I.; McLean, M.H. Current concepts in tumour-derived organoids. Br. J. Cancer 2020, 123, 1209–1218. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. 2017, 22, 456–472. [Google Scholar]
- Kronemberger, G.S.; Carneiro, F.A.; Rezende, D.F.; Baptista, L.S. Spheroids and organoids as humanized 3D scaffold-free engineered tissues for SARS-CoV-2 viral infection and drug screening. Artif. Organs. 2021, 45, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Vivarelli, S.; Candido, S.; Caruso, G.; Falzone, L.; Libra, M. Patient-derived tumor organoids for drug repositioning in cancer care: A promising approach in the era of tailored treatment. Cancers 2020, 12, 3636. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef]
- Datta, P.; Dey, M.; Ataie, Z.; Unutmaz, D.; Ozbolat, I.T. 3D bioprinting for reconstituting the cancer microenvironment. NPJ Precis. Oncol. 2020, 4, 18. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.H.; Karlsson, K.; Kuo, C.J. Applications of organoids for cancer biology and precision medicine. Nat. Cancer 2020, 1, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Nuciforo, S.; Heim, M.H. Organoids to model liver disease. JHEP Rep. 2021, 3, 100198. [Google Scholar] [CrossRef] [PubMed]
- Nuciforo, S.; Fofana, I.; Matter, M.S.; Blumer, T.; Calabrese, D.; Boldanova, T.; Piscuoglio, S.; Wieland, S.; Ringnalda, F.; Schwank, G.; et al. Organoid Models of Human Liver Cancers Derived from Tumor Needle Biopsies. Cell Rep. 2018, 24, 1363–1376. [Google Scholar] [CrossRef] [Green Version]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and animal models: Are 3D cultures the ideal tool to study cancer-microenvironment interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [Green Version]
- Augustine, R.; Kalva, S.N.; Ahmad, R.; Zahid, A.A.; Hasan, S.; Nayeem, A.; McClements, L.; Hasan, A. 3D Bioprinted cancer models: Revolutionizing personalized cancer therapy. Transl. Oncol. 2021, 14, 101015. [Google Scholar] [CrossRef]
- Bae, J.; Han, S.; Park, S. Recent Advances in 3D Bioprinted Tumor Microenvironment. Biochip J. 2020, 14, 137–147. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, X.; Zhou, X.; Liu, C. A 3D bioprinting liver tumor model for drug screening. World J. Pharm. Pharm. Sci. 2016, 5, 196–213. [Google Scholar]
- Wu, Q.; Liu, J.; Wang, X.; Feng, L.; Wu, J.; Zhu, X.; Wen, W.; Gong, X. Organ-on-a-chip: Recent breakthroughs and future prospects. Biomed. Eng. Online 2020, 19, 9. [Google Scholar] [CrossRef] [Green Version]
- Trujillo-de Santiago, G.; Flores-Garza, B.G.; Tavares-Negrete, J.A.; Lara-Mayorga, I.M.; González-Gamboa, I.; Zhang, Y.S.; Rojas-Martínez, A.; Ortiz-López, R.; Álvarez, M.M. The tumor-on-chip: Recent advances in the development of microfluidic systems to recapitulate the physiology of solid tumors. Materials 2019, 12, 2945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehrlich, A.; Duche, D.; Ouedraogo, G.; Nahmias, Y. Challenges and Opportunities in the Design of Liver-on-Chip Microdevices. Annu. Rev. Biomed. Eng. 2019, 21, 219–239. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Cuzzucoli, F.; Jiang, J.; Liang, L.G.; Wang, Y.; Kong, M.; Zhao, X.; Cui, W.; Li, J.; Wang, S.Q. Development of a biomimetic liver tumor-on-a-chip model based on decellularized liver matrix for toxicity testing. Lab Chip 2018, 18, 3379–3392. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, F.; Yesil-Celiktas, O.; Kazan, A.; Maharjan, S.; Saghazadeh, S.; Firoozbakhsh, K.; Firoozabadi, B.; Zhang, Y.S. A hepatocellular carcinoma–bone metastasis-on-a-chip model for studying thymoquinone-loaded anticancer nanoparticles. Bio-Des. Manuf. 2020, 3, 189–202. [Google Scholar] [CrossRef]
- Li, Z.; Zheng, W.; Wang, H.; Cheng, Y.; Fang, Y.; Wu, F.; Sun, G.; Sun, G.; Lv, C.; Hui, B. Application of animal models in cancer research: Recent progress and future prospects. Cancer Manag. Res. 2021, 13, 2455–2475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Moore, L.; Ji, P. Mouse models for cancer research. Chinese J. Cancer. 2007, 30, 149–152. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Meyer, C.; Xu, C.; Weng, H.; Hellerbrand, C.; ten Dijke, P.; Dooley, S. Animal models of chronic liver diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G449–G468. [Google Scholar] [CrossRef] [Green Version]
- Gargiulo, G. Next-generation in vivo modeling of human cancers. Front. Oncol. 2018, 8, 429. [Google Scholar] [CrossRef]
- Santos, N.P.; Colaço, A.A.; Oliveira, P.A. Animal models as a tool in hepatocellular carcinoma research: A Review. Tumor Biol. 2017, 39, 1010428317695923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.E.; Henderson, J.M.; Gorrell, M.D. Animal models for hepatocellular carcinoma. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 993–1002. [Google Scholar] [CrossRef]
- Guerin, M.V.; Finisguerra, V.; Van den Eynde, B.J.; Bercovici, N.; Trautmann, A. Preclinical murine tumor models: A structural and functional perspective. Elife 2020, 9, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Maronpot, R.R. Biological basis of differential susceptibility to hepatocarcinogenesis among mouse strains. J. Toxicol. Pathol. 2009, 22, 11–33. [Google Scholar] [CrossRef] [Green Version]
- Rogers, A.B. Stress of strains: Inbred mice in liver research. Gene Expr. 2018, 19, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Jilkova, Z.M.; Kurma, K.; Decaens, T. Animal Models of Hepatocellular Carcinoma: The Role of Immune System. Cancers 2019, 11, 1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heindryckx, F.; Colle, I.; Van Vlierberghe, H. Experimental mouse models for hepatocellular carcinoma research. Int. J. Exp. Pathol. 2009, 90, 367–386. [Google Scholar] [CrossRef]
- He, L.; Tian, D.A.; Li, P.Y.; He, X.X. Mouse models of liver cancer: Progress and recommendations. Oncotarget 2015, 6, 23306–23322. [Google Scholar] [CrossRef]
- Memon, A.; Pyao, Y.; Jung, Y.; Lee, J.I.; Lee, W.K. A modified protocol of diethylnitrosamine administration in mice to model hepatocellular carcinoma. Int. J. Mol. Sci. 2020, 21, 5461. [Google Scholar] [CrossRef]
- Da Costa, R.M.G.; Paula-Santos, N.; Rocha, A.F.; Colaç, A.; Lopes, C.; Oliveira, P.A. The N-nitrosodiethylamine mouse model: Sketching a timeline of evolution of chemically-induced hepatic lesions. Anticancer Res. 2014, 34, 7029–7037. [Google Scholar]
- Mohammed, E.S.; El-Beih, N.M.; El-Hussieny, E.A.; EL-Ahwany, E.; Hassan, M.; Zoheiry, M. Effects of free and nanoparticulate curcumin on chemically induced liver carcinoma in an animal model. Arch. Med. Sci. 2018, 17, 218–227. [Google Scholar] [CrossRef]
- Luo, M.; Yang, F.; Huang, S.X.; Kuang, Z.P.; Luo, X.L.; Li, Y.D.; Wu, J.N.; Xie, Y.A. Two-stage model of chemically induced hepatocellular carcinoma in mouse. Oncol. Res. 2013, 20, 517–528. [Google Scholar] [CrossRef]
- Brown, Z.J.; Heinrich, B.; Greten, T.F. Mouse models of hepatocellular carcinoma: An overview and highlights for immunotherapy research. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 536–554. [Google Scholar] [CrossRef]
- Kersten, K.; Visser, K.E.; Miltenburg, M.H.; Jonkers, J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 2017, 9, 137–153. [Google Scholar] [CrossRef]
- Jung, J. Human tumor xenograft models for preclinical assessment of anticancer drug development. Toxicol. Res. 2014, 30, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Schepers, E.; Avella, D.; Kimchi, E.T.; Kaifi, J.T.; Staveley-O’carroll, K.F.; Li, G. An oncogenic hepatocyte-induced orthotopic mouse model of hepatocellular cancer arising in the setting of hepatic inflammation and fibrosis. J. Vis. Exp. 2019, 2019, 2–9. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, J.; Liu, W.N.; Zhao, Y. Cancer Immunotherapies and Humanized Mouse Drug Testing Platforms. Transl. Oncol. 2019, 12, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Bresnahan, E.; Lindblad, K.E.; de Galarreta, M.R.; Lujambio, A. Mouse Models of Oncoimmunology in Hepatocellular Carcinoma. Clin. Cancer Res. 2020, 26, 5276–5286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiberger, T.; Chen, Y.; Ramjiawan, R.R.; Hato, T.; Fan, C.; Samuel, R.; Roberge, S.; Huang, P.; Lauwers, G.Y.; Zhu, A.X.; et al. An orthotopic mouse model of hepatocellular carcinoma with underlying liver cirrhosis. Nat. Protoc. 2015, 10, 1264–1274. [Google Scholar] [CrossRef] [Green Version]
- Walrath, J.C.; Hawes, J.J.; Van Dyke, T.; Reilly, K.M. Genetically engineered mouse models in cancer research. Adv. Cancer Res. 2010, 106, 113–164. [Google Scholar]
- Singh, M.; Murriel, C.L.; Johnson, L. Genetically engineered mouse models: Closing the gap between preclinical data and trial outcomes. Cancer Res. 2012, 72, 2695–2700. [Google Scholar] [CrossRef] [Green Version]
- Ju, H.L.; Han, K.H.; Lee, J.D.; Ro, S.W. Transgenic mouse models generated by hydrodynamic transfection for genetic studies of liver cancer and preclinical testing of anti-cancer therapy. Int. J. Cancer 2016, 138, 1601–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, J.; Dufour, J.F. Tumor suppressor and hepatocellular carcinoma. World J. Gastroenterol. 2008, 14, 1720–1733. [Google Scholar] [CrossRef] [Green Version]
- Chung, S.I.; Moon, H.; Kim, D.Y.; Cho, K.J.; Ju, H.L.; Kim, D.Y.; Ahn, S.H.; Han, K.H.; Ro, S.W. Development of a transgenic mouse model of hepatocellular carcinoma with a liver fibrosis background. BMC Gastroenterol. 2016, 16, 13. [Google Scholar] [CrossRef] [Green Version]
- Guil-Luna, S.; Sedlik, C.; Piaggio, E. Humanized Mouse Models to Evaluate Cancer Immunotherapeutics. Annu. Rev. Cancer Biol. 2020, 5, 119–136. [Google Scholar] [CrossRef]
- Tian, H.; Lyu, Y.; Yang, Y.G.; Hu, Z. Humanized Rodent Models for Cancer Research. Front. Oncol. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Akkina, R. New generation humanized mice for virus research: Comparative aspects and future prospects. Virology 2013, 23, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Wang, X.; Chen, D.; Liu, X.; Wang, X. Humanized mouse model: A review on preclinical applications for cancer immunotherapy. Am. J. Cancer Res. 2020, 10, 4568–4584. [Google Scholar] [PubMed]
- Maurice Morillon, Y.; Sabzevari, A.; Schlom, J.; Greiner, J.W. The development of next-generation PBMC humanized mice for preclinical investigation of cancer immunotherapeutic agents. Anticancer Res. 2020, 40, 5329–5341. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shuen, T.W.H.; Toh, T.B.; Chan, X.Y.; Liu, M.; Tan, S.Y.; Fan, Y.; Yang, H.; Lyer, S.G.; Bonney, G.K.; et al. Development of a new patient-derived xenograft humanised mouse model to study human-specific tumour microenvironment and immunotherapy. Gut 2018, 67, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Shi, J.; Li, S.; Wang, Q.; Wang, Q.; Wen, X.; Yang, F.; Duan, Z.; Yang, Y.; Zhang, X.; et al. A novel xenograft model of human HCC in immunocompetent mouse. bioRxiv 2019. [CrossRef]
- Trisilowati; Mallet, D.G. In Silico Experimental Modeling of Cancer Treatment. ISRN Oncol. 2012, 2012, 1–8. [Google Scholar]
- Thoolen, B.; ten Kate, F.J.W.; van Diest, P.J.; Malarkey, D.E.; Elmore, S.A.; Maronpot, R.R. Comparative histomorphological review of rat and human hepatocellular proliferative lesions. J. Toxicol. Pathol. 2012, 25, 189–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shankaraiah, R.C.; Gramantieri, L.; Fornari, F.; Sabbioni, S.; Callegari, E.; Negrini, M. Animal models of hepatocellular carcinoma prevention. Cancers 2019, 11, 1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Lu, S.; Zheng, H.; Xu, M.; Song, J.; Yang, W.; Weng, Q.; Zheng, L.; Fan, X.; Cheng, X.; et al. Identification of the Potential Metabolic Pathways Involved in the Hepatic Tumorigenesis of Rat Diethylnitrosamine-Induced Hepatocellular Carcinoma via 1 H NMR-Based Metabolomic Analysis. Biomed. Res. Int. 2019, 2019, 9367082. [Google Scholar] [CrossRef] [Green Version]
- Ciccarelli, O.; Colson, A.; De Saeger, C.; Reding, R.; Sempoux, C.; Leclercq, I.A.; Stärkel, P. Tumoral response and tumoral phenotypic changes in a rat model of diethylnitrosamine-induced hepatocellular carcinoma after salirasib and sorafenib administration. Onco. Targets. Ther. 2018, 11, 7143–7153. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.T.; Liu, C.J.; Su, T.H.; Cheng, H.R.; Jeng, Y.M.; Lin, H.L.; Wang, C.C.; Kao, J.H.; Chen, P.J.; Chen, D.S.; et al. Characterization of metastatic tumor antigen 1 and its interaction with hepatitis B virus X protein in NF-κB signaling and tumor progression in a woodchuck hepatocellular carcinoma model. Oncotarget 2016, 7, 47173–47185. [Google Scholar] [CrossRef] [Green Version]
- Kim, A.Y.; Yacoub, J.H.; Field, D.H.; Park, B.U.; Kallakury, B.; Korolowicz, K.E.; Menne, S. Suitability of the woodchuck HCC as a preclinical model for evaluation of intra-arterial therapies. Anim. Model. Exp. Med. 2020, 3, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.Y.; Ma, X.Z.; Ouyang, B.; Ings, D.P.; Marwah, S.; Liu, J.; Chen, A.Y.; Gupta, R.; Manuel, J.; Chen, X.C.; et al. Nanoparticle Uptake in a Spontaneous and Immunocompetent Woodchuck Liver Cancer Model. ACS Nano 2020, 14, 4698–4715. [Google Scholar] [CrossRef] [PubMed]
- Press, Z.W. Application of the woodchuck animal model for the treatment of hepatitis B virus-induced liver cancer. World J. Gastrointest Oncol. 2021, 13, 509–535. [Google Scholar]
- Blair, R. Transarterial Chemoembolization in a Woodchuck Model of Hepatocellular Carcinoma William. Physiol. Behav. 2017, 176, 139–148. [Google Scholar]
- Zhao, S.; Huang, J.; Ye, J. A fresh look at zebrafish from the perspective of cancer research. J. Exp. Clin. Cancer Res. 2015, 34, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Hason, M.; Bartůnĕk, P. Zebrafish models of cancer-new insights on modeling human cancer in a non-mammalian vertebrate. Genes 2019, 10, 935. [Google Scholar] [CrossRef] [Green Version]
- Huiting, L.; Laroche, F.; Feng, H. The Zebrafish as a Tool to Cancer Drug Discovery Current Challenges in Drug Discovery HHS Public Access. Austin J. Pharmacol. Ther. 2015, 3, 1069. [Google Scholar]
- Zhang, T.; Peterson, R.T. Zebrafish as a Platform for Drug Screening; Elsevier: Amsterdam, The Netherlands, 2019; ISBN 9780128124314. [Google Scholar]
- Cassar, S.; Adatto, I.; Freeman, J.L.; Gamse, J.T.; Lawrence, C.; Muriana, A.; Peterson, R.T.; Van Cruchten, S.; Zon, L.I. Use of Zebrafish in Drug Discovery Toxicology. Chem. Res. Toxicol. 2020, 33, 95–118. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Glasgow, E.; Agarwal, S. Zebrafish Xenografts for Drug Discovery and Personalized Medicine. Trends Cancer 2020, 6, 569–579. [Google Scholar] [CrossRef]
- Wrighton, P.J.; Oderberg, I.M.; Goessling, W. There Is Something Fishy About Liver Cancer: Zebrafish Models of Hepatocellular Carcinoma. CMGH 2019, 8, 347–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, J.; Gong, Z. Transgenic zebrafish for modeling hepatocellular carcinoma. MedComm 2020, 1, 140–156. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Emelyanov, A.; Koh, C.H.V.; Spitsbergen, J.M.; Parinov, S.; Gong, Z. An inducible kras V12 transgenic zebrafish model for liver tumorigenesis and chemical drug screening. DMM Dis. Model. Mech. 2012, 5, 63–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeanquartier, F.; Jean-Quartier, C.; Kotlyar, M.; Tokar, T.; Hauschild, A.C.; Jurisica, I.; Holzinger, A. Machine learning for In Silico modeling of tumor growth. In Machine Learning for Health Informatics; Holzinger, A., Ed.; Lecture Notes in Computer Science; Springer: Cham, Swithzerland, 2016; Volume 9605, pp. 415–434. [Google Scholar]
- Sacan, A.; Ekins, S.; Kortagere, S. Applications and limitations of in silico models in drug discovery. Methods Mol. Biol. 2012, 910, 87–124. [Google Scholar] [PubMed]
- Jeanquartier, F.; Jean-Quartier, C.; Cemernek, D.; Holzinger, A. In silico modeling for tumor growth visualization. BMC Syst. Biol. 2016, 10, 59. [Google Scholar] [CrossRef] [Green Version]
- Jean-Quartier, C.; Jeanquartier, F.; Jurisica, I.; Holzinger, A. In silico cancer research towards 3R. BMC Cancer 2018, 18, 408. [Google Scholar] [CrossRef]
- Liu, T.P.; Hong, Y.H.; Yang, P.M. In silico and in vitro identification of inhibitory activities of sorafenib on histone deacetylases in hepatocellular carcinoma cells. Oncotarget 2017, 8, 86168–86180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.N.; Li, H.; Yao, H.; Liu, X.; Li, L.; Leung, K.S.; Kung, H.; Lu, D.; Wong, M.H.; Lin, M.C.M. In Silico Identification and In Vitro and In Vivo Validation of Anti-Psychotic Drug Fluspirilene as a Potential CDK2 Inhibitor and a Candidate Anti-Cancer Drug Xi-Nan. PLoS ONE 2015, 10, e0132072. [Google Scholar]
- Sun, Y.; Zhang, Z. In Silico Identification of Crucial Genes and Specific Pathways in Hepatocellular Cancer. Genet. Test. Mol. Biomark. 2020, 24, 296–308. [Google Scholar] [CrossRef]
- Mabrouk, M.S. Discovering best candidates for Hepatocellular Carcinoma (HCC) by in-silico techniques and tools. Int. J. Bioinform. Res. Appl. 2012, 8, 141–152. [Google Scholar] [CrossRef]
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020, 471, 61–71. [Google Scholar] [CrossRef]
- Azuaje, F. Artificial intelligence for precision oncology: Beyond patient stratification. NPJ Precis. Oncol. 2019, 3, 1–5. [Google Scholar] [CrossRef]
- Nagy, M.; Radakovich, N.; Nazha, A. Machine Learning in Oncology: What Should Clinicians Know? JCO Clin. Cancer Inform. 2020, 4, 799–810. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Javed, Z.; Sadia, H.; Qureshi, I.A.; Irshad, A.; Ahmed, R.; Malik, K.; Raza, S.; Abbas, A.; Pezzani, R.; et al. Clinical applications of artificial intelligence and machine learning in cancer diagnosis: Looking into the future. Cancer Cell Int. 2021, 21, 1–11. [Google Scholar] [CrossRef]
- Ho, D. Artificial intelligence in cancer therapy. Science 2020, 367, 982–983. [Google Scholar] [CrossRef]
- Lai, Q.; Spoletini, G.; Mennini, G.; Laureiro, Z.L.; Tsilimigras, D.I.; Pawlik, T.M.; Rossi, M. Prognostic role of artificial intelligence among patients with hepatocellular cancer: A systematic review. World J. Gastroenterol. 2020, 26, 6679–6688. [Google Scholar] [CrossRef] [PubMed]
- Linton-Reid, K. Introduction: An Overview of AI in Oncology Drug Discovery and Development, Artificial Intelligence in Oncology Drug Discovery and Development; John, W.C., Belle, T., Eds.; IntechOpen: Rijeka, Croatia, 2020. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef] [Green Version]
- Rashidi, H.H.; Tran, N.K.; Betts, E.V.; Howell, L.P.; Green, R. Artificial Intelligence and Machine Learning in Pathology: The Present Landscape of Supervised Methods. Acad. Pathol. 2019, 6, 2374289519873088. [Google Scholar] [CrossRef]
- Roemer, E.J.; West, K.L.; Northrup, J.B.; Iverson, J.M. Supervised Machine Learning in Oncology: A Clinician’s Guide. Physiol. Behav. 2016, 176, 139–148. [Google Scholar]
- Zou, Z.M.; Chang, D.H.; Liu, H.; Xiao, Y.D. Current updates in machine learning in the prediction of therapeutic outcome of hepatocellular carcinoma: What should we know? Insights Imaging 2021, 12, 31. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning based multi-omics integration robustly predicts survival in liver cancer. Clin. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez, M.J.; Grande, R.G. Application of artificial intelligence in the diagnosis and treatment of hepatocellular carcinoma: A review. World J. Gastroenterol. 2020, 26, 5617–5628. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Morimoto, K.; Kajihara, S.; Tateishi, R.; Shiina, S.; Koike, K.; Yatomi, Y. Machine-learning Approach for the Development of a Novel Predictive Model for the Diagnosis of Hepatocellular Carcinoma. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abajian, A.; Murali, N.; Savic, L.J.; Laage-, F.M.; Nezami, N.; Duncan, J.S.; Schlachter, T.; Lin, M.; Geschwind, J.; Chapiro, J. Predicting Treatment Response to Intra-arterial Therapies of Hepatocellular Carcinoma using Supervised Machine Learning—An Artificial Intelligence Concept. J. Vasc. Interv. Radiol. 2018, 29, 850–857. [Google Scholar] [CrossRef]
- Morshid, A.; Elsayes, K.M.; Khalaf, A.M.; Elmohr, M.M.; Yu, J.; Kaseb, A.O.; Hassan, M.; Mahvash, A.; Wang, Z.; Hazle, J.D.; et al. A Machine Learning Model to Predict Hepatocellular Carcinoma Response to Transcatheter Arterial Chemoembolization. Radiol. Artif. Intell. 2019, 1, e180021. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, H.; Zeng, Y.; Liu, Z.; Ma, H.; Liu, J. Development and Validation of a Machine Learning Prognostic Model for Hepatocellular Carcinoma Recurrence After Surgical Resection. Front. Oncol. 2021, 10, 1–9. [Google Scholar] [CrossRef]
- Saito, A.; Toyoda, H.; Kobayashi, M.; Koiwa, Y.; Fujii, H.; Fujita, K.; Maeda, A.; Kaneoka, Y.; Hazama, S.; Nagano, H.; et al. Prediction of early recurrence of hepatocellular carcinoma after resection using digital pathology images assessed by machine learning. Mod. Pathol. 2021, 34, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Kaneko, S. Liver cancer stem cells: Recent progress in basic and clinical research. Regen. Ther. 2021, 17, 34–37. [Google Scholar] [CrossRef] [PubMed]





| Cell Line | Origin | Disease | Sensitivity to HCC Chemotherapy (Sorafenib) | Frequency (No. of PubMed Studies) | Applications |
|---|---|---|---|---|---|
| HepG2 | Homo sapiens, 15-year-old male [56] | HCC | IC50 = 6 µM [57]; IC50 = 7.42 µM [58] | 32,929 | 3D modeling; cancer research; toxicology studies; high-throughput screening [56] |
| Hep3B | Homo sapiens, 8-year-old juvenile male [56] | HCC | IC50 = 3.31 µM [58] | 2908 | 3D cell culture; high-throughput screening; cancer research; infectious and sexually transmitted disease research; toxicology evaluations [56] |
| HuH-7 | Homo sapiens, 57-year-old male [48] | Well-differentiated HCC | IC50 = 5.97 µM [58] | 2545 | 3D modeling [11]; drug testing [59,60] and repurposing [61]; drug metabolism studies [62] |
| C3A | Homo sapiens, 15-year-old male [48] | Differentiated HCC | - | 2070 | 3D cultures and cancer research [56] |
| SKHep1 | Homo sapiens, 52-year-old male [48] | Adenocarcinoma | IC50 = 5.3 ± 0.5 µM [63] | 976 | 3D modeling; cancer research; toxicology studies; high-throughput screening; cardiovascular disease research [56] |
| HepaRG | Homo sapiens, female patient [52] | Tumor from the liver of a female diagnosed with chronic hepatitis C and macronodular cirrhosis [54] | - | 835 | Bioartificial liver application [64]; in vitro drug metabolism and toxicology evaluations [65]; 3D model design [66] |
| Hepa1-6 | Mus musculus, C57L mouse strain [56] | Hepatoma | Effective concentrations = 10–50 µM [67] | 386 | 3D cultures and cancer research [56] |
| LMH | Gallus gallus, Leghorn strain chicken [56] | chemically induced HCC | - | 321 | 3D cultures and cancer research [56] |
| SNU-475 | Homo sapiens, 43-year-old male [56] | grade II–IV/V HCC | Effective concentrations = 20–50 µM [68] | 20 | 3D modeling; infectious disease research; sexually transmitted disease research; cancer research [56] |
| SNU-387 | Homo sapiens, 41-year-old female [56] | grade IV/V pleomorphic HCC | Effective concentrations = 10–50 µM [68] | 17 | 3D modeling; infectious disease research; sexually transmitted disease research; cancer research [56] |
| In Vitro 3D Model | Cell Line(s) | Observations | Reference |
|---|---|---|---|
| Co-culture on polycaprolactone electrospun scaffolds | HepG2 and patient-derived human healthy hepatocytes (HHH) | antiproliferative and antioxidant activities of the scaffold in the case of HepG2 cells and their co-culture with HHH | [73] |
| Co-culture on double-layered fibrous scaffolds incorporated with hydrogel micropatterns | HepG2 spheroids and fibroblasts | ↑ albumin secretion | [74] |
| Co-culture | HuH-7 and LX2 | induced drug (sorafenib) resistance in HCC cells by HGF/c-Met/Akt and Jak2/Stat3 signaling pathways | [72] |
| Co-culture | HepaRG and LX2 | increased expression of proinflammatory cytokines; ↑ VEGFA and matrix metalloproteinase-9 expression in hepatic stellate cells; permissive proangiogenic microenvironment | [75] |
| Co-culture | HuH-7 spheroids and human umbilical vein endothelial cells (HUVEC) | ↑ proliferation and gene expression of HCC-related genes; activation of the epithelial–mesenchymal transition (EMT) and angiogenic pathways; ↑ angiogenesis and vessel maturation | [76] |
| Spheroids | HuH-7 | activation of apoptotic and proliferative HIF-1α and ERK signals | [76] |
| Spheroids | HepG2 | experimental model for genotoxicity assessment | [77] |
| Organoid-like spheroids in porous alginate scaffolds | HuH-1, HuH-7, HepG2, Hep3B, SK-Hep-1 | ↑ sensitivity to TGF/β-induced EMT; ↑ in vivo tumorigenic and metastatic potential; ↑ resistance to chemotherapeutic drugs as compared to 2D cultures | [78] |
| Tumor Organoid System | HCCLM3, Hep3B, HUVEC, and human primary fibroblasts | similar features to human HCC observed in vivo; ↑ neo-angiogenesis-related markers (VEGFR2, VEGF, HIF-a), tumor-related inflammatory factors (CXCR4, CXCL12, TNF-a epithelial–mesenchymal transition markers (TGFb, Vimentin, MMP9) | [79] |
| Bioprinted Model | HepG2, NIH 3T3 | ↑ adhesion, viability, proliferation, function | [80] |
| Bioprinted Model | HepG2 | ↑ expression of tumor-related genes, differences in drug resistance genes as compared to 2D model | [81] |
| Cirrhotic decellularized ECM Scaffold Based Bioprinted Model | HepG2 | ↓ cell growth; ↑ invasion markers (matrix metalloproteinases MMP2 and MMP9, Twist-related protein 1) | [82] |
| Type of 3D In Vitro Model | Specific Features | Biomedical Applications | Reference(s) |
|---|---|---|---|
| Co-cultures |
|
| [89] |
| Spheroids |
|
| [90,91,92] |
| Organoids |
|
| [12,37,90,92,93,94,95] |
| Scaffold-based models |
|
| [92,96,97] |
| Bioprinted and 3D printed models |
|
| [97,98] |
| Organ-on-a-chip |
|
| [92,99] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blidisel, A.; Marcovici, I.; Coricovac, D.; Hut, F.; Dehelean, C.A.; Cretu, O.M. Experimental Models of Hepatocellular Carcinoma—A Preclinical Perspective. Cancers 2021, 13, 3651. https://doi.org/10.3390/cancers13153651
Blidisel A, Marcovici I, Coricovac D, Hut F, Dehelean CA, Cretu OM. Experimental Models of Hepatocellular Carcinoma—A Preclinical Perspective. Cancers. 2021; 13(15):3651. https://doi.org/10.3390/cancers13153651
Chicago/Turabian StyleBlidisel, Alexandru, Iasmina Marcovici, Dorina Coricovac, Florin Hut, Cristina Adriana Dehelean, and Octavian Marius Cretu. 2021. "Experimental Models of Hepatocellular Carcinoma—A Preclinical Perspective" Cancers 13, no. 15: 3651. https://doi.org/10.3390/cancers13153651