Immobilization of Jagged1 Enhances Vascular Smooth Muscle Cells Maturation by Activating the Notch Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Smooth Muscle Cell and 10T1/2 Cell Culture
2.2. HCASMC Serum Starvation
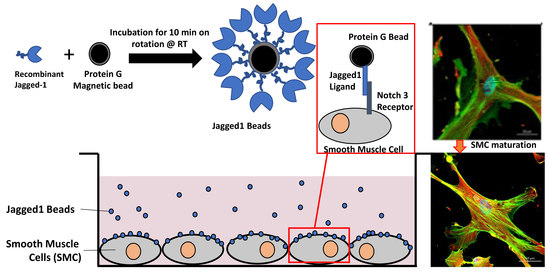
2.3. Jagged1/Fc Protein Bead Immobilization and Notch-Directed SMC Response
2.4. HCASMC-Jagged1-TGFβ1 Cross-Talk
2.5. Jagged1-Notch3 Magnetic Tweezer Assay
2.6. RNA Isolation and qPCR Analysis
2.7. Immunofluorescence Microscopy
2.8. Western Blotting
2.9. Statistical Analysis
3. Results
3.1. The Effect of Bead-Bound Jagged1 as a Surrogate for Endothelial-Bound Jagged1 for Notch Activation in HCASMCs
3.2. Co-Presentation of Jagged1 and TGFβ1 to Enhance SMC Maturity
3.3. Notch Activation in Embryonic Multipotent Mesenchymal Cells and Their Differentiation towards a Vascular Smooth Muscle Cell Lineage
3.4. Mechanosensitivity of Jagged1-Notch3 Signaling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mack, J.J.; Luisa Iruela-Arispe, M. NOTCH regulation of the endothelial cell phenotype. Curr. Opin. Hematol. 2018, 25, 212–218. [Google Scholar] [CrossRef]
- Hofmann, J.J.; Iruela-Arispe, M.L. Notch signaling in blood vessels: Who is talking to whom about what? Circ. Res. 2007, 100, 1556–1568. [Google Scholar] [CrossRef] [Green Version]
- Baeten, J.T.; Lilly, B. Notch Signaling in Vascular Smooth Muscle Cells. Adv. Pharmacol. 2017, 78, 351–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Campos, A.H.; Prince, C.Z.; Mou, Y.; Pollman, M.J. Coordinate Notch3-Hairy-related Transcription Factor Pathway Regulation in Response to Arterial Injury. J. Biol. Chem. 2002, 277, 23165–23171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis-Knowlton, J.; Turner, J.E.; Turner, A.; Damian-Loring, S.; Hagler, N.; Henderson, T.; Emery, I.F.; Bond, K.; Duarte, C.W.; Vary, C.P.H.; et al. Characterization of smooth muscle cells from human atherosclerotic lesions and their responses to Notch signaling. Lab. Investig. 2019, 99, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, P.; Ferrari, R. The Notch pathway: A new therapeutic target in atherosclerosis? Eur. Heart J. Suppl. 2015, 17, A74–A76. [Google Scholar] [CrossRef] [Green Version]
- Aquila, G.; Fortini, C.; Pannuti, A.; Delbue, S.; Pannella, M.; Morelli, M.B.; Caliceti, C.; Castriota, F.; De Mattei, M.; Ongaro, A.; et al. Distinct gene expression profiles associated with Notch ligands Delta-like 4 and Jagged1 in plaque material from peripheral artery disease patients: A pilot study. J. Transl. Med. 2017, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Lin, C.H.; Lilly, B. Notch signaling governs phenotypic modulation of smooth muscle cells. Vascul. Pharmacol. 2014, 63, 88–96. [Google Scholar] [CrossRef]
- Liu, H.; Kennard, S.; Lilly, B. Notch3 expression is induced in mural cells through an autoregulatory loop that requires endothelial-expressed Jagged-1. Vascular 2010, 104, 466–475. [Google Scholar] [CrossRef]
- Bhattacharyya, A.; Lin, S.; Sandig, M.; Mequanint, K. Regulation of Vascular Smooth Muscle Cell Phenotype in Three-Dimensional Coculture System by Jagged1-Selective Notch3 Signaling. Tissue Eng. Part A 2014, 20, 1175–1187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doi, H.; Iso, T.; Sato, H.; Yamazaki, M.; Matsui, H.; Tanaka, T.; Manabe, I.; Arai, M.; Nagai, R.; Kurabayashi, M. Jagged1-selective notch signaling induces smooth muscle differentiation via a RBP-Jκ-dependent pathway. J. Biol. Chem. 2006, 281, 28555–28564. [Google Scholar] [CrossRef] [Green Version]
- Zohorsky, K.; Mequanint, K. Designing Biomaterials to Modulate Notch Signaling in Tissue Engineering and Regenerative Medicine. Tissue Eng. Part B Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Abuammah, A.; Maimari, N.; Towhidi, L.; Frueh, J.; Chooi, K.Y.; Warboys, C.; Krams, R. New developments in mechanotransduction: Cross talk of the Wnt, TGF-β and Notch signalling pathways in reaction to shear stress. Curr. Opin. Biomed. Eng. 2018, 5, 96–104. [Google Scholar] [CrossRef]
- Luo, K. Signaling cross talk between TGF-β/Smad and other signaling pathways. Cold Spring Harb. Perspect. Biol. 2017, 9, a022137. [Google Scholar] [CrossRef] [Green Version]
- Langridge, P.D.; Struhl, G. Epsin-Dependent Ligand Endocytosis Activates Notch by Force. Cell 2017, 171, 1383–1396.e12. [Google Scholar] [CrossRef]
- Narui, Y.; Salaita, K. Membrane tethered delta activates notch and reveals a role for spatio-mechanical regulation of the signaling pathway. Biophys. J. 2013, 105, 2655–2665. [Google Scholar] [CrossRef] [Green Version]
- Vooijs, M.; Schroeter, E.H.; Pan, Y.; Blandford, M.; Kopan, R. Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins is not regulated through oligomerization. J. Biol. Chem. 2004, 279, 50864–50873. [Google Scholar] [CrossRef] [Green Version]
- Nandagopal, N.; Santat, L.A.; LeBon, L.; Sprinzak, D.; Bronner, M.E.; Elowitz, M.B. Dynamic Ligand Discrimination in the Notch Signaling Pathway. Cell 2018, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Ha, T. Defining single molecular forces required to activate integrin and Notch signaling. Nano Lett. 2013, 340, 991–994. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Rahil, Z.; Li, I.T.S.; Chowdhury, F.; Leckband, D.E.; Chemla, Y.R.; Ha, T. Constructing modular and universal single molecule tension sensor using protein G to study mechano-sensitive receptors. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, W.R.; Zimmerman, B.; He, L.; Miles, L.J.; Huang, J.; Tiyanont, K.; McArthur, D.G.; Aster, J.C.; Perrimon, N.; Loparo, J.J.; et al. Mechanical Allostery: Evidence for a Force Requirement in the Proteolytic Activation of Notch. Dev. Cell 2015, 33, 729–736. [Google Scholar] [CrossRef] [Green Version]
- Pedrosa, A.R.; Trindade, A.; Fernandes, A.C.; Carvalho, C.; Gigante, J.; Tavares, A.T.; Diéguez-Hurtado, R.; Yagita, H.; Adams, R.H.; Duarte, A. Endothelial jagged1 antagonizes Dll4 regulation of endothelial branching and promotes vascular maturation downstream of Dll4/Notch1. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1134–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boucher, J.; Gridley, T.; Liaw, L. Molecular Pathways of Notch Signaling in Vascular Smooth Muscle Cells. Front. Physiol. 2012, 3, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benedito, R.; Roca, C.; Sörensen, I.; Adams, S.; Gossler, A.; Fruttiger, M.; Adams, R.H. The Notch Ligands Dll4 and Jagged1 Have Opposing Effects on Angiogenesis. Cell 2009, 137, 1124–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, R.M.; Martins, M.C.L.; Almeida-Porada, G.; Barbosa, M.A. Induction of notch signaling by immobilization of jagged-1 on self-assembled monolayers. Biomaterials 2009, 30, 6879–6887. [Google Scholar] [CrossRef]
- Lin, J.; Lin, Y.; Su, L.; Su, Q.; Guo, W.; Huang, X.; Wang, C.; Lin, L. The role of Jagged1/Notch pathway-mediated angiogenesis of hepatocarcinoma cells in vitro, and the effects of the spleen-invigorating and blood stasis-removing recipe. Oncol. Lett. 2017, 14, 3616–3622. [Google Scholar] [CrossRef] [Green Version]
- Osathanon, T.; Manokawinchoke, J.; Sa-Ard-Iam, N.; Mahanonda, R.; Pavasant, P.; Suwanwela, J. Jagged1 promotes mineralization in human bone-derived cells. Arch. Oral Biol. 2019, 99, 134–140. [Google Scholar] [CrossRef]
- Putti, M.; de Jong, S.M.J.; Stassen, O.M.J.A.; Sahlgren, C.M.; Dankers, P.Y.W. A Supramolecular Platform for the Introduction of Fc-Fusion Bioactive Proteins on Biomaterial Surfaces. ACS Appl. Polym. Mater. 2019, 1, 2044–2054. [Google Scholar] [CrossRef]
- Li, H.; Yu, B.; Zhang, Y.; Pan, Z.; Xu, W.; Li, H. Jagged1 protein enhances the differentiation of mesenchymal stem cells into cardiomyocytes. Biochem. Biophys. Res. Commun. 2006, 341, 320–325. [Google Scholar] [CrossRef]
- Dishowitz, M.I.; Zhu, F.; Sundararaghavan, H.G.; Ifkovits, J.L.; Burdick, J.A.; Hankenson, K.D. Jagged1 immobilization to an osteoconductive polymer activates the Notch signaling pathway and induces osteogenesis. J. Biomed. Mater. Res. Part A 2014, 102, 1558–1567. [Google Scholar] [CrossRef] [PubMed]
- Blache, U.; Vallmajo-Martin, Q.; Horton, E.R.; Guerrero, J.; Djonov, V.; Scherberich, A.; Erler, J.T.; Martin, I.; Snedeker, J.G.; Milleret, V.; et al. Notch-inducing hydrogels reveal a perivascular switch of mesenchymal stem cell fate. EMBO Rep. 2018, 19, e45964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckstead, B.L.; Santosa, D.M.; Giachelli, C.M. Mimicking cell–cell interactions at the biomaterial–cell interface for control of stem cell differentiation. J. Biomed. Mater. Res. Part A 2006, 79A, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Sjöqvist, M.; Andersson, E.R. Do as I say, Not(ch) as I do: Lateral control of cell fate. Dev. Biol. 2019, 447, 58–70. [Google Scholar] [CrossRef]
- Hoglund, V.J.; Majesky, M.W. Patterning the artery wall by lateral induction of Notch signaling. Circulation 2012, 125, 212–215. [Google Scholar] [CrossRef] [Green Version]
- Han, M.; Wen, J.K.; Zheng, B.; Cheng, Y.; Zhang, C. Serum deprivation results in redifferentiation of human umbilical vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 2006, 291, 50–58. [Google Scholar] [CrossRef]
- Tsai, S.; Hollenbeck, S.T.; Ryer, E.J.; Edlin, R.; Yamanouchi, D.; Kundi, R.; Wang, C.; Liu, B.; Kent, C. TGF-B through Smad3 signaling stimulates vascular smooth muscle cell proliferation and neointimal formation. Am. J. Physiol. Heart Circ. Physiol. 2009, 297. [Google Scholar] [CrossRef] [Green Version]
- Low, E.L.; Baker, A.H.; Bradshaw, A.C. TGFβ smooth muscle cells and coronary artery disease: A review. Cell Signal. 2019, 53, 90–101. [Google Scholar] [CrossRef]
- Kurpinski, K.; Lam, H.; Chu, J.; Wang, A.; Kim, A.; Tsay, E.; Agrawal, S.; Schaffer, D.V.; Li, S. Transforming growth factor-β and notch signaling mediate stem cell differentiation into smooth muscle cells. Stem Cells 2010, 28, 734–742. [Google Scholar] [CrossRef]
- Blokzijl, A.; Dahlqvist, C.; Reissmann, E.; Falk, A.; Moliner, A.; Lendahl, U.; Ibáñez, C.F. Cross-talk between the Notch and TGF-β signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J. Cell Biol. 2003, 163, 723–728. [Google Scholar] [CrossRef]
- Dayekh, K.; Mequanint, K. The effects of progenitor and differentiated cells on ectopic calcification of engineered vascular tissues. Acta Biomater. 2020, 115, 288–298. [Google Scholar] [CrossRef]
- Fortini, M.E.; Bilder, D. Endocytic regulation of Notch signaling. Curr. Opin. Genet. Dev. 2009, 19, 323–328. [Google Scholar] [CrossRef] [Green Version]
- Meloty-Kapella, L.; Shergill, B.; Kuon, J.; Botvinick, E.; Weinmaster, G. Notch Ligand Endocytosis Generates Mechanical Pulling Force Dependent on Dynamin, Epsins, and Actin. Dev. Cell 2012, 22, 1299–1312. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Bhattacharyya, A.; Roszell, E.E.; Sandig, M.; Mequanint, K. The role of endothelial cell-bound Jagged1 in Notch3-induced human coronary artery smooth muscle cell differentiation. Biomaterials 2012, 33, 2462–2472. [Google Scholar] [CrossRef]
- Kibbie, J.; Teles, R.M.B.; Wang, Z.; Hong, P.; Montoya, D.; Krutzik, S.; Lee, S.; Kwon, O.; Modlin, R.L.; Cruz, D. Jagged1 Instructs Macrophage Differentiation in Leprosy. PLoS Pathog. 2016, 12, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Savary, E.; Sabourin, J.C.; Santo, J.; Hugnot, J.P.; Chabbert, C.; Van De Water, T.; Uziel, A.; Zine, A. Cochlear stem/progenitor cells from a postnatal cochlea respond to Jagged1 and demonstrate that notch signaling promotes sphere formation and sensory potential. Mech. Dev. 2008, 125, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Aho, S. Soluble form of Jagged1: Unique product of epithelial keratinocytes and a regulator of keratinocyte differentiation. J. Cell. Biochem. 2004, 92, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Luo, Z.; Wang, G.; Wang, Y.; Wang, Y.; Olmedo, M.; Morandi, M.M.; Barton, S.; Kevil, C.G.; Shu, B.; et al. Notch ligand Jagged1 promotes mesenchymal stromal cell-based cartilage repair. Exp. Mol. Med. 2018, 50, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caolo, V.; Schulten, H.M.; Zhuang, Z.W.; Murakami, M.; Wagenaar, A.; Verbruggen, S.; Molin, D.G.M.; Post, M.J. Soluble jagged-1 inhibits neointima formation by attenuating notch-herp2 signaling. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 1059–1065. [Google Scholar] [CrossRef] [Green Version]
- Urs, S.; Turner, B.; Tang, Y.; Rostama, B.; Small, D.; Liaw, L. Effect of soluble Jagged1-mediated inhibition of Notch signaling on proliferation and differentiation of an adipocyte progenitor cell model. Adipocyte 2012, 1, 46–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaya, O.; Binshtok, U.; Hersch, M.; Rivkin, D.; Weinreb, S.; Amir-Zilberstein, L.; Khamaisi, B.; Oppenheim, O.; Desai, R.A.; Goodyear, R.J.; et al. Cell-Cell Contact Area Affects Notch Signaling and Notch-Dependent Patterning. Dev. Cell 2017, 40, 505–511.e6. [Google Scholar] [CrossRef] [Green Version]
- Dayekh, K.; Mequanint, K. Comparative Studies of Fibrin-Based Engineered Vascular Tissues 2 and Notch Signaling from Progenitor Cells 1 3. ACS Biomater. Sci. Eng. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, V.K.; Andreadis, S.T. Stem Cell Sources for Vascular Tissue Engineering and Regeneration. Tissue Eng. Part B Rev. 2012, 18, 405–425. [Google Scholar] [CrossRef] [PubMed]
- Kiros, S.; Lin, S.; Xing, M.; Mequanint, K. Embryonic Mesenchymal Multipotent Cell Differentiation on Electrospun Biodegradable Poly(ester amide) Scaffolds for Model Vascular Tissue Fabrication. Ann. Biomed. Eng. 2020, 48, 980–991. [Google Scholar] [CrossRef] [PubMed]
- Hirschi, K.K.; Rohovsky, S.A.; D’Amore, P.A. PDGF, TGF-β, and heterotypic cell-cell interactions mediate endothelial cell-induced recruitment of 10T1/2 cells and their differentiation to a smooth muscle fate. J. Cell Biol. 1998, 141, 805–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndong, J.D.L.C.; Stephenson, Y.; Davis, M.E.; García, A.J.; Goudy, S. Controlled JAGGED1 delivery induces human embryonic palate mesenchymal cells to form osteoblasts. J. Biomed. Mater. Res. Part A 2018, 106, 552–560. [Google Scholar] [CrossRef]
- Boopathy, A.V.; Che, P.L.; Somasuntharam, I.; Fiore, V.F.; Cabigas, E.B.; Ban, K.; Brown, M.E.; Narui, Y.; Barker, T.H.; Yoon, Y.S.; et al. The modulation of cardiac progenitor cell function by hydrogel-dependent Notch1 activation. Biomaterials 2014, 35, 8103–8112. [Google Scholar] [CrossRef] [Green Version]
- Tung, J.C.; Paige, S.L.; Ratner, B.D.; Murry, C.E.; Giachelli, C.M. Engineered Biomaterials Control Differentiation and Proliferation of HumanEmbryonic-Stem-Cell-Derived Cardiomyocytes via Timed Notch Activation. Stem Cell Rep. 2014, 2, 271–281. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Mequanint, K. Bioreactor-induced mesenchymal progenitor cell differentiation and elastic fiber assembly in engineered vascular tissues. Acta Biomater. 2017, 59, 200–209. [Google Scholar] [CrossRef]
- Seo, D.; Southard, K.M.; Kim, J.; Cheon, J.; Gartner, Z.J.; Jun, Y.; Seo, D.; Southard, K.M.; Kim, J.; Lee, H.J.; et al. A Mechanogenetic Toolkit for Interrogating Cell Signaling in Space and Time. Cell 2016, 165, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mack, J.J.; Mosqueiro, T.S.; Archer, B.J.; Jones, W.M.; Sunshine, H.; Faas, G.C.; Briot, A.; Aragón, R.L.; Su, T.; Romay, M.C.; et al. NOTCH1 is a mechanosensor in adult arteries. Nat. Commun. 2017, 8, 1–18. [Google Scholar] [CrossRef]
- van Engeland, N.C.A.; Suarez Rodriguez, F.; Rivero-Müller, A.; Ristori, T.; Duran, C.L.; Stassen, O.M.J.A.; Antfolk, D.; Driessen, R.C.H.; Ruohonen, S.; Ruohonen, S.T.; et al. Vimentin regulates Notch signaling strength and arterial remodeling in response to hemodynamic stress. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Van Engeland, N.C.A.; Pollet, A.M.A.O.; Den Toonder, J.M.J.; Bouten, C.V.C.; Stassen, O.M.J.A.; Sahlgren, C.M. A biomimetic microfluidic model to study signalling between endothelial and vascular smooth muscle cells under hemodynamic conditions. Lab Chip 2018, 18, 1607–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loerakker, S.; Stassen, O.M.J.A.; Fleur, M.; Boareto, M.; Bouten, C.V.C. Mechanosensitivity of Jagged—Notch signaling can induce a switch-type behavior in vascular homeostasis. Proc. Natl. Acad. Sci. USA 2018, 115. [Google Scholar] [CrossRef] [Green Version]
- Antfolk, D.; Sjöqvist, M.; Cheng, F.; Isoniemi, K.; Duran, C.L.; Rivero-Muller, A.; Antila, C.; Niemi, R.; Landor, S.; Bouten, C.V.C.; et al. Selective regulation of Notch ligands during angiogenesis is mediated by vimentin. Proc. Natl. Acad. Sci. USA 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, E.R.; Sandberg, R.; Lendahl, U. Notch signaling: Simplicity in design, versatility in function. Development 2011, 138, 3593–3612. [Google Scholar] [CrossRef] [Green Version]
- Ong, C.T.; Cheng, H.T.; Chang, L.W.; Ohtsuka, T.; Kageyama, R.; Stormo, G.D.; Kopan, R. Target selectivity of vertebrate notch proteins: Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 2006, 281, 5106–5119. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zohorsky, K.; Lin, S.; Mequanint, K. Immobilization of Jagged1 Enhances Vascular Smooth Muscle Cells Maturation by Activating the Notch Pathway. Cells 2021, 10, 2089. https://doi.org/10.3390/cells10082089
Zohorsky K, Lin S, Mequanint K. Immobilization of Jagged1 Enhances Vascular Smooth Muscle Cells Maturation by Activating the Notch Pathway. Cells. 2021; 10(8):2089. https://doi.org/10.3390/cells10082089
Chicago/Turabian StyleZohorsky, Kathleen, Shigang Lin, and Kibret Mequanint. 2021. "Immobilization of Jagged1 Enhances Vascular Smooth Muscle Cells Maturation by Activating the Notch Pathway" Cells 10, no. 8: 2089. https://doi.org/10.3390/cells10082089