Inhibition of Mitochondrial Complex Function—The Hepatotoxicity Mechanism of Emodin Based on Quantitative Proteomic Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. CCK8 Cell Survival Assay
2.4. Quantitative Proteomic Assays
2.4.1. Protein Extraction and Quantification
2.4.2. Proteolysis and Tandem Mass Tag (TMT) Labeling
2.4.3. LC/MS/MS Analysis
2.4.4. Analysis of Proteomics Data
2.5. Western Blot Analysis
2.6. MMP Measurement
2.7. Mitochondrial Respiratory Chain Complexes Activity Assay
3. Results
3.1. Cell Counting Kit-8(CCK-8) Analysis
3.2. Protein Identification and Screening of Differentially Expressed Proteins
3.3. Gene Ontology (GO) and Pathway Enrichment Analysis
3.4. Western Blot Analysis
3.5. MMP and Complex Activity Measurement
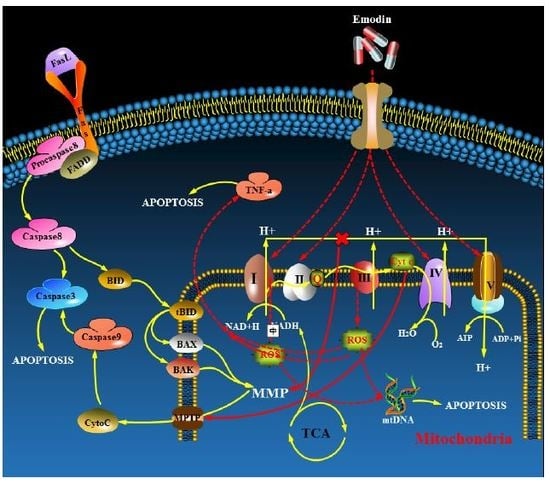
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BP | Biological processes |
| BSA | Bovine serum albumin |
| CC | Cellular component |
| CCCP | Carbonyl cyanide m-chlorophenyl hydrazine |
| CCK-8 | Cell Counting Kit-8 |
| CHAPS | 3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate) |
| COX | Cytochrome oxidase |
| Cyt C | Cytochrome C |
| DAVID | Database for Annotation, Visualization and Integrated Discovery |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FAD | Flavin adenine dinucleotide |
| GO | Gene ontology |
| JC-1 | Tetraethylbenzimidazolylcarbocyanine iodide |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MF | Molecular function |
| MMP | Mitochondrial membrane potential |
| MPTP | Mitochondrial permeability transition pore |
| mtDNA | Mitochondrial DNA |
| NADH | Nicotinamide adenine dinucleotide |
| NAFLD | Non-alcoholic fatty liver disease |
| NTP | National Toxicology Program |
| PVDF | Polyvinylidene fluoride |
| Q | Coenzyme Q |
| ROS | Reactive oxygen species |
| SDHA | Succinate dehydrogenase complex flavoprotein subunit A |
| TMT | Tandem Mass Tag |
| TNF-α | Tumor necrosis factor-α |
References
- Zhang, Y.X.; Li, J.S.; Peng, W.W.; Liu, X.; Yang, G.M.; Chen, L.H.; Cai, B.C. Comparative pharmacokinetics of aloe-emodin, rhein and emodin determined by liquid chromatography-mass spectrometry after oral administration of a rhubarb peony decoction and rhubarb extract to rats. Pharmazie 2013, 68, 333–339. [Google Scholar] [PubMed]
- Jin, J.H.; Ngoc, T.M.; Bae, K.; Kim, Y.S.; Kim, H.P. Inhibition of experimental atopic dermatitis by rhubarb (rhizomes of Rheum tanguticum) and 5-lipoxygenase inhibition of its major constituent, emodin. Phytother. Res. 2011, 25, 755–759. [Google Scholar] [CrossRef]
- Cai, J.; Razzak, A.; Hering, J.; Saed, A.; Babcock, T.A.; Helton, S.; Espat, N.J. Feasibility evaluation of emodin (rhubarb extract) as an inhibitor of pancreatic cancer cell proliferation in vitro. J. Parenter. Enter. Nutr. 2008, 32, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.M.; Kim, H.N.; Kim, Y.R.; Choi, Y.W.; Kim, C.M.; Shin, H.K.; Choi, B.T. Emodin from Polygonum multiflorum ameliorates oxidative toxicity in HT22 cells and deficits in photothrombotic ischemia. J. Ethnopharmacol. 2016, 188, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Huang, Y.Y.; Duh, P.D.; Wu, S.C. Hepatoprotection of emodin and Polygonum multiflorum against CCl(4)-induced liver injury. Pharm. Biol. 2012, 50, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Ni, B.; Lin, H.; Zhang, M.; Li, X.; Yin, X.; Qu, C.; Ni, J. Traditional usages, botany, phytochemistry, pharmacology and toxicology of Polygonum multiflorum Thunb.: A review. J. Ethnopharmacol. 2015, 159, 158–183. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.Z.; Sun, L.; Yuan, H.; Wu, A.Y.; Lu, J.G.; Ma, S.C. Simultaneous qualitative and quantitative analysis of 11 active compounds in rhubarb using two reference substances by UHPLC. J. Sep. Sci. 2018, 41. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.F.; Ni, B.R.; Lin, H.M.; Zhang, M.; Yan, L.; Qu, C.H.; Ni, J. Simultaneous determination of 14 constituents of Radix polygoni multiflori from different geographical areas by liquid chromatography-tandem mass spectrometry. Biomed. Chromatogr. 2015, 29, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.C.; Peng, Y.Y.; Ye, J.N. Determination of active ingredients of Polygonum cuspidatum Sied. et Zucc. by capillary electrophoresis with electrochemical detection. Electroanal 2004, 16, 1434–1438. [Google Scholar]
- He, D.; Chen, B.; Tian, Q.; Yao, S. Simultaneous determination of five anthraquinones in medicinal plants and pharmaceutical preparations by HPLC with fluorescence detection. J. Pharm. Biomed. Anal. 2009, 49, 1123–1127. [Google Scholar] [CrossRef]
- Dong, H.; Slain, D.; Cheng, J.; Ma, W.; Liang, W. Eighteen cases of liver injury following ingestion of Polygonum multiflorum. Complement. Ther. Med. 2014, 22, 70–74. [Google Scholar] [CrossRef]
- Li, Y.X.; Gong, X.H.; Liu, M.C.; Peng, C.; Li, P.; Wang, Y.T. Investigation of Liver Injury of Polygonum multiflorum Thunb. in Rats by Metabolomics and Traditional Approaches. Front. Pharm. 2017, 8, 791. [Google Scholar]
- Lin, L.; Li, H.; Lin, H.; Zhang, M.; Qu, C.; Yan, L.; Yin, X.; Ni, J. A New Perspective on Liver Injury by Traditional Chinese Herbs Such as Polygonum multiflorum: The Geographical Area of Harvest as an Important Contributory Factor. Front. Pharm. 2017, 8, 349. [Google Scholar] [CrossRef]
- Li, C.Y.; He, Q.; Gao, D.; Li, R.Y.; Zhu, Y.; Li, H.F.; Feng, W.W.; Yang, M.H.; Xiao, X.H.; Wang, J.B. Idiosyncratic drug-induced liver injury linked to Polygonum multiflorum: A case study by pharmacognosy. Chin. J. Integr. Med. 2017, 23, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; Kong, W.J.; Wang, H.J.; Zhao, H.P.; Xiao, H.Y.; Dai, C.M.; Xiao, X.H.; Zhao, Y.L.; Jin, C.; Zhang, L.; et al. Toxic effects caused by rhubarb (Rheum palmatum L.) are reversed on immature and aged rats. J. Ethnopharmacol. 2011, 134, 216–220. [Google Scholar]
- Lin, L.; Lin, H.; Zhang, M.; Ni, B.; Yin, X.; Qu, C.; Ni, J. A novel method to analyze hepatotoxic components in Polygonum multiflorum using ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. J. Hazard. Mater. 2015, 299, 249–259. [Google Scholar] [CrossRef]
- Tu, C.; Gao, D.; Li, X.F.; Li, C.Y.; Li, R.S.; Zhao, Y.L.; Li, N.; Jia, G.L.; Pang, J.Y.; Cui, H.R.; et al. Inflammatory stress potentiates emodin-induced liver injury in rats. Front. Pharm. 2015, 6, 233. [Google Scholar] [CrossRef] [PubMed]
- National Toxicology, P. NTP Toxicology and Carcinogenesis Studies of EMODIN (CAS NO. 518-82-1) Feed Studies in F344/N Rats and B6C3F1 Mice. Natl. Toxicol. Program. Tech. Rep. Ser. 2001, 493, 1–278. [Google Scholar]
- Lin, S.P.; Chu, P.M.; Tsai, S.Y.; Wu, M.H.; Hou, Y.C. Pharmacokinetics and tissue distribution of resveratrol, emodin and their metabolites after intake of Polygonum cuspidatum in rats. J. Ethnopharmacol. 2012, 144, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Song, Z.F.; Ma, C. Emodin Studies on Pharmacokinetics and Distribution in Rat Liver after Polygonum cuspidatum Sieb. et Zucc. Extract Administration. World Sci. Technol. 2008, 10, 64–67. [Google Scholar] [CrossRef]
- Gong, X.H.; Li, Y.; Zhang, R.Q.; Xie, X.F.; Peng, C.; Li, Y.X. The synergism mechanism of Rhubarb Anthraquinones on constipation elucidated by comparative pharmacokinetics of Rhubarb extract between normal and diseased rats. Eur. J. Drug Metab. Pharmacokinet. 2015, 40, 379–388. [Google Scholar] [CrossRef]
- Sharma, V.; Anderson, D.; Dhawan, A. Zinc oxide nanoparticles induce oxidative DNA damage and ROS-triggered mitochondria mediated apoptosis in human liver cells (HepG2). Apoptosis 2012, 17, 852–870. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Dudkina, N.V.; Folea, I.M.; Boekema, E.J. Towards structural and functional characterization of photosynthetic and mitochondrial supercomplexes. Micron 2015, 72, 39–51. [Google Scholar] [CrossRef]
- Fishbein, W.N.; Stowell, R.E. Studies on the mechanism of freezing damage to mouse liver using a mitochondrial enzyme assay. I. Temporal localization of the injury phase during slow freezing. Cryobiology 1968, 4, 283–289. [Google Scholar] [CrossRef]
- Dong, X.; Fu, J.; Yin, X.; Yang, C.; Ni, J. Aloe-emodin Induces Apoptosis in Human Liver HL-7702 Cells through Fas Death Pathway and the Mitochondrial Pathway by Generating Reactive Oxygen Species. Phytother. Res. 2017, 31, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Gu, L.Q.; Wu, J.Y. Apoptosis-inducing activity of new pyrazole emodin derivatives in human hepatocellular carcinoma HepG2 cells. Biol. Pharm. Bull. 2007, 30, 1113–1116. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Yang, J.; He, R.; Gao, F.; Sang, H.; Tang, X.; Ye, R.D. Emodin enhances arsenic trioxide-induced apoptosis via generation of reactive oxygen species and inhibition of survival signaling. Cancer Res. 2004, 64, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; He, D.; Li, K.; Liu, H.; Wang, B.; Zheng, L.; Li, J. Emodin targets mitochondrial cyclophilin D to induce apoptosis in HepG2 cells. Biomed. Pharmacother. Biomed. Pharmacother. 2017, 90, 222–228. [Google Scholar] [CrossRef]
- De Lonlay, P.; Valnot, I.; Barrientos, A.; Gorbatyuk, M.; Tzagoloff, A.; Taanman, J.W.; Benayoun, E.; Chretien, D.; Kadhom, N.; Lombes, A.; et al. A mutant mitochondrial respiratory chain assembly protein causes complex III deficiency in patients with tubulopathy, encephalopathy and liver failure. Nat. Genet. 2001, 29, 57–60. [Google Scholar] [CrossRef]
- Kwong, J.Q.; Henning, M.S.; Starkov, A.A.; Manfredi, G. The mitochondrial respiratory chain is a modulator of apoptosis. J. Cell Biol. 2007, 179, 1163–1177. [Google Scholar] [CrossRef] [Green Version]
- Vendemiale, G.; Grattagliano, I.; Caraceni, P.; Caraccio, G.; Domenicali, M.; Dall’Agata, M.; Trevisani, F.; Guerrieri, F.; Bernardi, M.; Altomare, E. Mitochondrial oxidative injury and energy metabolism alteration in rat fatty liver: Effect of the nutritional status. Hepatology 2001, 33, 808–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; He, L.; Lemasters, J.J. Mitochondrial permeability transition: A common pathway to necrosis and apoptosis. Biochem. Biophys. Res. Commun. 2003, 304, 463–470. [Google Scholar] [CrossRef]
- Schriewer, J.M.; Peek, C.B.; Bass, J.; Schumacker, P.T. ROS-mediated PARP activity undermines mitochondrial function after permeability transition pore opening during myocardial ischemia-reperfusion. J. Am. Heart Assoc. 2013, 2, e000159. [Google Scholar] [CrossRef]
- Higuchi, M.; Proske, R.J.; Yeh, E.T. Inhibition of mitochondrial respiratory chain complex I by TNF results in cytochrome c release, membrane permeability transition, and apoptosis. Oncogene 1998, 17, 2515–2524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spierings, D.; McStay, G.; Saleh, M.; Bender, C.; Chipuk, J.; Maurer, U.; Green, D.R. Connected to death: The (unexpurgated) mitochondrial pathway of apoptosis. Science 2005, 310, 66–67. [Google Scholar] [CrossRef]
- Trost, L.C.; Lemasters, J.J. Role of the mitochondrial permeability transition in salicylate toxicity to cultured rat hepatocytes: Implications for the pathogenesis of Reye’s syndrome. Toxicol. Appl. Pharm. 1997, 147, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Serviddio, G.; Bellanti, F.; Giudetti, A.M.; Gnoni, G.V.; Capitanio, N.; Tamborra, R.; Romano, A.D.; Quinto, M.; Blonda, M.; Vendemiale, G.; et al. Mitochondrial oxidative stress and respiratory chain dysfunction account for liver toxicity during amiodarone but not dronedarone administration. Free Radic. Biol. Med. 2011, 51, 2234–2242. [Google Scholar] [CrossRef]
- Van Houten, B.; Woshner, V.; Santos, J.H. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair 2006, 5, 145–152. [Google Scholar] [CrossRef]
- Wallace, D.C. Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagenesis 2010, 51, 440–450. [Google Scholar] [CrossRef]
- Kashimshetty, R.; Desai, V.G.; Kale, V.M.; Lee, T.; Moland, C.L.; Branham, W.S.; New, L.S.; Chan, E.C.; Younis, H.; Boelsterli, U.A. Underlying mitochondrial dysfunction triggers flutamide-induced oxidative liver injury in a mouse model of idiosyncratic drug toxicity. Toxicol. Appl. Pharm. 2009, 238, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Alston, C.L.; Davison, J.E.; Meloni, F.; van der Westhuizen, F.H.; He, L.; Hornig-Do, H.T.; Peet, A.C.; Gissen, P.; Goffrini, P.; Ferrero, I.; et al. Recessive germline SDHA and SDHB mutations causing leukodystrophy and isolated mitochondrial complex II deficiency. J. Med. Genet. 2012, 49, 569–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renkema, G.H.; Wortmann, S.B.; Smeets, R.J.; Venselaar, H.; Antoine, M.; Visser, G.; Ben-Omran, T.; van den Heuvel, L.P.; Timmers, H.J.; Smeitink, J.A.; et al. SDHA mutations causing a multisystem mitochondrial disease: Novel mutations and genetic overlap with hereditary tumors. Eur. J. Hum. Genet. 2015, 23, 202–209. [Google Scholar] [CrossRef] [PubMed]









| GO.ID | Term | Annotated | Significant | p-Value | p Adjust | Names (Uniport Accession) | GO Category |
|---|---|---|---|---|---|---|---|
| GO:0098573 | intrinsic component of mitochondrial membrane | 86 | 6 | 9.4 × 10−5 | 1.12 × 10−4 | A0A140TA86, P50416, Q16891, Q9Y3D6, P17152, Q99595 | Cellular Component |
| GO:0032592 | integral component of mitochondrial membrane | 85 | 6 | 8.8 × 10−5 | 1.06 × 10−4 | A0A140TA86, P50416, Q16891, Q9Y3D6, P17152, Q99595 | Cellular Component |
| GO:0032543 | mitochondrial translation | 130 | 18 | 5.9 × 10−16 | 3.02 × 10−15 | Q9BQC6, Q96EY7, Q9NYK5, Q9Y3D9, Q8N983, Q96A35, Q9BZE1, Q9H2W6, Q13084, O95182, Q8IXM3, Q969S9, Q9Y291, P82932, P82675, P46199, P42704, Q96DV4 | Biological Process |
| GO:0140053 | mitochondrial gene expression | 147 | 18 | 5.3 × 10−15 | 2.33 × 10−14 | Q9BQC6, Q96EY7, Q9NYK5, Q9Y3D9, Q8N983, Q96A35, Q9BZE1, Q9H2W6, Q13084, O95182, Q8IXM3, Q969S9, Q9Y291, P82932, P82675, P46199, P42704, Q96DV4 | Biological Process |
| GO:0005747 | mitochondrial respiratory chain complex I | 75 | 11 | 4.3 × 10−11 | 1.29 × 10−10 | P56556, O95182, O95139, O00217, Q16718, O00483, P28331, O43678, Q9NX14, O75306, P49821 | Cellular Component |
| GO:0005746 | mitochondrial respiratory chain | 138 | 17 | 4.2 × 10−15 | 1.57 × 10−14 | P56556, P31040, P13073, O95182, O95139, P21912, O00217, Q16718, O00483, H0UI06, P28331, O43678, P20674, Q7KZN9, Q9NX14, O75306, P49821 | Cellular Component |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 101 | 11 | 3.8 × 10−9 | 8.99 × 10−9 | P56556, Q9NPL8, O95182, O95139, O00217, Q16718, P28331, O43678, Q9NX14, O75306, P49821 | Biological Process |
| GO:0098800 | inner mitochondrial membrane protein complex | 400 | 20 | 3.6 × 10−10 | 9.53 × 10−10 | P56556, A0A140TA86, Q9Y5J7, P31040, P13073, Q16891, O95182, O95139, P00846, P21912, O00217, Q16718, O00483, P28331, O43678, P20674, Q9NX14, O75306, Q99595, P49821 | Cellular Component |
| GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | 20 | 4 | 3.5 × 10−5 | 3.9 × 10−5 | P13073, O00483, P20674, Q7KZN9 | Biological Process |
| GO:0098798 | mitochondrial protein complex | 430 | 22 | 3.0 × 10−11 | 9.43 × 10−10 | O76031, P56556, A0A140TA86, Q9Y5J7, P31040, P13073, Q16891, O95182, O95139, P00846, F8W8Z9, P21912, O00217, Q16718, O00483, P28331, O43678, P20674, Q9NX14, O75306, Q99595, P49821 | Cellular Component |
| GO:0006839 | mitochondrial transport | 316 | 14 | 2.6 × 10−6 | 3.88 × 10−6 | Q9Y5J7, P50416, Q9Y3D6, O00170, Q9H300, P00846, Q14790, Q99436, P40763, Q5HYI7, O00165, Q9NQZ5, Q9BZL1, Q99595 | Biological Process |
| GO:0070125 | mitochondrial translational elongation | 86 | 13 | 2.1 × 10−12 | 7.04 × 10−12 | Q96EY7, Q9NYK5, Q9Y3D9, Q8N983, Q96A35, Q9BZE1, Q9H2W6, Q13084, Q8IXM3, Q9Y291, P82932, P82675, Q96DV4 | Biological Process |
| GO:0032981 | mitochondrial respiratory chain complex I assembly | 62 | 11 | 1.7 × 10−11 | 5.13 × 10−11 | P56556, Q9NPL8, O95182, O95139, O00217, Q16718, P28331, O43678, Q9NX14, O75306, P49821 | Biological Process |
| GO:0097031 | mitochondrial respiratory chain complex I biogenesis | 62 | 11 | 1.7 × 10−11 | 5.13 × 10−11 | P56556, Q9NPL8, O95182, O95139, O00217, Q16718, P28331, O43678, Q9NX14, O75306, P49821 | Biological Process |
| GO:0005759 | mitochondrial matrix | 506 | 30 | 1.5 × 10−16 | 5.7 × 10−16 | O76031, P48735, Q9BQC6, Q96RQ3, Q9UNQ2, Q07820, Q9NYK5, Q32P41, Q6NVY1, Q8N983, Q9BZE1, Q9H2W6, Q16822, Q13084, O95182, P30038, Q8IXM3, Q969S9, Q9Y291, P11498, Q5U5X0, Q8NFF5, P82932, O00217, P82675, P28331, O75306, P42704, Q96AG4, Q13057 | Cellular Component |
| GO:0005761 | mitochondrial ribosome | 84 | 11 | 1.5 × 10−10 | 4.08 × 10−10 | Q9BQC6, Q9NYK5, Q8N983, Q9BZE1, Q9H2W6, Q13084, O95182, Q8IXM3, Q9Y291, P82932, P82675 | Cellular Component |
| GO:0005741 | mitochondrial outer membrane | 242 | 12 | 1.3 × 10−6 | 2.29 × 10−6 | Q9NUQ2, Q07820, P50416, Q9Y3D6, Q969Z3, P00387, Q14790, F8W8Z9, Q5HYI7, O00165, Q13057, A0A0C4DFN1 | Cellular Component |
| GO:0070126 | mitochondrial translational termination | 86 | 14 | 1.0 × 10−13 | 3.73 × 10−13 | Q96EY7, Q9NYK5, Q9Y3D9, Q8N983, Q96A35, Q9BZE1, Q9H2W6, Q13084, Q8IXM3, Q969S9, Q9Y291, P82932, P82675, Q96DV4 | Biological Process |
| Accession | Description | Complex | Log Ratio (3/1) | p-Value |
|---|---|---|---|---|
| P28331 | NADH:ubiquinone oxidoreductase core subunit S1 (NDUFS1) | Complex I | −0.7842 | 0.0002 |
| O75306 | NADH:ubiquinone oxidoreductase core subunit S2 (NDUFS2) | −0.4512 | 0.0017 | |
| O00217 | NADH:ubiquinone oxidoreductase core subunit S8 (NDUFS8) | −0.3746 | 0.0005 | |
| P49821 | NADH:ubiquinone oxidoreductase core subunit V1 (NDUFV1) | −0.5872 | 0.0008 | |
| E7EPT4 | NADH:ubiquinone oxidoreductase core subunit V2 (NDUFV2) | −0.4440 | 0.0063 | |
| O43678 | NADH:ubiquinone oxidoreductase subunit A2 (NDUFA2) | −0.2866 | 0.0001 | |
| Q16718 | NADH:ubiquinone oxidoreductase subunit A5 (NDUFA5) | −0.4784 | 0.0028 | |
| P56556 | NADH:ubiquinone oxidoreductase subunit A6 (NDUFA6) | −0.7647 | 0.0010 | |
| O95182 | NADH:ubiquinone oxidoreductase subunit A7 (NDUFA7) | 0.4626 | 0.0017 | |
| Q9NX14 | NADH:ubiquinone oxidoreductase subunit B11 (NDUFB11) | −0.5076 | 0.0004 | |
| O95139 | NADH:ubiquinone oxidoreductase subunit B6 (NDUFB6) | −0.3481 | 0.0001 | |
| O00483 | NDUFA4, mitochondrial complex associated (NDUFA4) | −0.4872 | 0.0004 | |
| P31040 | succinate dehydrogenase complex flavoprotein subunit A (SDHA) | Complex II | −0.7329 | 0.0004 |
| P21912 | succinate dehydrogenase complex iron sulfur subunit B (SDHB) | −0.5184 | 0.0046 | |
| Q7KZN9 | COX15, cytochrome c oxidase assembly homolog (COX15) | Complex IV | −0.4991 | 0.0001 |
| P13073 | cytochrome c oxidase subunit 4I1 (COX4I1) | −0.3085 | 0.0003 | |
| P20674 | cytochrome c oxidase subunit 5A (COX5A) | −0.5156 | 0.0011 | |
| H0UI06 | cytochrome c oxidase subunit 7A2 (COX7A2) | −0.7194 | 0.0061 | |
| P00846 | ATP synthase F0 subunit 6 (ATP6) | Complex V | −1.0014 | 0.0048 |
| Q9Y487 | ATPase H+ transporting V0 subunit a2 (ATP6V0A2) | 0.3322 | 0.0043 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, L.; Liu, Y.; Fu, S.; Qu, C.; Li, H.; Ni, J. Inhibition of Mitochondrial Complex Function—The Hepatotoxicity Mechanism of Emodin Based on Quantitative Proteomic Analyses. Cells 2019, 8, 263. https://doi.org/10.3390/cells8030263
Lin L, Liu Y, Fu S, Qu C, Li H, Ni J. Inhibition of Mitochondrial Complex Function—The Hepatotoxicity Mechanism of Emodin Based on Quantitative Proteomic Analyses. Cells. 2019; 8(3):263. https://doi.org/10.3390/cells8030263
Chicago/Turabian StyleLin, Longfei, Yuling Liu, Sai Fu, Changhai Qu, Hui Li, and Jian Ni. 2019. "Inhibition of Mitochondrial Complex Function—The Hepatotoxicity Mechanism of Emodin Based on Quantitative Proteomic Analyses" Cells 8, no. 3: 263. https://doi.org/10.3390/cells8030263