CDK-Independent and PCNA-Dependent Functions of p21 in DNA Replication
Abstract
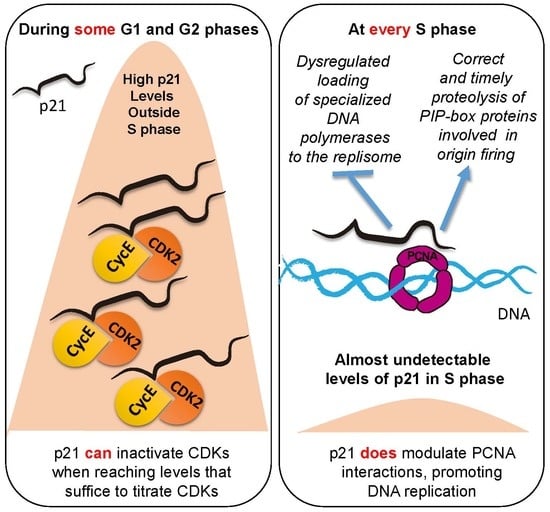
:1. A Broadly Accepted Notion: p21 Controls the Cell Cycle through Cyclin Kinases Inhibition
2. Can Cell-Cycle Arrest Be Achieved by p21–PCNA Interaction?
3. p21 Levels are Differentially Regulated during and by Cell Cycle
4. p21 Exerts Specific Functions at S-Phase-Specific Level, Regulating Genomic Stability through PCNA Interaction
5. Genotoxins Do Not Induce p21 Accumulation in S Phase
6. CDK-Independent Effect of p21 Overexpression on Damaged DNA Replication, Cell Death, and Genomic Stability
7. Therapeutic Opportunities for Cancer Treatment
8. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abbas, T.; Dutta, A. p21 in cancer: Intricate networks and multiple activities. Nat. Rev. Cancer 2009, 9, 400–414. [Google Scholar] [CrossRef] [PubMed]
- Karimian, A.; Ahmadi, Y.; Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 2016, 42, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Cmielova, J.; Rezacova, M. p21Cip1/Waf1 protein and its function based on a subcellular localization [corrected]. J. Cell Biochem. 2011, 112, 3502–3506. [Google Scholar] [CrossRef] [PubMed]
- Galea, C.A.; Wang, Y.; Sivakolundu, S.G.; Kriwacki, R.W. Regulation of cell division by intrinsically unstructured proteins: Intrinsic flexibility, modularity, and signaling conduits. Biochemistry 2008, 47, 7598–7609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, C.; Zhang, P.; Harper, J.W.; Elledge, S.J.; Leder, P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 1995, 82, 675–684. [Google Scholar] [CrossRef] [Green Version]
- Harper, J.W.; Adami, G.R.; Wei, N.; Keyomarsi, K.; Elledge, S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993, 75, 805–816. [Google Scholar] [CrossRef]
- Sherr, C.J.; Roberts, J.M. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999, 13, 1501–1512. [Google Scholar] [CrossRef] [Green Version]
- Ball, K.L. p21: Structure and functions associated with cyclin-CDK binding. Prog. Cell Cycle Res. 1997, 3, 125–134. [Google Scholar]
- Harper, J.W.; Elledge, S.J.; Keyomarsi, K.; Dynlacht, B.; Tsai, L.H.; Zhang, P.; Dobrowolski, S.; Bai, C.; Connell-Crowley, L.; Swindell, E.; et al. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell 1995, 6, 387–400. [Google Scholar] [CrossRef] [Green Version]
- Al Bitar, S.; Gali-Muhtasib, H. The Role of the Cyclin Dependent Kinase Inhibitor p21(cip1/waf1) in Targeting Cancer: Molecular Mechanisms and Novel Therapeutics. Cancers 2019, 11, 1475. [Google Scholar] [CrossRef] [Green Version]
- Cai, K.; Dynlacht, B.D. Activity and nature of p21(WAF1) complexes during the cell cycle. Proc. Natl. Acad. Sci. USA 1998, 95, 12254–12259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottifredi, V.; Prives, C. The S phase checkpoint: When the crowd meets at the fork. Semin. Cell Dev. Biol. 2005, 16, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Laptenko, O.; Beckerman, R.; Freulich, E.; Prives, C. p53 binding to nucleosomes within the p21 promoter in vivo leads to nucleosome loss and transcriptional activation. Proc. Natl. Acad. Sci. USA 2011, 108, 10385–10390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunz, F.; Dutriaux, A.; Lengauer, C.; Waldman, T.; Zhou, S.; Brown, J.P.; Sedivy, J.M.; Kinzler, K.W.; Vogelstein, B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 1998, 282, 1497–1501. [Google Scholar] [CrossRef] [PubMed]
- Brugarolas, J.; Chandrasekaran, C.; Gordon, J.I.; Beach, D.; Jacks, T.; Hannon, G.J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 1995, 377, 552–557. [Google Scholar] [CrossRef]
- Barr, A.R.; Cooper, S.; Heldt, F.S.; Butera, F.; Stoy, H.; Mansfeld, J.; Novak, B.; Bakal, C. DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat. Commun. 2017, 8, 14728. [Google Scholar] [CrossRef] [Green Version]
- Spencer, S.L.; Cappell, S.D.; Tsai, F.C.; Overton, K.W.; Wang, C.L.; Meyer, T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 2013, 155, 369–383. [Google Scholar] [CrossRef] [Green Version]
- Toettcher, J.E.; Loewer, A.; Ostheimer, G.J.; Yaffe, M.B.; Tidor, B.; Lahav, G. Distinct mechanisms act in concert to mediate cell cycle arrest. Proc. Natl. Acad. Sci. USA 2009, 106, 785–790. [Google Scholar] [CrossRef] [Green Version]
- Munoz-Espin, D.; Serrano, M. Cellular senescence: From physiology to pathology. Nat. Rev. Mol. Cell Biol. 2014, 15, 482–496. [Google Scholar] [CrossRef]
- Miyachi, K.; Fritzler, M.J.; Tan, E.M. Autoantibody to a nuclear antigen in proliferating cells. J. Immunol. 1978, 121, 2228–2234. [Google Scholar]
- Gulbis, J.M.; Kelman, Z.; Hurwitz, J.; O’Donnell, M.; Kuriyan, J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 1996, 87, 297–306. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Tan, C.K.; Zhou, J.Q.; You, M.; Carastro, L.M.; Downey, K.M.; So, A.G. Direct interaction of proliferating cell nuclear antigen with the small subunit of DNA polymerase delta. J. Biol. Chem. 2002, 277, 24340–24345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishna, T.S.; Kong, X.P.; Gary, S.; Burgers, P.M.; Kuriyan, J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 1994, 79, 1233–1243. [Google Scholar] [CrossRef]
- Choe, K.N.; Moldovan, G.L. Forging Ahead through Darkness: PCNA, Still the Principal Conductor at the Replication Fork. Mol. Cell 2017, 65, 380–392. [Google Scholar] [CrossRef] [Green Version]
- De Biasio, A.; Blanco, F.J. Proliferating cell nuclear antigen structure and interactions: Too many partners for one dancer? Adv. Protein Chem. Struct. Biol. 2013, 91, 1–36. [Google Scholar]
- Boehm, E.M.; Gildenberg, M.S.; Washington, M.T. The Many Roles of PCNA in Eukaryotic DNA Replication. Enzymes 2016, 39, 231–254. [Google Scholar]
- Xu, H.; Zhang, P.; Liu, L.; Lee, M.Y. A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry 2001, 40, 4512–4520. [Google Scholar] [CrossRef]
- Havens, C.G.; Walter, J.C. Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 2009, 35, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Prestel, A.; Wichmann, N.; Martins, J.M.; Marabini, R.; Kassem, N.; Broendum, S.S.; Otterlei, M.; Nielsen, O.; Willemoes, M.; Ploug, M.; et al. The PCNA interaction motifs revisited: Thinking outside the PIP-box. Cell Mol. Life Sci. 2019, 76, 4923–4943. [Google Scholar] [CrossRef] [Green Version]
- Bruning, J.B.; Shamoo, Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure 2004, 12, 2209–2219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oku, T.; Ikeda, S.; Sasaki, H.; Fukuda, K.; Morioka, H.; Ohtsuka, E.; Yoshikawa, H.; Tsurimoto, T. Functional sites of human PCNA which interact with p21 (Cip1/Waf1), DNA polymerase delta and replication factor C. Genes Cells 1998, 3, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Besson, A.; Dowdy, S.F.; Roberts, J.M. CDK inhibitors: Cell cycle regulators and beyond. Dev. Cell 2008, 14, 159–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, S.; Carr, A.M. Replication stress and genome rearrangements: Lessons from yeast models. Curr. Opin. Genet. Dev. 2013, 23, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Marotta, M.; Suzuki, R.; Diede, S.J.; Tapscott, S.J.; Niida, A.; Chen, X.; Mouakkad, L.; Kondratova, A.; Giuliano, A.E.; et al. Impediment of Replication Forks by Long Non-coding RNA Provokes Chromosomal Rearrangements by Error-Prone Restart. Cell Rep. 2017, 21, 2223–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoimenov, I.; Helleday, T. PCNA on the crossroad of cancer. Biochem. Soc. Trans. 2009, 37, 605–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naryzhny, S.N.; Lee, H. Characterization of proliferating cell nuclear antigen (PCNA) isoforms in normal and cancer cells: There is no cancer-associated form of PCNA. FEBS Lett. 2007, 581, 4917–4920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottifredi, V.; McKinney, K.; Poyurovsky, M.V.; Prives, C. Decreased p21 levels are required for efficient restart of DNA synthesis after S phase block. J. Biol. Chem. 2004, 279, 5802–5810. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Hurwitz, J.; Massague, J. Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature 1995, 375, 159–161. [Google Scholar] [CrossRef]
- Pan, Z.Q.; Reardon, J.T.; Li, L.; Flores-Rozas, H.; Legerski, R.; Sancar, A.; Hurwitz, J. Inhibition of nucleotide excision repair by the cyclin-dependent kinase inhibitor p21. J. Biol. Chem. 1995, 270, 22008–22016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivji, M.K.; Ferrari, E.; Ball, K.; Hubscher, U.; Wood, R.D. Resistance of human nucleotide excision repair synthesis in vitro to p21Cdn1. Oncogene 1998, 17, 2827–2838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, M.P.; Balajee, A.S.; Bohr, V.A. The C-terminal domain of p21 inhibits nucleotide excision repair In vitro and In vivo. Mol. Biol. Cell 1999, 10, 2119–2129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores-Rozas, H.; Kelman, Z.; Dean, F.B.; Pan, Z.Q.; Harper, J.W.; Elledge, S.J.; O’Donnell, M.; Hurwitz, J. Cdk-interacting protein 1 directly binds with proliferating cell nuclear antigen and inhibits DNA replication catalyzed by the DNA polymerase delta holoenzyme. Proc. Natl. Acad. Sci. USA 1994, 91, 8655–8659. [Google Scholar] [CrossRef] [Green Version]
- Podust, V.N.; Podust, L.M.; Goubin, F.; Ducommun, B.; Hubscher, U. Mechanism of inhibition of proliferating cell nuclear antigen-dependent DNA synthesis by the cyclin-dependent kinase inhibitor p21. Biochemistry 1995, 34, 8869–8875. [Google Scholar] [CrossRef] [PubMed]
- Waga, S.; Hannon, G.J.; Beach, D.; Stillman, B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature 1994, 369, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, M.; Robetorye, R.S.; Pereira-Smith, O.M.; Smith, J.R. The C-terminal region of p21SDI1/WAF1/CIP1 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J. Biol. Chem. 1995, 270, 17060–17063. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Reichner, C.; Wu, X.; Levine, A.J. Analysis of wild-type and mutant p21WAF-1 gene activities. Mol. Cell Biol. 1996, 16, 1786–1793. [Google Scholar] [CrossRef] [Green Version]
- Medema, R.H.; Klompmaker, R.; Smits, V.A.; Rijksen, G. p21waf1 can block cells at two points in the cell cycle, but does not interfere with processive DNA-replication or stress-activated kinases. Oncogene 1998, 16, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Ogryzko, V.V.; Wong, P.; Howard, B.H. WAF1 retards S-phase progression primarily by inhibition of cyclin-dependent kinases. Mol. Cell Biol. 1997, 17, 4877–4882. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Jackson, P.K.; Kirschner, M.W.; Dutta, A. Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature 1995, 374, 386–388. [Google Scholar] [CrossRef]
- Mansilla, S.F.; Soria, G.; Vallerga, M.B.; Habif, M.; Martinez-Lopez, W.; Prives, C.; Gottifredi, V. UV-triggered p21 degradation facilitates damaged-DNA replication and preserves genomic stability. Nucleic Acids Res. 2013, 41, 6942–6951. [Google Scholar] [CrossRef]
- Soria, G.; Speroni, J.; Podhajcer, O.L.; Prives, C.; Gottifredi, V. p21 differentially regulates DNA replication and DNA-repair-associated processes after UV irradiation. J. Cell Sci. 2008, 121, 3271–3282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cayrol, C.; Knibiehler, M.; Ducommun, B. p21 binding to PCNA causes G1 and G2 cell cycle arrest in p53-deficient cells. Oncogene 1998, 16, 311–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cazzalini, O.; Perucca, P.; Riva, F.; Stivala, L.A.; Bianchi, L.; Vannini, V.; Ducommun, B.; Prosperi, E. p21CDKN1A does not interfere with loading of PCNA at DNA replication sites, but inhibits subsequent binding of DNA polymerase delta at the G1/S phase transition. Cell Cycle 2003, 2, 596–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Waga, S.; Hannon, G.J.; Beach, D.; Stillman, B. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature 1994, 371, 534–537. [Google Scholar] [CrossRef]
- Perucca, P.; Cazzalini, O.; Mortusewicz, O.; Necchi, D.; Savio, M.; Nardo, T.; Stivala, L.A.; Leonhardt, H.; Cardoso, M.C.; Prosperi, E. Spatiotemporal dynamics of p21CDKN1A protein recruitment to DNA-damage sites and interaction with proliferating cell nuclear antigen. J. Cell Sci. 2006, 119, 1517–1527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adimoolam, S.; Lin, C.X.; Ford, J.M. The p53-regulated cyclin-dependent kinase inhibitor, p21 (cip1, waf1, sdi1), is not required for global genomic and transcription-coupled nucleotide excision repair of UV-induced DNA photoproducts. J. Biol. Chem. 2001, 276, 25813–25822. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.L.; Ford, J.M.; Hollander, M.C.; Bortnick, R.A.; Amundson, S.A.; Seo, Y.R.; Deng, C.X.; Hanawalt, P.C.; Fornace, A.J., Jr. p53-mediated DNA repair responses to UV radiation: Studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol. Cell Biol. 2000, 20, 3705–3714. [Google Scholar] [CrossRef]
- Wani, M.A.; Wani, G.; Yao, J.; Zhu, Q.; Wani, A.A. Human cells deficient in p53 regulated p21(waf1/cip1) expression exhibit normal nucleotide excision repair of UV-induced DNA damage. Carcinogenesis 2002, 23, 403–410. [Google Scholar] [CrossRef] [Green Version]
- Bendjennat, M.; Boulaire, J.; Jascur, T.; Brickner, H.; Barbier, V.; Sarasin, A.; Fotedar, A.; Fotedar, R. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell 2003, 114, 599–610. [Google Scholar] [CrossRef]
- Starostina, N.G.; Kipreos, E.T. Multiple degradation pathways regulate versatile CIP/KIP CDK inhibitors. Trends Cell Biol. 2012, 22, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Deng, T.; Yan, G.; Song, X.; Xie, L.; Zhou, Y.; Li, J.; Hu, X.; Li, Z.; Hu, J.; Zhang, Y.; et al. Deubiquitylation and stabilization of p21 by USP11 is critical for cell-cycle progression and DNA damage responses. Proc. Natl. Acad. Sci. USA 2018, 115, 4678–4683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bornstein, G.; Bloom, J.; Sitry-Shevah, D.; Nakayama, K.; Pagano, M.; Hershko, A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 2003, 278, 25752–25757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciznadija, D.; Zhu, X.H.; Koff, A. Hdm2- and proteasome-dependent turnover limits p21 accumulation during S phase. Cell Cycle 2011, 10, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Abbas, T.; Sivaprasad, U.; Terai, K.; Amador, V.; Pagano, M.; Dutta, A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008, 22, 2496–2506. [Google Scholar] [CrossRef] [Green Version]
- Coleman, K.E.; Grant, G.D.; Haggerty, R.A.; Brantley, K.; Shibata, E.; Workman, B.D.; Dutta, A.; Varma, D.; Purvis, J.E.; Cook, J.G. Sequential replication-coupled destruction at G1/S ensures genome stability. Genes Dev. 2015, 29, 1734–1746. [Google Scholar] [CrossRef] [Green Version]
- Stuart, S.A.; Wang, J.Y. Ionizing radiation induces ATM-independent degradation of p21Cip1 in transformed cells. J. Biol. Chem. 2009, 284, 15061–15070. [Google Scholar] [CrossRef] [Green Version]
- Sheng, C.; Mendler, I.H.; Rieke, S.; Snyder, P.; Jentsch, M.; Friedrich, D.; Drossel, B.; Loewer, A. PCNA-Mediated Degradation of p21 Coordinates the DNA Damage Response and Cell Cycle Regulation in Individual Cells. Cell Rep. 2019, 27, 48–58 e47. [Google Scholar] [CrossRef] [Green Version]
- Stewart-Ornstein, J.; Lahav, G. Dynamics of CDKN1A in Single Cells Defined by an Endogenous Fluorescent Tagging Toolkit. Cell Rep. 2016, 14, 1800–1811. [Google Scholar] [CrossRef] [Green Version]
- Vassilev, L.T. MDM2 inhibitors for cancer therapy. Trends Mol. Med. 2007, 13, 23–31. [Google Scholar] [CrossRef]
- Prives, C. Signaling to p53: Breaking the MDM2-p53 circuit. Cell 1998, 95, 5–8. [Google Scholar] [CrossRef] [Green Version]
- Galanos, P.; Vougas, K.; Walter, D.; Polyzos, A.; Maya-Mendoza, A.; Haagensen, E.J.; Kokkalis, A.; Roumelioti, F.M.; Gagos, S.; Tzetis, M.; et al. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat. Cell Biol. 2016, 18, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Mauro, M.; Rego, M.A.; Boisvert, R.A.; Esashi, F.; Cavallo, F.; Jasin, M.; Howlett, N.G. p21 promotes error-free replication-coupled DNA double-strand break repair. Nucleic Acids Res. 2012, 40, 8348–8360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottifredi, V.; Shieh, S.; Taya, Y.; Prives, C. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc. Natl. Acad. Sci. USA 2001, 98, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Duzdevich, D.; Warner, M.D.; Ticau, S.; Ivica, N.A.; Bell, S.P.; Greene, E.C. The dynamics of eukaryotic replication initiation: Origin specificity, licensing, and firing at the single-molecule level. Mol. Cell 2015, 58, 483–494. [Google Scholar] [CrossRef] [Green Version]
- Galanos, P.; Pappas, G.; Polyzos, A.; Kotsinas, A.; Svolaki, I.; Giakoumakis, N.N.; Glytsou, C.; Pateras, I.S.; Swain, U.; Souliotis, V.L.; et al. Mutational signatures reveal the role of RAD52 in p53-independent p21-driven genomic instability. Genome Biol. 2018, 19, 37. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Starostina, N.G.; Kipreos, E.T. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008, 22, 2507–2519. [Google Scholar] [CrossRef] [Green Version]
- Bertolin, A.P.; Mansilla, S.F.; Gottifredi, V. The identification of translesion DNA synthesis regulators: Inhibitors in the spotlight. DNA Repair 2015, 32, 158–164. [Google Scholar] [CrossRef] [Green Version]
- Soria, G.; Gottifredi, V. PCNA-coupled p21 degradation after DNA damage: The exception that confirms the rule? DNA Repair 2010, 9, 358–364. [Google Scholar] [CrossRef] [Green Version]
- Mansilla, S.F.; Bertolin, A.P.; Bergoglio, V.; Pillaire, M.J.; Gonzalez Besteiro, M.A.; Luzzani, C.; Miriuka, S.G.; Cazaux, C.; Hoffmann, J.S.; Gottifredi, V. Cyclin Kinase-independent role of p21(CDKN1A) in the promotion of nascent DNA elongation in unstressed cells. Elife 2016, 5, e18020. [Google Scholar] [CrossRef]
- Waters, L.S.; Minesinger, B.K.; Wiltrout, M.E.; D’Souza, S.; Woodruff, R.V.; Walker, G.C. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev.: MMBR 2009, 73, 134–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.J.; Colnaghi, L.; Huang, T.T. Dysregulation of DNA polymerase kappa recruitment to replication forks results in genomic instability. EMBO J. 2012, 31, 908–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bienko, M.; Green, C.M.; Sabbioneda, S.; Crosetto, N.; Matic, I.; Hibbert, R.G.; Begovic, T.; Niimi, A.; Mann, M.; Lehmann, A.R.; et al. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol. Cell 2010, 37, 396–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maya-Mendoza, A.; Moudry, P.; Merchut-Maya, J.M.; Lee, M.; Strauss, R.; Bartek, J. High speed of fork progression induces DNA replication stress and genomic instability. Nature 2018, 559, 279–284. [Google Scholar] [CrossRef]
- Liu, Y.; Kwiatkowski, D.J. Combined CDKN1A/TP53 mutation in bladder cancer is a therapeutic target. Mol. Cancer Ther. 2015, 14, 174–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Origanti, S.; Cai, S.R.; Munir, A.Z.; White, L.S.; Piwnica-Worms, H. Synthetic lethality of Chk1 inhibition combined with p53 and/or p21 loss during a DNA damage response in normal and tumor cells. Oncogene 2013, 32, 577–588. [Google Scholar] [CrossRef] [Green Version]
- Hauge, S.; Macurek, L.; Syljuasen, R.G. p21 limits S phase DNA damage caused by the Wee1 inhibitor MK1775. Cell Cycle 2019, 18, 834–847. [Google Scholar] [CrossRef]
- Garg, P.; Burgers, P.M. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 115–128. [Google Scholar] [CrossRef]
- Prives, C.; Gottifredi, V. The p21 and PCNA partnership: A new twist for an old plot. Cell Cycle 2008, 7, 3840–3846. [Google Scholar] [CrossRef] [Green Version]
- Mattia, M.; Gottifredi, V.; McKinney, K.; Prives, C. p53-Dependent p21 mRNA elongation is impaired when DNA replication is stalled. Mol. Cell Biol. 2007, 27, 1309–1320. [Google Scholar] [CrossRef] [Green Version]
- Beckerman, R.; Donner, A.J.; Mattia, M.; Peart, M.J.; Manley, J.L.; Espinosa, J.M.; Prives, C. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 2009, 23, 1364–1377. [Google Scholar] [CrossRef] [Green Version]
- Koumenis, C.; Alarcon, R.; Hammond, E.; Sutphin, P.; Hoffman, W.; Murphy, M.; Derr, J.; Taya, Y.; Lowe, S.W.; Kastan, M.; et al. Regulation of p53 by hypoxia: Dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol. Cell Biol. 2001, 21, 1297–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Zeng, S.X.; Lu, H. UV Induces p21 rapid turnover independently of ubiquitin and Skp2. J. Biol. Chem. 2006, 281, 26876–26883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.Y.; Yu, S.J.; Park, Y.G.; Kim, J.; Sohn, J. Glycogen synthase kinase 3beta phosphorylates p21WAF1/CIP1 for proteasomal degradation after UV irradiation. Mol. Cell Biol. 2007, 27, 3187–3198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soria, G.; Podhajcer, O.; Prives, C.; Gottifredi, V. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene 2006, 25, 2829–2838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savio, M.; Coppa, T.; Cazzalini, O.; Perucca, P.; Necchi, D.; Nardo, T.; Stivala, L.A.; Prosperi, E. Degradation of p21CDKN1A after DNA damage is independent of type of lesion, and is not required for DNA repair. DNA Repair 2009, 8, 778–785. [Google Scholar] [CrossRef]
- Nishitani, H.; Shiomi, Y.; Iida, H.; Michishita, M.; Takami, T.; Tsurimoto, T. CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 2008, 283, 29045–29052. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Budhavarapu, V.N.; Herrera, C.R.; Nam, H.W.; Kim, Y.S.; Yew, P.R. The CRL4Cdt2 ubiquitin ligase mediates the proteolysis of cyclin-dependent kinase inhibitor Xic1 through a direct association with PCNA. Mol. Cell Biol. 2010, 30, 4120–4133. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, A.R.; Niimi, A.; Ogi, T.; Brown, S.; Sabbioneda, S.; Wing, J.F.; Kannouche, P.L.; Green, C.M. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair 2007, 6, 891–899. [Google Scholar] [CrossRef]
- Ripley, B.M.; Gildenberg, M.S.; Washington, M.T. Control of DNA Damage Bypass by Ubiquitylation of PCNA. Genes 2020, 11, 138. [Google Scholar] [CrossRef] [Green Version]
- Masutani, C.; Kusumoto, R.; Iwai, S.; Hanaoka, F. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 2000, 19, 3100–3109. [Google Scholar] [CrossRef]
- Washington, M.T.; Johnson, R.E.; Prakash, L.; Prakash, S. Accuracy of lesion bypass by yeast and human DNA polymerase eta. Proc. Natl. Acad. Sci. USA 2001, 98, 8355–8360. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, A.R. Replication of damaged DNA by translesion synthesis in human cells. FEBS Lett. 2005, 579, 873–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendel, A.; Ziv, O.; Gueranger, Q.; Geacintov, N.; Livneh, Z. Reduced efficiency and increased mutagenicity of translesion DNA synthesis across a TT cyclobutane pyrimidine dimer, but not a TT 6-4 photoproduct, in human cells lacking DNA polymerase eta. DNA Repair 2008, 7, 1636–1646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avkin, S.; Sevilya, Z.; Toube, L.; Geacintov, N.; Chaney, S.G.; Oren, M.; Livneh, Z. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol. Cell 2006, 22, 407–413. [Google Scholar] [CrossRef] [PubMed]
- LaBaer, J.; Garrett, M.D.; Stevenson, L.F.; Slingerland, J.M.; Sandhu, C.; Chou, H.S.; Fattaey, A.; Harlow, E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997, 11, 847–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bloom, J.; Amador, V.; Bartolini, F.; DeMartino, G.; Pagano, M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 2003, 115, 71–82. [Google Scholar] [CrossRef] [Green Version]
- Tsanov, N.; Kermi, C.; Coulombe, P.; van der Laan, S.; Hodroj, D.; Maiorano, D. PIP degron proteins, substrates of CRL4Cdt2, and not PIP boxes, interfere with DNA polymerase eta and kappa focus formation on UV damage. Nucleic Acids Res. 2014, 42, 3692–3706. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Chatterjee, N.; Hemann, M.T.; Walker, G.C. Inhibition of mutagenic translesion synthesis: A possible strategy for improving chemotherapy? PLoS Genet. 2017, 13, e1006842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korzhnev, D.M.; Hadden, M.K. Targeting the Translesion Synthesis Pathway for the Development of Anti-Cancer Chemotherapeutics. J. Med. Chem. 2016, 59, 9321–9336. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Han, C.; Zhao, R.; Cui, T.; Dai, Y.; Mao, C.; Zhao, W.; Zhang, X.; Yu, J.; Wang, Q.E. Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells. Proc. Natl. Acad. Sci. USA 2015, 112, 4411–4416. [Google Scholar] [CrossRef] [Green Version]
- Lerner, L.K.; Francisco, G.; Soltys, D.T.; Rocha, C.R.; Quinet, A.; Vessoni, A.T.; Castro, L.P.; David, T.I.; Bustos, S.O.; Strauss, B.E.; et al. Predominant role of DNA polymerase eta and p53-dependent translesion synthesis in the survival of ultraviolet-irradiated human cells. Nucleic Acids Res. 2017, 45, 1270–1280. [Google Scholar] [CrossRef]
- Federico, M.B.; Siri, S.O.; Calzetta, N.L.; Paviolo, N.S.; de la Vega, M.B.; Martino, J.; Campana, M.C.; Wiesmuller, L.; Gottifredi, V. Unscheduled MRE11 activity triggers cell death but not chromosome instability in polymerase eta-depleted cells subjected to UV irradiation. Oncogene 2020. [Google Scholar] [CrossRef]
- Izuta, S. Inhibition of DNA polymerase eta by oxetanocin derivatives. Nucleic Acids Symp. Ser. (Oxf.) 2006, 269–270. [Google Scholar] [CrossRef] [Green Version]
- Mizushina, Y.; Motoshima, H.; Yamaguchi, Y.; Takeuchi, T.; Hirano, K.; Sugawara, F.; Yoshida, H. 3-O-methylfunicone, a selective inhibitor of mammalian Y-family DNA polymerases from an Australian sea salt fungal strain. Mar. Drugs 2009, 7, 624–639. [Google Scholar] [CrossRef] [Green Version]
- Vanarotti, M.; Evison, B.J.; Actis, M.L.; Inoue, A.; McDonald, E.T.; Shao, Y.; Heath, R.J.; Fujii, N. Small-molecules that bind to the ubiquitin-binding motif of REV1 inhibit REV1 interaction with K164-monoubiquitinated PCNA and suppress DNA damage tolerance. Bioorg. Med. Chem. 2018, 26, 2345–2353. [Google Scholar] [CrossRef]
- Yamanaka, K.; Dorjsuren, D.; Eoff, R.L.; Egli, M.; Maloney, D.J.; Jadhav, A.; Simeonov, A.; Lloyd, R.S. A comprehensive strategy to discover inhibitors of the translesion synthesis DNA polymerase kappa. PLoS ONE 2012, 7, e45032. [Google Scholar] [CrossRef] [Green Version]
- Punchihewa, C.; Inoue, A.; Hishiki, A.; Fujikawa, Y.; Connelly, M.; Evison, B.; Shao, Y.; Heath, R.; Kuraoka, I.; Rodrigues, P.; et al. Identification of small molecule proliferating cell nuclear antigen (PCNA) inhibitor that disrupts interactions with PIP-box proteins and inhibits DNA replication. J. Biol. Chem. 2012, 287, 14289–14300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sail, V.; Rizzo, A.A.; Chatterjee, N.; Dash, R.C.; Ozen, Z.; Walker, G.C.; Korzhnev, D.M.; Hadden, M.K. Identification of Small Molecule Translesion Synthesis Inhibitors That Target the Rev1-CT/RIR Protein-Protein Interaction. ACS Chem. Biol. 2017, 12, 1903–1912. [Google Scholar] [CrossRef] [PubMed]
- Actis, M.L.; Ambaye, N.D.; Evison, B.J.; Shao, Y.; Vanarotti, M.; Inoue, A.; McDonald, E.T.; Kikuchi, S.; Heath, R.; Hara, K.; et al. Identification of the first small-molecule inhibitor of the REV7 DNA repair protein interaction. Bioorg. Med. Chem. 2016, 24, 4339–4346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojtaszek, J.L.; Chatterjee, N.; Najeeb, J.; Ramos, A.; Lee, M.; Bian, K.; Xue, J.Y.; Fenton, B.A.; Park, H.; Li, D.; et al. A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy. Cell 2019, 178, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Villafanez, F.; Garcia, I.A.; Carbajosa, S.; Pansa, M.F.; Mansilla, S.; Llorens, M.C.; Angiolini, V.; Guantay, L.; Jacobs, H.; Madauss, K.P.; et al. AKT inhibition impairs PCNA ubiquitylation and triggers synthetic lethality in homologous recombination-deficient cells submitted to replication stress. Oncogene 2019, 38, 4310–4324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benamar, M.; Guessous, F.; Du, K.; Corbett, P.; Obeid, J.; Gioeli, D.; Slingluff, C.L.; Jr Abbas, T. Inactivation of the CRL4-CDT2-SET8/p21 ubiquitylation and degradation axis underlies the therapeutic efficacy of pevonedistat in melanoma. EBioMedicine 2016, 10, 85–100. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Xie, K.; Zhang, X.Q.; Pridgen, E.M.; Park, G.Y.; Cui, D.S.; Shi, J.; Wu, J.; Kantoff, P.W.; Lippard, S.J.; et al. Enhancing tumor cell response to chemotherapy through nanoparticle-mediated codelivery of siRNA and cisplatin prodrug. Proc. Natl. Acad. Sci. USA 2013, 110, 18638–18643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roninson, I.B. Oncogenic functions of tumour suppressor p21(Waf1/Cip1/Sdi1): Association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett. 2002, 179, 1–14. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansilla, S.F.; De La Vega, M.B.; Calzetta, N.L.; Siri, S.O.; Gottifredi, V. CDK-Independent and PCNA-Dependent Functions of p21 in DNA Replication. Genes 2020, 11, 593. https://doi.org/10.3390/genes11060593
Mansilla SF, De La Vega MB, Calzetta NL, Siri SO, Gottifredi V. CDK-Independent and PCNA-Dependent Functions of p21 in DNA Replication. Genes. 2020; 11(6):593. https://doi.org/10.3390/genes11060593
Chicago/Turabian StyleMansilla, Sabrina Florencia, María Belén De La Vega, Nicolás Luis Calzetta, Sebastián Omar Siri, and Vanesa Gottifredi. 2020. "CDK-Independent and PCNA-Dependent Functions of p21 in DNA Replication" Genes 11, no. 6: 593. https://doi.org/10.3390/genes11060593