Microbial and Plant-Assisted Bioremediation of Heavy Metal Polluted Environments: A Review
Abstract
:1. Introduction
2. Bioremediation
3. Effects of Heavy Metals on the Environment
4. Mechanism of Heavy Metal Remediation by Microorganisms
- (1)
- Sequestration of toxic metals by cell wall components or by intracellular metal binding proteins and peptides such as metallothioneins (MT) and phytochelatins along with compounds such as bacterial siderophores which are mostly catecholates, compared to fungi that produce hydroxamate siderophores.
- (2)
- Alteration of biochemical pathways to block metal uptake.
- (3)
- Conversion of metals to innocuous forms by enzymes.
- (4)
- Reduction of intracellular concentration of metals using precise efflux systems [36].
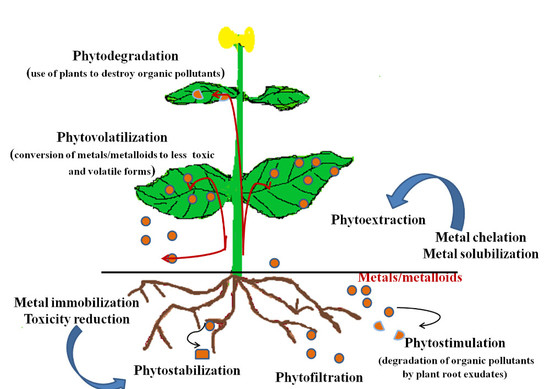
5. Phytoremediation
5.1. Phytoextraction/Phytoaccumulation
5.2. Phytofiltration
5.3. Phytostimulation
5.4. Phytostabilization
5.5. Phytovolatilization
5.6. Phytodegradation
5.7. Rhizofiltration
6. Plant Mechanisms for Metal Detoxification
7. Role of Plant Growth-Promoting Bacteria (PGPR) in Plant Growth under Abiotic Stress
7.1. Siderophore Production
7.2. Phosphate Solubilization
7.3. Aminoacyclopropane-1-Carboxylate Deaminase Production
7.4. Indole-3-Acetic Acid Production
8. Bioremediation Using Advanced Molecular Techniques and Genetically Engineered Organisms
9. Future Prospects for Bioremediation
10. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nematian, M.A.; Kazemeini, F. Accumulation of Pb, Zn, C and Fe in plants and hyperaccumulator choice in galali iron mine area, Iran. Int. J. Agric. Crop Sci. 2013, 5, 426. [Google Scholar]
- Kabata-Pendias, A. Trace Elements in Soils and Plants; CRC Press: New York, NY, USA, 2010. [Google Scholar]
- Dixit, R.; Malaviya, D.; Pandiyan, K.; Singh, U.B.; Sahu, A.; Shukla, R.; Singh, B.P.; Rai, J.P.; Sharma, P.K.; Lade, H. Bioremediation of heavy metals from soil and aquatic environment: An overview of principles and criteria of fundamental processes. Sustainability 2015, 7, 2189–2212. [Google Scholar] [CrossRef]
- Tak, H.I.; Ahmad, F.; Babalola, O.O. Advances in the application of plant growth-promoting rhizobacteria in phytoremediation of heavy metals. In Reviews of Environmental Contamination and Toxicology; Whitacre, D.M., Ed.; Springer: New York, NY, USA, 2013; Volume 223, pp. 33–52. [Google Scholar]
- Chandra, K.; Salman, A.S.; Mohd, A.; Sweety, R.; Ali, K.N. Protection against fca induced oxidative stress induced DNA damage as a model of arthritis and in vitro anti-arthritic potential of costus speciosus rhizome extract. Int. J. Pharm. Phytopharmacol. Res. 2015, 7, 383–389. [Google Scholar]
- Mani, S. Production of reactive oxygen species and its implication in human diseases. In Free Radicals in Human Health and Disease; Vibha, R., Umesh, C.S.Y., Eds.; Springer: New Delhi, India, 2015; pp. 3–15. [Google Scholar]
- Chibuike, G.; Obiora, S. Heavy metal polluted soils: Effect on plants and bioremediation methods. Appl. Environ. Soil Sci. 2014, 2014, doi–10. [Google Scholar] [CrossRef]
- Ekperusi, O.; Aigbodion, F. Bioremediation of petroleum hydrocarbons from crude oil-contaminated soil with the earthworm: Hyperiodrilus africanus. 3 Biotech 2015, 5, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Ayangbenro, A.S.; Babalola, O.O. A new strategy for heavy metal polluted environments: A review of microbial biosorbents. Int. J. Environ. Res. Public Health 2017, 14, 94. [Google Scholar] [CrossRef] [PubMed]
- Blaylock, M.J.; Salt, D.E.; Dushenkov, S.; Zakharova, O.; Gussman, C.; Kapulnik, Y.; Ensley, B.D.; Raskin, I. Enhanced accumulation of Pb in Indian mustard by soil-applied chelating agents. Environ. Sci. Technol. 1997, 31, 860–865. [Google Scholar] [CrossRef]
- Verma, J.P.; Jaiswal, D.K. Book review: Advances in biodegradation and bioremediation of industrial waste. Front. Microbiol. 2016, 6, 1555. [Google Scholar] [CrossRef]
- Jain, S.; Arnepalli, D. Biominerlisation as a remediation technique: A critical review. In Proceedings of the Indian Geotechnical Conference (IGC2016), Chennai, India, 15–17 December 2016. [Google Scholar]
- Mathialagan, T.; Viraraghavan, T. Biosorption of pentachlorophenol from aqueous solutions by a fungal biomass. Bioresour. Technol. 2009, 100, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Brandt, S.; Zeng, A.P.; Deckwer, W.D. Adsorption and desorption of pentachlorophenol on cells of mycobacterium chlorophenolicum PCP-1. Biotechnol. Bioeng. 1997, 55, 480–489. [Google Scholar] [CrossRef]
- Bosso, L.; Lacatena, F.; Cristinzio, G.; Cea, M.; Diez, M.C.; Rubilar, O. Biosorption of pentachlorophenol by anthracophyllum discolor in the form of live fungal pellets. New Biotechnol. 2015, 32, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Jianlong, W.; Yi, Q.; Horan, N.; Stentiford, E. Bioadsorption of pentachlorophenol (PCP) from aqueous solution by activated sludge biomass. Bioresour. Technol. 2000, 75, 157–161. [Google Scholar] [CrossRef]
- Yeung, A.T. Remediation technologies for contaminated sites. In Advances in Environmental Geotechnics; Chen, Y., Tang, X., Zhan, L., Eds.; Springer: New York, NY, USA, 2009; pp. 328–369. [Google Scholar]
- Tandon, P.K.; Singh, S.B. Redox processes in water remediation. Environ. Chem. Lett. 2016, 14, 15–25. [Google Scholar] [CrossRef]
- Gadd, G.M. Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 2010, 156, 609–643. [Google Scholar] [CrossRef] [PubMed]
- Rajapaksha, A.U.; Vithanage, M.; Ok, Y.S.; Oze, C. Cr (VI) formation related to Cr (III)-muscovite and birnessite interactions in ultramafic environments. Environ. Sci. Technol. 2013, 47, 9722–9729. [Google Scholar] [CrossRef] [PubMed]
- Bolan, N.S.; Kunhikrishnan, A.; Naidu, R. Carbon storage in a heavy clay soil landfill site after biosolid application. Sci. Total Environ. 2013, 465, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Beiyuan, J.; Awad, Y.M.; Beckers, F.; Tsang, D.C.; Ok, Y.S.; Rinklebe, J. Mobility and phytoavailability of As and Pb in a contaminated soil using pine sawdust biochar under systematic change of redox conditions. Chemosphere 2017, 178, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Alburquerque, J.; De La Fuente, C.; Bernal, M. Improvement of soil quality after “alperujo” compost application to two contaminated soils characterised by differing heavy metal solubility. J. Environ. Manag. 2011, 92, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Xu, P.; Zeng, G.; Yang, C.; Huang, D.; Zhang, J. Bioremediation of soils contaminated with polycyclic aromatic hydrocarbons, petroleum, pesticides, chlorophenols and heavy metals by composting: Applications, microbes and future research needs. Biotechnol. Adv. 2015, 33, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Ok, Y.S.; Uchimiya, S.M.; Chang, S.X.; Bolan, N. Biochar: Production, Characterization, and Applications; CRC Press: New York, NY, USA, 2015. [Google Scholar]
- Mohan, D.; Sarswat, A.; Ok, Y.S.; Pittman, C.U. Organic and inorganic contaminants removal from water with biochar, a renewable, low cost and sustainable adsorbent―A critical review. Bioresour. Technol. 2014, 160, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.B.; Zhou, J.L.; Ngo, H.H.; Guo, W.; Chen, M. Progress in the preparation and application of modified biochar for improved contaminant removal from water and wastewater. Bioresour. Technol. 2016, 214, 836–851. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Ali, S.; Qayyum, M.F.; Ibrahim, M.; Zia-ur-Rehman, M.; Abbas, T.; Ok, Y.S. Mechanisms of biochar-mediated alleviation of toxicity of trace elements in plants: A critical review. Environ. Sci. Pollut. Res. 2016, 23, 2230–2248. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Bolan, N.; Prévoteau, A.; Vithanage, M.; Biswas, J.K.; Ok, Y.S.; Wang, H. Applications of biochar in redox-mediated reactions. Bioresour. Technol. 2017, 246, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Klüpfel, L.; Keiluweit, M.; Kleber, M.; Sander, M. Redox properties of plant biomass-derived black carbon (biochar). Environ. Sci. Technol. 2014, 48, 5601–5611. [Google Scholar] [CrossRef] [PubMed]
- Saquing, J.M.; Yu, Y.-H.; Chiu, P.C. Wood-derived black carbon (biochar) as a microbial electron donor and acceptor. Environ. Sci. Technol. Lett. 2016, 3, 62–66. [Google Scholar] [CrossRef]
- Graber, E.; Tsechansky, L.; Lew, B.; Cohen, E. Reducing capacity of water extracts of biochars and their solubilization of soil Mn and Fe. Eur. J. Soil Sci. 2014, 65, 162–172. [Google Scholar] [CrossRef]
- Violante, A.; Cozzolino, V.; Perelomov, L.; Caporale, A.; Pigna, M. Mobility and bioavailability of heavy metals and metalloids in soil environments. J. Soil Sci. Plant Nutr. 2010, 10, 268–292. [Google Scholar] [CrossRef]
- Tandon, P.K.; Shukla, R.C.; Singh, S.B. Removal of arsenic (III) from water with clay-supported zerovalent iron nanoparticles synthesized with the help of tea liquor. Ind. Eng. Chem. Res. 2013, 52, 10052–10058. [Google Scholar] [CrossRef]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation techniques-classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 2016, 32, 180. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.T.; Azam, M.; Ali, A.; Haq, Q.M.R. Prospects for exploiting bacteria for bioremediation of metal pollution. Crit. Rev. Environ. Sci. Technol. 2014, 44, 519–560. [Google Scholar] [CrossRef]
- Rayu, S.; Karpouzas, D.G.; Singh, B.K. Emerging technologies in bioremediation: Constraints and opportunities. Biodegradation 2012, 23, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Mani, D.; Kumar, C. Biotechnological advances in bioremediation of heavy metals contaminated ecosystems: An overview with special reference to phytoremediation. Int. J. Environ. Sci. Technol. 2014, 11, 843–872. [Google Scholar] [CrossRef]
- Lu, L.; Huggins, T.; Jin, S.; Zuo, Y.; Ren, Z.J. Microbial metabolism and community structure in response to bioelectrochemically enhanced remediation of petroleum hydrocarbon-contaminated soil. Environ. Sci. Technol. 2014, 48, 4021–4029. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Thavamani, P.; Ramadass, K.; Naidu, R.; Srivastava, P.; Megharaj, M. Remediation trials for hydrocarbon-contaminated soils in arid environments: Evaluation of bioslurry and biopiling techniques. Int. Biodeterior. Biodegrad. 2015, 101, 56–65. [Google Scholar] [CrossRef]
- Kushwaha, A.; Rani, R.; Kumar, S.; Gautam, A. Heavy metal detoxification and tolerance mechanisms in plants: Implications for phytoremediation. Environ. Rev. 2015, 24, 39–51. [Google Scholar] [CrossRef]
- Gupta, A.; Joia, J.; Sood, A.; Sood, R.; Sidhu, C.; Kaur, G. Microbes as potential tool for remediation of heavy metals: A review. J. Microb. Biochem. Technol. 2016, 8, 364–372. [Google Scholar] [CrossRef]
- Ndeddy Aka, R.J.; Babalola, O.O. Effect of bacterial inoculation of strains of pseudomonas aeruginosa, alcaligenes feacalis and Bacillus subtilis on germination, growth and heavy metal (Cd, Cr, and Ni) uptake of Brassica juncea. Int. J. Phytorem. 2016, 18, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Skórzyńska-Polit, E.; Drążkiewicz, M.; Krupa, Z. Lipid peroxidation and antioxidative response in arabidopsis thaliana exposed to cadmium and copper. Acta Physiol. Plant. 2010, 32, 169. [Google Scholar] [CrossRef]
- Upadhyay, N.; Vishwakarma, K.; Singh, J.; Mishra, M.; Kumar, V.; Rani, R.; Mishra, R.K.; Chauhan, D.K.; Tripathi, D.K.; Sharma, S. Tolerance and reduction of chromium (VI) by Bacillus sp. Mnu16 isolated from contaminated coal mining soil. Front. Plant Sci. 2017, 8, 778. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.D.; Pal, D.; Penta, S.; Kumar, A. Ecotoxic heavy metals transformation by bacteria and fungi in aquatic ecosystem. World J. Microbiol. Biotechnol. 2015, 31, 1595–1603. [Google Scholar] [CrossRef] [PubMed]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol. 2014, 7, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Muszynska, E.; Hanus-Fajerska, E. Why are heavy metal hyperaccumulating plants so amazing? BioTechnol. J. Biotechnol. Comput. Biol. Bionanotechnol. 2015, 96, 265–271. [Google Scholar] [CrossRef]
- Pourrut, B.; Shahid, M.; Dumat, C.; Winterton, P.; Pinelli, E. Lead uptake, toxicity, and detoxification in plants. In Reviews of Environmental Contamination and Toxicology; Springer: New York, NY, USA, 2011; Volume 213, pp. 113–136. [Google Scholar]
- Jadia, C.D.; Fulekar, M. Phytoremediation of heavy metals: Recent techniques. Afr. J. Biotechnol. 2009, 8, 921–928. [Google Scholar]
- Gaur, N.; Flora, G.; Yadav, M.; Tiwari, A. A review with recent advancements on bioremediation-based abolition of heavy metals. Environ. Sci. Process. Impacts 2014, 16, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Flora, G.J. Arsenic toxicity and possible treatment strategies: Some recent advancement. Curr. Trends Biotechnol. Pharm. 2012, 6, 280–289. [Google Scholar]
- Dadzie, E. Assessment of Heavy Metal Contamination of the Densu River, Weija From Leachate. Master’s Thesis, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana, 2012. [Google Scholar]
- Tschirhart, C.; Handschumacher, P.; Laffly, D.; Bénéfice, E. Resource management, networks and spatial contrasts in human mercury contamination along the Rio Beni (Bolivian Amazon). Hum. Ecol. 2012, 40, 511–523. [Google Scholar] [CrossRef]
- Lakherwal, D. Adsorption of heavy metals: A review. Int. J. Environ. Res. Dev. 2014, 4, 41–48. [Google Scholar]
- Gupta, P.; Diwan, B. Bacterial exopolysaccharide mediated heavy metal removal: A review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 2016, 13, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Selatnia, A.; Boukazoula, A.; Kechid, N.; Bakhti, M.; Chergui, A.; Kerchich, Y. Biosorption of lead (II) from aqueous solution by a bacterial dead streptomyces rimosus biomass. Biochem. Eng. J. 2004, 19, 127–135. [Google Scholar] [CrossRef]
- Kang, C.-H.; Kwon, Y.-J.; So, J.-S. Bioremediation of heavy metals by using bacterial mixtures. Ecol. Eng. 2016, 89, 64–69. [Google Scholar] [CrossRef]
- Wang, J.; Chen, C. Biosorbents for heavy metals removal and their future. Biotechnol. Adv. 2009, 27, 195–226. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Anil Dwivedi, K. Biological wastes the tool for biosorption of arsenic. J. Bioremed. Biodegrad. 2015, 7, 2. [Google Scholar] [CrossRef]
- Coelho, L.M.; Rezende, H.C.; Coelho, L.M.; de Sousa, P.A.; Melo, D.F.; Coelho, N.M. Bioremediation of polluted waters using microorganisms. In Advances in Bioremediation of Wastewater and Polluted Soil; Shiomi, N., Ed.; InTech: Shanghai, China, 2015. [Google Scholar]
- Fomina, M.; Gadd, G.M. Biosorption: Current perspectives on concept, definition and application. Bioresour. Technol. 2014, 160, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Nayak, A.; Agarwal, S. Bioadsorbents for remediation of heavy metals: Current status and their future prospects. Environ. Eng. Res. 2015, 20, 1–18. [Google Scholar] [CrossRef]
- Srivastava, S.; Agrawal, S.; Mondal, M. A review on progress of heavy metal removal using adsorbents of microbial and plant origin. Environ. Sci. Pollut. Res. 2015, 22, 15386–15415. [Google Scholar] [CrossRef] [PubMed]
- Machado, M.D.; Soares, E.V.; Soares, H.M. Removal of heavy metals using a brewer’s yeast strain of Saccharomyces cerevisiae: Chemical speciation as a tool in the prediction and improving of treatment efficiency of real electroplating effluents. J. Hazard. Mater. 2010, 180, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.-Q.; Li, S.; Zhu, H.-Y.; Jiang, R.; Yin, L.-F. Biosorption of copper(II) from aqueous solution by mycelial pellets of rhizopus oryzae. Afr. J. Biotechnol. 2012, 11, 1403–1411. [Google Scholar]
- Abbas, S.H.; Ismail, I.M.; Mostafa, T.M.; Sulaymon, A.H. Biosorption of heavy metals: A review. J. Chem. Sci. Technol. 2014, 3, 74–102. [Google Scholar]
- Mustapha, M.U.; Halimoon, N. Microorganisms and biosorption of heavy metals in the environment: A review paper. J. Microb. Biochem. Technol. 2015, 7, 253–256. [Google Scholar] [CrossRef]
- Donot, F.; Fontana, A.; Baccou, J.; Schorr-Galindo, S. Microbial exopolysaccharides: Main examples of synthesis, excretion, genetics and extraction. Carbohydr. Polym. 2012, 87, 951–962. [Google Scholar] [CrossRef]
- Öner, E.T. Microbial production of extracellular polysaccharides from biomass. In Pretreatment Techniques for Biofuels and Biorefineries; Fang, Z., Ed.; Springer: New York, NY, USA, 2013; pp. 35–56. [Google Scholar]
- François, F.; Lombard, C.; Guigner, J.-M.; Soreau, P.; Brian-Jaisson, F.; Martino, G.; Vandervennet, M.; Garcia, D.; Molinier, A.-L.; Pignol, D. Isolation and characterization of environmental bacteria capable of extracellular biosorption of mercury. Appl. Environ. Microbiol. 2012, 78, 1097–1106. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Wang, Y.; Gong, L.; Wang, M.; Wang, H.; He, N.; Zheng, Y.; Li, Q. Formation of soluble Cr (III) end-products and nanoparticles during Cr (VI) reduction by bacillus cereus strain XMCr-6. Biochem. Eng. J. 2013, 70, 166–172. [Google Scholar] [CrossRef]
- Kanmani, P.; Aravind, J.; Preston, D. Remediation of chromium contaminants using bacteria. Int. J. Environ. Sci. Technol. 2012, 9, 183–193. [Google Scholar] [CrossRef]
- Achal, V.; Pan, X.; Fu, Q.; Zhang, D. Biomineralization based remediation of As (III) contaminated soil by Sporosarcina ginsengisoli. J. Hazard. Mater. 2012, 201, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.L.; Ceretti, H.M.; Daniel, M.A.; Ramírez, S.A.; Zalts, A. Cadmium, Zinc and Copper biosorption mediated by Pseudomonas veronii 2e. Bioresour. Technol. 2008, 99, 5574–5581. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, D.; Udayasooriyan, C.; Kamaladevi, B. Chromium (VI) reduction by Pseudomonas putida and Bacillus subtilis isolated from contaminated soils. Int. J. Environ. Sci. 2014, 5, 522. [Google Scholar]
- Rahman, A.; Nahar, N.; Nawani, N.N.; Jass, J.; Hossain, K.; Saud, Z.A.; Saha, A.K.; Ghosh, S.; Olsson, B.; Mandal, A. Bioremediation of hexavalent chromium (VI) by a soil-borne bacterium, enterobacter cloacae b2-dha. J. Environ. Sci. Health Part A 2015, 50, 1136–1147. [Google Scholar] [CrossRef] [PubMed]
- Taştan, B.E.; Ertuğrul, S.; Dönmez, G. Effective bioremoval of reactive dye and heavy metals by Aspergillus versicolor. Bioresour. Technol. 2010, 101, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Kumar Ramasamy, R.; Congeevaram, S.; Thamaraiselvi, K. Evaluation of isolated fungal strain from e-waste recycling facility for effective sorption of toxic heavy metal Pb (II) ions and fungal protein molecular characterization―A mycoremediation approach. Asian J. Exp. Biol. Sci. 2011, 2, 342–347. [Google Scholar]
- Achal, V.; Kumari, D.; Pan, X. Bioremediation of chromium contaminated soil by a brown-rot fungus, gloeophyllum sepiarium. Res. J. Microbiol. 2011, 6, 166. [Google Scholar] [CrossRef]
- Sukumar, M. Reduction of hexavalent chromium by rhizopus oryzae. Afr. J. Environ. Sci. Technol. 2010, 4, 412–418. [Google Scholar]
- Farhan, S.N.; Khadom, A.A. Biosorption of heavy metals from aqueous solutions by Saccharomyces cerevisiae. Int. J. Ind. Chem. 2015, 6, 119–130. [Google Scholar] [CrossRef]
- Lívia de CF, M.H.; Benedito, C. Potential application of modified Saccharomyces cerevisiae for removing lead and cadmium. J. Bioremed. Biodegrad. 2015, 6, 2. [Google Scholar]
- Lee, Y.-C.; Chang, S.-P. The biosorption of heavy metals from aqueous solution by Spirogyra and Cladophora filamentous macroalgae. Bioresour. Technol. 2011, 102, 5297–5304. [Google Scholar] [CrossRef] [PubMed]
- Mane, P.; Bhosle, A. Bioremoval of some metals by living algae Spirogyra sp. And Spirullina sp. From aqueous solution. Int. J. Environ. Res. 2012, 6, 571–576. [Google Scholar]
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef] [PubMed]
- Teschler, J.K.; Zamorano-Sánchez, D.; Utada, A.S.; Warner, C.J.; Wong, G.C.; Linington, R.G.; Yildiz, F.H. Living in the matrix: Assembly and control of Vibrio cholerae biofilms. Nat. Rev. Microbiol. 2015, 13, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Khan, E.; Sajad, M.A. Phytoremediation of heavy metals―Concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Jan, S.; Parray, J.A. Approaches to Heavy Metal Tolerance in Plants; Springer: New Delhi, India, 2016. [Google Scholar]
- Abbaszadeh-Dahaji, P.; Omidvari, M.; Ghorbanpour, M. Increasing phytoremediation efficiency of heavy metal-contaminated soil using PGPR for sustainable agriculture. In Plant-Microbe Interaction: An Approach to Sustainable Agriculture; Choudhary, D.K., Varma, A., Tuteja, N., Eds.; Springer: New Delhi, India, 2016; pp. 187–204. [Google Scholar]
- Choudhary, D.K.; Varma, A.; Tuteja, N. Plant-Microbe Interaction: An Approach to Sustainable Agriculture; Springer: New Delhi, India, 2017. [Google Scholar]
- Jutsz, A.M.; Gnida, A. Mechanisms of stress avoidance and tolerance by plants used in phytoremediation of heavy metals. Arch. Environ. Prot. 2015, 41, 104–114. [Google Scholar] [CrossRef]
- Jabeen, R.; Ahmad, A.; Iqbal, M. Phytoremediation of heavy metals: Physiological and molecular mechanisms. Bot. Rev. 2009, 75, 339–364. [Google Scholar] [CrossRef]
- Rascio, N.; Navari-Izzo, F. Heavy metal hyperaccumulating plants: How and why do they do it? And what makes them so interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Van der Ent, A.; Baker, A.J.; Reeves, R.D.; Pollard, A.J.; Schat, H. Hyperaccumulators of metal and metalloid trace elements: Facts and fiction. Plant Soil 2013, 362, 319–334. [Google Scholar] [CrossRef]
- Madejón, P.; Murillo, J.M.; Marañón, T.; Cabrera, F.; Soriano, M. Trace element and nutrient accumulation in sunflower plants two years after the Aznalcollar mine spill. Sci. Total Environ. 2003, 307, 239–257. [Google Scholar] [CrossRef]
- Lin, J.; Jiang, W.; Liu, D. Accumulation of copper by roots, hypocotyls, cotyledons and leaves of sunflower (Helianthus annuus L.). Bioresour. Technol. 2003, 86, 151–155. [Google Scholar] [CrossRef]
- Marchiol, L.; Fellet, G.; Perosa, D.; Zerbi, G. Removal of trace metals by Sorghum bicolor and Helianthus annuus in a site polluted by industrial wastes: A field experience. Plant Physiol. Biochem. 2007, 45, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Adesodun, J.K.; Atayese, M.O.; Agbaje, T.; Osadiaye, B.A.; Mafe, O.; Soretire, A.A. Phytoremediation potentials of sunflowers (Tithonia diversifolia and Helianthus annuus) for metals in soils contaminated with zinc and lead nitrates. Water Air Soil Pollut. 2010, 207, 195–201. [Google Scholar] [CrossRef]
- Herrero, E.; Lopez-Gonzalvez, A.; Ruiz, M.; Lucas-Garcia, J.; Barbas, C. Uptake and distribution of zinc, cadmium, lead and copper in Brassica napus var. Oleifera and Helianthus annus grown in contaminated soils. Int. J. Phytoremed. 2003, 5, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Angelova, V.R.; Perifanova-Nemska, M.; Uzunova, G.; Ivanov, K.; Lee, H. Potential of sunflower (Helianthus annuus L.) for phytoremediation of soils contaminated with heavy metals. World J. Sci. Eng. Technol. 2016, 10, 1–11. [Google Scholar]
- Ebbs, S.D.; Kochian, L.V. Toxicity of zinc and copper to brassica species: Implications for phytoremediation. J. Environ. Qual. 1997, 26, 776–781. [Google Scholar] [CrossRef]
- Islam, M.S.; Ueno, Y.; Sikder, M.T.; Kurasaki, M. Phytofiltration of arsenic and cadmium from the water environment using Micranthemum umbrosum (jf GMEL) sf blake as a hyperaccumulator. Int. J. Phytoremed. 2013, 15, 1010–1021. [Google Scholar] [CrossRef] [PubMed]
- Freitas, E.V.; Nascimento, C.W.; Souza, A.; Silva, F.B. Citric acid-assisted phytoextraction of lead: A field experiment. Chemosphere 2013, 92, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ferreiro, J.; Lu, H.; Fu, S.; Méndez, A.; Gascó, G. Use of phytoremediation and biochar to remediate heavy metal polluted soils: A review. Solid Earth 2014, 5, 65. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Zhu, J.; Zhou, Q.X.; Zhan, J. Fertilizer amendment for improving the phytoextraction of cadmium by a hyperaccumulator Rorippa globosa (turcz.) thell. J. Soils Sed. 2011, 11, 915. [Google Scholar] [CrossRef]
- Vassilev, A.; Schwitzguébel, J.-P.; Thewys, T.; Van Der Lelie, D.; Vangronsveld, J. The use of plants for remediation of metal-contaminated soils. Sci. World J. 2004, 4, 9–34. [Google Scholar] [CrossRef] [PubMed]
- Slatter, K.A. Nickel Accumulation and Tolerance in Berkheya Codii and Its Application in Phytoremediation. Master’s Thesis, University of Kwazulu, Natal, South Africa, 2013. [Google Scholar]
- Fulekar, M. Phytoremediation of heavy metals by Helianthus annuus in aquatic and soil environment. Int. J. Curr. Microbiol. App. Sci. 2016, 5, 392–404. [Google Scholar] [CrossRef]
- Mengoni, A.; Cecchi, L.; Gonnelli, C. Nickel hyperaccumulating plants and alyssum bertolonii: Model systems for studying biogeochemical interactions in serpentine soils. In Bio-Geo Interactions in Metal-Contaminated Soils; Kothe, E., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 31, pp. 279–296. [Google Scholar]
- Broadhurst, C.L.; Chaney, R.L. Growth and metal accumulation of an alyssum murale nickel hyperaccumulator ecotype co-cropped with alyssum montanum and perennial ryegrass in serpentine soil. Front. Plant Sci. 2016, 7, 451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wen, X.; Huang, Y.; Inoue, C.; Liang, Y. Higher accumulation capacity of cadmium than zinc by Arabidopsis halleri ssp. Germmifera in the field using different sowing strategies. Plant Soil 2017, 418, 1–12. [Google Scholar] [CrossRef]
- Claire-Lise, M.; Nathalie, V. The use of the model species arabidopsis halleri towards phytoextraction of cadmium polluted soils. New Biotechnol. 2012, 30, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Bothe, H. Plants in heavy metal soils. In Detoxification of Heavy Metals; Sherameti, I., Varma, A., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2011; Volume 30, pp. 35–57. [Google Scholar]
- Chen, B.; Ai, W.; Gong, H.; Gao, X.; Qiu, B. Cleaning up of heavy metals-polluted water by a terrestrial hyperaccumulator Sedum alfredii hance. Front. Biol. 2013, 8, 599–605. [Google Scholar] [CrossRef]
- Alford, É.R.; Pilon-Smits, E.A.; Fakra, S.C.; Paschke, M.W. Selenium hyperaccumulation by astragalus (Fabaceae) does not inhibit root nodule symbiosis. Am. J. Bot. 2012, 99, 1930–1941. [Google Scholar] [CrossRef] [PubMed]
- Nalla, S.; Hardaway, C.J.; Sneddon, J. Phytoextraction of selected metals by the first and second growth seasons of Spartina alterniflora. Instrum. Sci. Technol. 2012, 40, 17–28. [Google Scholar] [CrossRef]
- Rathinasabapathi, B. Arsenic hyperaccumulator fern pteris vittata: Utilities for arsenic phytoremediation and plant biotechnology. In Working with Ferns; Fernández, H., Kumar, A., Revilla, M.A., Eds.; Springer: New York, NY, USA, 2011; pp. 261–269. [Google Scholar]
- Xie, Q.-E.; Yan, X.-L.; Liao, X.-Y.; Li, X. The arsenic hyperaccumulator fern Pteris vittata L. Environ. Sci. Technol. 2009, 43, 8488–8495. [Google Scholar] [CrossRef] [PubMed]
- Datta, R.; Das, P.; Tappero, R.; Punamiya, P.; Elzinga, E.; Sahi, S.; Feng, H.; Kiiskila, J.; Sarkar, D. Evidence for exocellular arsenic in fronds of Pteris vittata. Sci. Rep. 2017, 7, 2839. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Han, F.X.; Chen, J.; Sridhar, B.M.; Monts, D.L. Phytoextraction and accumulation of mercury in three plant species: Indian mustard (Brassica juncea), beard grass (Polypogon monospeliensis), and chinese brake fern (Pteris vittata). Int. J. Phytoremed. 2008, 10, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, P.; Ye, Z.; Lan, C.; Xie, Z.; Shu, W. Chemically assisted phytoextraction of heavy metal contaminated soils using three plant species. Plant Soil 2005, 276, 153–162. [Google Scholar] [CrossRef]
- Mesjasz-Przybyłowicz, J.; Nakonieczny, M.; Migula, P.; Augustyniak, M.; Tarnawska, M.; Reimold, W.; Koeberl, C.; Przybyłowicz, W.; Głowacka, E. Uptake of cadmium, lead nickel and zinc from soil and water solutions by the nickel hyperaccumulator Berkheya coddii. Acta Biol. Crac. Ser. Bot. 2004, 46, 75–85. [Google Scholar]
- Rahman, M.A.; Reichman, S.M.; De Filippis, L.; Sany, S.B.T.; Hasegawa, H. Phytoremediation of toxic metals in soils and wetlands: Concepts and applications. In Environmental Remediation Technologies for Metal-Contaminated Soils; Hasegawa, H., Rahman, M.M., Rahman, I., Eds.; Springer: Tokyo, Japan, 2016; pp. 161–195. [Google Scholar]
- Gaiero, J.R.; McCall, C.A.; Thompson, K.A.; Day, N.J.; Best, A.S.; Dunfield, K.E. Inside the root microbiome: Bacterial root endophytes and plant growth promotion. Am. J. Bot. 2013, 100, 1738–1750. [Google Scholar] [CrossRef] [PubMed]
- Lone, M.I.; He, Z.-L.; Stoffella, P.J.; Yang, X.-E. Phytoremediation of heavy metal polluted soils and water: Progresses and perspectives. J. Zhejiang Univ. Sci. B 2008, 9, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Arora, K.; Sharma, S.; Monti, A. Bio-remediation of Pb and Cd polluted soils by switchgrass: A case study in india. Int. J. Phytoremed. 2016, 18, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, D.K.; Varma, A. Microbial-Mediated Induced Systemic Resistance in Plants; Springer: New York, NY, USA, 2016. [Google Scholar]
- Ma, Y.; Prasad, M.; Rajkumar, M.; Freitas, H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol. Adv. 2011, 29, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, M.; Sandhya, S.; Prasad, M.; Freitas, H. Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnol. Adv. 2012, 30, 1562–1574. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Maiti, S.K. Phytoremediation of metal mine waste. Appl. Ecol. Environ. Res. 2010, 8, 207–222. [Google Scholar]
- Favas, P.J.; Pratas, J.; Varun, M.; D’Souza, R.; Paul, M.S. Phytoremediation of soils contaminated with metals and metalloids at mining areas: Potential of native flora. In Environmental Risk Assessment of Soil Contamination; Maria, C., Hernandez, S., Eds.; InTech: Shanghai, China, 2014. [Google Scholar]
- Khanam, A. Phytoremediation: A green bio-engineering technology for cleanup the environmental contaminants. Int. J. Recent Sci. Res. 2016, 7, 9925–9928. [Google Scholar]
- Ogunmayowa, O.T. Coupling Bio/Phytoremediation with Switchgrass to Biofuel Feedstock Production in Mixed-Contaminant Soils. Ph.D. Thesis, Tennessee State University, Nashville, TN, USA, 2015. [Google Scholar]
- Babalola, O.O. Beneficial bacteria of agricultural importance. Biotechnol. Lett. 2010, 32, 1559–1570. [Google Scholar] [CrossRef] [PubMed]
- López-Chuken, U.J. Hydroponics and environmental clean-up. In Hydroponics―A Standard Methodology for Plant Biological Researches; Toshiki, A., Ed.; InTech: Shanghai, China, 2012. [Google Scholar]
- Emamverdian, A.; Ding, Y.; Mokhberdoran, F.; Xie, Y. Heavy metal stress and some mechanisms of plant defense response. Sci. World J. 2015, 2015, 756120. [Google Scholar] [CrossRef] [PubMed]
- Krumova, E.; Kostadinova, N.; Miteva-Staleva, J.; Gryshko, V.; Angelova, M. Cellular response to Cu-and Zn-induced oxidative stress in aspergillus fumigatus isolated from polluted soils in Bulgaria. CLEAN Soil Air Water 2016, 44, 657–666. [Google Scholar] [CrossRef]
- Hossain, M.A.; Piyatida, P.; da Silva, J.A.T.; Fujita, M. Molecular mechanism of heavy metal toxicity and tolerance in plants: Central role of glutathione in detoxification of reactive oxygen species and methylglyoxal and in heavy metal chelation. J. Bot. 2012, 2012, 872875. [Google Scholar] [CrossRef]
- Bücker-Neto, L.; Paiva, A.L.S.; Machado, R.D.; Arenhart, R.A.; Margis-Pinheiro, M. Interactions between plant hormones and heavy metals responses. Genet. Mol. Biol. 2017, 40 (Suppl. 1), 373–386. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.N.C.; de Souza, V.V.; da Silva Souza, T. Cytotoxic, genotoxic and mutagenic effects of sewage sludge on Allium cepa. Chemosphere 2016, 148, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Prasad, S.M.; Singh, S.; Singh, M. Phytoremediation potential of weed plants’ oxidative biomarker and antioxidant responses. Chem. Ecol. 2016, 32, 684–706. [Google Scholar] [CrossRef]
- Bielen, A.; Remans, T.; Vangronsveld, J.; Cuypers, A. The influence of metal stress on the availability and redox state of ascorbate, and possible interference with its cellular functions. Int. J. Mol. Sci. 2013, 14, 6382–6413. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.; Matos, M. Assessment of the impact of aluminum on germination, early growth and free proline content in Lactuca sativa L. Ecotoxicol. Environ. Saf. 2016, 131, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Rastgoo, L.; Alemzadeh, A.; Afsharifar, A. Isolation of two novel isoforms encoding zinc-and copper-transporting P1b-atpase from gouan (Aeluropus littoralis). Plant Omics J. 2011, 4, 377–383. [Google Scholar]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012. [Google Scholar] [CrossRef]
- Solanki, R.; Dhankhar, R. Biochemical changes and adaptive strategies of plants under heavy metal stress. Biologia 2011, 66, 195–204. [Google Scholar] [CrossRef]
- Gupta, D.; Huang, H.; Corpas, F. Lead tolerance in plants: Strategies for phytoremediation. Environ. Sci. Pollut. Res. 2013, 20, 2150–2161. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Yang, J.-L.; Li, C.-H. Advances in metallotionein studies in forest trees. Plant Omics 2012, 5, 46. [Google Scholar]
- Ehsanpour, A.A.; Zarei, S.; Abbaspour, J. The role of over expression of p5cs gene on proline, catalase, ascorbate peroxidase activity and lipid peroxidation of transgenic tobacco (Nicotiana tabacum L.) plant under in vitro drought stress. J. Cell Mol. Res. 2012, 4, 43–49. [Google Scholar]
- Saba, H.; Jyoti, P.; Neha, S. Mycorrhizae and phytochelators as remedy in heavy metal contaminated land remediation. Int. Res. J. Environ. Sci. 2013, 2, 74–78. [Google Scholar]
- Verkleij, J.; Sneller, F.; Schat, H. Metallothioneins and phytochelatins: Ecophysiological aspects. In Sulphur in Plants; Abrol, Y.P., Ahmad, A., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 163–176. [Google Scholar]
- Guo, J.; Xu, L.; Su, Y.; Wang, H.; Gao, S.; Xu, J.; Que, Y. Scmt2–1-3, a metallothionein gene of sugarcane, plays an important role in the regulation of heavy metal tolerance/accumulation. BioMed Res. Int. 2013, 2013, 904769. [Google Scholar] [CrossRef] [PubMed]
- Macovei, A.; Ventura, L.; Donà, M.; Faè, M.; Balestrazzi, A.; Carbonera, D. Effects of heavy metal treatments on metallothionein expression profiles in white poplar (Populus alba L.) cell suspension cultures. An. Univ. Oradea Fasc. Biol. 2010, 1, 194–198. [Google Scholar]
- Mishra, S.; Dubey, R. Heavy metal uptake and detoxification mechanisms in plants. Int. J. Agric. Res. 2006, 1, 122–141. [Google Scholar]
- Grennan, A.K. Metallothioneins, a diverse protein family. Plant Physiol. 2011, 155, 1750–1751. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Lade, H. Plant-growth-promoting rhizobacteria to improve crop growth in saline soils: A review. Agron. Sustain. Dev. 2014, 34, 737–752. [Google Scholar] [CrossRef]
- Nehra, V.; Choudhary, M. A review on plant growth promoting rhizobacteria acting as bioinoculants and their biological approach towards the production of sustainable agriculture. J. Appl. Nat. Sci. 2015, 7, 540–556. [Google Scholar]
- Rajkumar, M.; Ae, N.; Prasad, M.N.V.; Freitas, H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010, 28, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Ahemad, M.; Malik, A. Bioaccumulation of heavy metals by zinc resistant bacteria isolated from agricultural soils irrigated with wastewater. Bacteriol. J. 2011, 2, 12–21. [Google Scholar] [CrossRef]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ.-Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Ahemad, M. Implications of bacterial resistance against heavy metals in bioremediation: A review. J. Inst. Integr. Omics Appl. Biotechnol. 2012, 3, 3. [Google Scholar]
- Bhattacharyya, P.; Jha, D. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Viveros, O.; Jorquera, M.; Crowley, D.; Gajardo, G.; Mora, M. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant Nutr. 2010, 10, 293–319. [Google Scholar] [CrossRef]
- Ramadan, E.M.; AbdelHafez, A.A.; Hassan, E.A.; Saber, F.M. Plant growth promoting rhizobacteria and their potential for biocontrol of phytopathogens. Afr. J. Microbiol. Res. 2016, 10, 486–504. [Google Scholar]
- Ahmed, E.; Holmström, S.J. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nishizawa, N.K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of plant growth promoting rhizobacteria in agricultural sustainability—A review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.S.; Pandey, V.C.; Singh, D. Efficient soil microorganisms: A new dimension for sustainable agriculture and environmental development. Agric. Ecosyst. Environ. 2011, 140, 339–353. [Google Scholar] [CrossRef]
- Dell’mour, M.; Schenkeveld, W.; Oburger, E.; Fischer, L.; Kraemer, S.; Puschenreiter, M.; Lämmerhofer, M.; Koellensperger, G.; Hann, S. Analysis of iron-phytosiderophore complexes in soil related samples: LC-ESI-MS/MS versus CE-MS. Electrophoresis 2012, 33, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P.; Singh, E. Applications and mechanisms of plant growth-stimulating rhizobacteria. In Plant-Microbe Interaction: An Approach to Sustainable Agriculture; Choudhary, D., Varma, A., Tuteja, N., Eds.; Springer: Singapore, 2016; pp. 37–62. [Google Scholar]
- Milošević, N.A.; Marinković, J.B.; Tintor, B.B. Mitigating abiotic stress in crop plants by microorganisms. Zbornik Matice Srpske za Prirodne Nauke 2012, 123, 17–26. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Zahir, Z.A.; Naveed, M.; Nawaz, S. Mitigation of salinity-induced negative impact on the growth and yield of wheat by plant growth-promoting rhizobacteria in naturally saline conditions. Ann. Microbiol. 2013, 63, 225–232. [Google Scholar] [CrossRef]
- Ratna Kumar, P.; Raina, S.K.; Kumar, S.; Bhagat, K.P.; Singh, Y.; Bal, S.K. Adaptation and mitigation strategies of plant under drought and high-temperature stress. Clim. Chang. Plant Abiot. Stress Toler. 2013, 421–436. [Google Scholar] [CrossRef]
- Zafar-ul-Hye, M.; Farooq, H.M.; Zahir, Z.A.; Hussain, M.; Hussain, A. Application of acc-deaminase containing rhizobacteria with fertilizer improves maize production under drought and salinity stress. Int. J. Agric. Biol. 2014, 16, 591–596. [Google Scholar]
- Glick, B.R. Bacteria with acc deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Vanderleyden, J. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef] [PubMed]
- Wasilkowski, D.; Swędzioł, Ż.; Mrozik, A. Przydatność genetycznie modyfikowanych mikroorganizmów do bioremediacji zanieczyszczonych środowisk. Chemik 2012, 66, 817–826. (In Polish) [Google Scholar]
- Wolejko, E.; Wydro, U.; Loboda, T. The ways to increase efficiency of soil bioremediation. Ecol. Chem. Eng. 2016, 23, 155. [Google Scholar] [CrossRef]
- Verma, N.; Singh, M. Biosensors for heavy metals. Biometals 2005, 18, 121–129. [Google Scholar] [CrossRef] [PubMed]
- D’souza, S. Microbial biosensors. Biosens. Bioelectron. 2001, 16, 337–353. [Google Scholar] [CrossRef]
- Azad, M.A.K.; Amin, L.; Sidik, N.M. Genetically engineered organisms for bioremediation of pollutants in contaminated sites. Chin. Sci. Bull. 2014, 59, 703–714. [Google Scholar] [CrossRef]
- Kang, J.W. Removing environmental organic pollutants with bioremediation and phytoremediation. Biotechnol. Lett. 2014, 36, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wilson, D.B. Genetic engineering of bacteria and their potential for Hg2+ bioremediation. Biodegradation 1997, 8, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Barkay, T.; Miller, S.M.; Summers, A.O. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 2003, 27, 355–384. [Google Scholar] [CrossRef]
- Deckwer, W.-D.; Becker, F.; Ledakowicz, S.; Wagner-Döbler, I. Microbial removal of ionic mercury in a three-phase fluidized bed reactor. Environ. Sci. Technol. 2004, 38, 1858–1865. [Google Scholar] [CrossRef] [PubMed]
- Marconi, A.M.; Kieboom, J.; de Bont, J.A. Improving the catabolic functions in the toluene-resistant strain pseudomonas putidas12. Biotechnol. Lett. 1997, 19, 603–606. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, F.; Chen, J.; Sun, G. Arsenic removal from contaminated soil via biovolatilization by genetically engineered bacteria under laboratory conditions. J. Environ. Sci. 2011, 23, 1544–1550. [Google Scholar] [CrossRef]
- Rojas, L.A.; Yáñez, C.; González, M.; Lobos, S.; Smalla, K.; Seeger, M. Characterization of the metabolically modified heavy metal-resistant cupriavidus metallidurans strain MSR33 generated for mercury bioremediation. PLoS ONE 2011, 6, e17555. [Google Scholar] [CrossRef] [PubMed]
- Sone, Y.; Mochizuki, Y.; Koizawa, K.; Nakamura, R.; Pan-Hou, H.; Itoh, T.; Kiyono, M. Mercurial-resistance determinants in pseudomonas strain k-62 plasmid pmr68. AMB Express 2013, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Samanta, S.K.; Singh, O.V.; Jain, R.K. Polycyclic aromatic hydrocarbons: Environmental pollution and bioremediation. Trends Biotechnol. 2002, 20, 243–248. [Google Scholar] [CrossRef]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHS): A review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Dagar, V.K.; Khasa, Y.P.; Kuhad, R.C. Genetically modified microorganisms (GMOS) for bioremediation. In Biotechnology for Environmental Management and Resource Recovery; Kuhad, R., Singh, A., Eds.; Springer: New Delhi, India, 2013; pp. 191–218. [Google Scholar]
- Zaidi, A.; Wani, P.A.; Khan, M.S. Bioremediation: A natural method for the management of polluted environment. In Toxicity of Heavy Metals to Legumes and Bioremediation; Zaidi, A., Wani, P., Khan, M., Eds.; Springer: Vienna, Austria, 2012; pp. 101–114. [Google Scholar]
- Buermans, H.; Den Dunnen, J. Next generation sequencing technology: Advances and applications. Biochim. Biophys. Acta 2014, 1842, 1932–1941. [Google Scholar] [CrossRef] [PubMed]
- Divya, B.; Kumar, M.D. Plant-microbe interaction with enhanced bioremediation. Res. J. Biotechnol. 2011, 6, 72–79. [Google Scholar]
- Varsha, M.; Nidhi, M.; Anurag, M. Heavy metals in plants: Phytoremediation: Plants used to remediate heavy metal pollution. Agric. Biol. J. N. Am. 2010, 1, 40–46. [Google Scholar]
- Arshad, M.; Saleem, M.; Hussain, S. Perspectives of bacterial acc deaminase in phytoremediation. Trends Biotechnol. 2007, 25, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, J.D.; He, Z.; Zhou, J. Use of functional gene arrays for elucidating in situ biodegradation. Front. Microbiol. 2012, 3, 339. [Google Scholar] [CrossRef] [PubMed]
- Davison, J. Towards safer vectors for the field release of recombinant bacteria. Environ. Biosaf. Res. 2002, 1, 9–18. [Google Scholar] [CrossRef]



| Class of Microorganisms | Heavy Metal Removed | References |
|---|---|---|
| 1. Bacteria | ||
| Bacillus cereus strain XMCr-6 | Cr (VI) | [72] |
| Kocuria flava | Cu | [61] |
| Bacillus cereus | Cr (VI) | [61,73] |
| Sporosarcina ginsengisoli | As (III) | [61,74] |
| Pseudomonas veronii | Cd, Zn, Cu | [61,75] |
| Pseudomonas putida | Cr (VI) | [76] |
| Enterobacter cloacae B2-DHA | Cr (VI) | [77] |
| Bacillus subtilis | Cr (VI) | [76] |
| 2. Fungi | ||
| Aspergillus versicolor | Ni, Cu | [61,78] |
| Aspergillus fumigatus | Pb | [79] |
| Gloeophyllum sepiarium | Cr (VI) | [80] |
| Rhizopus oryzae (MPRO) | Cr (VI) | [81] |
| 3. Yeast | ||
| Sacharomyces cerevisiae | Pb, Cd | [82,83] |
| 4. Algae | ||
| Spirogyra spp. and Cladophora spp. | Pb (II), Cu (II) | [61,84] |
| Spirogyra spp. and Spirullina spp. | Cr Cu, Fe, Mn, Zn | [61,85] |
| Hydrodictylon, Oedogonium and Rhizoclonium spp. | As | [60,61] |
| Family | Species | Heavy Metals | References |
|---|---|---|---|
| Asteraceae | Berkheya coddii | Ni | [108] |
| Asteraceae | Helianthus annuus | Pb, Cd, Zn | [101,109] |
| Brassicaceae | Alyssum bertolonii | Ni | [110] |
| Brassicaceae | Alyssum murale | Ni | [111] |
| Brassicaceae | Arabidopsis halleri | Zn, Cd | [112] |
| Brassicaceae | Arabidopsis halleri | Cd Cd | [113] |
| Caryophyllaceae | Minuartia verna | Zn, Cd, Pb | [114] |
| Crassulaceae | Sedum alfredii | Pb | [7,115] |
| Euphorbiaceae | Euphorbia cheiradenia | Cu, Fe, Pb, Zn | [1] |
| Fabaceae | Astragalus racemosus | Se | [116] |
| Fabaceae | Medicago sativa | Pb | [7] |
| Poaceae | Spartina argentinensis | Cr | [117] |
| Pteridaceae | Pteris vittata | As | [118,119,120] |
| Pteridaceae | Pteris vittata | Hg | [121] |
| Violaceae | Viola boashanensis | Pb, Zn, Cd | [122] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ojuederie, O.B.; Babalola, O.O. Microbial and Plant-Assisted Bioremediation of Heavy Metal Polluted Environments: A Review. Int. J. Environ. Res. Public Health 2017, 14, 1504. https://doi.org/10.3390/ijerph14121504
Ojuederie OB, Babalola OO. Microbial and Plant-Assisted Bioremediation of Heavy Metal Polluted Environments: A Review. International Journal of Environmental Research and Public Health. 2017; 14(12):1504. https://doi.org/10.3390/ijerph14121504
Chicago/Turabian StyleOjuederie, Omena Bernard, and Olubukola Oluranti Babalola. 2017. "Microbial and Plant-Assisted Bioremediation of Heavy Metal Polluted Environments: A Review" International Journal of Environmental Research and Public Health 14, no. 12: 1504. https://doi.org/10.3390/ijerph14121504