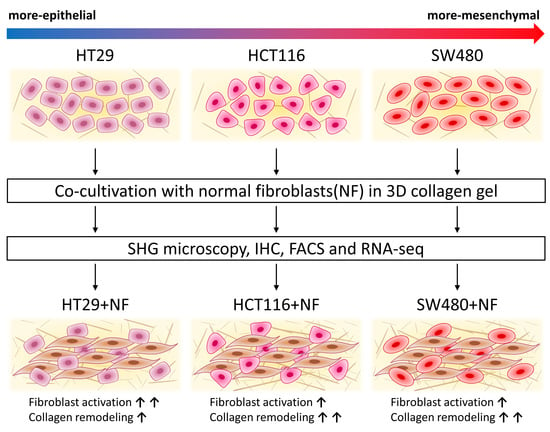
Expression of EMT-Related Genes in Hybrid E/M Colorectal Cancer Cells Determines Fibroblast Activation and Collagen Remodeling
Abstract
:1. Introduction
2. Results
2.1. Epithelial/Mesenchymal States of Cancer Cells
2.2. The Molecular Phenotype of NFs and CAFs and Activation of Normal Fibroblasts in Co-Culture
2.3. Collagen Remodeling
2.3.1. Collagen structure and Collagen-Related Gene Expression in Monocultures of Cancer Cells
2.3.2. Collagen Remodeling and Collagen-Related Gene Expression in Co-Cultures of Cancer Cells and Normal Fibroblasts
2.3.3. Collagen Organization and Collagen-Related Gene Expression in CAFs and Normal Fibroblasts
2.3.4. Collagen Structure in Tumor Xenografts
3. Discussion
3.1. Communication of Colorectal Cancer Cells and Normal Fibroblasts Induces Activation of the Normal Fibroblasts
3.2. Colorectal Cancer Cells, Normal Fibroblasts, and Their Co-Cultures Remodel Collagen in Different Modes
4. Materials and Methods
4.1. Cell Cultures
4.2. Co-Culturing Cancer Cells and Fibroblasts in a Collagen-Based 3D Model
4.3. Isolation of CAFs and Normal Fibroblasts
4.4. Immunofluorescence
4.5. Wound-Healing Assay
4.6. Spheroid Formation
4.7. Tumor Xenografts
4.8. Fluorescence-Activated Cell Sorting
4.9. RNA Isolation and Sequencing
4.10. Second Harmonic Generation (SHG) Microscopy
4.11. Quantitative Analysis of Collagen
4.12. Statistical Analysis
5. Conclusions
- Cancer cells impact on collagen structure and its change due to co-cultivation with fibroblasts
- Cancer cells, when cultured alone, differ in their collagen remodeling capacity. An increase in the density of collagen was observed only for the SW480 (invasive) cell line, but not for HT29. SW480 demonstrated the ability to create quite thick, oriented fibers.
- Co-culturing of colorectal cancer cell lines with normal fibroblasts resulted in a greater amount of fibrillar collagen during all periods of cultivation.
- Fibroblast-mediated remodeling of collagen led to the formation of highly ordered fiber networks, which are more favorable for cancer cell invasion. However, after prolonged cultivation in the absence of cancer cells, the CAFs lost the ability to organize collagen in ways that promote invasion, and they actually initiated matrix degradation.
- Expression of “collagen” genes in cancer cells and its change during co-cultivation with fibroblasts
- The genes related to collagen biosynthesis and catabolism tend to be more active in the more-mesenchymal and mobile SW480 cells than in the more-epithelial and less mobile HT29 cells. Moreover, the LOXL2 and LOXL3 genes, which are necessary for collagen fibril organization, are SW480 specific.
- The co-cultivation of colon cancer cell lines HT29 or SW480 with NFs leads to the activation of collagen biosynthesis and collagen fibril organization genes in both the fibroblasts and the cancer cells.
- The genes related to collagen biosynthesis and collagen fibril organization tend to be more active in skin fibroblasts (NFs) than in colon CAFs, whereas CAF marker genes are significantly up-regulated in colorectal CAFs compared to normal skin fibroblasts.
- The co-cultivation of the colorectal cancer cell lines HT29 or SW480 with skin fibroblasts leads to the activation of several CAF marker genes expression in the fibroblasts (ACTA2, POSTN, TNC, and DES). During co-cultivation, the HT29 line activates normal fibroblasts to a greater extent than does SW480.
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| EMT | epithelial–mesenchymal transition |
| ECM | extracellular matrix |
| CAF | cancer-associated fibroblast |
| NF | normal fibroblast |
| SHG | second harmonic generation |
| IHC | immunohistochemistry |
| FACS | fluorescence-activated cell sorting |
| RNA-seq | RNA sequencing |
References
- Hanahan, D.; Coussens, L.M. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sverdlov, E. Missed Druggable Cancer Hallmark: Cancer–Stroma Symbiotic Crosstalk as Paradigm and Hypothesis for Cancer Therapy. Bioessays 2018, 40, 1800079. [Google Scholar] [CrossRef]
- Valkenburg, K.C.; De Groot, A.E.; Pienta, K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018, 15, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Chopra, V.; Sangarappillai, R.M.; Romero-Canelón, I.; Jones, A.M. Lysyl Oxidase Like-2 (LOXL2): An Emerging Oncology Target. Adv. Ther. 2020, 3, 1900119. [Google Scholar] [CrossRef]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [Green Version]
- Nurmik, M.; Ullmann, P.; Rodriguez, F.; Haan, S.; Letellier, E. In search of definitions: Cancer-associated fibroblasts and their markers. Int. J. Cancer 2020, 146, 895–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nissen, N.I.; Karsdal, M.; Willumsen, N. Collagens and Cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J. Exp. Clin. Cancer Res. 2019, 38, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporale, A.; Vestri, A.R.; Benvenuto, E.; Mariotti, M.; Cosenza, U.M.; Scarpini, M.; Giuliani, A.; Mingazzini, P.; Angelico, F. Is desmoplasia a protective factor for survival in patients with colorectal carcinoma? Clin. Gastroenterol. Hepatol. 2005, 3, 370–375. [Google Scholar] [CrossRef]
- Sirica, A.E.; Gores, G.J. Desmoplastic stroma and cholangiocarcinoma: Clinical implications and therapeutic targeting. Hepatology 2014, 59, 2397–2402. [Google Scholar] [CrossRef] [Green Version]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef] [Green Version]
- Walker, C.; Mojares, E.; del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwager, S.C.; Taufalele, P.V.; Reinhart-King, C.A. Cell–Cell Mechanical Communication in Cancer. Cell. Mol. Bioeng. 2019, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Keely, P.J. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK–ERK linkage. Oncogene 2009, 28, 4326–4343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acerbi, I.; Cassereau, L.; Dean, I.; Shi, Q.; Au, A.; Park, C.; Chen, Y.Y.; Liphardt, J.; Hwang, E.S.; Weaver, V.M. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. (UK) 2015, 7, 1120–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elia, I.; Rossi, M.; Stegen, S.; Broekaert, D.; Doglioni, G.; van Gorsel, M.; Boon, R.; Escalona-Noguero, C.; Torrekens, S.; Verfaillie, C.; et al. Breast cancer cells rely on environmental pyruvate to shape the metastatic niche. Nature 2019, 568, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Anguiano, M.; Morales, X.; Castilla, C.; Pena, A.R.; Ederra, C.; Martínez, M.; Ariz, M.; Esparza, M.; Amaveda, H.; Mora, M.; et al. The use of mixed collagen-Matrigel matrices of increasing complexity recapitulates the biphasic role of cell adhesion in cancer cell migration: ECM sensing, remodeling and forces at the leading edge of cancer invasion. PLoS ONE 2020, 15, e0220019. [Google Scholar] [CrossRef]
- Choi, S.Y.; Sung, R.; Lee, S.J.; Lee, T.G.; Kim, N.; Yoon, S.M.; Lee, E.J.; Chae, H.B.; Youn, S.J.; Park, S.M. Podoplanin, α-smooth muscle actin or S100A4 expressing cancer-associated fibroblasts are associated with different prognosis in colorectal cancers. J. Korean Med. Sci. 2013, 28, 1293–1301. [Google Scholar] [CrossRef] [Green Version]
- Wei, B.; Zhou, X.; Liang, C.; Zheng, X.; Lei, P.; Fang, J.; Han, X.; Wang, L.; Qi, C.; Wei, H. Human colorectal cancer progression correlates with LOX-induced ECM stiffening. Int. J. Biol. Sci. 2017, 13, 1450–1457. [Google Scholar] [CrossRef]
- Stadler, M.; Scherzer, M.; Walter, S.; Holzner, S.; Pudelko, K.; Riedl, A.; Unger, C.; Kramer, N.; Weil, B.; Neesen, J.; et al. Exclusion from spheroid formation identifies loss of essential cell-cell adhesion molecules in colon cancer cells. Sci. Rep. 2018, 8, 1151. [Google Scholar] [CrossRef] [Green Version]
- Janiszewska, M.; Primi, M.C.; Izard, T. Cell adhesion in cancer: Beyond the migration of single cells. J. Biol. Chem. 2020, 295, 2495–2505. [Google Scholar] [CrossRef] [Green Version]
- Smyrek, I.; Mathew, B.; Fischer, S.C.; Lissek, S.M.; Becker, S.; Stelzer, E.H.K. E-cadherin, actin, microtubules and FAK dominate different spheroid formation phases and important elements of tissue integrity. Biol. Open 2019, 8, bio037051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Marck, V.L.; Bracke, M.E. Epithelial-Mesenchymal Transitions in Human Cancer. In Madame Curie Bioscience Database; Landes Bioscience: Austin, TX, USA, 2013; Available online: https://www.ncbi.nlm.nih.gov/books/NBK6362/.
- Zhang, B.; Zhang, Z.; Xia, S.; Xing, C.; Ci, X.; Li, X.; Zhao, R.; Tian, S.; Ma, G.; Zhu, Z.; et al. KLF5 Activates MicroRNA 200 Transcription To Maintain Epithelial Characteristics and Prevent Induced Epithelial-Mesenchymal Transition in Epithelial Cells. Mol. Cell. Biol. 2013, 33, 4919–4935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyun, K.A.; Goo, K.B.; Han, H.; Sohn, J.; Choi, W.; Kim, S.I.; Jung, H.I.; Kim, Y.S. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 2016, 7, 24677–24687. [Google Scholar] [CrossRef] [Green Version]
- Jędroszka, D.; Orzechowska, M.; Hamouz, R.; Górniak, K.; Bednarek, A.K. Markers of epithelial-to-mesenchymal transition reflect tumor biology according to patient age and Gleason score in prostate cancer. PLoS ONE 2017, 12, e0188842. [Google Scholar] [CrossRef] [Green Version]
- Orr, B.; Riddick, A.C.P.; Stewart, G.D.; Anderson, R.A.; Franco, O.E.; Hayward, S.W.; Thomson, A.A. Identification of stromally expressed molecules in the prostate by tag-profiling of cancer-associated fibroblasts, normal fibroblasts and fetal prostate. Oncogene 2012, 31, 1130–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiga, K.; Hara, M.; Nagasaki, T.; Sato, T.; Takahashi, H.; Takeyama, H. Cancer-associated fibroblasts: Their characteristics and their roles in tumor growth. Cancers 2015, 7, 2443–2458. [Google Scholar] [CrossRef] [PubMed]
- Christodoulou, I.; Goulielmaki, M.; Devetzi, M.; Panagiotidis, M.; Koliakos, G.; Zoumpourlis, V. Mesenchymal stem cells in preclinical cancer cytotherapy: A systematic review. Stem Cell Res. Ther. 2018, 9, 1–38. [Google Scholar] [CrossRef]
- Fukagawa, D.; Sugai, T.; Osakabe, M.; Suga, Y.; Nagasawa, T.; Itamochi, H.; Sugiyama, T. Protein expression patterns in cancer-associated fibroblasts and cells undergoing the epithelial-mesenchymal transition in ovarian cancers. Oncotarget 2018, 9, 27514–27524. [Google Scholar] [CrossRef]
- Su, S.; Chen, J.; Yao, H.; Liu, J.; Yu, S.; Lao, L.; Wang, M.; Luo, M.; Xing, Y.; Chen, F.; et al. CD10+GPR77+ Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 2018, 172, 841.e16–856.e16. [Google Scholar] [CrossRef]
- Yeo, S.Y.; Lee, K.W.; Shin, D.; An, S.; Cho, K.H.; Kim, S.H. A positive feedback loop bi-stably activates fibroblasts. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Wong, P.F.; Wei, W.; Gupta, S.; Smithy, J.W.; Zelterman, D.; Kluger, H.M.; Rimm, D.L. Multiplex quantitative analysis of cancer-associated fibroblasts and immunotherapy outcome in metastatic melanoma. J. Immunother. Cancer 2019, 7, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtis, M.; Kenny, H.A.; Ashcroft, B.; Mukherjee, A.; Johnson, A.; Zhang, Y.; Helou, Y.; Batlle, R.; Liu, X.; Gutierrez, N.; et al. Fibroblasts Mobilize Tumor Cell Glycogen to Promote Proliferation and Metastasis. Cell Metab. 2019, 29, 141.e9–155.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavón, M.A.; Arroyo-Solera, I.; Céspedes, M.V.; Casanova, I.; León, X.; Mangues, R. uPA/uPAR and SERPINE1 in head and neck cancer: Role in tumor resistance, metastasis, prognosis and therapy. Oncotarget 2016, 7, 57351–57366. [Google Scholar] [CrossRef] [Green Version]
- Alekseenko, I.V.; Chernov, I.P.; Kostrov, S.V.; Sverdlov, E.D. Are synapse-like structures a possible way for crosstalk of cancer with its microenvironment? Cancers 2020, 12, 806. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef] [PubMed]
- LeBleu, V.S.; Kalluri, R. A peek into cancer-associated fibroblasts: Origins, functions and translational impact. DMM Dis. Model. Mech. 2018, 11, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Cortez, E.; Roswall, P.; Pietras, K. Functional subsets of mesenchymal cell types in the tumor microenvironment. Semin. Cancer Biol. 2014, 25, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Arina, A.; Idel, C.; Hyjek, E.M.; Alegre, M.L.; Wang, Y.; Bindokas, V.P.; Weichselbaum, R.R.; Schreiber, H. Tumor-associated fibroblasts predominantly come from local and not circulating precursors. Proc. Natl. Acad. Sci. USA 2016, 113, 7551–7556. [Google Scholar] [CrossRef] [Green Version]
- Ishii, G.; Ochiai, A.; Neri, S. Phenotypic and functional heterogeneity of cancer-associated fibroblast within the tumor microenvironment. Adv. Drug Deliv. Rev. 2016, 99, 186–196. [Google Scholar] [CrossRef]
- Darby, I.A.; Laverdet, B.; Bonté, F.; Desmoulière, A. Fibroblasts and myofibroblasts in wound healing. Clin. Cosmet. Investig. Dermatol. 2014, 7, 301–311. [Google Scholar]
- Wessels, D.J.; Pradhan, N.; Park, Y.N.; Klepitsch, M.A.; Lusche, D.F.; Daniels, K.J.; Conway, K.D.; Voss, E.R.; Hegde, S.V.; Conway, T.P.; et al. Reciprocal signaling and direct physical interactions between fibroblasts and breast cancer cells in a 3D environment. PLoS ONE 2019, 14, e0218854. [Google Scholar] [CrossRef] [PubMed]
- Koh, B.; Jeon, H.; Kim, D.; Kang, D.; Kim, K.R. Effect of fibroblast co-culture on the proliferation, viability and drug response of colon cancer cells. Oncol. Lett. 2019, 17, 2409–2417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, S.Y.; Lee, J.H.; Shin, Y.; Chung, S.; Kuh, H.J. Co-culture of tumor spheroids and fibroblasts in a collagen matrix-incorporated microfluidic chip mimics reciprocal activation in solid tumor microenvironment. PLoS ONE 2016, 11, e0159013. [Google Scholar] [CrossRef] [Green Version]
- Foster, D.S.; Jones, R.E.; Ransom, R.C.; Longaker, M.T.; Norton, J.A. The evolving relationship of wound healing and tumor stroma. JCI Insight 2018, 3, e99911. [Google Scholar] [CrossRef] [Green Version]
- Schnittert, J.; Bansal, R.; Storm, G.; Prakash, J. Integrins in wound healing, fibrosis and tumor stroma: High potential targets for therapeutics and drug delivery. Adv. Drug Deliv. Rev. 2018, 129, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Henriksson, M.L.; Edin, S.; Dahlin, A.M.; Oldenborg, P.A.; Öberg, Å.; Van Guelpen, B.; Rutegård, J.; Stenling, R.; Palmqvist, R. Colorectal cancer cells activate adjacent fibroblasts resulting in FGF1/FGFR3 signaling and increased invasion. Am. J. Pathol. 2011, 178, 1387–1394. [Google Scholar] [CrossRef]
- Peng, C.; Zou, X.; Xia, W.; Gao, H.; Li, Z.; Liu, N.; Xu, Z.; Gao, C.; He, Z.; Niu, W.; et al. Integrin αvβ6 plays a bi-directional regulation role between colon cancer cells and cancer-associated fibroblasts. Biosci. Rep. 2018, 38, BSR20180243. [Google Scholar] [CrossRef] [Green Version]
- Eberlein, C.; Rooney, C.; Ross, S.J.; Farren, M.; Weir, H.M.; Barry, S.T. E-Cadherin and EpCAM expression by NSCLC tumour cells associate with normal fibroblast activation through a pathway initiated by integrin αvβ6 and maintained through TGFβ signalling. Oncogene 2015, 34, 704–716. [Google Scholar] [CrossRef]
- Gao, M.-Q.; Kim, B.G.; Kang, S.; Choi, Y.P.; Park, H.; Kang, K.S.; Cho, N.H. Stromal fibroblasts from the interface zone of human breast carcinomas induce an epithelial-mesenchymal transition-like state in breast cancer cells in vitro. J. Cell Sci. 2010, 123, 3507–3514. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Zhou, L.; Li, D.; Andl, T.; Zhang, Y. Cancer-Associated Fibroblasts Build and Secure the Tumor Microenvironment. Front. Cell Dev. Biol. 2019, 7, 60. [Google Scholar] [CrossRef] [Green Version]
- Kubouchi, Y.; Yurugi, Y.; Wakahara, M.; Sakabe, T.; Haruki, T.; Nosaka, K.; Miwa, K.; Araki, K.; Taniguchi, Y.; Shiomi, T.; et al. Podoplanin expression in cancer-associated fibroblasts predicts unfavourable prognosis in patients with pathological stage IA lung adenocarcinoma. Histopathology 2018, 72, 490–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yurugi, Y.; Wakahara, M.; Matsuoka, Y.; Sakabe, T.; Kubouchi, Y.; Haruki, T.; Nosaka, K.; Miwa, K.; Araki, K.; Taniguchi, Y.; et al. Podoplanin expression in cancer-associated fibroblasts predicts poor prognosis in patients with squamous cell carcinoma of the lung. Anticancer Res. 2017, 37, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Shindo, K.; Aishima, S.; Ohuchida, K.; Fujiwara, K.; Fujino, M.; Mizuuchi, Y.; Hattori, M.; Mizumoto, K.; Tanaka, M.; Oda, Y. Podoplanin expression in cancer-associated fibroblasts enhances tumor progression of invasive ductal carcinoma of the pancreas. Mol. Cancer 2013, 12, 168. [Google Scholar] [CrossRef] [Green Version]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mierke, C.T.; Sauer, F.; Grosser, S.; Puder, S.; Fischer, T.; Käs, J.A. The two faces of enhanced stroma: Stroma acts as a tumor promoter and a steric obstacle. NMR Biomed. 2018, 31, e3831. [Google Scholar] [CrossRef]
- Malik, R.; Lelkes, P.I.; Cukierman, E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 2015, 33, 230–236. [Google Scholar] [CrossRef] [Green Version]
- Santi, A.; Kugeratski, F.G.; Zanivan, S. Cancer Associated Fibroblasts: The Architects of Stroma Remodeling. Proteomics 2018, 18, 1700167. [Google Scholar] [CrossRef]
- Case, A.; Brisson, B.K.; Durham, A.C.; Rosen, S.; Monslow, J.; Buza, E.; Salah, P.; Gillem, J.; Ruthel, G.; Veluvolu, S.; et al. Identification of prognostic collagen signatures and potential therapeutic stromal targets in canine mammary gland carcinoma. PLoS ONE 2017, 12, e0180448. [Google Scholar] [CrossRef] [Green Version]
- Emon, B.; Bauer, J.; Jain, Y.; Jung, B.; Saif, T. Biophysics of Tumor Microenvironment and Cancer Metastasis—A Mini Review. Comput. Struct. Biotechnol. J. 2018, 16, 279–287. [Google Scholar] [CrossRef]
- Sander, L.M. Modeling Contact Guidance and Invasion by Cancer Cells. Cancer Res. 2014, 74, 4588–4596. [Google Scholar] [CrossRef] [Green Version]
- Chiu, C.-L.; Digman, M.A.; Gratton, E. Cell Matrix Remodeling Ability Shown by Image Spatial Correlation. J. Biophys. 2013, 2013, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Libring, S.; Shinde, A.; Chanda, M.K.; Nuru, M.; George, H.; Saleh, A.M.; Abdullah, A.; Kinzer-Ursem, T.L.; Calve, S.; Wendt, M.K.; et al. The dynamic relationship of breast cancer cells and fibroblasts in fibronectin accumulation at primary and metastatic tumor sites. Cancers 2020, 12, 1270. [Google Scholar] [CrossRef]
- Jun, B.H.; Guo, T.; Libring, S.; Chanda, M.K.; Paez, J.S.; Shinde, A.; Wendt, M.K.; Vlachos, P.P.; Solorio, L. Fibronectin-expressing mesenchymal tumor cells promote breast cancer metastasis. Cancers 2020, 12, 2553. [Google Scholar] [CrossRef] [PubMed]
- Shinde, A.; Paez, J.S.; Libring, S.; Hopkins, K.; Solorio, L.; Wendt, M.K. Transglutaminase-2 facilitates extracellular vesicle-mediated establishment of the metastatic niche. Oncogenesis 2020, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harjanto, D.; Maffei, J.S.; Zaman, M.H. Quantitative analysis of the effect of cancer invasiveness and collagen concentration on 3D matrix remodeling. PLoS ONE 2011, 6, e24891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmelzer, C.E.H.; Heinz, A.; Troilo, H.; Lockhart-Cairns, M.P.; Jowitt, T.A.; Marchand, M.F.; Bidault, L.; Bignon, M.; Hedtke, T.; Barret, A.; et al. Lysyl oxidase-like 2 (LOXL2)-mediated cross-linking of tropoelastin. FASEB J. 2019, 33, 5468–5481. [Google Scholar] [CrossRef] [Green Version]
- Hanley, C.J.; Noble, F.; Ward, M.; Bullock, M.; Drifka, C.; Mellone, M.; Manousopoulou, A.; Johnston, H.E.; Hayden, A.; Thirdborough, S.; et al. A subset of myofibroblastic cancer-associated fibroblasts regulate collagen fiber elongation, which is prognostic in multiple cancers. Oncotarget 2016, 7, 6159–6174. [Google Scholar] [CrossRef] [Green Version]
- Dipersio, L.P. Regional growth differences of human tumour xenografts in nude mice. Lab. Anim. 1981, 15, 179–180. [Google Scholar] [CrossRef] [PubMed]
- De Both, N.J.; Vermey, M.; Dinjens, W.N.; Bosman, F.T. A comparative evaluation of various invasion assays testing colon carcinoma cell lines. Br. J. Cancer 1999, 81, 934–941. [Google Scholar] [CrossRef] [Green Version]
- Rajan, N.; Habermehl, J.; Coté, M.-F.; Doillon, C.J.; Mantovani, D. Preparation of ready-to-use, storable and reconstituted type I collagen from rat tail tendon for tissue engineering applications. Nat. Protoc. 2006, 1, 2753–2758. [Google Scholar] [CrossRef]
- Herrera, M.; Herrera, A.; Larriba, M.; Ferrer-Mayorga, G.; Herreros, A.; Bonilla, F.; Baulida, J.; Peña, C. Colon Cancer-associated Fibroblast Establishment and Culture Growth. Bio-protocol 2016, 6, e1773. [Google Scholar] [CrossRef] [Green Version]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Campagnola, P.J.; Dong, C.-Y. Second harmonic generation microscopy: Principles and applications to disease diagnosis. Laser Photon. Rev. 2011, 5, 13–26. [Google Scholar] [CrossRef]
- Dudenkova, V.V.; Shirmanova, M.V.; Lukina, M.M.; Feldshtein, F.I.; Virkin, A.; Zagainova, E.V. Examination of Collagen Structure and State by the Second Harmonic Generation Microscopy. Biochemistry 2019, 84, 89–107. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Druzhkova, I.; Shirmanova, M.; Ignatova, N.; Dudenkova, V.; Lukina, M.; Zagaynova, E.; Safina, D.; Kostrov, S.; Didych, D.; Kuzmich, A.; et al. Expression of EMT-Related Genes in Hybrid E/M Colorectal Cancer Cells Determines Fibroblast Activation and Collagen Remodeling. Int. J. Mol. Sci. 2020, 21, 8119. https://doi.org/10.3390/ijms21218119
Druzhkova I, Shirmanova M, Ignatova N, Dudenkova V, Lukina M, Zagaynova E, Safina D, Kostrov S, Didych D, Kuzmich A, et al. Expression of EMT-Related Genes in Hybrid E/M Colorectal Cancer Cells Determines Fibroblast Activation and Collagen Remodeling. International Journal of Molecular Sciences. 2020; 21(21):8119. https://doi.org/10.3390/ijms21218119
Chicago/Turabian StyleDruzhkova, Irina, Marina Shirmanova, Nadezhda Ignatova, Varvara Dudenkova, Maria Lukina, Elena Zagaynova, Dina Safina, Sergey Kostrov, Dmitry Didych, Alexey Kuzmich, and et al. 2020. "Expression of EMT-Related Genes in Hybrid E/M Colorectal Cancer Cells Determines Fibroblast Activation and Collagen Remodeling" International Journal of Molecular Sciences 21, no. 21: 8119. https://doi.org/10.3390/ijms21218119