Pharmacological Modulation of Ubiquitin-Proteasome Pathways in Oncogenic Signaling
Abstract
:1. Introduction
Methodology
2. Molecular Biology of the Components in Ubiquitin-Proteasome System
2.1. Ubiquitin
2.2. Proteasome
2.3. Deubiquitinase
2.4. Ubiquitination
2.4.1. E1 (Ubiquitin-Activating Enzyme)
2.4.2. E2 (Ubiquitin-Conjugating Enzyme)
2.4.3. E3 (Ubiquitin-Ligase Enzyme)
- (i)
- Really interesting new gene [RING] finger domain-containing ligases (BIRC7, Brca1, Cb1-b, cIAP1, IDOL, mdm2, SIAH1, RAD18, RNF4, TRAF6): The RING E3 enzymes are differentiated due to their RING or Ubox (CHIP) fold catalytic-domain which encourages the direct linking of ubiquitin from E2 with the substrate [44,45,46]. There are more than 600 RING finger E3s encoded in the mammalian genome. Structurally, it is a zinc coordinating domain that consists of spaced cysteine and histidine residues which promote E2 dependent ubiquitylation [47].
- (ii)
- Homologous to E6-associated protein C terminus [HECT] domain-containing ligases (SMURF1, NEDD4.1, HUWE1, E6AP): It consists of 28 members in human and based on similarities in the N- terminus domains, 15 members out of 28 can be divided into two subfamilies. The most prominent and well-studied category is the NEDD4 subfamily consisting of nine members, which are characterized by the presence of C2 and WW domains. Another subfamily is the HERC E3 ligase enzyme consisting of six members and shares one commonality, i.e., one or more RCC-like domains (regulators of chromatin condensation 1-like domains) [48].
- (iii)
- RING between RING (RBR) domain-containing ligases: Structurally, the RBR module has three Zn2+ binding motifs, a RING1 domain which interacts with E2 then followed by IBR, which is in-between RING1 finger domain and the RING2 domain of which catalytic cysteine is involved. There are 14 RBRs in humans; among these, only three members that are well understood are Parkin, HHAR1, and HOIP [49,50].
3. Physiological and Pathological Role of UPS in Human Body
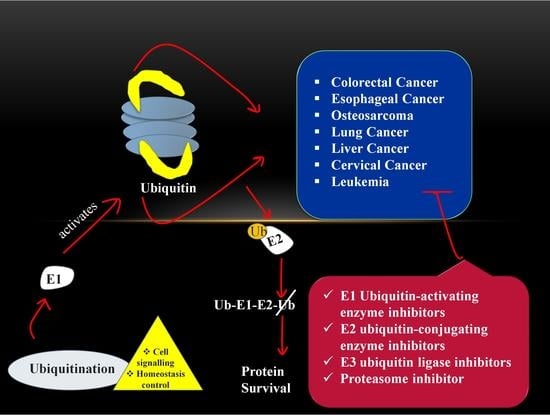
4. Role of Ubiquitin-Proteasome Pathway in Cancer
4.1. Colorectal Cancer
4.2. Esophageal Cancer
4.3. Osteosarcoma
4.4. Lung Cancer
4.5. Liver Cancer
4.6. Cervical Cancer
4.7. Leukemia
5. Preclinical Studies
5.1. In Vivo Studies
5.2. In Vitro Studies
6. Clinical Trial Drugs with Ongoing Stage and Category
7. Cancer Therapeutic Strategy via Targeting UPS
7.1. E1 Ubiquitin-Activating Enzyme Inhibitors
7.2. E2 Ubiquitin-Conjugating Enzyme Inhibitors
7.3. E3 Ubiquitin Ligase Inhibitors
8. Proteasome Inhibitor
9. Conclusions and Future Perspective
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| BMP-2 | Bone morphogenetic protein-2 |
| BTRC | β-transducin repeat containing |
| Cdc20 | Cell division cycle 20 |
| DDR | DNA damage response |
| DSBs | DNA double-strand breaks |
| FBA | F-box associated region |
| FBW7 | F box/WD repeat-containing protein 7 |
| FOXN | A member of the Forkhead box transcription factors |
| MARCH8 | Membrane-associated RING-CH |
| NAE | Neddylation activation enzyme |
| NAE1 | NEDD8 activating enzyme E1 |
| NEDD4L | Neural precursor cell expressed, developmentally down-regulated 4-like |
| OTUB | Ovarian tumor domain-containing ubiquitin aldehyde binding protein |
| Pirh2 | p53-Induced ring-H2 protein |
| PPAR | Peroxisome proliferator-activated receptors |
| TSC | Tuberous sclerosis complex |
| UBCH | Ubiquitin-conjugating enzyme H |
| Ube | Ubiquitin-conjugating enzyme |
| UCH | Ubiquitin carboxyl-terminal hydrolase |
| UCP | Ubiquitin carrier protein |
| UHRF | Ubiquitin-like with plant homeodomain and ring finger domains |
| UPP | Ubiquitin proteasome pathway |
| UPS | Ubiquitin proteasome system |
| USP | Ubiquitin specific peptidase |
| VHL | Von Hippel-Lindau |
| β-Trcp | β-transducin repeat-containing protein |
References
- Martin, G.S. Cell signaling and cancer. Cancer Cell 2003, 4, 167–174. [Google Scholar] [CrossRef] [Green Version]
- Sever, R.; Brugge, J.S. Signaling in cancer. Cold Spring Harb. Perspect. Med. 2014, 5, a006098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huangfu, W.C.; Fuchs, S.Y. Ubiquitination-dependent regulation of signaling receptors in cancer. Genes Cancer 2010, 1, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Nepal, S.; Shrestha, A.; Park, P.H. Ubiquitin specific protease 2 acts as a key modulator for the regulation of cell cycle by adiponectin and leptin in cancer cells. Mol. Cell. Endocrinol. 2015, 412, 44–55. [Google Scholar] [CrossRef]
- Spataro, V.; Norbury, C.; Harris, A.L. The ubiquitin-proteasome pathway in cancer. Br. J. Cancer 1998, 77, 448–455. [Google Scholar] [CrossRef] [Green Version]
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef]
- Dikic, I.; Wakatsuki, S.; Walters, K.J. Ubiquitin-binding domains—from structures to functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 659–671. [Google Scholar] [CrossRef]
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct. Target. Ther. 2020, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef] [Green Version]
- Weinert, B.T.; Schölz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef] [Green Version]
- Ferreira de Freitas, R.; Harding, R.J.; Franzoni, I.; Ravichandran, M.; Mann, M.K.; Ouyang, H.; Lautens, M.; Santhakumar, V.; Arrowsmith, C.H.; Schapira, M. Identification and structure–activity relationship of HDAC6 zinc-finger ubiquitin binding domain inhibitors. J. Med. Chem. 2018, 61, 4517–4527. [Google Scholar] [CrossRef]
- Scott, D.; Oldham, N.J.; Strachan, J.; Searle, M.S.; Layfield, R. Ubiquitin-binding domains: Mechanisms of ubiquitin recognition and use as tools to investigate ubiquitin-modified proteomes. Proteomics 2015, 15, 844–861. [Google Scholar] [CrossRef]
- Randles, L.; Walters, K.J. Ubiquitin and its binding domains. Front. Biosci. 2012, 17, 2140. [Google Scholar] [CrossRef] [Green Version]
- Hurley, J.H.; Lee, S.; Prag, G. Ubiquitin-binding domains. Biochem. J. 2006, 399, 361–372. [Google Scholar] [CrossRef]
- Baumeister, W.; Walz, J.; Zühl, F.; Seemüller, E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell 1998, 92, 367–380. [Google Scholar] [CrossRef] [Green Version]
- DeMartino, G.N.; Gillette, T.G. Proteasomes: Machines for all reasons. Cell 2007, 129, 659–662. [Google Scholar] [CrossRef] [Green Version]
- Saeki, Y.; Tanaka, K. Assembly and function of the proteasome. Ubiquitin Fam. Modif. Proteasome 2012, 832, 315–337. [Google Scholar]
- Tian, G.; Park, S.; Lee, M.J.; Huck, B.; McAllister, F.; Hill, C.P.; Gygi, S.P.; Finley, D. An asymmetric interface between the regulatory and core particles of the proteasome. Nat. Struct. Mol. Biol. 2011, 18, 1259–1267. [Google Scholar] [CrossRef] [Green Version]
- Gillette, T.G.; Kumar, B.; Thompson, D.; Slaughter, C.A.; DeMartino, G.N. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J. Biol. Chem. 2008, 283, 31813–31822. [Google Scholar] [CrossRef] [Green Version]
- Bard, J.A.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and function of the 26S proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B 2009, 85, 12–36. [Google Scholar] [CrossRef] [Green Version]
- Marshall, R.S.; Vierstra, R.D. Dynamic regulation of the 26S proteasome: From synthesis to degradation. Front. Mol. Biosci. 2019, 6, 40. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; Fried, V.A.; Finley, D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 1998, 94, 615–623. [Google Scholar] [CrossRef] [Green Version]
- Scheel, H.; Hofmann, K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinform. 2005, 6, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharyya, S.; Yu, H.; Mim, C.; Matouschek, A. Regulated protein turnover: Snapshots of the proteasome in action. Nat. Rev. Mol. Cell Biol. 2014, 15, 122–133. [Google Scholar] [CrossRef] [Green Version]
- Nijman, S.M.; Luna-Vargas, M.P.; Velds, A.; Brummelkamp, T.R.; Dirac, A.M.; Sixma, T.K.; Bernards, R. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005, 123, 773–786. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef] [Green Version]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbé, S. Deubiquitylases from genes to organism. Physiological. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar]
- Mevissen, T.E.; Komander, D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishna, S.; Suresh, B.; Baek, K.H. The role of deubiquitinating enzymes in apoptosis. Cell. Mol. Life Sci. 2011, 68, 15–26. [Google Scholar] [CrossRef]
- Hermanns, T.; Hofmann, K. Bacterial DUBs: Deubiquitination beyond the seven classes. Biochem. Soc. Trans. 2019, 47, 1857–1866. [Google Scholar] [CrossRef] [Green Version]
- Tsakiri, E.N.; Trougakos, I.P. The amazing ubiquitin-proteasome system: Structural components and implication in aging. Int. Rev. Cell Mol. Biol. 2015, 314, 171–237. [Google Scholar]
- Pelzer, C.; Kassner, I.; Matentzoglu, K.; Singh, R.K.; Wollscheid, H.P.; Scheffner, M.; Schmidtke, G.; Groettrup, M. UBE1L2, a Novel E1 Enzyme Specific for Ubiquitin. J. Biol. Chem. 2007, 282, 23010–23014. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, M.; Smith, H.E. E1 ubiquitin-activating enzyme UBA-1 plays multiple roles throughout C. elegans development. PLoS Genet. 2008, 4, e1000131. [Google Scholar]
- Lee, I.; Schindelin, H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 2008, 134, 268–278. [Google Scholar] [CrossRef] [Green Version]
- Allan, D.C.; Phillips, J.C. Evolution of the ubiquitin-activating enzyme Uba1 (E1). Phys. A Stat. Mech. Appl. 2017, 483, 456–461. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.J.; Chen, F.Z.; Chen, H.C. Ubiquitination involved enzymes and cancer. Med. Oncol. 2014, 31, 93. [Google Scholar] [CrossRef]
- Van Wijk, S.J.; Timmers, H.M. The family of ubiquitin-conjugating enzymes (E2s): Deciding between life and death of proteins. FASEB J. 2010, 24, 981–993. [Google Scholar] [CrossRef]
- Stewart, M.D.; Ritterhoff, T.; Klevit, R.E.; Brzovic, P.S. E2 enzymes: More than just middle men. Cell Res. 2016, 26, 423–440. [Google Scholar] [CrossRef] [Green Version]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Zheng, N.; Shabek, N. Ubiquitin ligases: Structure, function, and regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef]
- Balaji, V.; Hoppe, T. Regulation of E3 ubiquitin ligases by homotypic and heterotypic assembly. F1000Research 2020, 9, 88. [Google Scholar] [CrossRef]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 2012, 125, 531–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sluimer, J.; Distel, B. Regulating the human HECT E3 ligases. Cell. Mol. Life Sci. 2018, 75, 3121–3141. [Google Scholar] [CrossRef] [Green Version]
- Martino, L.; Brown, N.R.; Masino, L.; Esposito, D.; Rittinger, K. Determinants of E2-ubiquitin conjugate recognition by RBR E3 ligases. Sci. Rep. 2018, 8, 68. [Google Scholar] [CrossRef] [Green Version]
- Walden, H.; Rittinger, K. RBR ligase–mediated ubiquitin transfer: A tale with many twists and turns. Nat. Struct. Mol. Biol. 2018, 25, 440–445. [Google Scholar] [CrossRef]
- Khan, H.; Gupta, A.; Singh, T.G.; Kaur, A. Mechanistic insight on the role of leukotriene receptors in ischemic–reperfusion injury. Pharmacol. Rep. 2021, 73, 1240–1254. [Google Scholar] [CrossRef] [PubMed]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef] [PubMed]
- Rehni, A.K.; Singh, T.G.; Behl, N.; Arora, S. Possible involvement of ubiquitin proteasome system and other proteases in acute and delayed aspects of ischemic preconditioning of brain in mice. Biol. Pharm. Bull. 2010, 33, 1953–1957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldeira, M.V.; Salazar, I.L.; Curcio, M.; Canzoniero, L.M.; Duarte, C.B. Role of the ubiquitin–proteasome system in brain ischemia: Friend or foe? Prog. Neurobiol. 2014, 112, 50–69. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.; Huang, T.; Zhang, L.; Zhou, Y.; Luo, H.; Xu, H.; Wang, X. Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Front. Aging Neurosci. 2016, 8, 303. [Google Scholar] [CrossRef]
- Kostin, S.; Pool, L.; Elsässer, A.; Hein, S.; Drexler, H.C.; Arnon, E.; Hayakawa, Y.; Zimmermann, R.; Bauer, E.; Klövekorn, W.P.; et al. Myocytes die by multiple mechanisms in failing human hearts. Circ. Res. 2003, 92, 715–724. [Google Scholar] [CrossRef] [Green Version]
- Eefting, F.; Rensing, B.; Wigman, J.; Pannekoek, W.J.; Liu, W.M.; Cramer, M.J.; Lips, D.J.; Doevendans, P.A. Role of apoptosis in reperfusion injury. Cardiovasc. Res. 2004, 61, 414–426. [Google Scholar] [CrossRef]
- Shukla, S.K.; Rafiq, K. Proteasome biology and therapeutics in cardiac diseases. Transl. Res. 2019, 205, 64–76. [Google Scholar] [CrossRef]
- Attaix, D.; Ventadour, S.; Codran, A.; Béchet, D.; Taillandier, D.; Combaret, L. The ubiquitin-proteasome system and skeletal muscle wasting. Essays Biochem. 2005, 41, 173–186. [Google Scholar] [CrossRef]
- Lv, C.; Wang, X.; Guo, Y.; Yuan, S. Role of Selective Autophagy in Spermatogenesis and Male Fertility. Cells 2020, 9, 2523. [Google Scholar] [CrossRef]
- Hauser, P.V.; Perco, P.; Mühlberger, I.; Pippin, J.; Blonski, M.; Mayer, B.; Alpers, C.E.; Oberbauer, R.; Shankland, S.J. Microarray and bioinformatics analysis of gene expression in experimental membranous nephropathy. Nephron Exp. Nephrol. 2009, 112, e43–e58. [Google Scholar] [CrossRef] [Green Version]
- Kortenoeven, M.L.; Fenton, R.A. Renal aquaporins and water balance disorders. Biochim. Biophys. Acta Gen. Subj. 2014, 1840, 1533–1549. [Google Scholar] [CrossRef]
- Sharma, V.; Kaur, A.; Singh, T.G. Counteracting role of nuclear factor erythroid 2-related factor 2 pathway in Alzheimer’s disease. Biomed. Pharmacother. 2020, 129, 110373. [Google Scholar] [CrossRef]
- Gao, C.; Huang, W.; Kanasaki, K.; Xu, Y. The role of ubiquitination and sumoylation in diabetic nephropathy. BioMed Res. Int. 2014, 2014, 160692. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [Green Version]
- Williams, G.H.; Stoeber, K. The cell cycle and cancer. J. Pathol. 2012, 226, 352–364. [Google Scholar] [CrossRef]
- Thanikachalam, K.; Khan, G. Colorectal cancer and nutrition. Nutrients 2019, 11, 164. [Google Scholar] [CrossRef] [Green Version]
- Sporn, M.B. TGF-β: 20 years and counting. Microbes Infect. 1999, 1, 1251–1253. [Google Scholar] [CrossRef]
- Shi, Y.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef] [Green Version]
- Glasgow, E.; Mishra, L. Transforming growth factor-β signaling and ubiquitinators in cancer. Endocr. Relat. Cancer 2008, 15, 59–72. [Google Scholar] [CrossRef] [Green Version]
- Stolz, A.; Neufeld, K.; Ertych, N.; Bastians, H. Wnt-mediated protein stabilization ensures proper mitotic microtubule assembly and chromosome segregation. EMBO Rep. 2015, 16, 490–499. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, Y.; Xue, J.; Gong, A.; Yu, G.; Zhou, A.; Lin, K.; Zhang, S.; Zhang, N.; Gottardi, C.J.; et al. Wnt-induced deubiquitination FoxM1 ensures nucleus β-catenin transactivation. EMBO J. 2016, 35, 668–684. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Ou, B.; Zhao, S.; Liu, X.; Song, L.; Liu, X.; Wang, R.; Peng, Z. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signaling pathway. EBioMedicine 2019, 48, 236–247. [Google Scholar] [CrossRef] [Green Version]
- Yun, S.I.; Hong, H.K.; Yeo, S.Y.; Kim, S.H.; Cho, Y.B.; Kim, K.K. Ubiquitin-specific protease 21 promotes colorectal cancer metastasis by acting as a Fra-1 deubiquitinase. Cancers 2020, 12, 207. [Google Scholar] [CrossRef] [Green Version]
- Liang, Q.; Tang, C.; Tang, M.; Zhang, Q.; Gao, Y.; Ge, Z. TRIM47 is up-regulated in colorectal cancer, promoting ubiquitination and degradation of SMAD4. J. Exp. Clin. Cancer Res. 2019, 38, 159. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Liu, G.; Liu, R.; He, J.; Wang, G.; Zhang, H.; Liu, T.; Bai, J.; Cheng, N.; Qiu, J. Expression of ubiquitin conjugating enzyme E2T in colorectal cancers and clinical implications. Oncol. Lett. 2020, 20, 275. [Google Scholar] [CrossRef]
- Hao, Z.; Zhang, H.; Cowell, J. Ubiquitin-conjugating enzyme UBE2C: Molecular biology, role in tumorigenesis, and potential as a biomarker. Tumor Biol. 2012, 33, 723–730. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, W.; Qiao, G.; Zhao, B.; Liu, X.; Zhu, F. The Expression of Tripartite Motif Protein 36 and β-Catenin Correlates with the Prognosis of Esophageal Cancer. Gastroenterol. Res. Pract. 2020, 2020, 7641761. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Komatsu, S.; Ichikawa, D.; Hirajima, S.; Nishimura, Y.; Konishi, H.; Shiozaki, A.; Fujiwara, H.; Okamoto, K.; Tsuda, H.; et al. Overexpression of TRIM44 is related to invasive potential and malignant outcomes in esophageal squamous cell carcinoma. Tumor Biol. 2017, 39, 1010428317700409. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Zhao, Z.; Rong, L.; Xue, L.; Song, Y. RASSF6-TRIM16 axis promotes cell proliferation, migration and invasion in esophageal squamous cell carcinoma. J. Genet. Genom. 2019, 46, 477–488. [Google Scholar] [CrossRef]
- Wang, L.; Hou, Z.; Hasim, A.; Abuduerheman, A.; Zhang, H.; Niyaz, M.; Awut, I.; Upur, H.; Sheyhidin, I. RNF113A promotes the proliferation, migration and invasion, and is associated with a poor prognosis of esophageal squamous cell carcinoma. Int. J. Oncol. 2018, 52, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Saraya, A.; Das, P.; Sharma, R. Increased expression of MARCH8, an E3 ubiquitin ligase, is associated with growth of esophageal tumor. Cancer Cell Int. 2017, 17, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Shah, K.; Oza, M.J.; Behl, T. Reactivation of p53 gene by MDM2 inhibitors: A novel therapy for cancer treatment. Biomed. Pharmacother. 2019, 109, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fu, D.; Xi, J.; Ji, Z.; Liu, T.; Ma, Y.; Zhao, Y.; Dong, L.; Wang, Q.; Shen, X. Expression and clinical significance of UCH37 in human esophageal squamous cell carcinoma. Dig. Dis. Sci. 2012, 57, 2310–2317. [Google Scholar] [CrossRef]
- Zhang, W.; Zhuang, Y.; Zhang, Y.; Yang, X.; Zhang, H.; Wang, G.; Yin, W.; Wang, R.; Zhang, Z.; Xiao, W. Uev1A facilitates osteosarcoma differentiation by promoting Smurf1-mediated Smad1 ubiquitination and degradation. Cell Death Dis. 2017, 8, e2974. [Google Scholar] [CrossRef]
- Kim, B.G.; Lee, J.H.; Yasuda, J.; Ryoo, H.M.; Cho, J.Y. Phospho-Smad1 modulation by nedd4 e3 ligase in BMP/TGF-β signaling. J. Bone Miner. Res. 2011, 26, 1411–1424. [Google Scholar] [CrossRef]
- Georges, A.; Marcon, E.; Greenblatt, J.; Frappier, L. Identification and characterization of USP7 targets in cancer cells. Sci. Rep. 2018, 8, 15833. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.A.; Maecker, H.L.; French, D.M.; Liu, J.; Gregg, A.; Silverstein, L.B.; Cao, T.C.; Carano, R.A.; Dixit, V.M. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 2011, 146, 918–930. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Yang, Y.; Nolo, R.; Zweidler-McKay, P.A.; Hughes, D.P. Regulation of NOTCH signaling by reciprocal inhibition of HES1 and Deltex 1 and its role in osteosarcoma invasiveness. Oncogene 2010, 29, 2916–2926. [Google Scholar] [CrossRef] [Green Version]
- Yi, X.; Deng, X.; Zhao, Y.; Deng, B.; Deng, J.; Fan, H.; Du, Y.; Hao, L. Ubiquitin-like protein FAT10 promotes osteosarcoma growth by modifying the ubiquitination and degradation of YAP1. Exp. Cell Res. 2020, 387, 111804. [Google Scholar] [CrossRef]
- Gan, Z.; Han, K.; Lin, S.; Hu, H.; Shen, Z.; Min, D. Knockdown of ubiquitin-specific peptidase 39 inhibited the growth of osteosarcoma cells and induced apoptosis in vitro. Biol. Res. 2017, 50, 15. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Cai, X.; Xu, T.; Liu, K.; Yang, D.; Fan, L.; Li, G.; Yu, X. Tripartite Motif-Containing 46 Promotes Viability and Inhibits Apoptosis of Osteosarcoma Cells by Activating NF-κB Signaling Through Ubiquitination of PPARα. Oncol. Res. 2020, 28, 409. [Google Scholar] [CrossRef]
- Liang, J.; Xing, D.; Li, Z.; Shen, J.; Zhao, H.; Li, S. TRIM59 is upregulated and promotes cell proliferation and migration in human osteosarcoma. Mol. Med. Rep. 2016, 13, 5200–5206. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, Z.; Zhu, X.; Qian, G.; Zhou, Y.; Sun, Y.; Yu, W.; Wang, J.; Lu, H.; Lin, F.; et al. N6-Methyladenosine modification of the TRIM7 positively regulates tumorigenesis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. EBioMedicine 2020, 59, 102955. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, F.; Wang, X.; Li, G. Downregulation of Ubiquitin-Specific Protease 22 Inhibits Proliferation, Invasion, and Epithelial–Mesenchymal Transition in Osteosarcoma Cells. Oncol. Res. 2017, 25, 743. [Google Scholar] [CrossRef]
- Wang, Y.; Leng, H.; Chen, H.; Wang, L.; Jiang, N.; Huo, X.; Yu, B. Knockdown of UBE2T inhibits osteosarcoma cell proliferation, migration, and invasion by suppressing the PI3K/Akt signaling pathway. Oncol. Res. 2016, 24, 361. [Google Scholar] [CrossRef]
- Khan, H.; Singh, A.; Thapa, K.; Garg, N.; Grewal, A.K.; Singh, T.G. Therapeutic modulation of the phosphatidylinositol 3-kinases (PI3K) pathway in cerebral ischemic injury. Brain Res. 2021, 1761, 147399. [Google Scholar] [CrossRef]
- Shao, G.; Wang, R.; Sun, A.; Wei, J.; Peng, K.; Dai, Q.; Yang, W.; Lin, Q. The E3 ubiquitin ligase NEDD4 mediates cell migration signaling of EGFR in lung cancer cells. Mol. Cancer. 2018, 17, 24. [Google Scholar] [CrossRef] [Green Version]
- Gamell, C.; Gulati, T.; Levav-Cohen, Y.; Young, R.J.; Do, H.; Pilling, P.; Takano, E.; Watkins, N.; Fox, S.B.; Russell, P.; et al. Reduced abundance of the E3 ubiquitin ligase E6AP contributes to decreased expression of the INK4/ARF locus in non–small cell lung cancer. Sci. Signal. 2017, 10, eaaf8223. [Google Scholar] [CrossRef]
- Liu, L.; Yu, L.; Zeng, C.; Long, H.; Duan, G.; Yin, G.; Dai, X.; Lin, Z. E3 ubiquitin ligase HRD1 promotes lung tumorigenesis by promoting sirtuin 2 ubiquitination and degradation. Mol. Cell. Biol. 2020, 40, e00257-19. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Deng, Q.; Jiang, C.; Wang, X.; Niu, T.; Li, H.; Chen, T.; Jin, J.; Pan, W.; Cai, X.; et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene 2015, 34, 3957–3967. [Google Scholar] [CrossRef]
- Singh, S.; Singh, T.G. Role of nuclear factor kappa b (Nf-κb) signalling in neurodegenerative diseases: An mechanistic approach. Curr. Neuropharmacol. 2020, 18, 918–935. [Google Scholar] [CrossRef]
- Ren, H.; Xu, Y.; Wang, Q.; Jiang, J. E3 ubiquitin ligase tripartite motif-containing 71 promotes the proliferation of non-small cell lung cancer through the inhibitor of kappaB-α/nuclear factor kappaB pathway. Oncotarget 2018, 9, 10880. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Liu, P.; Wang, J.; Gong, T.; Zhang, F.; Ma, J.; Han, N. Ubiquitin-conjugating enzyme E2C regulates apoptosis-dependent tumor progression of non-small cell lung cancer via ERK pathway. Med. Oncol. 2015, 32, 149. [Google Scholar] [CrossRef]
- Gu, J.; Mao, W.; Ren, W.; Xu, F.; Zhu, Q.; Lu, C.; Lin, Z.; Zhang, Z.; Chu, Y.; Liu, R.; et al. Ubiquitin-protein ligase E3C maintains non-small-cell lung cancer stemness by targeting AHNAK-p53 complex. Cancer Lett. 2019, 443, 125–134. [Google Scholar] [CrossRef]
- Zhan, W.; Han, T.; Zhang, C.; Xie, C.; Gan, M.; Deng, K.; Fu, M.; Wang, J.B. TRIM59 promotes the proliferation and migration of non-small cell lung cancer cells by upregulating cell cycle related proteins. PLoS ONE 2015, 10, e0142596. [Google Scholar]
- Ning, J.; Zhang, J.; Liu, W.; Lang, Y.; Xue, Y.; Xu, S. Overexpression of ubiquitin-specific protease 22 predicts poor survival in patients with early-stage non-small cell lung cancer. Eur. J. Histochem. 2012, 56, e46. [Google Scholar] [CrossRef]
- Chan, D.W.; Chan, C.Y.; Yam, J.W.; Ching, Y.P.; Ng, I.O. Prickle-1 negatively regulates Wnt/β-catenin pathway by promoting Dishevelled ubiquitination/degradation in liver cancer. Gastroenterology 2006, 131, 1218–1227. [Google Scholar] [CrossRef]
- Guo, P.; Ma, X.; Zhao, W.; Huai, W.; Li, T.; Qiu, Y.; Zhang, Y.; Han, L. TRIM31 is upregulated in hepatocellular carcinoma and promotes disease progression by inducing ubiquitination of TSC1–TSC2 complex. Oncogene 2018, 37, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Qin, C.; Li, T.; Ma, X.; Qiu, Y.; Lin, Y.; Ma, D.; Qin, Z.; Sun, C.; Shen, X.; et al. The E3 ubiquitin ligase TRIM7 suppressed hepatocellular carcinoma progression by directly targeting Src protein. Cell Death Differ. 2020, 27, 1819–1831. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, C.; Wang, X.L.; Ly, P.; Belyi, V.; Xu-Monette, Z.Y.; Young, K.H.; Hu, W.; Feng, Z. E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ. 2014, 21, 1792–1804. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.F.; Zhang, M.F.; Tian, Q.H.; Zhang, C.Z. TRIM65 triggers β-catenin signaling via ubiquitylation of Axin1 to promote hepatocellular carcinoma. J. Cell Sci. 2017, 130, 3108–3115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, N.N.; Zhang, Z.Z.; Ren, J.H.; Zhang, J.; Zhou, Y.J.; Wong, V.K.; Law, B.Y.; Cheng, S.T.; Zhou, H.Z.; Chen, W.X.; et al. Overexpression of ubiquitin-conjugating enzyme E2 L3 in hepatocellular carcinoma potentiates apoptosis evasion by inhibiting the GSK3β/p65 pathway. Cancer Lett. 2020, 481, 1–4. [Google Scholar] [CrossRef]
- Liu, L.P.; Yang, M.; Peng, Q.Z.; Li, M.Y.; Zhang, Y.S.; Guo, Y.H.; Chen, Y.; Bao, S.Y. UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53Biochem. Biophys. Res. Commun. 2017, 493, 20–27. [Google Scholar] [CrossRef]
- Urbanik, T.; Köhler, B.C.; Boger, R.J.; Woerns, M.A.; Heeger, S.; Otto, G.; Hövelmeyer, N.; Galle, P.R.; Schuchmann, M.; Waisman, A.; et al. Down-regulation of CYLD as a trigger for NF-κB activation and a mechanism of apoptotic resistance in hepatocellular carcinoma cells. Int. J. Oncol. 2011, 38, 121–131. [Google Scholar]
- Seliger, B.; Fedorushchenko, A.; Brenner, W.; Ackermann, A.; Atkins, D.; Hanash, S.; Lichtenfels, R. Ubiquitin COOH-Terminal Hydrolase 1: A Biomarker of Renal Cell Carcinoma Associated with Enhanced Tumor Cell Proliferation and Migration. Clin. Cancer Res. 2007, 13, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Nanok, C.; Jearanaikoon, P.; Proungvitaya, S.; Limpaiboon, T. Aberrant methylation of HTATIP2 and UCHL1 as a predictive biomarker for cholangiocarcinoma. Mol. Med. Rep. 2018, 17, 4145–4153. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.J.; Zhu, J.J.; Yang, X.Y.; Biskup, E. NEDD4 promotes cell growth and migration via PTEN/PI3K/AKT signaling in hepatocellular carcinoma. Oncol. Lett. 2017, 14, 2649–2656. [Google Scholar] [CrossRef] [Green Version]
- Yuan, R.; Wang, K.; Hu, J.; Yan, C.; Li, M.; Yu, X.; Liu, X.; Lei, J.; Guo, W.; Wu, L.; et al. Ubiquitin-like protein FAT10 promotes the invasion and metastasis of hepatocellular carcinoma by modifying β-catenin degradation. Cancer Res. 2014, 74, 5287–5300. [Google Scholar] [CrossRef] [Green Version]
- Su, D.; Ma, S.; Shan, L.; Wang, Y.; Wang, Y.; Cao, C.; Liu, B.; Yang, C.; Wang, L.; Tian, S.; et al. Ubiquitin-specific protease 7 sustains DNA damage response and promotes cervical carcinogenesis. J. Clin. Investig. 2018, 128, 4280–4296. [Google Scholar] [CrossRef]
- Liang, J.; Nishi, H.; Bian, M.L.; Higuma, C.; Sasaki, T.; Ito, H.; Isaka, K. The ubiquitin-conjugating enzyme E2-EPF is overexpressed in cervical cancer and associates with tumor growth. Oncol. Rep. 2012, 28, 1519–1525. [Google Scholar] [CrossRef]
- Hu, J.; Meng, Y.; Zeng, J.; Zeng, B.; Jiang, X. Ubiquitin E3 Ligase MARCH7 promotes proliferation and invasion of cervical cancer cells through VAV2-RAC1-CDC42 pathway. Oncol. Lett. 2018, 16, 2312–2318. [Google Scholar] [CrossRef] [Green Version]
- Bai, M.; Che, Y.; Lu, K.; Fu, L. Analysis of deubiquitinase OTUD5 as a biomarker and therapeutic target for cervical cancer by bioinformatic analysis. PeerJ 2020, 8, e9146. [Google Scholar] [CrossRef]
- Zhang, Q.; Qiao, L.; Wang, X.; Ding, C.; Chen, J.J. UHRF1 epigenetically down-regulates UbcH8 to inhibit apoptosis in cervical cancer cells. Cell Cycle. 2018, 17, 300–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, W.; Guo, Q.; Zhu, C.; Song, Y.; Feng, H.; Cao, Y.; Du, M.; Chen, H. USP18 promotes cell proliferation and suppressed apoptosis in cervical cancer cells via activating AKT signaling pathway. BMC Cancer 2020, 20, 741. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martinez, E.; Morrisroe, C.; Sharrocks, A.D. The ubiquitin ligase UBE3A dampens ERK pathway signalling in HPV E6 transformed HeLa cells. PLoS ONE 2015, 10, e0119366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, M.; Zhao, C.; Wei, N.; Wu, X.; Cui, J.; Xing, Y. High expression of ubiquitin-specific protease 8 (USP8) is associated with poor prognosis in patients with cervical squamous cell carcinoma. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2018, 24, 4934. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zhao, W.; Sun, B.; Wang, X.; Liu, Q. Overexpression of TRIM24 is correlated with the progression of human cervical cancer. Am. J. Transl. Res. 2017, 9, 620. [Google Scholar]
- Aierken, G.; Seyiti, A.; Alifu, M.; Kuerban, G. Knockdown of tripartite-59 (TRIM59) inhibits cellular proliferation and migration in human cervical cancer cells. Oncol. Res. 2017, 25, 381. [Google Scholar] [CrossRef]
- Song, Y.; Guo, Q.; Gao, S.; Hua, K. Tripartite motif-containing protein 3 plays a role of tumor inhibitor in cervical cancer. Biochem. Biophys. Res. Commun. 2018, 498, 686–692. [Google Scholar] [CrossRef]
- Lu, X.; Zhuang, H.; Yu, Q.; Zhang, X.; Wu, Z.; Zhang, L.; Xu, Y.; Wu, B.; Yang, L.; Ma, A.; et al. Identification of the UBA2-WTIP fusion gene in acute myeloid leukemia. Exp. Cell Res. 2018, 371, 409–416. [Google Scholar] [CrossRef]
- Seghatoleslam, A.; Monabbati, A.; Bozorg-Ghalati, F.; Nikseresht, M.; Bordbar, M.R.; Rahvar, M.; Owji, A.A. Expression of UBE2Q2, a putative member of the ubiquitin-conjugating enzyme family in pediatric acute lymphoblastic leukemia. Arch. Iran. Med. 2012, 15, 352–355. [Google Scholar]
- Luo, H.; Qin, Y.; Reu, F.; Ye, S.; Dai, Y.; Huang, J.; Wang, F.; Zhang, D.; Pan, L.; Zhu, H.; et al. Microarray-based analysis and clinical validation identify ubiquitin-conjugating enzyme E2E1 (UBE2E1) as a prognostic factor in acute myeloid leukemia. J. Hematol. Oncol. 2016, 9, 125. [Google Scholar] [CrossRef] [Green Version]
- King, B.; Trimarchi, T.; Reavie, L.; Xu, L.; Mullenders, J.; Ntziachristos, P.; Aranda-Orgilles, B.; Perez-Garcia, A.; Shi, J.; Vakoc, C.; et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 2013, 153, 1552–1566. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Bei, L.; Shah, C.A.; Huang, W.; Platanias, L.C.; Eklund, E.A. The E3 ubiquitin ligase Triad1 influences development of Mll-Ell-induced acute myeloid leukemia. Oncogene 2018, 37, 2532–2544. [Google Scholar] [CrossRef] [Green Version]
- Wang, E.; Kawaoka, S.; Yu, M.; Shi, J.; Ni, T.; Yang, W.; Zhu, J.; Roeder, R.G.; Vakoc, C.R. Histone H2B ubiquitin ligase RNF20 is required for MLL-rearranged leukemia. Proc. Natl. Acad. Sci. USA 2013, 110, 3901–3906. [Google Scholar] [CrossRef] [Green Version]
- Shan, H.; Li, X.; Xiao, X.; Dai, Y.; Huang, J.; Song, J.; Liu, M.; Yang, L.; Lei, H.; Tong, Y.; et al. USP7 deubiquitinates and stabilizes NOTCH1 in T-cell acute lymphoblastic leukemia. Signal. Transduct. Target Ther. 2018, 3, 29. [Google Scholar] [CrossRef] [Green Version]
- Qiu, G.Z.; Mao, X.Y.; Ma, Y.; Gao, X.C.; Wang, Z.; Jin, M.Z.; Sun, W.; Zou, Y.X.; Lin, J.; Fu, H.L.; et al. Ubiquitin-specific protease 22 acts as an oncoprotein to maintain glioma malignancy through deubiquitinating B cell-specific Moloney murine leukemia virus integration site 1 for stabilization. Cancer Sci. 2018, 109, 2199–2210. [Google Scholar] [CrossRef] [Green Version]
- Quintás-Cardama, A.; Zhang, N.; Qiu, Y.H.; Post, S.; Creighton, C.J.; Cortes, J.; Coombes, K.R.; Kornblau, S.M. Loss of TRIM62 expression is an independent adverse prognostic factor in acute myeloid leukemia. Clin. Lymphoma Myeloma Leuk. 2015, 15, 115–127. [Google Scholar] [CrossRef] [Green Version]
- Schneider, D.; Chua, R.L.; Molitor, N.; Hamdan, F.H.; Rettenmeier, E.M.; Prokakis, E.; Mishra, V.K.; Kari, V.; Wegwitz, F.; Johnsen, S.A.; et al. The E3 ubiquitin ligase RNF40 suppresses apoptosis in colorectal cancer cells. Clin. Epigenet. 2019, 11, 98. [Google Scholar] [CrossRef] [Green Version]
- Thrift, A.P. The epidemic of oesophageal carcinoma: Where are we now? Cancer Epidemiol. 2016, 41, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Lagergren, J.; Fitzgerald, R.C.; Lordick, F.; Shah, M.A.; Lagergren, P.; Cunningham, D. Oesophageal cancer. Nat. Rev. Dis Primers 2017, 3, 17048. [Google Scholar] [CrossRef] [PubMed]
- Lagergren, J.; Smyth, E.; Cunningham, D.; Lagergren, P. Oesophageal cancer. Lancet 2017, 390, 2383–2396. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Li, X.; Wang, W.; Gao, T.; Shi, Z. UBE2C is involved in the functions of ECRG4 on esophageal squamous cell carcinoma. Biomed. Pharmacother. 2018, 98, 201–206. [Google Scholar] [CrossRef]
- Gatti, V.; Bernassola, F.; Talora, C.; Melino, G.; Peschiaroli, A. The impact of the ubiquitin system in the pathogenesis of squamous cell carcinomas. Cancers 2020, 12, 1595. [Google Scholar] [CrossRef]
- Kuwano, H.; Saeki, H.; Kawaguchi, H.; Sonoda, K.; Kitamura, K.; Nakashima, H.; Toh, Y.; Sugimachi, K. Proliferative activity of cancer cells in front and center areas of carcinoma in situ and invasive sites of esophageal squamous-cell carcinoma. Int. J. Cancer 1998, 78, 149–152. [Google Scholar] [CrossRef]
- Park, H.B.; Kim, J.W.; Baek, K.H. Regulation of Wnt signaling through ubiquitination and deubiquitination in cancers. Int. J. Mol. Sci. 2020, 21, 3904. [Google Scholar] [CrossRef]
- Moghbeli, M.; Abbaszadegan, M.R.; Golmakani, E.; Forghanifard, M.M. Correlation of Wnt and NOTCH pathways in esophageal squamous cell carcinoma. Cell Commun. Signal. 2016, 10, 129–135. [Google Scholar] [CrossRef] [Green Version]
- Moore, D.D.; Luu, H.H. Osteosarcoma. Orthop. Oncol. 2014, 162, 65–92. [Google Scholar]
- Kansara, M.; Thomas, D.M. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007, 26, 959248. [Google Scholar] [CrossRef]
- Ohata, Y.; Ozono, K. Bone and stem cells. The mechanism of osteogenic differentiation from mesenchymal stem cell. Clin. Calcium 2014, 24, 501–508. [Google Scholar]
- Vriend, J.; Reiter, R.J. Melatonin, bone regulation and the ubiquitin-proteasome connection: A review. Life Sci. 2016, 145, 152–160. [Google Scholar] [CrossRef]
- Franceschi, R.T.; Ge, C.; Xiao, G.; Roca, H.; Jiang, D. Transcriptional regulation of osteoblasts. Cells Tissues Organs 2009, 189, 144–152. [Google Scholar] [CrossRef]
- Wang, Z.; Kang, W.; You, Y.; Pang, J.; Ren, H.; Suo, Z.; Liu, H.; Zheng, Y. USP7: Novel drug target in cancer therapy. Front. Pharmacol. 2019, 10, 427. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, D.; Tian, Z.; Nicholson, B.; Kumar, K.S.; Zhou, B.; Carrasco, R.; McDermott, J.L.; Leach, C.A.; Fulcinniti, M.; Kodrasov, M.P.; et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 2012, 22, 345–358. [Google Scholar] [CrossRef] [Green Version]
- Ying, M.; Zhang, L.; Zhou, Q.; Shao, X.; Cao, J.; Zhang, N.; Li, W.; Zhu, H.; Yang, B.; He, Q. The E3 ubiquitin protein ligase MDM2 dictates all-trans retinoic acid-induced osteoblastic differentiation of osteosarcoma cells by modulating the degradation of RARα. Oncogene 2016, 35, 4358–4367. [Google Scholar] [CrossRef]
- De Groot, P.M.; Wu, C.C.; Carter, B.W.; Munden, R.F. The epidemiology of lung cancer. Transl. Lung Cancer Res. 2018, 7, 220. [Google Scholar] [CrossRef]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Zhao, X.C.; Wang, G.Z.; Wen, Z.S.; Zhou, Y.C.; Hu, Q.; Zhang, B.; Qu, L.W.; Gao, S.H.; Liu, J.; Ma, L.; et al. Systematic identification of CDC34 that functions to stabilize EGFR and promote lung carcinogenesis. EBioMedicine 2020, 53, 102689. [Google Scholar] [CrossRef]
- Jeon, H.S.; Jen, J. TGF-2 Signaling and the Role of Inhibitory Smads in Non-small Cell Lung Cancer. J. Thorac. Oncol. 2010, 5, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Li, D.; Zhou, P.; Li, W. Smurf1-positive expression indicates favorable survival for resected non-small cell lung cancer patients. Int. J. Clin. Exp. Pathol. 2018, 11, 399. [Google Scholar]
- Xie, C.; Powell, C.; Yao, M.; Wu, J.; Dong, Q. Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int. J. Biochem. Cell Biol. 2014, 47, 113–117. [Google Scholar] [CrossRef]
- Gupta, A.; Khan, H.; Kaur, A.; Singh, T.G. Novel Targets Explored in the Treatment of Alcohol Withdrawal Syndrome. CNS Neurol. Disord. Drug Targets 2020, 20, 158–173. [Google Scholar] [CrossRef]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef]
- Roura, E.; Castellsagué, X.; Pawlita, M.; Travier, N.; Waterboer, T.; Margall, N.; Bosch, F.X.; De Sanjosé, S.; Dillner, J.; Gram, I.T.; et al. Smoking as a major risk factor for cervical cancer and pre-cancer: Results from the EPIC cohort. Int. J. Cancer 2014, 135, 453–466. [Google Scholar] [CrossRef]
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute myeloid leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [Green Version]
- Pui, C.H.; Relling, M.; Downing, J.R. Acute lymphoblastic leukemia. N. Engl. J. Med. 2004, 350, 1535–1548. [Google Scholar] [CrossRef] [Green Version]
- Melo, J.V.; Hughes, T.P.; Apperley, J.F. Chronic myeloid leukemia. ASH Educ. Program Book 2003, 1, 132–152. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.W.; Ali, M.; Wood, T.E.; Wong, D.; Maclean, N.; Wang, X.; Gronda, M.; Skrtic, M.; Li, X.; Hurren, R.; et al. The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood Am Soc Hemat. 2010, 115, 2251–2259. [Google Scholar] [CrossRef] [Green Version]
- Eliseeva, E.; Pati, D.; Diccinanni, M.B.; Yu, A.L.; Mohsin, S.K.; Margolin, J.F.; Plon, S.E. Expression and localization of the CDC34 ubiquitin-conjugating enzyme in pediatric acute lymphoblastic leukemia. Cell Growth Differ. Publ. Am. J. Cancer Res. 2001, 12, 427–434. [Google Scholar]
- Cui, X.; Shen, W.; Wang, G.; Huang, Z.; Wen, D.; Yang, Y.; Liu, Y.; Cui, L. Ring finger protein 152 inhibits colorectal cancer cell growth and is a novel prognostic biomarker. Am. J. Transl. Res. 2018, 10, 3701. [Google Scholar] [PubMed]
- Peyser, B.D.; Hermone, A.; Salamoun, J.M.; Burnett, J.C.; Hollingshead, M.G.; McGrath, C.F.; Gussio, R.; Wipf, P. Specific RITA modification produces hyperselective cytotoxicity while maintaining in vivo antitumor efficacy. Mol. Cancer Ther. 2019, 18, 1765–1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coughlin, K.; Anchoori, R.; Iizuka, Y.; Meints, J.; MacNeill, L.; Vogel, R.I.; Orlowski, R.Z.; Lee, M.K.; Roden, R.B.; Bazzaro, M. Small-molecule RA-9 inhibits proteasome-associated DUBs and ovarian cancer in vitro and in vivo via exacerbating unfolded protein responses. Clin. Cancer Res. 2014, 20, 3174–3186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, H.; Jing, B.; Xia, Y.; Zhang, Y.; Hu, M.; Cai, H.; Tong, Y.; Zhou, L.; Yang, L.; Yang, J.; et al. WP1130 reveals USP24 as a novel target in T-cell acute lymphoblastic leukemia. Cancer Cell Int. 2019, 19, 56. [Google Scholar] [CrossRef]
- Soong, R.S.; Anchoori, R.K.; Roden, R.B.; Cho, R.L.; Chen, Y.C.; Tseng, S.C.; Huang, Y.L.; Liao, P.C.; Shyu, Y.C. Bis-benzylidine Piperidone RA190 treatment of hepatocellular carcinoma via binding RPN13 and inhibiting NF-κB signaling. BMC Cancer 2020, 20, 386. [Google Scholar] [CrossRef]
- Song, Y.; Li, S.; Ray, A.; Das, D.S.; Qi, J.; Samur, M.K.; Tai, Y.T.; Munshi, N.; Carrasco, R.D.; Chauhan, D.; et al. Blockade of deubiquitylating enzyme Rpn11 triggers apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Oncogene 2017, 36, 5631–5638. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Fang, L.; Zhao, H.; Xiang, T.; Wang, D. MDM2 inhibitor Nutlin-3a suppresses proliferation and promotes apoptosis in osteosarcoma cells. Acta Biochim. Biophys. Sin. 2012, 44, 685–691. [Google Scholar] [CrossRef] [Green Version]
- Tchoghandjian, A.; Soubéran, A.; Tabouret, E.; Colin, C.; Denicolaï, E.; Jiguet-Jiglaire, C.; El-Battari, A.; Villard, C.; Baeza-Kallee, N.; Figarella-Branger, D. Inhibitor of apoptosis protein expression in glioblastomas and their in vitro and in vivo targeting by SMAC mimetic GDC-0152. Cell Death Dis. 2016, 7, e2325. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; Sun, H.; Peng, Y.; Lu, J.; Nikolovska-Coleska, Z.; McEachern, D.; Liu, L.; Qiu, S.; Yang, C.Y.; Miller, R.; et al. A potent and orally active antagonist (SM-406/AT-406) of multiple inhibitor of apoptosis proteins (IAPs) in clinical development for cancer treatment. J. Med. Chem. 2011, 54, 2714–2726. [Google Scholar] [CrossRef] [Green Version]
- Xia, S.; Zhang, X.; Li, C.; Guan, H. Oridonin inhibits breast cancer growth and metastasis through blocking the Notch signaling. Saudi Pharm. J. 2017, 25, 638–643. [Google Scholar] [CrossRef]
- Shi, C.S.; Kuo, K.L.; Lin, W.C.; Chen, M.S.; Liu, S.H.; Liao, S.M.; Hsu, C.H.; Chang, Y.W.; Chang, H.C.; Huang, K.H. Neddylation inhibitor, MLN4924 suppresses angiogenesis in huvecs and solid cancers: In vitro and in vivo study. Am. J. Cancer Res. 2020, 10, 953. [Google Scholar]
- Fu, C.; Zhu, X.; Xu, P.; Li, Y. Pharmacological inhibition of USP7 promotes antitumor immunity and contributes to colon cancer therapy. OncoTargets Ther. 2019, 12, 609. [Google Scholar] [CrossRef] [Green Version]
- Fukui, S.; Nagasaka, K.; Miyagawa, Y.; Kikuchi-Koike, R.; Kawata, Y.; Kanda, R.; Ichinose, T.; Sugihara, T.; Hiraike, H.; Wada-Hiraike, O.; et al. The proteasome deubiquitinase inhibitor bAP15 downregulates TGF-β/Smad signaling and induces apoptosis via UCHL5 inhibition in ovarian cancer. Oncotarget 2019, 10, 5932. [Google Scholar] [CrossRef] [Green Version]
- Kuo, K.L.; Liu, S.H.; Lin, W.C.; Chow, P.M.; Chang, Y.W.; Yang, S.P.; Shi, C.S.; Hsu, C.H.; Liao, S.M.; Chang, H.C.; et al. The deubiquitinating enzyme inhibitor PR-619 enhances the cytotoxicity of cisplatin via the suppression of anti-apoptotic bcl-2 protein: In vitro and in vivo study. Cells 2019, 8, 1268. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Nam, S.; Lee, C.S.; Li, B.; Coppola, D.; Hamilton, A.D.; Dou, Q.P.; Sebti, S.M. CEP1612, a dipeptidyl proteasome inhibitor, induces p21WAF1 and p27KIP1 expression and apoptosis and inhibits the growth of the human lung adenocarcinoma A-549 in nude mice. Cancer Res. 2001, 61, 1280–1284. [Google Scholar]
- Milacic, V.; Banerjee, S.; Landis-Piwowar, K.R.; Sarkar, F.H.; Majumdar, A.P.; Dou, Q.P. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer Res. 2008, 68, 7283–7292. [Google Scholar] [CrossRef] [Green Version]
- Li, L.W.; Yu, X.Y.; Yang, Y.; Zhang, C.P.; Guo, L.P.; Lu, S.H. Expression of esophageal cancer related gene 4 (ECRG4), a novel tumor suppressor gene, in esophageal cancer and its inhibitory effect on the tumor growth in vitro and in vivo. Int. J. Cancer Res. 2009, 125, 1505–1513. [Google Scholar] [CrossRef]
- Jung, Y.; Yi, Y.S.; Yoo, D.S.; Kim, J.H.; Yang, W.S.; Lee, J.; Park, K.W.; Kweon, D.H.; Hong, S.; Cho, J.Y. 8-(Tosylamino) quinoline inhibits tumour progression through targeting phosphoinositide-3-kinase/Akt pathway. Pharm. Int. J. Pharm. Sci. 2013, 68, 146–152. [Google Scholar]
- Wang, X.; Mazurkiewicz, M.; Hillert, E.K.; Olofsson, M.H.; Pierrou, S.; Hillertz, P.; Gullbo, J.; Selvaraju, K.; Paulus, A.; Akhtar, S.; et al. The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Sci. Rep. 2016, 6, 26979. [Google Scholar] [CrossRef]
- Jiang, L.; Sun, Y.; Wang, J.; He, Q.; Chen, X.; Lan, X.; Chen, J.; Dou, Q.P.; Shi, X.; Liu, J. Proteasomal cysteine deubiquitinase inhibitor b-AP15 suppresses migration and induces apoptosis in diffuse large B cell lymphoma. J. Exp. Clin. Cancer Res. 2019, 38, 453. [Google Scholar] [CrossRef]
- Ungermannova, D.; Parker, S.J.; Nasveschuk, C.G.; Wang, W.; Quade, B.; Zhang, G.; Kuchta, R.D.; Phillips, A.J.; Liu, X. Largazole and its derivatives selectively inhibit ubiquitin activating enzyme (E1). PLoS ONE 2012, 7, e29208. [Google Scholar]
- Tsukamoto, S.; Hirota, H.; Imachi, M.; Fujimuro, M.; Onuki, H.; Ohta, T.; Yokosawa, H. Himeic acid A: A new ubiquitin-activating enzyme inhibitor isolated from a marine-derived fungus, Aspergillus sp. Bioorg. Med. Chem. Lett. 2005, 15, 191–194. [Google Scholar] [CrossRef]
- Hashimoto, M.; Kato, H.; Katsuki, A.; Tsukamoto, S.; Fujii, I. Identification of the Biosynthetic Gene Cluster for Himeic Acid A: A Ubiquitin-Activating Enzyme (E1) Inhibitor in Aspergillus japonicus MF275. ChemBioChem 2018, 19, 535–539. [Google Scholar] [CrossRef]
- Hann, Z.S.; Ji, C.; Olsen, S.K.; Lu, X.; Lux, M.C.; Tan, D.S.; Lima, C.D. Structural basis for adenylation and thioester bond formation in the ubiquitin E1. Proc. Natl. Acad. Sci. USA 2019, 116, 15475–15484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, Y.; Suzuki, M. Design, synthesis, and biological evaluation of novel ubiquitin-activating enzyme inhibitors. Bioorg. Med. Chem. Lett. 2018, 28, 2723–2727. [Google Scholar] [CrossRef]
- Chen, J.; Dexheimer, T.S.; Ai, Y.; Liang, Q.; Villamil, M.A.; Inglese, J.; Maloney, D.J.; Jadhav, A.; Simeonov, A.; Zhuang, Z. Selective and cell-active inhibitors of the USP1/UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem. Biol. 2011, 18, 1390–1400. [Google Scholar] [CrossRef] [Green Version]
- Tsukamoto, S.; Takeuchi, T.; Rotinsulu, H.; Mangindaan, R.E.; van Soest, R.W.; Ukai, K.; Kobayashi, H.; Namikoshi, M.; Ohta, T.; Yokosawa, H. Leucettamol A: A new inhibitor of Ubc13-Uev1A interaction isolated from a marine sponge, Leucetta aff. microrhaphis. Bioorg. Med. Chem. Lett. 2008, 18, 6319–6320. [Google Scholar] [CrossRef]
- Pulvino, M.; Liang, Y.; Oleksyn, D.; DeRan, M.; Van Pelt, E.; Shapiro, J.; Sanz, I.; Chen, L.; Zhao, J. Inhibition of proliferation and survival of diffuse large B-cell lymphoma cells by a small-molecule inhibitor of the ubiquitin-conjugating enzyme Ubc13-Uev1A. Blood Am. Soc. Hemat. 2012, 120, 1668–1677. [Google Scholar] [CrossRef] [PubMed]
- Ushiyama, S.; Umaoka, H.; Kato, H.; Suwa, Y.; Morioka, H.; Rotinsulu, H.; Losung, F.; Mangindaan, R.E.; de Voogd, N.J.; Yokosawa, H.; et al. Manadosterols A and B, sulfonated sterol dimers inhibiting the Ubc13–Uev1A interaction, isolated from the marine sponge Lissodendryx fibrosa. J. Nat. Prod. 2012, 75, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Eun, J.S.; Kim, B.G.; Kim, S.Y.; Jeon, H.; Soh, Y. Vitexin, an HIF-1alpha inhibitor, has anti-metastatic potential in PC12 cells. Mol. Cells 2006, 22, 291–299. [Google Scholar] [PubMed]
- Yang, Y.; Ludwig, R.L.; Jensen, J.P.; Pierre, S.A.; Medaglia, M.V.; Davydov, I.V.; Safiran, Y.J.; Oberoi, P.; Kenten, J.H.; Phillips, A.C.; et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 2005, 7, 547–559. [Google Scholar] [CrossRef] [Green Version]
- Matsuzawa, M.; Kakeya, H.; Yamaguchi, J.; Shoji, M.; Onose, R.; Osada, H.; Hayashi, Y. Enantio-and Diastereoselective Total Synthesis of (+)-Panepophenanthrin, a Ubiquitin-Activating Enzyme Inhibitor, and Biological Properties of Its New Derivatives. Chem. Asian J. 2006, 1, 845–851. [Google Scholar] [CrossRef]
- Vandenberghe, I.; Créancier, L.; Vispé, S.; Annereau, J.P.; Barret, J.M.; Pouny, I.; Samson, A.; Aussagues, Y.; Massiot, G.; Ausseil, F.; et al. Physalin B, a novel inhibitor of the ubiquitin-proteasome pathway, triggers NOXA-associated apoptosis. Biochem. Pharmacol. 2008, 76, 453–462. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, D.D.; Wu, Y.P.; Su, D.; Zhou, T.Y.; Gai, R.H.; Fu, Y.Y.; Zheng, L.; He, Q.J.; Zhu, H.; et al. MDM2 promotes epithelial–mesenchymal transition and metastasis of ovarian cancer SKOV3 cells. Br. J. Cancer 2017, 117, 1192–1201. [Google Scholar] [CrossRef] [Green Version]
- Downey-Kopyscinski, S.; Daily, E.W.; Gautier, M.; Bhatt, A.; Florea, B.I.; Mitsiades, C.S.; Richardson, P.G.; Driessen, C.; Overkleeft, H.S.; Kisselev, A.F. An inhibitor of proteasome β2 sites sensitizes myeloma cells to immunoproteasome inhibitors. Blood Adv. 2018, 2, 2443–2451. [Google Scholar] [CrossRef]
- Niewerth, D.; van Meerloo, J.; Jansen, G.; Assaraf, Y.G.; Hendrickx, T.C.; Kirk, C.J.; Anderl, J.L.; Zweegman, S.; Kaspers, G.J.; Cloos, J. Anti-leukemic activity and mechanisms underlying resistance to the novel immunoproteasome inhibitor PR-924. Biochem. Pharmacol. 2014, 89, 43–51. [Google Scholar] [CrossRef]
- Li, J.; Yakushi, T.; Parlati, F.; Mackinnon, A.L.; Perez, C.; Ma, Y.; Carter, K.P.; Colayco, S.; Magnuson, G.; Brown, B.; et al. Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat. Chem. Bio. 2017, 13, 486–493. [Google Scholar] [CrossRef] [Green Version]
- Hu, S.; Jin, Y.; Liu, Y.; Ljungman, M.; Neamati, N. Synthesis and mechanistic studies of quinolin-chlorobenzothioate derivatives with proteasome inhibitory activity in pancreatic cancer cell lines. Eur. J. Med. Chem. 2018, 158, 884–895. [Google Scholar] [CrossRef]
- Han, K.H.; Kwak, M.; Lee, T.H.; Park, M.S.; Jeong, I.H.; Kim, M.J.; Jin, J.O.; Lee, P.C. USP14 inhibition regulates tumorigenesis by inducing autophagy in lung cancer in vitro. Int. J. Mol. Sci. 2019, 20, 5300. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Wang, J.; Yuan, X.; Yang, S.; Xu, X.; Li, K.; He, Y.; Wei, L.; Zhang, J.; Tian, Y. IU1 suppresses proliferation of cervical cancer cells through MDM2 degradation. Int. J. Mol. Sci. 2020, 16, 2951. [Google Scholar] [CrossRef]
- Reinstein, E.; Ciechanover, A. Narrative review: Protein degradation and human diseases: The ubiquitin connection. Ann. Intern. Med. 2006, 145, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Micel, L.N.; Tentler, J.J.; Smith, P.G.; Eckhardt, G.S. Role of ubiquitin ligases and the proteasome in oncogenesis: Novel targets for anticancer therapies. J. Clin. Oncol. 2013, 31, 1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Kitagaki, J.; Dai, R.M.; Tsai, Y.C.; Lorick, K.L.; Ludwig, R.L.; Pierre, S.A.; Jensen, J.P.; Davydov, I.V.; Oberoi, P.; et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007, 67, 9472–9481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barghout, S.H.; Patel, P.S.; Wang, X.; Xu, G.W.; Kavanagh, S.; Halgas, O.; Zarabi, S.F.; Gronda, M.; Hurren, R.; Jeyaraju, D.V.; et al. Preclinical evaluation of the selective small-molecule UBA1 inhibitor, TAK-243, in acute myeloid leukemia. Leukemia 2019, 33, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Shirazi, F.; Singh, R.K.; Kuiatse, I.; Wang, H.; Lee, H.C.; Berkova, Z.; Berger, A.; Hyer, M.; Chattopadhyay, N.; et al. Ubiquitin-activating enzyme inhibition induces an unfolded protein response and overcomes drug resistance in myeloma. Blood Am. Soc. Hematol. 2019, 133, 1572–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyer, M.L.; Milhollen, M.A.; Ciavarri, J.; Fleming, P.; Traore, T.; Sappal, D.; Huck, J.; Shi, J.; Gavin, J.; Brownell, J.; et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 2018, 24, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Best, S.; Hashiguchi, T.; Kittai, A.; Bruss, N.; Paiva, C.; Okada, C.; Liu, T.; Berger, A.; Danilov, A.V. Targeting ubiquitin-activating enzyme induces ER stress–mediated apoptosis in B-cell lymphoma cells. Blood Adv. 2019, 3, 51–62. [Google Scholar] [CrossRef] [Green Version]
- Shen, M.; Schmitt, S.; Buac, D.; Dou, Q.P. Targeting the ubiquitin–proteasome system for cancer therapy. Expert Opin. Ther. Targets. 2013, 17, 1091–1108. [Google Scholar] [CrossRef] [Green Version]
- Sekizawa, R.; Ikeno, S.; Nakamura, H.; Naganawa, H.; Matsui, S.; Iinuma, H.; Takeuchi, T. Panepophenanthrin, from a mushroom strain, a novel inhibitor of the ubiquitin-activating enzyme. J. Nat. Prod. 2002, 65, 1491–1493. [Google Scholar] [CrossRef]
- Mehta, G.; Ramesh, S.S. Enantioselective total synthesis of (+)-panepophenanthrin, a novel inhibitor of the ubiquitin-activating enzyme. Tetrahedron Lett. 2004, 45, 1985–1987. [Google Scholar] [CrossRef] [Green Version]
- Mori, N.; Yamada, Y.; Ikeda, S.; Yamasaki, Y.; Tsukasaki, K.; Tanaka, Y.; Tomonaga, M.; Yamamoto, N.; Fujii, M. Bay 11-7082 inhibits transcription factor NF-κB and induces apoptosis of HTLV-I–infected T-cell lines and primary adult T-cell leukemia cells. Blood Am. Soc. Hematol. 2002, 100, 1828–1834. [Google Scholar]
- Strickson, S.; Campbell, D.G.; Emmerich, C.H.; Knebel, A.; Plater, L.; Ritorto, M.S.; Shpiro, N.; Cohen, P. The anti-inflammatory drug BAY 11-7082 suppresses the MyD88-dependent signalling network by targeting the ubiquitin system. Biochem. J. 2013, 451, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Fan, Y.H.; Xu, X.; Zhang, H.; Dou, J.; Tang, Y.; Zhong, X.; Rojas, Y.; Yu, Y.; Zhao, Y.; et al. A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways. Cell Death Dis. 2014, 5, e1079. [Google Scholar] [CrossRef] [Green Version]
- Garg, P.; Ceccarelli, D.F.; Keszei, A.F.; Kurinov, I.; Sicheri, F.; Sidhu, S.S. Structural and functional analysis of ubiquitin-based inhibitors that target the backsides of E2 enzymes. J. Mol. Biol. 2020, 432, 952–966. [Google Scholar] [CrossRef]
- Ceccarelli, D.F.; Tang, X.; Pelletier, B.; Orlicky, S.; Xie, W.; Plantevin, V.; Neculai, D.; Chou, Y.C.; Ogunjimi, A.; Al-Hakim, A.; et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell 2011, 145, 1075–1087. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Sun, Y. Targeting CDC34 E2 ubiquitin conjugating enzyme for lung cancer therapy. EBioMedicine 2020, 54, 102718. [Google Scholar] [CrossRef]
- Sanders, M.A.; Brahemi, G.; Nangia-Makker, P.; Balan, V.; Morelli, M.; Kothayer, H.; Westwell, A.D.; Shekhar, M.P. Novel inhibitors of Rad6 ubiquitin conjugating enzyme: Design, synthesis, identification, and functional characterization. Mol. Cancer Ther. 2013, 12, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Kothayer, H.; Spencer, S.M.; Tripathi, K.; Westwell, A.D.; Palle, K. Synthesis and in vitro anticancer evaluation of some 4, 6-diamino-1, 3, 5-triazine-2-carbohydrazides as Rad6 ubiquitin conjugating enzyme inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 2030–2034. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Nag, A. CUL4A ubiquitin ligase: A promising drug target for cancer and other human diseases. Open Biol. 2014, 4, 130217. [Google Scholar] [CrossRef] [Green Version]
- Al-Ghabkari, A.; Narendran, A. In vitro characterization of a potent p53-MDM2 inhibitor, RG7112 in neuroblastoma cancer cell lines. Cancer Biother. Radiopharm. 2019, 34, 252–257. [Google Scholar] [CrossRef]
- Pan, W.W.; Zhou, J.J.; Yu, C.; Xu, Y.; Guo, L.J.; Zhang, H.Y.; Zhou, D.; Song, F.Z.; Fan, H.Y. Ubiquitin E3 ligase CRL4CDT2/DCAF2 as a potential chemotherapeutic target for ovarian surface epithelial cancer. J. Biol. Chem. 2013, 288, 29680–29691. [Google Scholar] [CrossRef] [Green Version]
- Azmi, A.S.; Philip, P.A.; Beck, F.W.; Wang, Z.; Banerjee, S.; Wang, S.; Yang, D.; Sarkar, F.H.; Mohammad, R.M. MI-219-zinc combination: A new paradigm in MDM2 inhibitor-based therapy. Oncogene 2011, 30, 117–126. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.; Sigoillot, F.; Gaur, S.; Choi, S.; Pfaff, K.L.; Oh, D.C.; Hathaway, N.; Dimova, N.; Cuny, G.D.; King, R.W. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell 2010, 18, 382–395. [Google Scholar] [CrossRef] [Green Version]
- You, L.; Liu, H.; Huang, J.; Xie, W.; Wei, J.; Ye, X.; Qian, W. The novel anticancer agent JNJ-26854165 is active in chronic myeloid leukemic cells with unmutated BCR/ABL and T315I mutant BCR/ABL through promoting proteosomal degradation of BCR/ABL proteins. Oncotarget 2017, 8, 7777. [Google Scholar] [CrossRef] [Green Version]
- Makii, C.; Oda, K.; Ikeda, Y.; Sone, K.; Hasegawa, K.; Uehara, Y.; Nishijima, A.; Asada, K.; Koso, T.; Fukuda, T.; et al. MDM2 is a potential therapeutic target and prognostic factor for ovarian clear cell carcinomas with wild type TP53. Oncotarget 2016, 7, 75328. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Ando, H.; Suzuki, T.; Ogura, T.; Hotta, K.; Imamura, Y.; Yamaguchi, Y.; Handa, H. Identification of a primary target of thalidomide teratogenicity. Science 2010, 327, 1345–1350. [Google Scholar] [CrossRef] [Green Version]
- Asatsuma-Okumura, T.; Ito, T.; Handa, H. Molecular mechanisms of cereblon-based drugs. Pharmacol. Ther. 2019, 202, 132–139. [Google Scholar] [CrossRef]
- Azmi, A.S.; Aboukameel, A.; Banerjee, S.; Wang, Z.; Mohammad, M.; Wu, J.; Wang, S.; Yang, D.; Philip, P.A.; Sarkar, F.H.; et al. MDM2 inhibitor MI-319 in combination with cisplatin is an effective treatment for pancreatic cancer independent of p53 function. Eur. J. Cancer 2010, 46, 1122–1131. [Google Scholar] [CrossRef] [Green Version]
- Kisselev, A.F.; Callard, A.; Goldberg, A.L. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J. Biol. Chem. 2006, 281, 8582–8590. [Google Scholar] [CrossRef] [Green Version]
- Lawasut, P.; Chauhan, D.; Laubach, J.; Hayes, C.; Fabre, C.; Maglio, M.; Mitsiades, C.; Hideshima, T.; Anderson, K.C.; Richardson, P.G. New proteasome inhibitors in myeloma. Curr. Hematol. Malig. Rep. 2012, 7, 258–266. [Google Scholar] [CrossRef]
- McHugh, A.; Fernandes, K.; South, A.P.; Mellerio, J.E.; Salas-Alanís, J.C.; Proby, C.M.; Leigh, I.M.; Saville, M.K. Preclinical comparison of proteasome and ubiquitin E1 enzyme inhibitors in cutaneous squamous cell carcinoma: The identification of mechanisms of differential sensitivity. Oncotarget 2018, 9, 20265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, L.; Mohan, R.; Kwok, B.H.; Elofsson, M.; Sin, N.; Crews, C.M. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 1999, 96, 10403–10408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidor-Kaczmarek, J.; Cichorek, M.; Spodnik, J.H.; Wójcik, S.; Moryś, J. Proteasome inhibitors against amelanotic melanoma. Cell Biol. Toxicol. 2017, 33, 557–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Wang, K.; Zhen, S.; Wang, R.; Luo, W. Carfilzomib induces G2/M cell cycle arrest in human endometrial cancer cells via upregulation of p21Waf1/Cip1 and p27Kip1. Taiwan J. Obstet. Gynecol. 2016, 55, 847–851. [Google Scholar] [CrossRef]
- Vogl, D.T.; Martin, T.G.; Vij, R.; Hari, P.; Mikhael, J.R.; Siegel, D.; Wu, K.L.; Delforge, M.; Gasparetto, C. Phase I/II study of the novel proteasome inhibitor delanzomib (CEP-18770) for relapsed and refractory multiple myeloma. Leuk. Lymphoma. 2017, 58, 1872–1879. [Google Scholar] [CrossRef]







| S. No. | Type of Enzyme | Enzymes Involved | Modulation of Pathways Involved | Cancer Type | References |
|---|---|---|---|---|---|
| 1. | E3 ligase | Ectodermin, TRIM 47 | ↓ Smad 4 in TGF-β signaling | Promotes colorectal cancer | [75] |
| 2. | E3 ligase | FBXW7 | ↓ Wnt/β-catenin | Inhibits colorectal cancer | [71] |
| 3. | Deubiquitinating enzymes | USP5 | ↑ Wnt/β-catenin | Promotes colorectal cancer | [72] |
| 4. | Deubiquitinating enzymes | USP11, USP21 | ↑ERK/MAPK | Promotes colorectal cancer | [73,74] |
| 5. | Deubiquitinating enzymes | UBE2T | ↓ p53 pathway, ↑pentose phosphate pathway, etc., | Promotes colorectal cancer | [76] |
| 6. | E3 ligase | TRIM 67 | ↓ p53 degradation | Inhibits colorectal cancer | [75] |
| 7. | Ubiquitin-conjugating enzyme E2C | UBE2C | Pro-apoptotic | Inhibits esophageal cancer | [77] |
| 8. | E3 ligase | TRIM36 | ↑ Wnt/β-catenin | Promotes esophageal cancer | [78] |
| 9. | E3 ligase | TRIM44 | ↑ mTOR | Promotes esophageal cancer | [79] |
| 10. | E3 ligase | TRIM16 | ↑ TGF β/Snail | Promotes esophageal cancer | [80] |
| 11. | E3 ligase | RNF113A | ---- | Promotes esophageal cancer | [81] |
| 12. | E3 ligase | MARCH 8 | ---- | Promotes esophageal cancer | [82] |
| 13. | E3 ligase | Gankyrin | ↑ p53 degradation | Promotes esophageal cancer | [83] |
| 14. | Deubiquitinating enzymes | Ubiquitin carboxyl-terminal hydrolase 37 | ↑ TGF-β signaling | Promotes esophageal cancer | [84] |
| 15. | E2 ligase | Uev1A | bone morphogenetic protein signaling | Inhibits osteosarcoma | [85] |
| 16. | E3 ligase | Nedd4 | ↑ TGF-β signaling | Promotes osteosarcoma | [86] |
| 17. | E3 ligase | USP7 | ↑ Wnt/β-catenin | Promotes osteosarcoma | [87] |
| 18. | Deubiquitinating enzymes | USP1 | stabilize “inhibitors of DNA binding.” | Promotes osteosarcoma | [88] |
| 19. | E3 ligase | Deltex1 | ↓ NOTCH/HES1 | Inhibits osteosarcoma | [89] |
| 20. | E2 ligase | FAT10 | ↓ Hippo/YAP1 | Inhibits osteosarcoma | [90] |
| 21. | Deubiquitinating enzymes | USP39 | ↑ p21 | Inhibits osteosarcoma | [91] |
| 22. | E3 ligases | TRIM46, TRIM21, TRIM14, and TRIM23 | ↑NF-κB | Promotes osteosarcoma | [92] |
| 23. | E3 ligases | TRIM59 and TRIM7 | ↓ p53 and E-Cadherin | Promotes osteosarcoma | [93,94] |
| 24. | Deubiquitinating enzymes and E2 ligases | USP22 and UBE2T | ↑ PI3K/AKT | Promotes osteosarcoma | [95,96] |
| 25. | E3 ligases | Smurf1 | ↑ TGF-β signaling | Promotes lung cancer | [70] |
| 26. | E3 ligases | NEDD4-1 | ↑ PI3K/PTEN | Promotes lung cancer | [97] |
| 27. | E3 ligases | NEDD4 | EGFR mutation | Promotes lung cancer | [98] |
| 28. | E3 ligases | UBE3A | ↓ p16INK4a | Promotes lung cancer | [99] |
| 29. | E3 ligases | HRD1 | ↓ Sirtuin 2 | Promotes lung cancer | [100] |
| 30. | Deubiquitinating enzymes | USP37 | ↑ c-Myc | Promotes lung cancer | [101] |
| 31. | E3 ligases | TRIM7, TRIM71 | ↑ NF-κB | Promotes lung cancer | [102,103] |
| 32. | E2C ligases | UBE2C | ↑ ERK | Promotes Lung cancer | [104] |
| 33. | E3 ligases | UBE3C, TRIM59 | ↓ p53 | Promotes Lung cancer | [105,106] |
| 34. | Deubiquitinating enzymes | USP22 | ↑ ERK/AKT | Promotes lung cancer | [107] |
| 35. | E3 ligases | Prickle-1 | ↓ Wnt/β-catenin | Inhibits liver cancer | [108] |
| 36. | E3 ligases | TRIM31 | ↑ mTOR | Promotes liver cancer | [109] |
| 37. | E3 ligases | TRIM7 | ↓ PI3K | Inhibits liver cancer | [110] |
| 38. | E3 ligases | TRIM32 | ↑ mutated p53 | Promotes liver cancer | [111] |
| 39. | E3 ligases | TRIM65 | ↑ β-catenin | Promotes liver cancer | [112] |
| 40. | E2 ligases | UBE2L3 | ↓ p65 | Promotes liver cancer | [113] |
| 41. | E2 ligases | UBE2T | ↓ p53, p21, and noxa | Promotes liver cancer | [114] |
| 42. | Deubiquitinating enzymes | CYLD | ↓ NF-κB | Inhibits liver cancer | [115] |
| 43. | Deubiquitinating enzymes | UCHL1 | Apoptotic resistance | Promotes liver cancer | [116,117] |
| 44. | E3 ligases | NEDD4 | ↑ PTEN/PI3K/AKT | Promotes liver cancer | [118] |
| 45. | E3 ligases | FAT10 | ----- | Promotes liver cancer | [119] |
| 46. | E3 ligases | USP7 | Facilitates DNA repair by stabilizing MDC1 | Promotes cervical cancer | [120] |
| 47. | E2 ligases | E2-EPF | ↑ pVHL-HIF | Promotes cervical cancer | [121] |
| 48. | E3 ligases | MARCH 7 | ↑ VAV1/RAC1/CDC42 | Promotes cervical cancer | [122] |
| 49. | Deubiquitinating enzymes | Ovarian-tumor proteases deubiquitinase 5 | ↑ PI3K-AKT | Promotes cervical cancer | [123] |
| 50. | E3 ligases and E2 ligases | UHRF1, UBE2L6 | Promotes hypermethylation | Promotes cervical cancer | [124] |
| 51. | Deubiquitinating enzymes | USP18 | ↑ PI3K/AKT | Promotes cervical cancer | [125] |
| 52. | E3 ligases | UBE3A | ↓ ERK | Inhibits cervical cancer | [126] |
| 53. | Deubiquitinating enzymes | USP8 | Stabilizes FLIPL and EGFR signaling | Promotes cervical cancer | [127] |
| 54. | E3 ligases | TRIM 24 | ↑NF-κB/AKT | Promotes cervical cancer | [128] |
| 55. | E3 ligases | TRIM59 | ↓ p53 pathway, ↑ Ras/Rad, ↑ ERK | Promotes cervical cancer | [129] |
| 56. | E3 ligases | TRIM3 | ↑ p53 pathway, ↑ Caspase 3 | Inhibits cervical cancer | [130] |
| 57. | E1 ligases | UBA2 | ↑ ERK1/2, STAT3, and STAT5 | Promotes leukemia | [131] |
| 58. | E2 ligases and E2R1 | UBE2Q2 and CDC34 | ↓ IκB | Promotes leukemia | [132] |
| 59. | E2 ligases | UBE2E1 | ↓ HOX gene (HOXA9 and HOXA10) | Inhibits leukemia | [133] |
| 60. | E3 ligase | Fbxw7 | ↑ c-Myc, Notch1 | Promotes leukemia | [134] |
| 61. | E3 ligases | Triad1 | ↓HOX genes | Inhibits leukemia | [135] |
| 62. | E3 ligases | RNF20 | Interacts with histone H3 lys79 (H3K79) methyltransferase DOT1L | Promotes leukemia | [136] |
| 63. | E3 ligase | USP7 | ↑ NOTCH1 | Promotes leukemia | [137] |
| 64. | Deubiquitinating enzymes | USP22 | Stabilize BMI1 | Promotes leukemia | [138] |
| 65. | E3 ligases | TRIM62 | ↑ NOTCH and β-catenin signaling | Promotes leukemia | [139] |
| S. No. | Drug | Cancer | Signaling Pathway | Animal Models | Reference |
|---|---|---|---|---|---|
| 1. | RNF152 | Colorectal cancer | It is inactivating mTORC1 to induce autophagy and apoptotic cell death. | Immunodeficient nude mice | [171] |
| 2. | RITA (2,5-bis[5-hydroxymethyl2-thienyl] furan, NSC 652287) | Renal carcinoma | Block TP53–mdm2 complex and reactivation of p53 and Induction of Tumor cell Apoptosis | Mouse xenograft model | [172] |
| 3. | RA-9 | Ovarian cancer | Apoptosis and proteotoxic stress | Mice xenograft model | [173] |
| 4. | WP1130 | T-cell acute lymphoblastic leukemia | Induces apoptosis by accelerating the collapse of mitochondrial transmembrane potential via USP24-Mcl-1 axis | Tumor xenografts in NOD-SCID mice | [174] |
| 5. | The bis-benzylidine piperidone RA190 | Hepatocellular carcinoma | Nuclear factor κB (NF-κB) signaling | Male nude mice CAnN.Cg-Foxn1nu/CrlNarl | [175] |
| 6. | O-phenanthroline (OPA) | Multiple myeloma | Caspase cascade and endoplasmic stress response signaling | Murine xenograft model of human MM | [176] |
| 7. | Nutlin-3a | Osteosarcoma | Competitively binds the mdm2-p53 interacting site, activates p53 pathway | Human xenograft OS animal model with SAOS-2, U2OS, MG63 cell lines in SCID mice | [177] |
| 8. | GDC-0152 | Glioblastomas | Antagonists of the inhibitor of IAPs Postponed tumor formation and slowed down tumor growth | U87MG- iRFP cell grafted mice | [178] |
| 9. | SM-406/AT-406 | Human cancer cell (ovarian and breast cancer) | Antagonizes XIAP BIR3 induces rapid degradation of cellular cIAP1 protein | SCID mice bearing MDA-MB-231 xenograft tumors | [179] |
| 10. | Oridonin | Breast cancer | Tumor suppressive effect via inhibiting Notch receptors expression | Male BALB/C athymic nude mice | [180] |
| 11. | MLN4924 | Human urothelial cell carcinoma, cervical cancer, renal carcinoma, pharyngeal squamous carcinoma | Inhibits cell viability and induced apoptosis in HUVECs (human umbilical vascular endothelial cells) | Xenograft SCID mice | [181] |
| 12. | P5091 | Colorectal cancer | Elevated mRNA level of IFN-γ and TNF-α | Female BALB/c mice (CT26 xenograft model) | [182] |
| 13. | bAP15 | Ovarian cancer | Regulating TGF-β signaling, dephosphorylating Smad2, inducing apoptosis | Mice xenograft models of SKOV3 | [183] |
| 14. | PR-619 | Bladder urothelial carcinoma (UC) | Suppression of the Bcl-2 level | Nude mice Xenograft Matrigel culture | [184] |
| 15. | CEP1612 [phthalimide-(CH2)8CH-(cyclopentyl) CO-Arg(NO2)-Leu-H] | Human lung adenocarcinoma | Accumulation of p21WAF1 and p27KIP1, inducing apoptosis | A-549 tumor-bearing nude mice | [185] |
| 16. | Curcumin | Human colon cancer | Inhibit the proteasome and induce apoptosis | HCT-116 tumor-bearing ICR SCID mice | [186] |
| 17. | P5091 | Multiple Myeloma Cells | Inhibited USP7 activity, decreased HDM2, and increased p21 levels, induces apoptosis | Human plasmacytoma xenograft and SCID-hu mouse models | [155] |
| 18. | ECRG4 | Esophageal cancer | Inhibits NF-κB expression and nuclear translocation, attenuates NF-κB target gene COX-2 expression | BALB/c nude mice | [187] |
| 19. | 8-(tosylamino) quinoline (8-TQ) | Human cancer cells | Inhibition of molecular signaling machineries composed of phosphoinositide 3-kinase (PI3K)/phosphoinositide-dependent kinase-1 (PDK1)/Akt and extracellular-signal-regulated kinase (ERK) | murine T-cell lymphoma RMA cells in mice | [188] |
| 20. | VLX1570 | Multiple myeloma | Decrease in ERK phosphorylation; USP14 inhibitor | Xenograft model in immunocompromised mice | [189] |
| 21. | b-AP15 | Large B cell lymphoma | Inhibits Wnt/β-catenin and TGFβ/Smad pathways; USP14 and UCHL5 deubiquitinases | Mouse xenograft models of SU-DHL-4 and SU-DHL-2 cells | [190] |
| S. No. | Drugs | Category | Cell Lines | Reference |
|---|---|---|---|---|
| 1. | Largazole | Ubiquitin activating enzyme (UAE) inhibitor | Kip16, a GFP-p27 expressing Cell Line | [191] |
| 2. | Himeic acid A | UAE inhibitor | Western blotting with anti-Flag antibody | [192,193] |
| 3. | Ub-vinylsulfonamide (Ub-AVSN) | UAE inhibitor | N597A variant and the WT assay | [194,195] |
| 4. | Pimozide or GW7647 | USP1/UAF1 inhibitor | H596 and H460 cell lines | [196] |
| 5. | Leucettamol A | Ubc13-Uev1A inhibitor | Escherichia coli BL21 cells | [197] |
| 6. | NSC697923 | Ubc13-Uev1A inhibitor | ABC (activated B cell-like)-DLBCL cells and GCB (germinal center B cell-like)-DLBCL cells | [198] |
| 7. | Manadosterols A and B | Ubc13-Uev1A inhibitor | Escherichia coli BL21 cells | [199] |
| 8. | Vitexin | ubiquitin-conjugating enzyme E2-25K inhibitor | Rat pheochromocytoma PC12 cells, HepG2 (human hepatocellular carcinoma), and HOS (human osteosarcoma) cells | [200] |
| 9. | HLI98 family (C, D, E) | Ubiquitin ligase enzyme inhibitor | SAOS cells | [201] |
| 10. | RKTS-80, -81, -82 | E1 inhibitors | human breast cancer MCF-7 cells | [202] |
| 11. | Physalin B | proteasome inhibitors | human DLD-1 colon cancer cells | [203] |
| 12. | HLI-373 | E3 ligase inhibitor | ovarian SKOV3 cells | [204] |
| 13. | ONX-0914 | Immunoproteasome inhibitors | KMS-11 cells | [205] |
| 14. | PR-924 | Immunoproteasome inhibitor | Human T-cell ALL CCRF-CEM cells, human myeloid leukemic THP1 cells, and human multiple myeloma RPMI-8226 cells | [206] |
| 15. | Capzimin | Proteasome inhibitor | HCT116 cell lines | [207] |
| 16. | QCBT7 | Proteasome inhibitor | colon carcinoma cell line HCT 116. | [208] |
| 17. | IU1-47 | USP14 inhibitor | A549 and H1299 cell lines | [209] |
| 18. | IU1 | USP14 selective inhibitor | HeLa and SiHa cells (cervical cancer cells) | [210] |
| S. No. | Drug in Clinical Trials | Category | Cancer | Phase | NCT Number |
|---|---|---|---|---|---|
| 1. | TAK-243 (formerly known as MLN7243) | UAE (ubiquitin-activating enzyme) inhibitor | Advanced Malignant Solid tumors | Phase I (terminated) | NCT02045095 |
| 2. | Disulfiram and Cooper | Zinc fingers and RING-finger ubiquitin E3 ligases inhibitors | Breast Neoplasm Female, Metastatic Breast Cancer | Phase 2 | NCT03323346 |
| 3. | KPG-818 | Ubiquitin ligase modulator | Selected hematological malignancies (multiple myeloma, mantle cell lymphoma, follicular lymphoma, diffuse large B-cell lymphoma, indolent lymphoma, adult T-cell leukemia-lymphoma, or chronic lymphocytic leukemia | Phase 1 | NCT04283097 |
| 4. | Vorinostat (MK-0683) + Bortezomib | HDAC (Histone deacetylases) inhibitors + proteasome inhibitor | Multiple Myeloma | Phase 3 | NCT00773747 |
| 5. | MLN4924 | Nedd8 activating enzyme inhibitor | lymphoma or multiple myeloma | Phase 1 (completed) | NCT00722488 |
| 6. | Bortezomib + Doxorubicin | Proteasome inhibitor | Advanced, Recurrent, or Metastatic Adenoid Cystic Carcinoma of the Head and Neck | Phase 2 (completed) | NCT00077428 |
| 7. | NPI-0052 | Proteasome inhibitor | Solid tumors, lymphomas, leukemias, and multiple myeloma. | Phase 1 (completed) | NCT00629473 |
| 8. | Marizomib(NPI-0052) + Vorinostat | Proteasome inhibitor + HDAC (Histone deacetylases) inhibitors | Non-Small Cell Lung cancer, Pancreatic cancer, Melanoma, Lymphoma Multiple Myeloma | Phase 1 (completed) | NCT00667082 |
| 9. | JNJ-26854165 | E3 ligase inhibitors | Neoplasms | Phase 1 (completed) | NCT00676910 |
| 10. | Bortezomib (PS-341) | proteasome inhibitor | Squamous cell carcinomas of the head and neck (SCCHN) | Phase 1 (completed) | NCT00011778 |
| 11. | TAK-981 + Pembrolizumab | SUMOylation inhibitor + immunosuppressant | Advanced or Metastatic Solid tumors | Phase 1 Phase 2 | NCT04381650 |
| 12. | Oprozomib | Proteasome inhibitor | Advanced Refractory or Recurrent Solid tumors | Phase 1 (completed) | NCT01129349 |
| 13. | Carfilzomib | Proteasome inhibitor | Neuroendocrine cancer | Phase 2 | NCT02318784 |
| 14. | MLN9708 | Proteasome inhibitor | Advanced non-hematologic malignancies | Phase 1 (completed) | NCT00830869 |
| 15. | MLN9708 + Vorinostat | Proteasome inhibitor + HDAC inhibitor | Advanced p53 Mutant malignancies | Phase 1 | NCT02042989 |
| 16. | GSK2110183 | Proteasome inhibitor | Multiple myeloma | Phase 1 (completed) | NCT01445587 |
| 17. | Trastuzumab + PS-341 | Proteasome inhibitor | Breast cancer, Stage 4 | Phase 1 (completed) | NCT00199212 |
| 18. | Finasteride | Ubiquitin-conjugating enzyme inhibitor | Adenocarcinoma of the ProstateStage II Prostate cancer | Phase 2 | NCT00438464 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, A.; Khan, H.; Singh, T.G.; Grewal, A.K.; Najda, A.; Kawecka-Radomska, M.; Kamel, M.; Altyar, A.E.; Abdel-Daim, M.M. Pharmacological Modulation of Ubiquitin-Proteasome Pathways in Oncogenic Signaling. Int. J. Mol. Sci. 2021, 22, 11971. https://doi.org/10.3390/ijms222111971
Sharma A, Khan H, Singh TG, Grewal AK, Najda A, Kawecka-Radomska M, Kamel M, Altyar AE, Abdel-Daim MM. Pharmacological Modulation of Ubiquitin-Proteasome Pathways in Oncogenic Signaling. International Journal of Molecular Sciences. 2021; 22(21):11971. https://doi.org/10.3390/ijms222111971
Chicago/Turabian StyleSharma, Anmol, Heena Khan, Thakur Gurjeet Singh, Amarjot Kaur Grewal, Agnieszka Najda, Małgorzata Kawecka-Radomska, Mohamed Kamel, Ahmed E. Altyar, and Mohamed M. Abdel-Daim. 2021. "Pharmacological Modulation of Ubiquitin-Proteasome Pathways in Oncogenic Signaling" International Journal of Molecular Sciences 22, no. 21: 11971. https://doi.org/10.3390/ijms222111971