Coenzyme Q Biosynthesis: An Update on the Origins of the Benzenoid Ring and Discovery of New Ring Precursors
Abstract
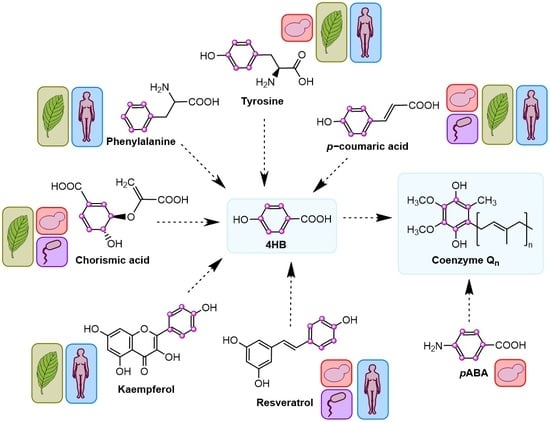
:1. Introduction
2. 4-Hydroxybenzoic Acid (4HB)
2.1. Classic Ring Precursor Identification
2.1.1. Radiolabeling
2.1.2. Stable Isotope
2.2. Source of 4HB
2.2.1. E. coli
2.2.2. S. cerevisiae
2.2.3. Mammals
2.2.4. Plants
3. Other Ring Precursors
3.1. pABA in S. cerevisiae
3.2. Natural Products
4. Closing Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Aussel, L.; Pierrel, F.; Loiseau, L.; Lombard, M.; Fontecave, M.; Barras, F. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1837, 1004–1011. [Google Scholar] [CrossRef]
- Wang, Y.; Hekimi, S. The Complexity of Making Ubiquinone. Trends Endocrinol. Metab. 2019, 30, 929–943. [Google Scholar] [CrossRef]
- Awad, A.M.; Bradley, M.C.; Fernández-Del-Río, L.; Nag, A.; Tsui, H.S.; Clarke, C.F. Coenzyme Q10 deficiencies: Pathways in yeast and humans. Essays Biochem. 2018, 62, 361–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turunen, M.; Olsson, J.; Dallner, G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta (BBA)-Bioenerg. 2004, 1660, 171–199. [Google Scholar] [CrossRef] [Green Version]
- Alcázar-Fabra, M.; Rodríguez-Sánchez, F.; Trevisson, E.; Brea-Calvo, G. Primary coenzyme Q deficiencies: A literature review and online platform of clinical features to uncover genotype-phenotype correlations. Free Radic. Biol. Med. 2021, 167, 141–180. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hekimi, S. Understanding Ubiquinone. Trends Cell Biol. 2016, 26, 367–378. [Google Scholar] [CrossRef]
- Stefely, J.A.; Pagliarini, D.J. Biochemistry of Mitochondrial Coenzyme Q Biosynthesis. Trends Biochem. Sci. 2017, 42, 824–843. [Google Scholar] [CrossRef]
- Lombard, J.; Moreira, D. Origins and Early Evolution of the Mevalonate Pathway of Isoprenoid Biosynthesis in the Three Domains of Life. Mol. Biol. Evol. 2011, 28, 87–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boronat, A.; Rodríguez-Concepción, M. Terpenoid biosynthesis in prokaryotes. In Biotechnology of Isoprenoids; Schrader, J., Bohlmann, J., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer Nature: Basingstoke, UK, 2015; Volume 148, pp. 3–18. [Google Scholar]
- Lohr, M.; Schwender, J.; Polle, J.E. Isoprenoid biosynthesis in eukaryotic phototrophs: A spotlight on algae. Plant Sci. 2012, 185–186, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Bentlage, B.; Rogers, T.S.; Bachvaroff, T.R.; Delwiche, C.F. Complex Ancestries of Isoprenoid Synthesis in Dinoflagellates. J. Eukaryot. Microbiol. 2016, 63, 123–137. [Google Scholar] [CrossRef]
- Abby, S.S.; Kazemzadeh, K.; Vragniau, C.; Pelosi, L.; Pierrel, F. Advances in bacterial pathways for the biosynthesis of ubiqui-none. Biochim. Biophys. Acta (BBA)-Bioenerg. 2020, 1861, 148259. [Google Scholar] [CrossRef]
- Hayashi, K.; Ogiyama, Y.; Yokomi, K.; Nakagawa, T.; Kaino, T.; Kawamukai, M. Functional conservation of coenzyme Q bio-synthetic genes among yeasts, plants, and humans. PLoS ONE 2014, 9, e99038. [Google Scholar] [CrossRef] [Green Version]
- Kawamukai, M. Biosynthesis of coenzyme Q in eukaryotes. Biosci. Biotechnol. Biochem. 2016, 80, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Tsui, H.S.; Clarke, C.F. Ubiquinone Biosynthetic Complexes in Prokaryotes and Eukaryotes. Cell Chem. Biol. 2019, 26, 465–467. [Google Scholar] [CrossRef] [PubMed]
- Chehade, M.H.; Pelosi, L.; Fyfe, C.D.; Loiseau, L.; Rascalou, B.; Brugière, S.; Kazemzadeh, K.; Ciccone, L.; Aussel, L.; Couté, Y.; et al. A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone. Cell Chem. Biol. 2019, 26, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Marbois, B.; Xie, L.X.; Choi, S.; Hirano, K.; Hyman, K.; Clarke, C.F. para-Aminobenzoic acid is a precursor in coenzyme Q6 bio-synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 27827–27838. [Google Scholar] [CrossRef] [Green Version]
- Pierrel, F.; Hamelin, O.; Douki, T.; Kieffer-Jaquinod, S.; Mühlenhoff, U.; Ozeir, M.; Lill, R.; Fontecave, M. Involvement of mito-chondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 2010, 17, 449–459. [Google Scholar] [CrossRef]
- Nishida, I.; Yanai, R.; Matsuo, Y.; Kaino, T.; Kawamukai, M. Benzoic acid inhibits Coenzyme Q biosynthesis in Schizosaccharo-myces pombe. PLoS ONE 2020, 15, e0242616. [Google Scholar] [CrossRef] [PubMed]
- Ozeir, M.; Mühlenhoff, U.; Webert, H.; Lill, R.; Fontecave, M.; Pierrel, F. Coenzyme Q Biosynthesis: Coq6 Is Required for the C5-Hydroxylation Reaction and Substrate Analogs Rescue Coq6 Deficiency. Chem. Biol. 2011, 18, 1134–1142. [Google Scholar] [CrossRef]
- Ozeir, M.; Pelosi, L.; Ismail, A.; Mellot-Draznieks, C.; Fontecave, M.; Pierrel, F. Coq6 Is Responsible for the C4-deamination Reac-tion in Coenzyme Q Biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 24140–24151. [Google Scholar] [CrossRef] [Green Version]
- Lapointe, C.P.; Stefely, J.A.; Jochem, A.; Hutchins, P.D.; Wilson, G.M.; Kwiecien, N.W.; Coon, J.J.; Wickens, M.; Pagliarini, D.J. Multi-omics Reveal Specific Targets of the RNA-Binding Protein Puf3p and Its Orchestration of Mitochondrial Biogenesis. Cell Syst. 2018, 6, 125–135. [Google Scholar] [CrossRef] [Green Version]
- Veling, M.T.; Reidenbach, A.G.; Freiberger, E.C.; Kwiecien, N.W.; Hutchins, P.D.; Drahnak, M.J.; Jochem, A.; Ulbrich, A.; Rush, M.J.; Russell, J.D.; et al. Multi-omic Mitoprotease Profiling Defines a Role for Oct1p in Coenzyme Q Production. Mol. Cell 2017, 68, 970–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vögtle, F.N.; Prinz, C.; Kellermann, J.; Lottspeich, F.; Pfanner, N.; Meisinger, C. Mitochondrial protein turnover: Role of the pre-cursor intermediate peptidase Oct1 in protein stabilization. Mol. Biol. Cell 2011, 22, 2135–2143. [Google Scholar] [CrossRef]
- Acosta Lopez, M.J.; Trevisson, E.; Canton, M.; Vazquez-Fonseca, L.; Morbidoni, V.; Baschiera, E.; Frasson, C.; Pelosi, L.; Ras-calou, B.; Desbats, M.A.; et al. Vanillic Acid Restores Coenzyme Q Biosynthesis and ATP Production in Human Cells Lacking COQ6. Oxid. Med. Cell. Longev. 2019, 2019, 3904905. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.X.; Ozeir, M.; Tang, J.Y.; Chen, J.Y.; Jaquinod, S.K.; Fontecave, M.; Clarke, C.F.; Pierrel, F. Overexpression of the Coq8 ki-nase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosyn-thetic pathway. J. Biol. Chem. 2012, 287, 23571–23581. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.X.; Williams, K.J.; He, C.H.; Weng, E.; Khong, S.; Rose, T.E.; Kwon, O.; Bensinger, S.J.; Marbois, B.N.; Clarke, C.F. Resveratrol and para-coumarate serve as ring precursors for coenzyme Q biosynthesis. J. Lipid Res. 2015, 56, 909–919. [Google Scholar] [CrossRef] [Green Version]
- Gibson, F. Chemical and Genetic Studies on the Biosynthesis of Ubiquinone by Escherichia coli. Biochem. Soc. Trans. 1973, 1, 317–326. [Google Scholar] [CrossRef]
- Poon, W.W.; Do, T.Q.; Marbois, B.N.; Clarke, C.F. Sensitivity to treatment with polyunsaturated fatty acids is a general charac-teristic of the ubiquinone-deficient yeast coq mutants. Mol. Asp. Med. 1997, 18, 121–127. [Google Scholar] [CrossRef]
- Poon, W.W.; Marbois, B.N.; Faull, K.F.; Clarke, C.F. 3-Hexaprenyl-4-hydroxybenzoic acid forms a predominant intermediate pool in ubiquinone biosynthesis in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 1995, 320, 305–314. [Google Scholar] [CrossRef]
- Marbois, B.N.; Clarke, C.F. The COQ7 gene encodes a protein in saccharomyces cerevisiae necessary for ubiquinone biosynthe-sis. J. Biol. Chem. 1996, 271, 2995–3004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sippel, C.J.; Goewert, R.R.; Slachman, F.N.; Olson, R.E. The regulation of ubiquinone-6 biosynthesis by Saccharomyces cere-visiae. J. Biol. Chem. 1983, 258, 1057–1061. [Google Scholar] [CrossRef]
- Goewert, R.R.; Sippel, C.J.; Olson, R.E. Identification of 3,4-dihydroxy-5-hexaprenylbenzoic acid as an intermediate in the bio-synthesis of ubiquinone-6 by Saccharomyces cerevisiae. Biochemistry 1981, 20, 4217–4223. [Google Scholar] [CrossRef]
- Bradley, M.C.; Yang, K.; Fernández-Del-Río, L.; Ngo, J.; Ayer, A.; Tsui, H.S.; Novales, N.A.; Stocker, R.; Shirihai, O.S.; Barros, M.H.; et al. COQ11 deletion mitigates respiratory deficiency caused by mutations in the gene encoding the coenzyme Q chaperone protein Coq10. J. Biol. Chem. 2020, 295, 6023–6042. [Google Scholar] [CrossRef] [Green Version]
- Allan, C.M.; Awad, A.M.; Johnson, J.S.; Shirasaki, D.I.; Wang, C.; Blaby-Haas, C.E.; Merchant, S.S.; Loo, J.A.; Clarke, C.F. Identification of Coq11, a New Coenzyme Q Biosynthetic Protein in the CoQ-Synthome in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 7517–7534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, U.C.; Clarke, C.F. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 2007, 7, S62–S71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C.H.; Xie, L.X.; Allan, C.M.; Tran, U.C.; Clarke, C.F. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2014, 1841, 630–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefely, J.A.; Licitra, F.; Laredj, L.; Reidenbach, A.G.; Kemmerer, Z.A.; Grangeray, A.; Jaeg-Ehret, T.; Minogue, C.E.; Ulbrich, A.; Hutchins, P.D.; et al. Cerebellar Ataxia and Coenzyme Q Deficiency through Loss of Unorthodox Kinase Activity. Mol. Cell 2016, 63, 608–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohman, D.C.; Forouhar, F.; Beebe, E.T.; Stefely, M.S.; Minogue, C.E.; Ulbrich, A.; Stefely, J.A.; Sukumar, S.; Luna-Sánchez, M.; Jochem, A.; et al. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosyn-thesis. Proc. Natl. Acad. Sci. USA 2014, 111, E4697–E4705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floyd, B.J.; Wilkerson, E.M.; Veling, M.T.; Minogue, C.E.; Xia, C.; Beebe, E.T.; Wrobel, R.L.; Cho, H.; Kremer, L.S.; Alston, C.L.; et al. Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function. Mol. Cell 2016, 63, 621–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashraf, S.; Gee, H.Y.; Woerner, S.; Xie, L.X.; Vega-Warner, V.; Lovric, S.; Fang, H.; Song, X.; Cattran, D.C.; Avila-Casado, C.; et al. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J. Clin. Investig. 2013, 123, 5179–5189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, K.; Jochem, A.; Le Vasseur, M.; Lewis, S.; Paulson, B.R.; Reddy, T.R.; Russell, J.D.; Coon, J.J.; Pagliarini, D.J.; Nunnari, J. Coenzyme Q biosynthetic proteins assemble in a substrate-dependent manner into domains at ER–mitochondria contacts. J. Cell Biol. 2019, 218, 1353–1369. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg-Bord, M.; Tsui, H.S.; Antunes, D.; Fernandez-Del-Rio, L.; Bradley, M.C.; Dunn, C.D.; Nguyen, T.P.T.; Rapaport, D.; Clarke, C.F.; Schuldiner, M. The Endoplasmic Reticulum-Mitochondria Encounter Structure Complex Coordinates Coenzyme Q Biosynthesis. Contact 2019, 2, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Tsui, H.S.; Pham, N.V.B.; Amer, B.R.; Bradley, M.C.; Gosschalk, J.E.; Gallagher-Jones, M.; Ibarra, H.; Clubb, R.T.; Blaby-Haas, C.E.; Clarke, C.F. Human COQ10A and COQ10B are distinct lipid-binding START domain proteins required for coenzyme Q function. J. Lipid Res. 2019, 60, 1293–1310. [Google Scholar] [CrossRef]
- Fino, C.; Vestergaard, M.; Ingmer, H.; Pierrel, F.; Gerdes, K.; Harms, A. PasT of Escherichia coli sustains antibiotic tolerance and aerobic respiration as a bacterial homolog of mitochondrial Coq10. MicrobiologyOpen 2020, 9, e1064. [Google Scholar] [CrossRef] [PubMed]
- Alcázar-Fabra, M.; Trevisson, E.; Brea-Calvo, G. Clinical syndromes associated with Coenzyme Q10 deficiency. Essays Biochem. 2018, 62, 377–398. [Google Scholar] [CrossRef]
- Fernández-Del-Río, L.; Kelly, M.E.; Contreras, J.; Bradley, M.C.; James, A.M.; Murphy, M.P.; Payne, G.S.; Clarke, C.F. Genes and lipids that impact uptake and assimilation of exogenous coenzyme Q in Saccharomyces cerevisiae. Free Radic. Biol. Med. 2020, 154, 105–118. [Google Scholar] [CrossRef]
- Gutierrez-Mariscal, F.M.; Yubero-Serrano, E.M.; Villalba, J.M.; Lopez-Miranda, J. Coenzyme Q10: From bench to clinic in aging diseases, a translational review. Crit. Rev. Food Sci. Nutr. 2019, 59, 2240–2257. [Google Scholar] [CrossRef] [PubMed]
- Yubero, D.; Montero, R.; Santos-Ocaña, C.; Salviati, L.; Navas, P.; Artuch, R. Molecular diagnosis of coenzyme Q10 deficiency: An update. Expert Rev. Mol. Diagn. 2018, 18, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Rudney, H.; Parson, W.W. The conversion of p-hydroxybenzaldehyde to the benzoquinone ring of ubiquinone in Rhodospiril-lum rubrum. J. Biol. Chem. 1963, 238, 3137–3138. [Google Scholar] [CrossRef]
- Parson, W.W.; Rudney, H. The biosynthesis of the benzoquinone ring of ubiquinone from p-hydroxybenzaldehyde and p-hydroxybenzoic acid in rat kidney, Azotobacter vinelandii, and baker’s yeast. Proc. Natl. Acad. Sci. USA 1964, 51, 444–450. [Google Scholar] [CrossRef] [Green Version]
- Olson, R.E.; Rudney, H. Biosynthesis of Ubiquinone. Vitam. Horm. 1983, 40, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Elliott, W.H.; Waller, G.R. Vitamins and Cofactors. In Biochemical Applications of Mass Spectromery; Waller, G.R., Ed.; Wiley-Interscience: New York, NY, USA, 1973; pp. 499–536. [Google Scholar]
- Olson, R.E. Biosynthesis of Ubiquinones in Animals. Vitam. Horm. 1967, 24, 551–574. [Google Scholar] [CrossRef]
- Olson, R.E.; Dialamieh, G.H.; Bentley, R.; Springer, C.M.; Ramsey, V.G. Studies on coenzyme Q. Pattern of labeling in coenzyme Q9 after administration of isotopic acetate and aromatic amino acids to rats. J. Biol. Chem. 1965, 240, 514–523. [Google Scholar] [CrossRef]
- Siebert, M.; Severin, K.; Heide, L. Formation of 4-hydroxybenzoate in Escherichia coli: Characterization of the ubiC gene and its encoded enzyme chorismate pyruvate-lyase. Microbiology 1994, 140, 897–904. [Google Scholar] [CrossRef] [Green Version]
- Nichols, B.P.; Green, J.M. Cloning and sequencing of Escherichia coli ubiC and purification of chorismate lyase. J. Bacteriol. 1992, 174, 5309–5316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Payet, L.A.; Leroux, M.; Willison, J.C.; Kihara, A.; Pelosi, L.; Pierrel, F. Mechanistic details of early steps in coenzyme Q biosyn-thesis pathway in yeast. Cell Chem. Biol. 2016, 23, 1241–1250. [Google Scholar] [CrossRef] [Green Version]
- Stefely, J.A.; Kwiecien, N.W.; Freiberger, E.C.; Richards, A.L.; Jochem, A.; Rush, M.J.P.; Ulbrich, A.; Robinson, K.P.; Hutchins, P.D.; Veling, M.T.; et al. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat. Biotechnol. 2016, 34, 1191–1197. [Google Scholar] [CrossRef] [Green Version]
- Valera, M.J.; Boido, E.; Ramos, J.C.; Manta, E.; Radi, R.; Dellacassa, E.; Carrau, F. The Mandelate Pathway, an Alternative to the Phenylalanine Ammonia Lyase Pathway for the Synthesis of Benzenoids in Ascomycete Yeasts. Appl. Environ. Microbiol. 2020, 86, e00701-20. [Google Scholar] [CrossRef] [PubMed]
- Valera, M.J.; Zeida, A.; Boido, E.; Beltran, G.; Torija, M.J.; Mas, A.; Radi, R.; Dellacassa, E.; Carrau, F. Genetic and transcriptomic evidences suggest ARO10 genes are involved in benzenoid biosynthesis by yeast. Yeast 2020, 37, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.P.; Jochem, A.; Johnson, S.E.; Reddy, T.R.; Russell, J.D.; Coon, J.J.; Pagliarini, D.J. Defining intermediates and redun-dancies in coenzyme Q precursor biosynthesis. J. Biol. Chem. 2021, 296, 100643. [Google Scholar] [CrossRef]
- Luengo, A.; Li, Z.; Gui, D.Y.; Sullivan, L.B.; Zagorulya, M.; Do, B.T.; Ferreira, R.; Naamati, A.; Ali, A.; Lewis, C.A.; et al. Increased demand for NAD+ relative to ATP drives aerobic glycolysis. Mol. Cell 2021, 81, 691–707.e6. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.E.; Bentley, R.; Aiyar, A.S.; Dialameh, G.H.; Gold, P.H.; Ramsey, V.G.; Springer, C.M. Benzoate derivates as intermedi-ates in the biosynthesis of the coenzyme Q in the rat. J. Biol. Chem. 1963, 238, 3146–3148. [Google Scholar] [CrossRef]
- Enríquez, J.A.; Sánchez-Cabo, F.; Vázquez, J. Hypothesis Driven versus Hypothesis-free: Filling the Gaps in CoQ Biosynthesis. Cell Metab. 2016, 24, 525–526. [Google Scholar] [CrossRef] [Green Version]
- Mehere, P.; Han, Q.; Lemkul, J.A.; Vavricka, C.J.; Robinson, H.; Bevan, D.R.; Li, J. Tyrosine aminotransferase: Biochemical and structural properties and molecular dynamics simulations. Protein Cell 2010, 1, 1023–1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soubeyrand, E.; Kelly, M.; Keene, S.A.; Bernert, A.C.; Latimer, S.; Johnson, T.S.; Elowsky, C.; Colquhoun, T.A.; Block, A.K.; Basset, G.J. Arabidopsis 4-COUMAROYL-COA LIGASE 8 contributes to the biosynthesis of the benzenoid ring of coenzyme Q in pe-roxisomes. Biochem. J. 2019, 476, 3521–3532. [Google Scholar] [CrossRef]
- Block, A.; Widhalm, J.R.; Fatihi, A.; Cahoon, R.E.; Wamboldt, Y.; Elowsky, C.; Mackenzie, S.A.; Cahoon, E.B.; Chapple, C.; Dudareva, N.; et al. The Origin and Biosynthesis of the Benzenoid Moiety of Ubiquinone (Coenzyme Q) in Arabidopsis. Plant Cell 2014, 26, 1938–1948. [Google Scholar] [CrossRef] [Green Version]
- Soubeyrand, E.; Johnson, T.S.; Latimer, S.; Block, A.; Kim, J.; Colquhoun, T.A.; Butelli, E.; Martin, C.; Wilson, M.A.; Basset, G.J. The Peroxidative Cleavage of Kaempferol Contributes to the Biosynthesis of the Benzenoid Moiety of Ubiquinone in Plants. Plant Cell 2018, 30, 2910–2921. [Google Scholar] [CrossRef] [Green Version]
- Tsao, R. Chemistry and Biochemistry of Dietary Polyphenols. Nutrients 2010, 2, 1231–1246. [Google Scholar] [CrossRef]
- Zhang, H.; Tsao, R. Dietary polyphenols, oxidative stress and antioxidant and anti-inflammatory effects. Curr. Opin. Food Sci. 2016, 8, 33–42. [Google Scholar] [CrossRef]
- Hatia, S.; Septembre-Malaterre, A.; Le Sage, F.; Badiou-Bénéteau, A.; Baret, P.; Payet, B.; Lefebvre d’Hellencourt, C.; Gonthier, M.P. Evaluation of antioxidant properties of major dietary polyphenols and their protective effect on 3T3-L1 preadipocytes and red blood cells exposed to oxidative stress. Free Radic. Res. 2014, 48, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, D.; Rodriguez-Mateos, A.; Spencer, J.P.; Tognolini, M.; Borges, G.; Crozier, A. Dietary (Poly) phenolics in Human Health: Structures, Bioavailability, and Evidence of Protective Effects Against Chronic Diseases. Antiox. Redox Signal. 2013, 18, 1818–1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuršvietienė, L.; Stanevičienė, I.; Mongirdienė, A.; Bernatonienė, J. Multiplicity of effects and health benefits of resveratrol. Medicina 2016, 52, 148–155. [Google Scholar] [CrossRef]
- Fernández-del-Río, L.; Nag, A.; Casado, E.G.; Ariza, J.; Awad, A.M.; Joseph, A.I.; Kwon, O.; Verdin, E.; de Cabo, R.; Schneider, C.; et al. Kaempferol increases levels of coenzyme Q in kidney cells and serves as a biosynthetic ring precursor. Free Radic. Biol. Med. 2017, 110, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Kozłowska, A.; Szostak-Wegierek, D. Flavonoids—food sources and health benefits. Rocz. Państw. Zakładu Hig. 2014, 65, 79–85. [Google Scholar]
- Soubeyrand, E.; Latimer, S.; Bernert, A.C.; Keene, S.A.; Johnson, T.S.; Shin, D.; Block, A.K.; Colquhoun, T.A.; Schäffner, A.R.; Kim, J.; et al. 3-O-glycosylation of kaempferol restricts the supply of the benzenoid precursor of ubiquinone (Coenzyme Q) in Arabidopsis thaliana. Phytochemistry 2021, 186, 112738. [Google Scholar] [CrossRef]
- Fernández-del-Río, L.; Soubeyrand, E.; Basset, G.J.; Clarke, C.F. Metabolism of the Flavonol Kaempferol in Kidney Cells Liber-ates the B-ring to Enter Coenzyme Q Biosynthesis. Molecules 2020, 25, 2955. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-del-Río, L.; Clarke, C.F. Coenzyme Q Biosynthesis: An Update on the Origins of the Benzenoid Ring and Discovery of New Ring Precursors. Metabolites 2021, 11, 385. https://doi.org/10.3390/metabo11060385
Fernández-del-Río L, Clarke CF. Coenzyme Q Biosynthesis: An Update on the Origins of the Benzenoid Ring and Discovery of New Ring Precursors. Metabolites. 2021; 11(6):385. https://doi.org/10.3390/metabo11060385
Chicago/Turabian StyleFernández-del-Río, Lucía, and Catherine F. Clarke. 2021. "Coenzyme Q Biosynthesis: An Update on the Origins of the Benzenoid Ring and Discovery of New Ring Precursors" Metabolites 11, no. 6: 385. https://doi.org/10.3390/metabo11060385