Scalable Extraction of Big Macromolecular Data in Azure Data Lake Environment
Abstract
:1. Introduction
2. Related Works
3. Methods and Technologies
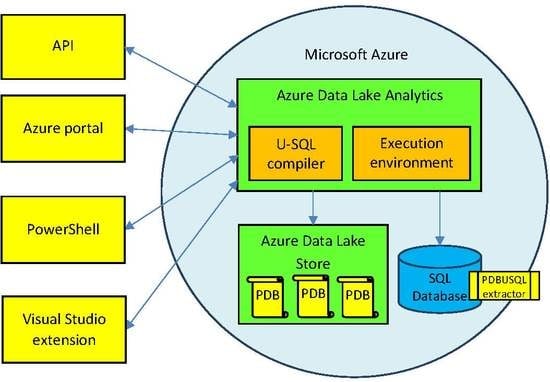
3.1. Extensions to the Azure Data Lake Environment
- Data Lake Store (DLS), which constitutes a petabyte scale, unlimited storage for a domain-related data lake, in which large collections of data located in various files are distributed across many storage servers. This enables performing read operations in parallel and improves the performance of data read operations.
- Data Lake Analytics (DLA), which enables efficient and scalable analysis of data stored in Big Data Lakes by parallelizing the analysis on a distributed infrastructure in the Azure cloud. It provides the U-SQL compiler and distributed execution environment for declarative processing and analysis of data stored in the Data Lake Store.
3.2. Setting Up the Azure Data Lake for Scalable Extraction of Macromolecular Data
- set up the Azure Data Lake environment,
- upload PDB files with macromolecular structures into the Azure Data Lake Store,
- register the PDBUSQLExtractor library in the Azure Data Lake Analytics.
3.3. Modules and Methods for Parsing PDB Files
- ATOM that presents the atomic coordinates for standard amino acids and nucleotides. It also presents the occupancy and temperature factor for each atom.
- HETATM that presents non-polymer or other non-standard chemical coordinates, such as water molecules or atoms presented in HET groups, together with the occupancy and temperature factor for each atom.
- SHEET that is used to identify the position of -sheets in protein molecules.
- HELIX that is used to identify the position of -helices in protein molecules.
- SEQRES that contains a listing of the consecutive chemical components (amino acids for proteins) covalently linked in a linear fashion to form a polymer.


3.4. Optimizations
- decompressed PDB macromolecular files,
- compressed PDB macromolecular files.
- individual macromolecular data files that are extracted in parallel,
- joined, sequential macromolecular data files that are also extracted in parallel.

4. Results
4.1. Experimental Setup
- data set DS1 containing 1475 files,
- data set DS2 containing 14,750 files.
- ATOM,
- SHEET,
- SEQRES.
- decompressed-individual files (DI),
- compressed-individual files (CI),
- decompressed-sequential files (DS),
- compressed-sequential files (CS).
4.2. Execution Times
4.3. Sample Calculations

5. Discussion
6. Availability
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ADL | Azure Data Lake |
| API | Application Programming Interface |
| AUs | allocation units |
| CI | compressed-individual files |
| CLR | common language runtime |
| CS | compressed-sequential files |
| CSV | comma-separated values |
| DI | decompressed-individual files |
| DLA | Data Lake Analytics |
| DLL | Dynamic-Link Library |
| DLS | Data Lake Store |
| DS | decompressed-sequential files |
| DS1 | data set 1 |
| DS2 | data set 2 |
| IMPSMS | In-Memory Protein Structure Management System |
| OS | operating system |
| PDB | Protein Data Bank |
| SDK | Software Development Kit |
| SQL | Structured Query Language |
| TSV | tab-separated values |
| UDO | user-defined operator |
| VMs | virtual machines |
References
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westbrook, J.; Fitzgerald, P. The PDB format, mmCIF, and other data formats. Methods Biochem. Anal. 2003, 44, 161–179. [Google Scholar] [PubMed]
- Bourne, P.E.; Berman, H.M.; McMahon, B.; Watenpaugh, K.D.; Westbrook, J.D.; Fitzgerald, P.M. The macromolecular Crystallographic Information File (mmCIF). Methods Enzymol. 1997, 277, 571–590. [Google Scholar] [PubMed]
- Wesbrook, J.; Ito, N.; Nakamura, H.; Henrick, K.; Berman, H. PDBML: The representation of archival macromolecular structure data in XML. Bioinformatics 2005, 21, 988–992. [Google Scholar] [CrossRef] [PubMed]
- Mrozek, D.; Brozek, M.; Małysiak-Mrozek, B. Parallel implementation of 3D protein structure similarity searches using a GPU and the CUDA. J Mol Model 2014, 20, 2067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, C.; Zuo, Y.; Zou, Q. O-GlcNAcPRED-II: An integrated classification algorithm for identifying O-GlcNAcylation sites based on fuzzy undersampling and a K-means PCA oversampling technique. Bioinformatics 2018, 34, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Masseroli, M.; Canakoglu, A.; Ceri, S. Integration and Querying of Genomic and Proteomic Semantic Annotations for Biomedical Knowledge Extraction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016, 13, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Ceri, S.; Kaitoua, A.; Masseroli, M.; Pinoli, P.; Venco, F. Data Management for Heterogeneous Genomic Datasets. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 14, 1251–1264. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Tang, J.; Zou, Q. Local-DPP: An improved DNA-binding protein prediction method by exploring local evolutionary information. Inf. Sci. 2017, 384, 135–144. [Google Scholar] [CrossRef]
- Hung, C.L.; Lin, C.Y. Open Reading Frame Phylogenetic Analysis on the Cloud. Int. J. Genom. 2013, 2013. [Google Scholar] [CrossRef]
- Macalino, S.J.Y.; Basith, S.; Clavio, N.A.B.; Chang, H.; Kang, S.; Choi, S. Evolution of In Silico Strategies for Protein-Protein Interaction Drug Discovery. Molecules 2018, 23, 1963. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, C.; Lu, K.; Fang, L.; Zhang, Y.; Li, S.; Guo, G.; Du, Y. An Interface for Biomedical Big Data Processing on the Tianhe-2 Supercomputer. Molecules 2017, 22, 2116. [Google Scholar] [CrossRef] [PubMed]
- Mrozek, D. Scalable Big Data Analytics for Protein Bioinformatics Computational Biology; Springer: Berlin, Germany, 2018; Volume 28. [Google Scholar]
- White, T. Hadoop—The Definitive Guide: Storage and Analysis at Internet Scale, 3rd ed.; O-Reilly: Sebastopol, CA, USA, 2012. [Google Scholar]
- Zaharia, M.; Xin, R.S.; Wendell, P.; Das, T.; Armbrust, M.; Dave, A.; Meng, X.; Rosen, J.; Venkataraman, S.; Franklin, M.J.; Ghodsi, A.; Gonzalez, J.; Shenker, S.; Stoica, I. Apache Spark: A Unified Engine for Big Data Processing. Commun. ACM 2016, 59, 56–65. [Google Scholar] [CrossRef]
- Mell, P.; Grance, T. The NIST Definition of Cloud Computing. Special Publication 800-145; 2011. Available online: http://nvlpubs.nist.gov/nistpubs/Legacy/SP/nistspecialpublication800-145.pdf (accessed on 10 October 2017).
- Tina, K.G.; Bhadra, R.; Srinivasan, N. PIC: Protein Interactions Calculator. Nucleic Acids Res. 2007, 35, W473–W476. [Google Scholar] [CrossRef] [PubMed]
- Chourasia, M.; Sastry, G.M.; Sastry, G.N. Aromatic–Aromatic Interactions Database, A2ID: An analysis of aromatic Π-networks in proteins. Int. J. Biol. Macromol. 2011, 48, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Bhattacharyya, R.; Dasgupta, M.; Mandal, S.; Chakrabarti, P. IntGeom: A Server for the Calculation of the Interaction Geometry between Planar Groups in Proteins. J. Proteom. Bioinform. 2009, 2, 60–63. [Google Scholar] [CrossRef]
- Hazelhurst, S. PH2: An Hadoop-based framework for mining structural properties from the PDB database. In Proceedings of the 2010 Annual Research Conference of the South African Institute of Computer Scientists and Information Technologists, Bela Bela, South Africa, 11–13 October 2010; pp. 104–112. [Google Scholar]
- Date, C. An Introduction to Database Systems, 8th ed.; Addison-Wesley: Boston, MA, USA, 2003. [Google Scholar]
- Robillard, D.E.; Mpangase, P.T.; Hazelhurst, S.; Dehne, F. SpeeDB: Fast structural protein searches. Bioinformatics 2015, 31, 3027–3034. [Google Scholar] [CrossRef] [PubMed]
- Małysiak-Mrozek, B.; Żur, K.; Mrozek, D. In-Memory Management System for 3D Protein Macromolecular Structures. Curr. Proteom. 2018, 15, 175–189. [Google Scholar] [CrossRef]
- Stephens, S.M.; Chen, J.Y.; Davidson, M.G.; Thomas, S.; Trute, B.M. Oracle Database 10g: A platform for BLAST search and Regular Expression pattern matching in life sciences. Nucleic Acids Res. 2005, 33, D675–D679. [Google Scholar] [CrossRef] [PubMed]
- BioSQL Homepage. Available online: http://biosql.org/ (accessed on 2 November 2018).
- Prlić, A.; Yates, A.; Bliven, S.E.; Rose, P.W.; Jacobsen, J.; Troshin, P.V.; Chapman, M.; Gao, J.; Koh, C.H.; Foisy, S.; et al. BioJava: An open-source framework for bioinformatics in 2012. Bioinformatics 2012, 28, 2693–2695. [Google Scholar] [PubMed]
- Mrozek, D.; Wieczorek, D.; Małysiak-Mrozek, B.; Kozielski, S. PSS-SQL: Protein Secondary Structure—Structured Query Language. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 1073–1076. [Google Scholar]
- Mrozek, D.; Socha, B.; Kozielski, S.; Małysiak-Mrozek, B. An efficient and flexible scanning of databases of protein secondary structures. J. Intell. Inf. Syst. 2016, 46, 213–233. [Google Scholar] [CrossRef]
- Hammel, L.; Patel, J.M. Searching on the Secondary Structure of Protein Sequences. In VLDB’02: Proceedings of the 28th International Conference on Very Large Databases, Hong Kong, China, 20–23 August 2002; Bernstein, P.A., Ioannidis, Y.E., Ramakrishnan, R., Papadias, D., Eds.; Morgan Kaufmann: San Francisco, CA, USA, 2002; pp. 634–645. [Google Scholar] [Green Version]
- Tata, S.; Friedman, J.S.; Swaroop, A. Declarative Querying for Biological Sequences. In Proceedings of the 22nd International Conference on Data Engineering (ICDE’06), Atlanta, GA, USA, 3–7 April 2006; pp. 87–98. [Google Scholar]
- Mrozek, D.; Małysiak-Mrozek, B.; Adamek, R. P3D-SQL: Extending Oracle PL/SQL Capabilities Towards 3D Protein Structure Similarity Searching. In Bioinformatics and Biomedical Engineering; Ortuño, F., Rojas, I., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; Volume 9043, pp. 548–556. [Google Scholar]
- Hung, C.L.; Lin, Y.L. Implementation of a Parallel Protein Structure Alignment Service on Cloud. Int. J. Genom. 2013. [Google Scholar] [CrossRef]
- Holm, L.; Kaariainen, S.; Rosenstrom, P.; Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 2008, 24, 2780–2781. [Google Scholar] [CrossRef] [Green Version]
- Gibrat, J.; Madej, T.; Bryant, S. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 1996, 6, 377–385. [Google Scholar] [CrossRef]
- Mrozek, D.; Daniłowicz, P.; Małysiak-Mrozek, B. HDInsight4PSi: Boosting performance of 3D protein structure similarity searching with HDInsight clusters in Microsoft Azure cloud. Inf. Sci. 2016, 349, 77–101. [Google Scholar] [CrossRef]
- Mrozek, D.; Suwała, M.; Małysiak-Mrozek, B. High-throughput and scalable protein function identification with Hadoop and Map-only pattern of the MapReduce processing model. J. Knowl. Inf. Syst. 2018, 1–34. [Google Scholar] [CrossRef]
- Małysiak-Mrozek, B.; Daniłowicz, P.; Mrozek, D. Efficient 3D Protein Structure Alignment on Large Hadoop Clusters in Microsoft Azure Cloud. Beyond Databases, Architectures and Structures. In Facing the Challenges of Data Proliferation and Growing Variety; Kozielski, S., Mrozek, D., Kasprowski, P., Małysiak-Mrozek, B., Kostrzewa, D., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 33–46. [Google Scholar]
- Mrozek, D.; Małysiak-Mrozek, B.; Kłapciński, A. Cloud4Psi: Cloud computing for 3D protein structure similarity searching. Bioinformatics 2014, 30, 2822–2825. [Google Scholar] [CrossRef]
- Mrozek, D. High-Performance Computational Solutions in Protein Bioinformatics; SpringerBriefs in Computer Science; Springer: Cham, Switzerland, 2014. [Google Scholar]
- Mrozek, D.; Kutyła, T.; Małysiak-Mrozek, B. Accelerating 3D Protein Structure Similarity Searching on Microsoft Azure Cloud with Local Replicas of Macromolecular Data. In Parallel Processing and Applied Mathematics—PPAM 2015; Wyrzykowski, R., Ed.; Lecture Notes in Computer Science; Springer: Heidelberg, Germany, 2016; Volume 9574, pp. 1–12. [Google Scholar]
- Hung, C.L.; Hua, G.J. Cloud Computing for Protein-Ligand Binding Site Comparison. Biomed. Res. Int. 2013. [Google Scholar] [CrossRef]
- Mrozek, D.; Gosk, P.; Małysiak-Mrozek, B. Scaling Ab Initio Predictions of 3D Protein Structures in Microsoft Azure Cloud. J. Grid Comput. 2015, 13, 561–585. [Google Scholar] [CrossRef]
- Zou, Q.; Wan, S.; Ju, Y.; Tang, J.; Zeng, X. Pretata: Predicting TATA binding proteins with novel features and dimensionality reduction strategy. BMC Syst. Biol. 2016, 10, 114. [Google Scholar] [CrossRef]
- Microsoft Azure. Overview of Microsoft Azure Data Lake Analytics. Available online: https://docs.microsoft.com/en-us/azure/data-lake-analytics/data-lake-analytics-overview (accessed on 7 November 2018).
- Microsoft Azure. Azure Data Lake Analytics Documentation. Available online: https://docs.microsoft.com/en-us/azure/data-lake-analytics/ (accessed on 18 December 2018).
- Protein Data Bank Contents Guide. Atomic Coordinate Entry Format Description, Version 3.3. Available online: http://www.wwpdb.org/documentation/file-format-content/format33/v3.3.html (accessed on 7 November 2018).










| Azure Data Lake | PC | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| # AUs | 1 | 2 | 8 | 32 | 128 | 512 | 1024 | 1475 | 1 |
| DI-ATOM | 12,400 | 6225 | 1606 | 459 | 171 | 100 | 94 | 94 | 464 |
| CI-ATOM | 11,740 | 5892 | 1526 | 441 | 170 | 103 | 97 | 97 | 516 |
| DI-SHEET | 12,225 | 6115 | 1548 | 412 | 128 | 60 | 60 | 60 | – |
| CI-SHEET | 11,793 | 5898 | 1484 | 391 | 120 | 60 | 56 | 56 | – |
| DI-SEQRES | 11,831 | 5922 | 1494 | 393 | 129 | 68 | 61 | 61 | – |
| CI-SEQRES | 11,746 | 5879 | 1482 | 396 | 124 | 57 | 57 | 57 | – |
| ATOM | SHEET | SEQRES | |
|---|---|---|---|
| DS | 116 | 46 | 51 |
| CS | 116 | 46 | 51 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mrozek, D.; Dąbek, T.; Małysiak-Mrozek, B. Scalable Extraction of Big Macromolecular Data in Azure Data Lake Environment. Molecules 2019, 24, 179. https://doi.org/10.3390/molecules24010179
Mrozek D, Dąbek T, Małysiak-Mrozek B. Scalable Extraction of Big Macromolecular Data in Azure Data Lake Environment. Molecules. 2019; 24(1):179. https://doi.org/10.3390/molecules24010179
Chicago/Turabian StyleMrozek, Dariusz, Tomasz Dąbek, and Bożena Małysiak-Mrozek. 2019. "Scalable Extraction of Big Macromolecular Data in Azure Data Lake Environment" Molecules 24, no. 1: 179. https://doi.org/10.3390/molecules24010179