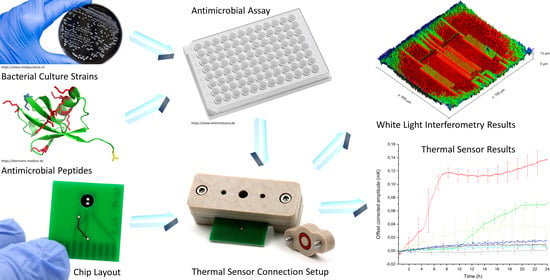
A Real-Time Thermal Sensor System for Quantifying the Inhibitory Effect of Antimicrobial Peptides on Bacterial Adhesion and Biofilm Formation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Culture Strains and Antibiotic Susceptibility Testing
2.2. Antimicrobial Peptides (AMPs)
2.3. Antimicrobial Assay—Determination of Minimal Inhibitory Concentrations (MICs) of Different AMPs
2.4. Whole Genome Sequencing
2.5. Thermal Sensor Fabrication Process
2.6. Chip Layout
2.7. Sample Preparation for Sensor Application
2.8. Cleaning and Sterilization Protocol of the Thermal Sensor and the Sensor Connection Setup
2.9. White Light Interferometry
2.10. Thermal Sensor Connection Setup
3. Results and Discussion
3.1. Results of MIC Experiments and Whole-Genome Sequencing
3.2. White Light Interferometry Results of Biofilm Thickness Measurements
3.3. Thermal Sensor Results
3.3.1. Thermal Sensor Results of Inhibition of Protamine
3.3.2. Thermal Sensor Results of Inhibition of OH-CATH-30
4. Conclusions
5. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaynes, R. The Discovery of Penicillin—New Insights After More Than 75 Years of Clinical Use. Emerg. Infect. Dis. 2017, 23, 849–853. [Google Scholar] [CrossRef]
- Bowler, P.G. Antibiotic resistance and biofilm tolerance: A combined threat in the treatment of chronic infections. J. Wound Care 2018, 27, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; E Kretzschmar, M.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Marshall, B.M.; Levy, S.B. Food Animals and Antimicrobials: Impacts on Human Health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antão, E.-M.; Vincze, S.; Hanke, R.; Klimmek, L.; Suchecka, K.; Lübke-Becker, A.; Wieler, L.H. Antibiotic resistance, the 3As and the road ahead. Gut Pathog. 2018, 10, 1–9. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Genet. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- da Silva Araújo, P.A. Biofilm Control with Antimicrobial Agents: The Role of the Exopolymeric Matrix; University of Porto: Porto, Portugal, 2014. [Google Scholar]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef]
- Mah, T.-F.C.; O’Toole, A.G. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as Complex Differentiated Communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, K. Riddle of Biofilm Resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adlhart, C.; Verran, J.; Azevedo, N.F.; Olmez, H.; Keinänen-Toivola, M.M.; Gouveia, I.; Melo, L.F.; Crijns, F. Surface modifications for antimicrobial effects in the healthcare setting: A critical overview. J. Hosp. Infect. 2018, 99, 239–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Melchior, M.; Vaarkamp, H.; Fink-Gremmels, J. Biofilms: A role in recurrent mastitis infections? Veter. J. 2006, 171, 398–407. [Google Scholar] [CrossRef]
- Mainau, E.; Temple, D.; Manteca, X. Welfare issues related to mastitis in dairy cows. Farm Anim. Welf. Educ. Cent. 2014, 10, 2. [Google Scholar]
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012, 65, 183–195. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, K. Why and how are peptide-lipid interactions utilized for self defence? Biochem. Soc. Trans. 2001, 29, 598–601. [Google Scholar] [CrossRef] [Green Version]
- Reddy, K.; Yedery, R.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef]
- Rapsch, K.; Bier, F.F.; Tadros, M.; Von Nickisch-Rosenegk, M. Identification of Antimicrobial Peptides and Immobilization Strategy Suitable for a Covalent Surface Coating with Biocompatible Properties. Bioconjugate Chem. 2014, 25, 308–319. [Google Scholar] [CrossRef]
- Cole, A.M.; Ganz, T. Human Antimicrobial Peptides: Analysis and Application. Biotechniques 2000, 29, 822–831. [Google Scholar] [CrossRef]
- Hancock, R.E.; Diamond, G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000, 8, 402–410. [Google Scholar] [CrossRef]
- Chung, P.Y.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 2017, 50, 405–410. [Google Scholar] [CrossRef]
- Ebenhan, T.; Gheysens, O.; Kruger, H.G.; Zeevaart, J.R.; Sathekge, M.M. Antimicrobial Peptides: Their Role as Infection-Selective Tracers for Molecular Imaging. BioMed. Res. Int. 2014, 2014, 1–15. [Google Scholar] [CrossRef]
- Park, S.-C.; Park, Y.; Hahm, K.-S. The Role of Antimicrobial Peptides in Preventing Multidrug-Resistant Bacterial Infections and Biofilm Formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolas, P. Multifunctional host defense peptides: Intracellular-targeting antimicrobial peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nat. Cell Biol. 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, e2437. [Google Scholar] [CrossRef]
- Neu, T.R.; Lawrence, J.R. Innovative techniques, sensors, and approaches for imaging biofilms at different scales. Trends Microbiol. 2015, 23, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.W.; Meyer, M.T.; Berkovich, A.; Subramanian, S.; Iliadis, A.A.; Bentley, W.E.; Ghodssi, R. A surface acoustic wave biofilm sensor integrated with a treatment method based on the bioelectric effect. Sens. Actuators A Phys. 2016, 238, 140–149. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.W.; Sardari, S.E.; Meyer, M.T.; Iliadis, A.A.; Wu, H.C.; Bentley, W.E.; Ghodssi, R. An ALD aluminum oxide passivated Surface Acoustic Wave sensor for early biofilm detection. Sens. Actuators B Chem. 2012, 163, 136–145. [Google Scholar] [CrossRef]
- Schmid, L.; Franke, T. Real-time size modulation and synchronization of a microfluidic dropmaker with pulsed surface acoustic waves (SAW). Sci. Rep. 2018, 8, 4541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pires, L.; Sachsenheimer, K.; Kleintschek, T.; Waldbaur, A.; Schwartz, T.; Rapp, B.E. Online monitoring of biofilm growth and activity using a combined multi-channel impedimetric and amperometric sensor. Biosens. Bioelectron. 2013, 47, 157–163. [Google Scholar] [CrossRef]
- Ghafar-Zadeh, E.; Sawan, M.; Shabani, A.; Zourob, M.; Chodavarapu, V. Bacteria growth monitoring through an on-chip capacitive sensor. In Proceedings of the 2008 IEEE 14th International Mixed-Signals Sensors, and Systems Test Workshop, Vancouver, BC, Canada, 18–20 June 2008; pp. 1–4. [Google Scholar]
- Reipa, V.; Almeida, J.; Cole, K.D. Long-term monitoring of biofilm growth and disinfection using a quartz crystal microbalance and reflectance measurements. J. Microbiol. Methods 2006, 66, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Lerchner, J.; Wolf, A.; Buchholz, F.; Mertens, F.; Neu, T.; Harms, H.; Maskow, T. Miniaturized calorimetry—A new method for real-time biofilm activity analysis. J. Microbiol. Methods 2008, 74, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Cui, H.; Xiong, X.; Chen, J.; Wang, Y.; Shen, J.; Luo, Y.; Chen, L. QCM-nanomagnetic beads biosensor for lead ion detection. Analyst 2017, 143, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Mariana, F.; Buchholz, F.; Lerchner, J.; Neu, T.R.; Harms, H.; Maskow, T. Chip-calorimetric monitoring of biofilm eradication with antibiotics provides mechanistic information. Int. J. Med. Microbiol. 2013, 303, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, M.; Stemme, G.; Kittilsland, G.; Pedersen, K. A silicon sensor for measurement of liquid flow and thickness of fouling biofilms. Sens. Actuators 1988, 13, 203–221. [Google Scholar] [CrossRef]
- Fischer, M.; Wahl, M.; Friedrichs, G. Design and field application of a UV-LED based optical fiber biofilm sensor. Biosens. Bioelectron. 2012, 33, 172–178. [Google Scholar] [CrossRef]
- Kwak, Y.H.; Lee, J.; Lee, J.; Kwak, S.H.; Oh, S.; Paek, S.-H.; Ha, U.-H.; Seo, S. A simple and low-cost biofilm quantification method using LED and CMOS image sensor. J. Microbiol. Methods 2014, 107, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Kristich, C.J.; Li, Y.-H.; Cvitkovitch, D.G.; Dunny, G.M. Esp-Independent Biofilm Formation by Enterococcus faecalis. J. Bacteriol. 2004, 186, 154–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zibaii, M.I.; Kazemi, A.; Latifi, H.; Azar, M.K.; Hosseini, S.M.; Ghezelaiagh, M.H. Measuring bacterial growth by refractive index tapered fiber optic biosensor. J. Photochem. Photobiol. B Biol. 2010, 101, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Romero, D.; Behrmann, O.; Dame, G.; Urban, G. Dynamic thermal sensor for biofilm monitoring. Sens. Actuators A Phys. 2014, 213, 43–51. [Google Scholar] [CrossRef]
- Ernst, H.; Jachimowicz, A.; Urban, G. Dynamic thermal sensor—Principles in MEMS for fluid characterization. IEEE Sens. J. 2001, 1, 361–367. [Google Scholar] [CrossRef]
- Ernst, H.; Jachimowicz, A.; Urban, G.A. High resolution flow characterization in Bio-MEMS. Sens. Actuators A Phys. 2002, 100, 54–62. [Google Scholar] [CrossRef]
- Çubukçu, A.S.; Romero, D.F.R.; Urban, G.A. A dynamic thermal flow sensor for simultaneous measurement of thermal conductivity and flow velocity of gases. Sens. Actuators A Phys. 2014, 208, 73–87. [Google Scholar] [CrossRef]
- Kuntner, J.; Kohl, F.; Jakoby, B. Simultaneous thermal conductivity and diffusivity sensing in liquids using a micromachined device. Sens. Actuators A Phys. 2006, 130, 62–67. [Google Scholar] [CrossRef]
- Beigelbeck, R.; Nachtnebel, H.; Kohl, F.; Jakoby, B. A novel measurement method for the thermal properties of liquids by utilizing a bridge-based micromachined sensor. Meas. Sci. Technol. 2011, 22. [Google Scholar] [CrossRef]
- Romero, D.R.; Kogan, K.; Cubukcu, A.S.; Urban, G.A. Simultaneous flow and thermal conductivity measurement of gases utilizing a calorimetric flow sensor. Sens. Actuators A Phys. 2013, 203, 225–233. [Google Scholar] [CrossRef]
- Watts, J.L. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals: Approved Standard; Clinical and Laboratory Standards Institute: Annapolis Junction, MD, USA, 2013. [Google Scholar]
- Feßler, A.; Böttner, A.; Fehr, M.; Kasper, H.; Kehrenberg, C.; Kietzmann, M. Mikrotiterplattenlayouts für Kleintiere, Großtiere und Mastitis: Aktualisierung der Layouts des DVG-Arbeitskreises Antibiotikaresistenz. Dtsch. Tierärzteblatt 2017, 65, 472–481. [Google Scholar]
- Miller, B.F.; Abrams, R.; Dorfman, A.; Klein, M. Antibacterial Properties of Protamine and Histone. Am. Assoc. Adv. Sci. Stable 1942, 96, 428–430. [Google Scholar] [CrossRef]
- Tadros, M. Method for the Production of Protamine. U.S. Patent 7,544,483 B2, 9 June 2009. [Google Scholar]
- Zhang, Y.; Zhao, H.; Yu, G.-Y.; Liu, X.-D.; Shen, J.-H.; Lee, W.-H.; Zhang, Y. Structure–function relationship of king cobra cathelicidin. Peptides 2010, 31, 1488–1493. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Munby, M.; Fujiki, J.; Aoki, K.; Kawaguchi, C.; Nakamura, K.; Nakamura, T.; Sasaki, M.; Sato, T.; Usui, M.; Sawa, H.; et al. Whole-Genome Sequence of Fluoroquinolone-Resistant Escherichia coli HUE1, Isolated in Hokkaido, Japan. Microbiol. Resour. Announc. 2020, 9, 3491–3500. [Google Scholar] [CrossRef]
- Manson, J.-A.E.; Seferis, J.C. Autoclave Processing of PEEK/Carbon Fiber Composites. J. Thermoplast. Compos. Mater. 1989, 2, 34–49. [Google Scholar] [CrossRef]
- Udachan, L.A.; Shivaprasad, S.M.; Ashrit, P.V.; Angadi, M.A. Electrical resistivity and temperature coefficient of resistance of vacuum evaporated thin chromium films. Phys. Status Solidi 1980, 60, K191–K194. [Google Scholar] [CrossRef]
- Gottlieb, C.T.; Thomsen, L.E.; Ingmer, H.; Mygind, P.H.; Kristensen, H.-H.; Gram, L. Antimicrobial peptides effectively kill a broad spectrum of Listeria monocytogenes and Staphylococcus aureus strains independently of origin, sub-type, or virulence factor expression. BMC Microbiol. 2008, 8, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, L.T.; Gill, T. Solubility and antimicrobial efficacy of protamine on Listeria monocytogenes and Escherichia coli as influenced by pH. J. Appl. Microbiol. 2000, 88, 1049–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinho, M.G.; De Lencastre, H.; Tomasz, A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. USA 2001, 98, 10886–10891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peschel, A.; Jack, R.W.; Otto, M.; Collins, L.V.; Staubitz, P.; Nicholson, G.; Kalbacher, H.; Nieuwenhuizen, W.F.; Jung, G.; Tarkowski, A.; et al. Staphylococcus aureus Resistance to Human Defensins and Evasion of Neutrophil Killing via the Novel Virulence Factor Mprf Is Based on Modification of Membrane Lipids with l-Lysine. J. Exp. Med. 2001, 193, 1067–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes Romero, D.F. Development of a Medium Independent Flow Measurement Technique Based on Oscillatory Thermal Excitation; Universität Freiburg: Freiburg, Germany, 2014. [Google Scholar]
- Steinhart, J.S.; Hart, S.R. Calibration curves for thermistors. Deep Sea Res. Oceanogr. Abstr. 1968, 15, 497–503. [Google Scholar] [CrossRef]
- Characklis, W.G.; Nevimons, M.J.; Picologlou, B.F. Influence of Fouling Biofilms on Heat Transfer. Heat Transf. Eng. 1981, 3, 23–37. [Google Scholar] [CrossRef]
- Li, M.; Cha, D.J.; Lai, Y.; Villaruz, A.E.; Sturdevant, D.E.; Otto, M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 2007, 66, 1136–1147. [Google Scholar] [CrossRef] [PubMed]
- Stumpe, S.; Schmid, R.; Stephens, D.L.; Georgiou, G.; Bakker, E.P. Identification of OmpT as the Protease That Hydrolyzes the Antimicrobial Peptide Protamine before It Enters Growing Cells of Escherichia coli. J. Bacteriol. 1998, 180, 4002–4006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, B.-Y.; Varadarajan, N.; Li, H.; Rodriguez, S.; Iverson, B.L.; Georgiou, G. Substrate Specificity of the Escherichia coli Outer Membrane Protease OmpP. J. Bacteriol. 2006, 189, 522–530. [Google Scholar] [CrossRef] [Green Version]
- Koo, S.-P.; Bayer, A.S.; Yeaman, M.R. Diversity in Antistaphylococcal Mechanisms among Membrane-Targeting Antimicrobial Peptides. Infect. Immun. 2001, 69, 4916–4922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correa, W.; Heinbockel, L.; Behrends, J.; Kaconis, Y.; Barcena-Varela, S.; Gutsmann, T.; Mauss, K.; Schürholz, T.; Schromm, A.B.; De Tejada, G.M.; et al. Antibacterial action of synthetic antilipopolysaccharide peptides (SALP) involves neutralization of both membrane-bound and free toxins. FEBS J. 2019, 286, 1576–1593. [Google Scholar] [CrossRef]
- Li, Y.; Wei, S.; Wu, J.; Jasensky, J.; Xi, C.; Li, H.; Xu, Y.; Wang, Q.; Marsh, E.N.G.; Brooks, C.L.; et al. Effects of Peptide Immobilization Sites on the Structure and Activity of Surface-Tethered Antimicrobial Peptides. J. Phys. Chem. C 2015, 119, 7146–7155. [Google Scholar] [CrossRef]
- Assoni, L.; Milani, B.; Carvalho, M.R.; Nepomuceno, L.N.; Waz, N.T.; Guerra, M.E.S.; Converso, T.R.; Darrieux, M. Resistance Mechanisms to Antimicrobial Peptides in Gram-Positive Bacteria. Front. Microbiol. 2020, 11, 593215. [Google Scholar] [CrossRef] [PubMed]
- Maria-Neto, S.; De Almeida, K.C.; Macedo, M.L.R.; Franco, O.L. Understanding bacterial resistance to antimicrobial peptides: From the surface to deep inside. Biochim. Biophys. Acta (BBA) Biomembr. 2015, 1848, 3078–3088. [Google Scholar] [CrossRef] [Green Version]
- Henderson, J.C.; Fage, C.D.; Cannon, J.R.; Brodbelt, J.S.; Keatinge-Clay, A.T.; Trent, M.S. Antimicrobial Peptide Resistance of Vibrio cholerae Results from an LPS Modification Pathway Related to Nonribosomal Peptide Synthetases. ACS Chem. Biol. 2014, 9, 2382–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aspedon, A.; Groisman, E.A. The antibacterial action of protamine: Evidence for disruption of cytoplasmic membrane energization in Salmonella typhimurium. Microbiology 1996, 142, 3389–3397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marín-Medina, N.; Mescola, A.; Alessandrini, A. Effects of the peptide Magainin H2 on Supported Lipid Bilayers studied by different biophysical techniques. Biochim. Biophys. Acta (BBA) Biomembr. 2018, 1860, 2635–2643. [Google Scholar] [CrossRef]
- Mescola, A.; Marín-Medina, N.; Ragazzini, G.; Accolla, M.; Alessandrini, A. Magainin-H2 effects on the permeabilization and mechanical properties of giant unilamellar vesicles. J. Colloid Interface Sci. 2019, 553, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, V. Impact of environmental biofilms: Industrial components and its remediation. J. Basic Microbiol. 2020, 60, 198–206. [Google Scholar] [CrossRef]







| Measurement Method | + Advantages | − Disadvantages | Ref. |
|---|---|---|---|
| Surface acoustic waves (SAWs) | + high sensitivity + easy to produce + low cost | − limited height − measurement − limited reusability | [30,31,32] |
| Electrochemical impedance spectroscopy (EIS) | + non-invasive + easy to integrate | − no long-term stability − limited reusability − baseline drift − low sensitivity | [34,35,36] |
| Quartz crystal microbalance (QCM) | + high sensitivity + label free | − temperature sensitive − affected by medium turbidity | [35,37] |
| Isothermal micro-calorimetry (IMC) | + metabolic information + non-invasive + non-destructive | − long settling time − heat source indistinguishable | [36,38] |
| Time-invariant heat transfer | + easy to prepare | − requires knowledge about thermal properties | [39] |
| Optical detection | + easy to handle | − no thickness measurement − time consuming − affected by medium turbidity | [35,40,41,42] |
| Antibiotic | Concentration in µg/mL |
|---|---|
| Penicillin G | 0.125–8 |
| Ampicillin | 4–16 |
| Cefazolin | 4–32 |
| Cefoperazone | 2–16 |
| Cefquinome | 1–8 |
| Oxacillin | 1–4 |
| Pirlimycin | 1–4 |
| Erythromycin | 0.125–4 |
| Marbofloxacin | 0.25–2 |
| Amoxicillin/clavulanic acid | 4/2–32/16 |
| Kanamycin/cefalexin | 4/0.4–32/3.2 |
| AMP | Sequence (Primary Structure) Molecular Weight (g/mol) Degree of Purity (%) |
|---|---|
| Protamine | MPRRRRSSSRPVRRRRRPRVSRRRRRRGGRRRR |
| 4381.24 | |
| 96.94 | |
| OH-CATH-30 | KFFKKLKNSVKKRAKKFFKKPRVIGVSIPF |
| 3595.55 | |
| 99.44 |
| Bacterial Strains | Protamine MIC in µg/mL | OH-CATH-30 MIC in µg/mL |
|---|---|---|
| E. coli (ATCC25922) | 31.25 | 15.63 |
| S. aureus (ATCC29213) | 15.63 | ≥1000 |
| E. coli (IMT37453) | 62.5 | 31.25 |
| S. aureus (IMT37556) | 31.25 | 250 |
| Bacterial Strains | Max. Value (Z-axis) in µm | Mean Value (Z-axis) in µm |
|---|---|---|
| E. coli (ATCC25922) | 12.14 ± 1.80 | 2.66 ± 1.12 |
| S. aureus (ATCC29213) | 16.58 ± 7.12 | 8.61 ± 5.07 |
| E. coli (IMT37453) | 21.48 ± 2.06 | 11.70 ± 1.43 |
| S. aureus (IMT37556) | 22.31 ± 1.00 | 12.97 ± 0.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wieland, T.; Assmann, J.; Bethe, A.; Fidelak, C.; Gmoser, H.; Janßen, T.; Kotthaus, K.; Lübke-Becker, A.; Wieler, L.H.; Urban, G.A. A Real-Time Thermal Sensor System for Quantifying the Inhibitory Effect of Antimicrobial Peptides on Bacterial Adhesion and Biofilm Formation. Sensors 2021, 21, 2771. https://doi.org/10.3390/s21082771
Wieland T, Assmann J, Bethe A, Fidelak C, Gmoser H, Janßen T, Kotthaus K, Lübke-Becker A, Wieler LH, Urban GA. A Real-Time Thermal Sensor System for Quantifying the Inhibitory Effect of Antimicrobial Peptides on Bacterial Adhesion and Biofilm Formation. Sensors. 2021; 21(8):2771. https://doi.org/10.3390/s21082771
Chicago/Turabian StyleWieland, Tobias, Julia Assmann, Astrid Bethe, Christian Fidelak, Helena Gmoser, Traute Janßen, Krishan Kotthaus, Antina Lübke-Becker, Lothar H. Wieler, and Gerald A. Urban. 2021. "A Real-Time Thermal Sensor System for Quantifying the Inhibitory Effect of Antimicrobial Peptides on Bacterial Adhesion and Biofilm Formation" Sensors 21, no. 8: 2771. https://doi.org/10.3390/s21082771