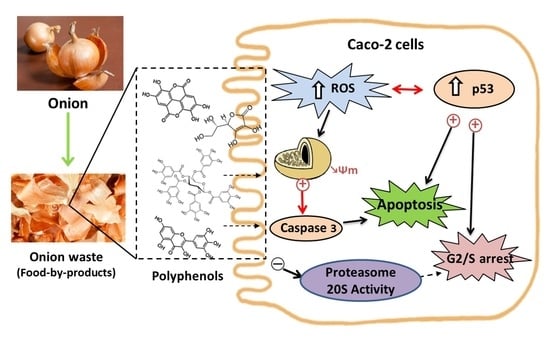
Valorization of Onion Waste by Obtaining Extracts Rich in Phenolic Compounds and Feasibility of Its Therapeutic Use on Colon Cancer
Abstract
:1. Introduction
2. Material and Methods
2.1. Extracts
2.2. Characterization of Phenolic Compounds of Onions Extracts by HPLC-DAD
2.3. Antioxidant Capacity of Onion Extract by DPPH, ABTS and FRAP
2.4. Determination of Total Phenolic and Total Flavonoids Content
2.5. Biological Studies
2.5.1. Cell Proliferation Assay and IC50Values
2.5.2. Propidium Iodide Staining of DNA Content and Cell Cycle Analysis
2.5.3. Measurements of Apoptosis
2.5.4. Mitochondrial Membrane Potential Assay
2.5.5. Determination of P53 and Caspase 3 Activity
2.5.6. Determination of Proteasome Activity
2.5.7. Determination of Intracellular Levels of Reactive Oxygen Species (ROS)
2.5.8. Theoretical Absorption Percentage and Caco-2 Permeability of Individual Phenolic Compounds
2.6. Statistical Analysis
3. Results and Discussion
3.1. Phenolic Composition and Antioxidant Activity in Onion WasteExtracts
3.2. Biological Studies with OnionWasteExtract
3.2.1. Antiproliferative Effect and Cell Cycle Arrest
3.2.2. Type of Cell Death
3.2.3. Activity of Proteasome 20 S Subunit
3.2.4. Redox Activity on Colon Cancer Cells
3.2.5. Redox Activity on a Model of Intestinal Barrier
3.3. Theoretical Absorption of Individual Phenolic Compounds (Based on Lipinnski Parameters)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ren, F.; Nian, Y.; Perussello, C.A. Effect of Storage, Food Processing and Novel Extraction Technologies on Onions Flavonoid Content: A Review. Food Res. Int. 2020, 132, 108953. [Google Scholar] [CrossRef] [PubMed]
- González-de-Peredo, A.V.; Vázquez-Espinosa, M.; Espada-Bellido, E.; Carrera, C.; Ferreiro-González, M.; Barbero, G.F.; Palma, M. Flavonol Composition and Antioxidant Activity of Onions (Allium cepa L.) Based on the Development of New Analytical Ultrasound-assisted Extraction Methods. Antioxidants 2021, 10, 273. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.S.; Almeida, D.P.F.; Simal-Gándara, J.; Pérez-Gregorio, M.R. Onions: A source of flavonoids. In Flavonoids—From Biosynthesis to Human Health; Justino, G.C., Ed.; IntechOpen: London, UK, 2017; pp. 439–471. [Google Scholar] [CrossRef] [Green Version]
- Campone, L.; Celano, R.; Piccinelli, A.L.; Pagano, I.; Carabetta, S.; di Sanzo, R.; Russo, M.; Ibañez, E.; Cifuentes, A.; Rastrelli, L. Response Surface Methodology to Optimize Supercritical Carbon Dioxide/Co-Solvent Extraction of Brown Onion Skin by-Product as Source of Nutraceutical Compounds. Food Chem. 2018, 269, 495–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benito-Román, Ó.; Blanco, B.; Sanz, M.T.; Beltrán, S. Subcritical Water Extraction of Phenolic Compounds from Onion Skin Wastes (Allium cepa cv. Horcal): Effect of Temperature and Solvent Properties. Antioxidants 2020, 9, 233. [Google Scholar] [CrossRef]
- Akdeniz, B.; Sumnu, G.; Sahin, S. Microencapsulation of Phenolic Compounds Extracted from Onion (Allium cepa) Skin. J. Food Process. Preserv. 2018, 42, e13648. [Google Scholar] [CrossRef]
- Yoo, K.S.; Lee, E.J.; Patil, B.S. Changes in flavor precursor, pungency, and sugar contents in short-day onion bulbs during five month storage at various temperatures and controlled-atmosphere. J. Food Sci. 2012, 77, C216–C221. [Google Scholar] [CrossRef]
- Hossain, M.B.; Lebelle, J.; Birsan, R.; Rai, D.K. Enrichment and Assessment of the Contributions of the Major Polyphenols to the Total Antioxidant Activity of Onion Extracts: A Fractionation by Flash Chromatography Approach. Antioxidants 2018, 7, 175. [Google Scholar] [CrossRef] [Green Version]
- Ren, F.; Reilly, K.; Gaffney, M.; Kerry, J.P.; Hossain, M.; Rai, D.K. Evaluation of Polyphenolic Content and Antioxidant Activity in Two Onion Varieties Grown under Organic and Conventional Production Systems. J. Sci. Food Agric. 2017, 97, 2982–2990. [Google Scholar] [CrossRef]
- Slimestad, R.; Fossen, T.; Vågen, I.M. Onions: A Source of Unique Dietary Flavonoids. J. Agric. Food Chem. 2007, 55, 10067–10080. [Google Scholar] [CrossRef]
- Søltoft, M.; Christensen, J.H.; Nielsen, J.; Knuthsen, P. Pressurised Liquid Extraction of Flavonoids in Onions. Method Development and Validation. Talanta 2009, 80, 269–278. [Google Scholar] [CrossRef]
- Brglez Mojzer, E.; Knez Hrnčič, M.; Škerget, M.; Knez, Ž.; Bren, U. Polyphenols: Extraction Methods, Antioxidative Action, Bioavailability and Anticarcinogenic Effects. Molecules 2016, 21, 901. [Google Scholar] [CrossRef]
- Assefa, A.D.; Jeong, Y.J.; Kim, D.J.; Jeon, Y.A.; Lee, J.R.; Ko, H.C.; Baek, H.J.; Sung, J.S. Assessing Phenolic Content and Antioxidant Potential Diversity in Allium Plants Using Multivariate Data Analysis. Hortic. Environ. Biotechnol. 2018, 59, 759–773. [Google Scholar] [CrossRef]
- Wang, T.Y.; Li, Q.; Bi, K. Bioactive Flavonoids in Medicinal Plants: Structure, Activity and Biological Fate. Asian J. Pharm. Sci. 2018, 13, 12–23. [Google Scholar] [CrossRef]
- Sunil, C.; Xu, B. An Insight into the Health-Promoting Effects of Taxifolin (Dihydroquercetin). Phytochemistry 2019, 166, 112066. [Google Scholar] [CrossRef]
- Polera, N.; Badolato, M.; Perri, F.; Carullo, G.; Aiello, F. Quercetin and Its Natural Sources in Wound Healing Management. Curr. Med. Chem. 2019, 26, 5825–5848. [Google Scholar] [CrossRef]
- Carullo, G.; Cappello, A.R.; Frattaruolo, L.; Badolato, M.; Armentano, B.; Aiello, F. Quercetin and Derivatives: Useful Tools in Inflammation and Pain Management. Future Med. Chem. 2017, 9, 79–93. [Google Scholar] [CrossRef]
- Harris, S.; Brunton, N.; Tiwari, U.; Cummins, E. Human Exposure Modelling of Quercetin in Onions (Allium cepa L.) Following Thermal Processing. Food Chem. 2015, 187, 135–139. [Google Scholar] [CrossRef]
- Pang, B.; Xu, X.; Lu, Y.; Jin, H.; Yang, R.; Jiang, C.; Shao, D.; Liu, Y.; Shi, J. Prediction of New Targets and Mechanisms for Quercetin in the Treatment of Pancreatic Cancer, Colon Cancer, and Rectal Cancer. Food Funct. 2019, 10, 5339–5349. [Google Scholar] [CrossRef]
- Ulusoy, H.G.; Sanlier, N. A Minireview of Quercetin: From Its Metabolism to Possible Mechanisms of Its Biological Activities. Crit. Rev. Food Sci. Nutr. 2020, 60, 3290–3303. [Google Scholar] [CrossRef]
- Tang, S.M.; Deng, X.T.; Zhou, J.; Li, Q.P.; Ge, X.X.; Miao, L. Pharmacological Basis and New Insights of Quercetin Action in Respect to Its Anti-Cancer Effects. Biomed. Pharmacother. 2020, 121, 109604. [Google Scholar] [CrossRef]
- Zhang, X.A.; Zhang, S.; Yin, Q.; Zhang, J. Quercetin Induces Human Colon Cancer Cells Apoptosis by Inhibiting the Nuclear Factor-Kappa B Pathway. Pharmacogn. Mag. 2015, 11, 404–409. [Google Scholar] [CrossRef] [Green Version]
- Chou, C.C.; Yang, J.S.; Lu, H.F.; Ip, S.W.; Lo, C.; Wu, C.C.; Lin, J.P.; Tang, N.Y.; Chung, J.G.; Chou, M.J.; et al. Quercetin-Mediated Cell Cycle Arrest and Apoptosis Involving Activation of a Caspase Cascade through the Mitochondrial Pathway in Human Breast Cancer MCF-7 Cells. Arch. Pharm. Res. 2010, 33, 1181–1191. [Google Scholar] [CrossRef]
- Metrani, R.; Singh, J.; Archaya, P.; Jayaprakasha, G.; Patil, B. Comparative Metabolomics Profiling of Polyphenols, Nutrients and Antioxidant Activities of Two Red Onion (Allium cepa L.) Cultivars. Plants 2020, 9, 1077. [Google Scholar] [CrossRef]
- Hithamani, G.; Kizhakayil, D.; Srinivasan, K. Uptake of Phenolic Compounds from Plant Foods in Human Intestinal Caco-2 Cells. J. Biosci. 2017, 42, 603–611. [Google Scholar] [CrossRef]
- Lee, S.U.; Lee, J.H.; Choi, S.H.; Lee, J.S.; Ohnisi-Kameyama, M.; Kozukue, N.; Levin, C.E.; Friedman, M. Flavonoid Content in Fresh, Home-Processed, and Light-Exposed Onions and in Dehydrated Commercial Onion Products. J. Agric. Food Chem. 2008, 56, 8541–8548. [Google Scholar] [CrossRef]
- Quero, J.; Mármol, I.; Cerrada, E.; Rodríguez-Yoldi, M.J. Insight into the Potential Application of Polyphenol-Rich Dietary Intervention in Degenerative Disease Management. Food Funct. 2020, 11, 2805–2825. [Google Scholar] [CrossRef]
- Esparza, I.; Moler, J.A.; Arteta, M.; Jiménez-Moreno, N.; Ancín-Azpilicueta, C. Phenolic Composition of Grape Stems from Different Spanish Varieties and Vintages. Biomolecules 2021, 11, 1221. [Google Scholar] [CrossRef]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventós, R.M. Analysis of Total Phenols and Other Oxidation Substrates and Antioxidants by Means of Folin-Ciocalteu Reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar] [CrossRef]
- Chandra, S.; Khan, S.; Avula, B.; Lata, H.; Yang, M.H.; Elsohly, M.A.; Khan, I.A. Assessment of Total Phenolic and Flavonoid Content, Antioxidant Properties, and Yield of Aeroponically and Conventionally Grown Leafy Vegetables and Fruit Crops: A Comparative Study. Evid. Based. Complement. Alternat. Med. 2014, 2014, 253875. [Google Scholar] [CrossRef]
- Sánchez-de-Diego, C.; Mármol, I.; Pérez, R.; Gascón, S.; Rodríguez-Yoldi, M.J.; Cerrada, E. The Anticancer Effect Related to Disturbances in Redox Balance on Caco-2 Cells Caused by an Alkynyl Gold (I) Complex. J. Inorg. Biochem. 2017, 166, 108–121. [Google Scholar] [CrossRef] [Green Version]
- Velderrain-Rodríguez, G.R.; Quero, J.; Osada, J.; Martín-Belloso, O.; Rodríguez-Yoldi, M.J. Phenolic-Rich Extracts from Avocado Fruit Residues as Functional Food Ingredients with Antioxidant and Antiproliferative Properties. Biomolecules 2021, 11, 977. [Google Scholar] [CrossRef] [PubMed]
- Bonaccorsi, P.; Caristi, C.; Gargiulli, C.; Leuzzi, U. Flavonol Glucoside Profile of Southern Italian Red Onion (Allium cepa L.). J. Agric. Food Chem. 2005, 53, 2733–2740. [Google Scholar] [CrossRef] [PubMed]
- Celano, R.; Docimo, T.; Piccinelli, A.L.; Gazzerro, P.; Tucci, M.; Di Sanzo, R.; Carabetta, S.; Campone, L.; Russo, M.; Rastrelli, L. Onion Peel: Turning a Food Waste into a Resource. Antioxidants 2021, 10, 304. [Google Scholar] [CrossRef] [PubMed]
- Nile, A.; Gansukh, E.; Park, G.S.; Kim, D.H.; Hariram Nile, S. Novel Insights on the Multi-Functional Properties of Flavonol Glucosides from Red Onion (Allium cepa L.) Solid Waste—In Vitro and In Silico Approach. Food Chem. 2021, 335, 127650. [Google Scholar] [CrossRef]
- Hernández-Aquino, E.; Muriel, P. Beneficial Effects of Naringenin in Liver Diseases: Molecular Mechanisms. World J. Gastroenterol. 2018, 24, 1679. [Google Scholar] [CrossRef]
- Adebooye, O.C.; Alashi, A.M.; Aluko, R.E. A Brief Review on Emerging Trends in Global Polyphenol Research. J. Food Biochem. 2018, 42, e12519. [Google Scholar] [CrossRef]
- Chen, X.X.; Lam, K.H.; Chen, Q.X.; Leung, G.P.H.; Tang, S.C.W.; Sze, S.C.W.; Xiao, J.B.; Feng, F.; Wang, Y.; Zhang, K.Y.B.; et al. Ficus Virens Proanthocyanidins Induced Apoptosis in Breast Cancer Cells Concomitantly Ameliorated 5-Fluorouracil Induced Intestinal Mucositis in Rats. Food Chem. Toxicol. 2017, 110, 49–61. [Google Scholar] [CrossRef]
- Chojnacka, K.; Lewandowska, U. Chemopreventive Effects of Polyphenol-Rich Extracts against Cancer Invasiveness and Metastasis by Inhibition of Type IV Collagenases Expression and Activity. J. Funct. Foods 2018, 46, 295–311. [Google Scholar] [CrossRef]
- Pan, P.; Huang, Y.W.; Oshima, K.; Yearsley, M.; Zhang, J.; Yu, J.; Arnold, M.; Wang, L.S. An Immunological Perspective for Preventing Cancer with Berries. J. Berry Res. 2018, 8, 163. [Google Scholar] [CrossRef]
- Rady, H.M.; Hemmaid, K.Z.; Esmaeil, N.N.; Eid, M.M.; Elshat, A.A. Sidr Kashmiry Honey and Its Fractions Induced Apoptosis in Hepatocellular Carcinoma in Vitro. Med. J. Nutr. Metab. 2018, 11, 343–351. [Google Scholar] [CrossRef]
- Reboredo-Rodríguez, P. Potential Roles of Berries in the Prevention of Breast Cancer Progression. J. Berry Res. 2018, 8, 307–323. [Google Scholar] [CrossRef]
- Quero, J.; Jiménez-Moreno, N.; Esparza, I.; Osada, J.; Cerrada, E.; Ancín-Azpilicueta, C.; Rodríguez-Yoldi, M.J. Grape Stem Extracts with Potential Anticancer and Antioxidant Properties. Antioxidants 2021, 10, 243. [Google Scholar] [CrossRef]
- Tanikawa, C.; Zhang, Y.Z.; Yamamoto, R.; Tsuda, Y.; Tanaka, M.; Funauchi, Y.; Mori, J.; Imoto, S.; Yamaguchi, R.; Nakamura, Y.; et al. The Transcriptional Landscape of P53 Signalling Pathway. EBioMedicine 2017, 20, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Kruiswijk, F.; Labuschagne, C.F.; Vousden, K.H. P53 in Survival, Death and Metabolic Health: A Lifeguard with a Licence to Kill. Nat. Rev. Mol. Cell Biol. 2015, 16, 393–405. [Google Scholar] [CrossRef]
- Niazi, S.; Purohit, M.; Niazi, J.H. Role of P53 Circuitry in Tumorigenesis: A Brief Review. Eur. J. Med. Chem. 2018, 158, 7–24. [Google Scholar] [CrossRef]
- Cipolletti, M.; Fernandez, V.S.; Montalesi, E.; Marino, M.; Fiocchetti, M. Beyond the Antioxidant Activity of Dietary Polyphenols in Cancer: The Modulation of Estrogen Receptors (ERs) Signaling. Int. J. Mol. Sci. 2018, 19, 2624. [Google Scholar] [CrossRef] [Green Version]
- Suvorova, I.I.; Knyazeva, A.R.; Pospelov, V.A. Resveratrol-Induced P53 Activation Is Associated with Autophagy in Mouse EmbryonicStem Cells. Biochem. Biophys. Res. Commun. 2018, 503, 2180–2185. [Google Scholar] [CrossRef]
- Vitkeviciene, A.; Baksiene, S.; Borutinskaite, V.; Navakauskiene, R. Epigallocatechin-3-Gallate and BIX-01294 Have Different Impact on Epigenetics and Senescence Modulation in Acute and Chronic Myeloid Leukemia Cells. Eur. J. Pharmacol. 2018, 838, 32–40. [Google Scholar] [CrossRef]
- Arul, D.; Subramanian, P. Naringenin (Citrus flavonone) Induces Growth Inhibition, Cell Cycle Arrest and Apoptosis in Human Hepatocellular Carcinoma Cells. Pathol. Oncol. Res. 2013, 19, 763–770. [Google Scholar] [CrossRef]
- Rao, S.; Chinkwo, K.; Santhakumar, A.; Johnson, S.; Blanchard, C. Apoptosis Induction Pathway in Human Colorectal Cancer Cell Line SW480 Exposed to Cereal Phenolic Extracts. Molecules 2019, 24, 2465. [Google Scholar] [CrossRef] [Green Version]
- Carvajal, L.A.; Manfredi, J.J. Another Fork in the Road-Life or Death Decisions by the Tumour Suppressor P53. EMBO Rep. 2013, 14, 414–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, H.; Reale, M.; Ullah, H.; Sureda, A.; Tejada, S.; Wang, Y.; Zhang, Z.J.; Xiao, J. Anti-Cancer Effects of Polyphenols via Targeting P53 Signaling Pathway: Updates and Future Directions. Biotechnol. Adv. 2020, 38, 107385. [Google Scholar] [CrossRef] [PubMed]
- Curti, V.; Di Lorenzo, A.; Dacrema, M.; Xiao, J.; Nabavi, S.M.; Daglia, M. In Vitro Polyphenol Effects on Apoptosis: An Update of Literature Data. Semin. Cancer Biol. 2017, 46, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Thakur, V.S.; Bhaskaran, N.; Nawab, A.; Babcook, M.A.; Jackson, M.W.; Gupta, S. Green Tea Polyphenols Induce P53-Dependent and P53-Independent Apoptosis in Prostate Cancer Cells through Two Distinct Mechanisms. PLoS ONE 2012, 7, e52572. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Yi, S.M.; Yun, J.W.; Jung, J.H.; Kim, D.H.; Kim, H.J.; Chang, S.-H.; Kim, G.; Ryu, C.H.; Shin, S.C.; et al. Polyphenols Isolated from Allium cepa L. Induces Apoptosis by Induction of P53 and Suppression of Bcl-2 through Inhibiting PI3K/Akt Signaling Pathway in AGS Human Cancer Cells. J. Cancer Prev. 2014, 19, 14–22. [Google Scholar] [CrossRef] [Green Version]
- Jacquemin, G.; Granci, V.; Gallouet, A.S.; Lalaoui, N.; Morlé, A.; Iessi, E.; Morizot, A.; Garrido, C.; Guillaudeux, T.; Micheau, O. Quercetin-Mediated Mcl-1 and Survivin Downregulation Restores TRAIL-Induced Apoptosis in Non-Hodgkin’s Lymphoma B Cells. Haematologica 2012, 97, 38–46. [Google Scholar] [CrossRef] [Green Version]
- Park, J.H.; Darvin, P.; Lim, E.J.; Joung, Y.H.; Hong, D.Y.; Park, E.U.; Park, S.H.; Choi, S.K.; Moon, E.S.; Cho, B.W.; et al. Hwanggeumchal sorghum Induces Cell Cycle Arrest, and Suppresses Tumor Growth and Metastasis through Jak2/STAT Pathways in Breast Cancer Xenografts. PLoS ONE 2012, 7, e40531. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The Ubiquitin System. Ann. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Arlt, A.; Bauer, I.; Schafmayer, C.; Tepel, J.; Müerköster, S.S.; Brosch, M.; Röder, C.; Kalthoff, H.; Hampe, J.; Moyer, M.P.; et al. Increased Proteasome Subunit Protein Expression and Proteasome Activity in Colon Cancer Relate to an Enhanced Activation of Nuclear Factor E2-Related Factor 2 (Nrf2). Oncogene 2009, 28, 3983–3996. [Google Scholar] [CrossRef] [Green Version]
- Milacic, V.; Banerjee, S.; Landis-Piwowar, K.R.; Sarkar, F.H.; Majumdar, A.P.N.; Dou, Q.P. Curcumin Inhibits the Proteasome Activity in Human Colon Cancer Cells in Vitro and in Vivo. Cancer Res. 2008, 68, 7283. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Qu, W.; Cheng, Y.; Sun, Y.; Jiang, Y.; Zou, T.; Wang, Z.; Xu, Y.; Zhao, H. The Inhibitory Effect of Intravesical Fisetin against Bladder Cancer by Induction of P53 and Down-Regulation of NF-Kappa B Pathways in a Rat Bladder Carcinogenesis Model. Basic Clin. Pharmacol. Toxicol. 2014, 115, 321–329. [Google Scholar] [CrossRef]
- Roy, P.; George, J.; Srivastava, S.; Tyagi, S.; Shukla, Y. Correction to: Inhibitory Effects of Tea Polyphenols by Targeting Cyclooxygenase-2 through Regulation of Nuclear Factor Kappa B, Akt and P53 in Rat Mammary Tumors. Investig. New Drugs 2019, 37, 1310–1311. [Google Scholar] [CrossRef]
- Shao, J.; Fujiwara, T.; Kadowaki, Y.; Fukazawa, T.; Waku, T.; Itoshima, T.; Yamatsuji, T.; Nishizaki, M.; Roth, J.A.; Tanaka, N. Overexpression of the Wild-Type P53 Gene Inhibits NF-Kappa B Activity and Synergizes with Aspirin to Induce Apoptosis in Human Colon Cancer Cells. Oncogene 2000, 19, 726–736. [Google Scholar] [CrossRef] [Green Version]
- Manda, G.; Isvoranu, G.; Comanescu, M.V.; Manea, A.; Debelec Butuner, B.; Korkmaz, K.S. The Redox Biology Network in Cancer Pathophysiology and Therapeutics. Redox Biol. 2015, 5, 347–357. [Google Scholar] [CrossRef] [Green Version]
- Budanov, A.V. The Role of Tumor Suppressor P53 in the Antioxidant Defense and Metabolism. Subcell. Biochem. 2014, 85, 337–358. [Google Scholar] [CrossRef] [Green Version]
- Kabe, Y.; Ando, K.; Hirao, S.; Yoshida, M.; Handa, H. Redox Regulation of NF-Kappa B Activation: Distinct Redox Regulation between the Cytoplasm and the Nucleus. Antioxid. Redox Signal. 2005, 7, 395–403. [Google Scholar] [CrossRef]
- Morgan, M.J.; Liu, Z.G. Crosstalk of Reactive Oxygen Species and NF-ΚB Signaling. Cell Res. 2011, 21, 103–115. [Google Scholar] [CrossRef] [Green Version]
- Roleira, F.M.F.; Tavares-Da-Silva, E.J.; Varela, C.L.; Costa, S.C.; Silva, T.; Garrido, J.; Borges, F. Plant Derived and Dietary Phenolic Antioxidants: Anticancer Properties. Food Chem. 2015, 183, 235–258. [Google Scholar] [CrossRef]
- León-González, A.J.; Auger, C.; Schini-Kerth, V.B. Pro-Oxidant Activity of Polyphenols and Its Implication on Cancer Chemoprevention and Chemotherapy. Biochem. Pharmacol. 2015, 98, 371–380. [Google Scholar] [CrossRef]
- Almeer, R.S.; Mahmoud, S.M.; Amin, H.K.; Abdel Moneim, A.E. Ziziphus Spina-Christi Fruit Extract Suppresses Oxidative Stress and P38 MAPK Expression in Ulcerative Colitis in Rats via Induction of Nrf2 and HO-1 Expression. Food Chem. Toxicol. 2018, 115, 49–62. [Google Scholar] [CrossRef]
- Kumar, V.L.; Pandey, A.; Verma, S.; Das, P. Protection Afforded by Methanol Extract of Calotropis procera Latex in Experimental Model of Colitis Is Mediated through Inhibition of Oxidative Stress and Pro-Inflammatory Signaling. Biomed. Pharmacother. 2019, 109, 1602–1609. [Google Scholar] [CrossRef]
- Chalouati, H.; Boutet, E.; Metais, B.; Fouche, E.; Ben Sâad, M.M.; Gamet-Payrastre, L. DNA Damage and Oxidative Stress Induced at Low Doses by the Fungicide Hexachlorobenzene in Human Intestinal Caco-2 Cells. Toxicol. Mech. Methods 2015, 25, 448–458. [Google Scholar] [CrossRef]
- Lee, S.I.; Kang, K.S. N-Acetylcysteine Modulates Lipopolysaccharide-Induced Intestinal Dysfunction. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Catanzaro, D.; Rancan, S.; Orso, G.; Dall’acqua, S.; Brun, P.; Giron, M.C.; Carrara, M.; Castagliuolo, I.; Ragazzi, E.; Caparrotta, L.; et al. Boswellia serrata Preserves Intestinal Epithelial Barrier from Oxidative and Inflammatory Damage. PLoS ONE 2015, 10, e0125375. [Google Scholar] [CrossRef] [Green Version]
- Lea, T. Caco-2 cell line. In The Impact of Food Bioactives on Health In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Cham, Switzerland, 2015; pp. 103–111. [Google Scholar] [CrossRef] [Green Version]
- Andreu, V.; Larrea, A.; Rodriguez-Fernandez, P.; Alfaro, S.; Gracia, B.; Luciá, A.; Usón, L.; Gomez, A.C.; Mendoza, G.; Lacoma, A.; et al. Matryoshka-Type Gastro-Resistant Microparticles for the Oral Treatment of Mycobacterium tuberculosis. Nanomedicine 2019, 14, 707–726. [Google Scholar] [CrossRef] [Green Version]
- Hardjono, S.; Siswandono, S.; Andayani, R. Evaluation of N-benzoylthiourea derivatives as possible analgesic agents by predicting their physicochemical and pharmacokinetic properties, toxicity, and analgesic activity. Indones. J. Biotechnol. 2017, 22, 76–85. [Google Scholar] [CrossRef] [Green Version]







| Phenolic Compound | Concentration (mg/g Extract) |
|---|---|
| Protocatechuic acid | 11.5 ± 0.3 |
| Ellagic acid | 0.10 ± 0.01 |
| Vanillic acid | 0.33 ± 0.03 |
| Quercetin | 10.2 ± 0.4 |
| Quercetin 3-glucoside | 1.16 ± 0.08 |
| Kaempferol | 0.44 ± 0.01 |
| Isorhamnetin | 0.24 ± 0.01 |
| Unknown flavonoid 1 1 | 2.57 ± 0.09 |
| Unknown flavonoid 2 1 | 12.3 ± 0.3 |
| Unknown flavonoid 3 1 | 0.87 ± 0.03 |
| Assays | Concentration |
|---|---|
| Antioxidant capacity by ABTS (mmol Trolox/g extract) | 1.11 ± 0.08 |
| Antioxidant capacity by FRAP (mmol Trolox/g extract) | 0.83 ± 0.01 |
| Antioxidant capacity by DPPH (mmol Trolox/g extract) | 0.49 ± 0.08 |
| Total Flavonoids (mg quercetin/g extract) | 64 ± 3 |
| Total Phenolic Content (mg gallic acid/g extract) | 177 ± 9 |
| Identified Compound | MW | TPSA | Log P | No. Atoms | Hydrogen Bonds Acceptors | Hydrogen Bonds Donors | Rotatable Bonds | Molecular Volume (Å3) | Violations to LIRF | % ABS | log Papp (10−6 cm/s) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Protocatechuic acid | 154.12 | 77.75 | 0.88 | 11 | 4 | 3 | 1 | 127.08 | 0 | 89.15 | 1.15 |
| 2 | Ellagic acid | 302.19 | 141.33 | 0.94 | 22 | 8 | 4 | 0 | 221.78 | 0 | 60.24 | 0.33 |
| 3 | Vanillic acid | 168.15 | 66.76 | 1.19 | 12 | 4 | 2 | 2 | 144.61 | 0 | 85.97 | 0.33 |
| 4 | Quercetin | 302.24 | 131.35 | 1.68 | 22 | 11 | 7 | 1 | 240.08 | 0 | 63.68 | −0.23 |
| 5 | Quercetin 3-glucoside | 464.38 | 210.50 | −0.36 | 33 | 12 | 8 | 4 | 372.21 | 2 | 36.38 | 0.24 |
| 6 | Kaempferol | 286.24 | 111.12 | 2.17 | 21 | 6 | 4 | 1 | 232.07 | 0 | 70.66 | 0.03 |
| 7 | Isorhamnetin | 316.26 | 120.36 | 1.99 | 23 | 7 | 4 | 2 | 257.61 | 0 | 67.48 | −0.003 |
| Compounds of matching spectra for the unknown flavonoids | ||||||||||||
| Quercetin 3,7,4′-triglucoside | 788.66 | 368.81 | −4.16 | 55 | 22 | 14 | 0 | 636.45 | 3 | −18.24 | −1.14 | |
| Isorhamnetin 3,4′-diglucoside | 640.55 | 278.66 | −2.07 | 45 | 17 | 10 | 8 | 521.86 | 3 | 12.86 | −1.10 | |
| Quercetin 4’-glucoside | 464.38 | 210.50 | −0.33 | 33 | 12 | 8 | 4 | 372.21 | 2 | 36.38 | 0.27 | |
| Isorhamnetin 4’-glucoside | 478.41 | 199.51 | −0.03 | 34 | 12 | 7 | 5 | 389.73 | 2 | 40.17 | 0.34 | |
| Quercetin 7,4’-diglucoside | 626.52 | 289.65 | −2.12 | 44 | 17 | 11 | 7 | 504.33 | 3 | 9.07 | −1.22 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paesa, M.; Nogueira, D.P.; Velderrain-Rodríguez, G.; Esparza, I.; Jiménez-Moreno, N.; Mendoza, G.; Osada, J.; Martin-Belloso, O.; Rodríguez-Yoldi, M.J.; Ancín-Azpilicueta, C. Valorization of Onion Waste by Obtaining Extracts Rich in Phenolic Compounds and Feasibility of Its Therapeutic Use on Colon Cancer. Antioxidants 2022, 11, 733. https://doi.org/10.3390/antiox11040733
Paesa M, Nogueira DP, Velderrain-Rodríguez G, Esparza I, Jiménez-Moreno N, Mendoza G, Osada J, Martin-Belloso O, Rodríguez-Yoldi MJ, Ancín-Azpilicueta C. Valorization of Onion Waste by Obtaining Extracts Rich in Phenolic Compounds and Feasibility of Its Therapeutic Use on Colon Cancer. Antioxidants. 2022; 11(4):733. https://doi.org/10.3390/antiox11040733
Chicago/Turabian StylePaesa, Mónica, Danielle Pires Nogueira, Gustavo Velderrain-Rodríguez, Irene Esparza, Nerea Jiménez-Moreno, Gracia Mendoza, Jesús Osada, Olga Martin-Belloso, María Jesús Rodríguez-Yoldi, and Carmen Ancín-Azpilicueta. 2022. "Valorization of Onion Waste by Obtaining Extracts Rich in Phenolic Compounds and Feasibility of Its Therapeutic Use on Colon Cancer" Antioxidants 11, no. 4: 733. https://doi.org/10.3390/antiox11040733