Large-Size Subunit Catalases Are Chimeric Proteins: A H2O2 Selecting Domain with Catalase Activity Fused to a Hsp31-Derived Domain Conferring Protein Stability and Chaperone Activity
Abstract
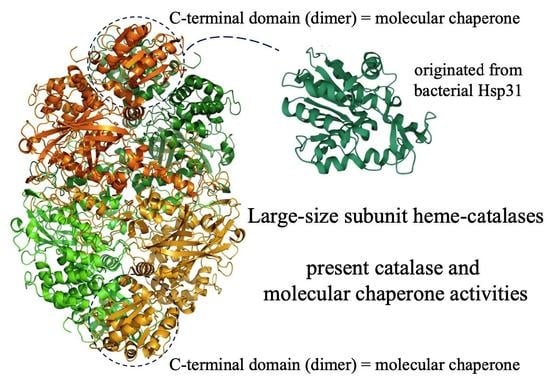
:1. Introduction
2. Materials and Methods
2.1. Purification of CAT-1
2.2. Optimal Temperature
2.3. Treatment of CAT-1 with Subtilisin
2.4. Saturation Kinetics of the CAT-1 Catalytic Domain
2.5. Determination of t1/2
2.6. Treatment of CAT-1 with Denaturation Agents
2.7. Expression of Proteins in Escherichia coli
2.8. Structural Alignment
2.9. Heat Stability and Molecular Chaperone Activity
2.10. Dimerization Modes of CT
2.11. Phylogeny of Bacterial Hsp31 and MC_CT
3. Results
3.1. Amino Acid Composition of the Catalase Domains and CTs
3.2. The CT Confers Overall Protein Stability and Chaperone Activity
3.3. Structural Alignment of the CT with the Members of the DJ-1/PfpI Superfamily
3.4. CT of LSCs Represents a New Dimer Conformation of the DJ-1/PfpI Superfamily
3.5. Possible Origin of the CT from a Bacterial Hsp31-like Protein
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klotz, M.G.; Loewen, P.C. The molecular evolution of catalatic hydroperoxidases: Evidence for multiple lateral transfer of genes between prokaryota and from bacteria into eukaryota. Mol. Biol. Evol. 2003, 20, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Zamocky, M.; Gasselhuber, B.; Furtmuller, P.G.; Obinger, C. Molecular evolution of hydrogen peroxide degrading enzymes. Arch. Biochem. Biophys. 2012, 525, 131–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansberg, W.; Salas-Lizana, R.; Domínguez, L. Fungal catalases: Function, phylogenetic origin and structure. Arch. Biochem. Biophys. 2012, 525, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Michán, S.; Lledías, F.; Baldwin, J.D.; Natvig, D.O.; Hansberg, W. Regulation and oxidation of two large monofunctional catalases. Free Radic. Biol. Med. 2002, 33, 521–532. [Google Scholar] [CrossRef]
- Díaz, A.; Rangel, P.; Montes de Oca, Y.; Lledías, F.; Hansberg, W. Molecular and kinetic study of catalase-1, a durable large catalase of Neurospora crassa. Free Radic. Biol. Med. 2001, 31, 1323–1333. [Google Scholar] [CrossRef]
- Díaz, A.; Valdés, V.J.; Rudiño-Piñera, E.; Horjales, E.; Hansberg, W. Structure-function relationships in fungal large-subunit catalases. J. Mol. Biol. 2009, 386, 218–232. [Google Scholar] [CrossRef]
- Maddi, A.; Bowman, S.M.; Free, S.J. Trifluoromethanesulfonic acid-based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans. Fungal Genet. Biol. 2009, 46, 768–781. [Google Scholar] [CrossRef] [Green Version]
- Michán, S.; Lledías, F.; Hansberg, W. Asexual development is increased in Neurospora crassa cat-3-null mutant strains. Eukaryot. Cell 2003, 2, 798–808. [Google Scholar] [CrossRef] [Green Version]
- Lledías, F.; Rangel, P.; Hansberg, W. Singlet oxygen is part of a hyperoxidant state generated during spore germination. Free Radic. Biol. Med. 1999, 26, 1396–1404. [Google Scholar] [CrossRef]
- Hansberg, W.; Aguirre, J. Hyperoxidant states cause microbial cell differentiation by cell isolation from dioxygen. J. Theor. Biol. 1990, 142, 201–221. [Google Scholar] [CrossRef]
- Aguirre, J.; Rios-Momberg, M.; Hewitt, D.; Hansberg, W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005, 13, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Hansberg, W.; Aguirre, J.; Rios-Momberg, M.; Rangel, P.; Peraza, L.; Montes de Oca, Y.; Cano-Domínguez, N. Cell Differentiation as a Response to Oxidative Stress; Elsevier: London, UK, 2008. [Google Scholar]
- Díaz, A.; Muñoz-Clares, R.A.; Rangel, P.; Valdés, V.J.; Hansberg, W. Functional and structural analysis of catalase oxidized by singlet oxygen. Biochimie 2005, 87, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Nava-Ramírez, T.; Hansberg, W. Chaperone activity of large-size subunit catalases. Free Radic. Biol. Med. 2020, 156, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Holm, L. Using Dali for protein structure comparison. Methods Mol. Biol. 2020, 2112, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Natarajan, P.; Ye, Y.; Hrabe, T.; Godzik, A. POSA: A user-driven, interactive multiple protein structure alignment server. Nucleic Acids Res. 2014, 42, W240–W245. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Vallve, S.; Palau, J.; Romeu, A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol. Biol. Evol. 1999, 16, 1125–1134. [Google Scholar] [CrossRef] [Green Version]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Erdos, G.; Pajkos, M.; Dosztanyi, Z. IUPred3: Prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 2021, 49, W297–W303. [Google Scholar] [CrossRef]
- Di Rienzo, L.; Miotto, M.; Bo, L.; Ruocco, G.; Raimondo, D.; Milanetti, E. Characterizing hydropathy of amino acid side chain in a protein environment by investigating the structural changes of áter molecules network. Front. Mol. Biosci. 2021, 8, 626837. [Google Scholar] [CrossRef]
- Shimizu, K.; Muraoka, Y.; Hirose, S.; Tomii, K.; Noguchi, T. Predicting mostly disordered proteins by using structure-unknown protein data. BMC Bioinformatics 2007, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Chelikani, P.; Donald, L.J.; Duckworth, H.W.; Loewen, P.C. Hydroperoxidase II of Escherichia coli exhibits enhanced resistance to proteolytic cleavage compared to other catalases. Biochemistry 2003, 42, 5729–5735. [Google Scholar] [CrossRef] [PubMed]
- Sevinc, M.S.; Switala, J.; Bravo, J.; Fita, I.; Loewen, P.C. Truncation and heme pocket mutations reduce production of functional catalase HPII in Escherichia coli. Protein Eng. 1998, 11, 549–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teo, J.W.; Kum, S.; Jureen, R.; Lin, R.T. Molecular characterization of a catalase-negative Staphylococcus aureus blood culture isolate. J. Clin. Microbiol. 2015, 53, 3699–3701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nava-Ramírez, T.; Hansberg, W. Mechanism for the molecular chaperone of large-size subunit catalases. Free Radic. Biol. Med. 2022, 156, 99–106. [Google Scholar] [CrossRef]
- Jung, H.J.; Kim, S.; Kim, Y.J.; Kim, M.K.; Kang, S.G.; Lee, J.H.; Kim, W.; Cha, S.S. Dissection of the dimerization modes in the DJ-1 superfamily. Mol. Cells 2012, 33, 163–171. [Google Scholar] [CrossRef]
- Andreeva, A.; Bekkhozhin, Z.; Omertassova, N.; Baizhumanov, T.; Yeltay, G.; Akhmetali, M.; Toibazar, D.; Utepbergenov, D. The apparent deglycase activity of DJ-1 results from the conversion of free methylglyoxal present in fast equilibrium with hemithioacetals and hemiaminals. J. Biol. Chem. 2019, 294, 18863–18872. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Cookson, M.R. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol. Biol. 2004, 4, 6. [Google Scholar] [CrossRef] [Green Version]
- Deiana, A.S.; Shimizu, K.; Giansanti, A. Amino acid composition and thermal stability of protein structures: The free energy geography of the Protein Data Bank. arXiv preprint 2010, arXiv:1012.5916 [q-bio.GN]. [Google Scholar]
- Aussel, L.; Ezraty, B. Methionine redox homeostasis in protein quality control. Front. Mol. Biosci. 2021, 8, 665492. [Google Scholar] [CrossRef]
- Garcia-Santamarina, S.; Boronat, S.; Hidalgo, E. Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction. Biochemistry 2014, 53, 2560–2580. [Google Scholar] [CrossRef] [Green Version]
- Vega-García, V.; Díaz-Vilchis, A.; Saucedo-Vázquez, J.P.; Solano-Peralta, A.; Rudiño-Piñera, E.; Hansberg, W. Structure, kinetics, molecular and redox properties of a cytosolic and developmentally regulated fungal catalase-peroxidase. Arch. Biochem. Biophys. 2018, 640, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, L.; Sosa-Peinado, A.; Hansberg, W. Catalase evolved to concentrate H2O2 at its active site. Arch. Biochem. Biophys. 2010, 500, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Jha, V.; Chelikani, P.; Carpena, X.; Fita, I.; Loewen, P.C. Influence of main channel structure on H2O2 access to the heme cavity of catalase KatE of Escherichia coli. Arch. Biochem. Biophys. 2012, 526, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, L.; Sosa-Peinado, A.; Hansberg, W. How catalase recognizes H2O2 in a sea of water. Proteins 2014, 82, 45–56. [Google Scholar] [CrossRef]
- Szilagyi, A.; Gyorffy, D.; Zavodszky, P. The twilight zone between protein order and disorder. Biophys. J. 2008, 95, 1612–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Liu, J.; Wang, G.; Cha, J.Y.; Li, G.; Chen, S.; Li, Z.; Guo, J.; Zhang, C.; Yang, Y.; et al. A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 2015, 27, 908–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Cui, L.; Xie, Z.; Zhang, Z.; Liu, E.; Peng, X. Two NCA1 isoforms interact with catalase in a mutually exclusive manner to redundantly regulate its activity in rice. BMC Plant Biol. 2019, 19, 105. [Google Scholar] [CrossRef]
- Du, H.; Kim, S.; Hur, Y.S.; Lee, M.S.; Lee, S.H.; Cheon, C.I. A cytosolic thioredoxin acts as a molecular chaperone for peroxisome matrix proteins as well as antioxidant in peroxisome. Mol. Cells 2015, 38, 187–194. [Google Scholar] [CrossRef] [Green Version]
- Yuan, F.; Yin, S.; Xu, Y.; Xiang, L.; Wang, H.; Li, Z.; Fan, K.; Pan, G. The richness and diversity of catalases in bacteria. Front. Microbiol. 2021, 12, 645477. [Google Scholar] [CrossRef]








| PDB | CAT | CT c | |
|---|---|---|---|
| hpho a | 41.53 | 35.28 | 49.14 |
| hphi | 43.49 | 43.82 | 37.22 |
| hcy/aro | 14.88 | 20.73 | 13.35 |
| D/E a | 0.81 | 1.21 | 0.80 |
| N/Q a | 1.03 | 1.16 | 1.07 |
| T/S a | 0.81 | 1.05 | 0.78 |
| R/K a | 0.95 | 1.22 | 0.83 |
| (F + W)/P a | 1.05 | 1.16 | 1.56 |
| IF b | 3.87 | 4.11 | −0.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hansberg, W.; Nava-Ramírez, T.; Rangel-Silva, P.; Díaz-Vilchis, A.; Mendoza-Oliva, A. Large-Size Subunit Catalases Are Chimeric Proteins: A H2O2 Selecting Domain with Catalase Activity Fused to a Hsp31-Derived Domain Conferring Protein Stability and Chaperone Activity. Antioxidants 2022, 11, 979. https://doi.org/10.3390/antiox11050979
Hansberg W, Nava-Ramírez T, Rangel-Silva P, Díaz-Vilchis A, Mendoza-Oliva A. Large-Size Subunit Catalases Are Chimeric Proteins: A H2O2 Selecting Domain with Catalase Activity Fused to a Hsp31-Derived Domain Conferring Protein Stability and Chaperone Activity. Antioxidants. 2022; 11(5):979. https://doi.org/10.3390/antiox11050979
Chicago/Turabian StyleHansberg, Wilhelm, Teresa Nava-Ramírez, Pablo Rangel-Silva, Adelaida Díaz-Vilchis, and Aydé Mendoza-Oliva. 2022. "Large-Size Subunit Catalases Are Chimeric Proteins: A H2O2 Selecting Domain with Catalase Activity Fused to a Hsp31-Derived Domain Conferring Protein Stability and Chaperone Activity" Antioxidants 11, no. 5: 979. https://doi.org/10.3390/antiox11050979