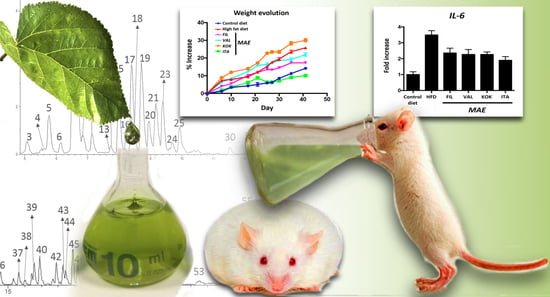
Comparative Study of the Antioxidant and Anti-Inflammatory Effects of Leaf Extracts from Four Different Morus alba Genotypes in High Fat Diet-Induced Obesity in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Plant Materials and Extraction Procedure of Bioactive Compounds from M. alba
2.3. RP-HPLC-ESI-TOF-MS Analysis
2.4. DPPH•Scavenging Activity
2.5. Oxidative Stress Assays
2.6. Effects of MAEs on High Fat Diet Fed Mice
2.6.1. Animals and Treatments
2.6.2. Glucose Tolerance Test
2.6.3. Plasma Determinations
2.6.4. Morphological Variables
2.6.5. Analysis of Gene Expression by RT-qPCR
2.6.6. Histological Studies
2.7. Statistics
3. Results and Discussion
3.1. Analytical Characterization of Phytochemicals in MAEs Yielded by PLE Conditions
3.2. Quantitation of Polar Compounds in MAEs
3.3. Radical Scavenging Activity of the M. alba Extracts
3.4. Antioxidant Activity of the M. alba Extracts on C. elegans
3.5. Effects of MAEs on Body Weight, Glucose Tolerance Test, Fat Accumulation, and Plasma Biochemical Profile
3.6. Effects of MAEs on Systemic Inflammatory Response
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; Naghavi, M.; et al. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [CrossRef] [PubMed]
- García-Ruiz, I.; Solís-Muñoz, P.; Fernandez-Moreira, D.; Grau, M.; Muñoz-Yagüe, M.T.; Solis-Herruzo, J.A. Omentectomy Prevents Metabolic Syndrome By Reducing Appetite and Body Weight In A Diet-Induced Obesity Rat Model. Sci. Rep. 2018, 8, 1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregor, M.F.; Hotamisligil, G.S. Inflammatory Mechanisms in Obesity. Annu. Rev. Immunol. 2011, 29, 415–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herranz-López, M.; Barrajón-Catalán, E.; Segura-Carretero, A.; Menendez, J.A.; Joven, J.; Micol, V. Lemon verbena (Lippia citriodora) polyphenols alleviate obesity-related disturbances in hypertrophic adipocytes through AMPK-dependent mechanisms. Phytomedicine 2015, 22, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Dugo, P.; Donato, P.; Cacciola, F.; Germanò, M.P.; Rapisarda, A.; Mondello, L. Characterization of the polyphenolic fraction of Morus alba leaves extracts by HPLC coupled to a hybrid IT-TOF MS system. J. Sep. Sci. 2009, 32, 3627–3634. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, K.T.A.; Zhao, W.G.; Nair, C.V.; Ercisli, S.; Tsou, C.H. Morus. In Wild Crop Relatives: Genomic and Breeding Resources—Tropical and Subtropical Fruits; Springer: Berlin/Heidelberg, Germany, 2011; pp. 75–95. [Google Scholar]
- Harauma, A.; Murayama, T.; Ikeyama, K.; Sano, H.; Arai, H.; Takano, R.; Kita, T.; Hara, S.; Kamei, K.; Yokode, M. Mulberry leaf powder prevents atherosclerosis in apolipoprotein E-deficient mice. Biochem. Biophys. Res. Commun. 2007, 358, 751–756. [Google Scholar] [CrossRef]
- Hu, X.-Q.; Thakur, K.; Chen, G.-H.; Hu, F.; Zhang, J.-G.; Zhang, H.-B.; Wei, Z.-J. Metabolic Effect of 1-Deoxynojirimycin from Mulberry Leaves on db/db Diabetic Mice Using Liquid Chromatography–Mass Spectrometry Based Metabolomics. J. Agric. Food Chem. 2017, 65, 4658–4667. [Google Scholar] [CrossRef]
- Ercisli, S.; Orhan, E. Chemical composition of white (Morus alba), red (Morus rubra) and black (Morus nigra) mulberry fruits. Food Chem. 2007, 103, 1380–1384. [Google Scholar] [CrossRef]
- Sheng, Y.; Liu, J.; Zheng, S.; Liang, F.; Luo, Y.; Huang, K.; Xu, W.; He, X. Mulberry leaves ameliorate obesity through enhancing brown adipose tissue activity and modulating gut microbiota. Food Funct. 2019, 10, 4771–4781. [Google Scholar] [CrossRef]
- Gunjal, S.; Ankola, A.V.; Bhat, K. In vitro antibacterial activity of ethanolic extract of Morus alba leaf against periodontal pathogens. Indian J. Dent. Res. 2015, 26, 533–536. [Google Scholar] [CrossRef]
- He, X.; Fang, J.; Ruan, Y.; Wang, X.; Sun, Y.; Wu, N.; Zhao, Z.; Chang, Y.; Ning, N.; Guo, H.; et al. Structures, bioactivities and future prospective of polysaccharides from Morus alba (white mulberry): A review. Food Chem. 2018, 245, 899–910. [Google Scholar] [CrossRef] [PubMed]
- Gui, Z.; Raman, S.T.; Ganeshan, A.K.P.G.; Chen, C.; Jin, C.; Li, S.-H.; Chen, H.-J. In vitro and In vivo antioxidant activity of flavonoid extracted from mulberry fruit (Morus alba L.). Pharmacogn. Mag. 2016, 12, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Tan, Y.-X.; Chen, R.-Y.; Kang, J. The latest review on the polyphenols and their bioactivities of ChineseMorusplants. J. Asian Nat. Prod. Res. 2014, 16, 690–702. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Chen, X.; Yu, H.; Chen, L.; Shen, M. Seasonal variations of iminosugars in mulberry leaves detected by hydrophilic interaction chromatography coupled with tandem mass spectrometry. Food Chem. 2018, 251, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.-Z.; Li, N.; Tuo, Z.-D.; Li, J.-L.; Xing, S.-S.; Li, B.-B.; Zhang, L.; Lee, H.-S.; Chen, J.-G.; Cui, L. Effects of Morus root bark extract and active constituents on blood lipids in hyperlipidemia rats. J. Ethnopharmacol. 2016, 180, 54–59. [Google Scholar] [CrossRef]
- Leyva-Jiménez, F.J.; Lozano-Sánchez, J.; Borrás-Linares, I.; Arráez-Román, D.; Segura-Carretero, A. Comparative study of conventional and pressurized liquid extraction for recovering bioactive compounds from Lippia citriodora leaves. Food Res. Int. 2018, 109, 213–222. [Google Scholar] [CrossRef]
- Garcia-Gómez, B.; González-Alvarez, H.; Martínez-Mora, C.; Cenis, L.J.; Perez-Hernandez, C.D.M.; Martínez-Zubiaur, Y.; Martínez-Gómez, P. The molecular characterization of an extended mulberry germplasm by SSR markers. Genetika 2019, 51, 389–403. [Google Scholar] [CrossRef] [Green Version]
- Gross, J.H. Mass Spectrometry; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Yen, G.-C.; Der Duh, P. Scavenging Effect of Methanolic Extracts of Peanut Hulls on Free-Radical and Active-Oxygen Species. J. Agric. Food Chem. 1994, 42, 629–632. [Google Scholar] [CrossRef]
- Martorell, P.; Bataller, E.; Llopis, S.; González, N.; Álvarez, B.; Monton, F.; Ortiz, P.; Ramón, D.; Genovés, S. A Cocoa Peptide Protects Caenorhabditis elegans from Oxidative Stress and β-Amyloid Peptide Toxicity. PLoS ONE 2013, 8, e63283. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Salcedo, E.M.; Tassotti, M.; Del Rio, D.; Hernández, F.; Martínez, J.J.; Mena, P. (Poly)phenolic fingerprint and chemometric analysis of white (Morus alba L.) and black (Morus nigra L.) mulberry leaves by using a non-targeted UHPLC–MS approach. Food Chem. 2016, 212, 250–255. [Google Scholar] [CrossRef]
- Shao, S.-Y.; Feng, Z.-M.; Yang, Y.-N.; Jiang, J.-S.; Zhang, P. Eight new phenylethanoid glycoside derivatives possessing potential hepatoprotective activities from the fruits of Forsythia suspensa. Fitoterapia 2017, 122, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Nastić, N.; Borrás-Linares, I.; Lozano-Sánchez, J.; Švarc-Gajić, J.; Segura-Carretero, A. Optimization of the extraction of phytochemicals from black mulberry (Morus nigra L.) leaves. J. Ind. Eng. Chem. 2018, 68, 282–292. [Google Scholar] [CrossRef]
- Cho, N.; Kim, H.W.; Kim, T.B.; Ransom, T.T.; Beutler, J.A.; Sung, S.H. Preparative Purification of Anti-Proliferative Diarylheptanoids from Betula platyphylla by High-Speed Counter-Current Chromatography. Molecules 2016, 21, 700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fakhfakh, N.; Jdir, H.; Jridi, M.; Rateb, M.; Belbahri, L.; Ayadi, M.A.; Nasri, M.; Zouari, N. The mallow, Malva aegyptiaca L. (Malvaceae): Phytochemistry analysis and effects on wheat dough performance and bread quality. LWT 2017, 75, 656–662. [Google Scholar] [CrossRef]
- Katsube, T.; Yamasaki, M.; Shiwaku, K.; Ishijima, T.; Matsumoto, I.; Abe, K.; Yamasaki, Y. Effect of flavonol glycoside in mulberry (Morus alba L.) leaf on glucose metabolism and oxidative stress in liver in diet-induced obese mice. J. Sci. Food Agric. 2010, 90, 2386–2392. [Google Scholar] [CrossRef]
- Hotamisligil, G.; Shargill, N.; Spiegelman, B. Adipose expression of tumor necrosis factor-alpha: Direct role in obesity-linked insulin resistance. Science 1993, 259, 87–91. [Google Scholar] [CrossRef]
- Ouchi, N.; Parker, J.L.; Lugus, J.J.; Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011, 11, 85–97. [Google Scholar] [CrossRef]
- Greenberg, A.S.; Obin, M.S. Obesity and the role of adipose tissue in inflammation and metabolism. Am. J. Clin. Nutr. 2006, 83, 461S–465S. [Google Scholar] [CrossRef] [Green Version]
- Pal, M.; Febbraio, M.A.; Lancaster, G.I. The roles of c-Jun NH2-terminal kinases (JNKs) in obesity and insulin resistance. J. Physiol. 2015, 594, 267–279. [Google Scholar] [CrossRef] [Green Version]
- Stienstra, R.; Duval, C.; Muller, M.; Kersten, S. PPARs, Obesity, and Inflammation. PPAR Res. 2006, 2007, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Imam, M.U.; Yida, Z.; Wang, F.; Wang, F. Peroxisome Proliferator-Activated Receptor Gamma (PPARγ) as a Target for Concurrent Management of Diabetes and Obesity-Related Cancer. Curr. Pharm. Des. 2017, 23, 3677–3688. [Google Scholar] [CrossRef] [PubMed]
- Haluzík, M.M.; Haluzík, M. PPAR-alpha and insulin sensitivity. Physiol. Res. 2005, 55, 115–122. [Google Scholar] [PubMed]
- Cypess, A.M.; Kahn, C.R. Brown fat as a therapy for obesity and diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 2010, 17, 143–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nedergaard, J.; Cannon, B. The Browning of White Adipose Tissue: Some Burning Issues. Cell Metab. 2014, 20, 396–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrovic, N.; Walden, T.B.; Shabalina, I.G.; Timmons, J.A.; Cannon, B.; Nedergaard, J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 2010, 285, 7153–7164. [Google Scholar] [CrossRef] [Green Version]
- Kershaw, E.E.; Flier, J.S. Adipose Tissue as an Endocrine Organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef]
- Acedo, S.C.; Gambero, S.; Cunha, F.G.P.; Lorand-Metze, I.; Gambero, A. Participation of leptin in the determination of the macrophage phenotype: An additional role in adipocyte and macrophage crosstalk. Vitr. Cell. Dev. Boil. Anim. 2013, 49, 473–478. [Google Scholar] [CrossRef]
- Al-Qahtani, S.M.; Bryzgalova, G.; Valladolid-Acebes, I.; Korach-André, M.; Dahlman-Wright, K.; Efendić, S.; Berggren, P.-O.; Portwood, N. 17β-Estradiol suppresses visceral adipogenesis and activates brown adipose tissue-specific gene expression. Horm. Mol. Boil. Clin. Investig. 2016, 29, 13–26. [Google Scholar] [CrossRef]
- Gavito, A.L.; Bautista, D.; Suarez, J.D.; Badran, S.; Arco, R.; Pavón, F.J.; Serrano, A.; Rivera, P.; Decara, J.; Cuesta, A.L.; et al. Chronic IL-6 Administration Desensitizes IL-6 Response in Liver, Causes Hyperleptinemia and Aggravates Steatosis in Diet-Induced-Obese Mice. PLoS ONE 2016, 11, e0157956. [Google Scholar] [CrossRef]
- Choi, H.M.; Doss, H.M.; Kim, K.S. Multifaceted Physiological Roles of Adiponectin in Inflammation and Diseases. Int. J. Mol. Sci. 2020, 21, 1219. [Google Scholar] [CrossRef] [Green Version]
- Kola, B.; Grossman, A.B.; Korbonits, M. The role of AMP-activated protein kinase in obesity. Front. Horm. Res. 2008, 36, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Miyazawa, M.; Kamei, A.; Abe, K.; Kojima, T. Ameliorative Effects of Mulberry (Morus alba L.) Leaves on Hyperlipidemia in Rats Fed a High-Fat Diet: Induction of Fatty Acid Oxidation, Inhibition of Lipogenesis, and Suppression of Oxidative Stress. Biosci. Biotechnol. Biochem. 2010, 74, 2385–2395. [Google Scholar] [CrossRef] [PubMed]
- Abu-Farha, M.; Al-Khairi, I.; Cherian, P.; Chandy, B.; Sriraman, D.; Al-Hubail, A.; Al-Refaei, F.H.; AlTerki, A.; Abubaker, J. Increased ANGPTL3, 4 and ANGPTL8/betatrophin expression levels in obesity and T2D. Lipids Heal. Dis. 2016, 15, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shepherd, P.R.; Kahn, B.B. Glucose Transporters and Insulin Action—Implications for Insulin Resistance and Diabetes Mellitus. N. Engl. J. Med. 1999, 341, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Jangra, S.; Raja Shekar, K.; Sharma, R.K.; Pothuraju, R.; Mohanty, A.K.; Kuruva, R.S. Ameliorative effect of fermentable fibres on adiposity and insulin resistance in C57BL/6 mice fed a high-fat and sucrose diet. Food Funct. 2019, 10, 3696–3705. [Google Scholar] [CrossRef]
- Nagata, N.; Xu, L.; Kohno, S.; Ushida, Y.; Aoki, Y.; Umeda, R.; Fuke, N.; Zhuge, F.; Ni, Y.; Nagashimada, M.; et al. Glucoraphanin Ameliorates Obesity and Insulin Resistance Through Adipose Tissue Browning and Reduction of Metabolic Endotoxemia in Mice. Diabetes 2017, 66, 1222–1236. [Google Scholar] [CrossRef] [Green Version]
- Kalis, C.; Kanzler, B.; Lembo, A.; Poltorak, A.; Galanos, C.; Freudenberg, M.A. Toll-like receptor 4 expression levels determine the degree of LPS-susceptibility in mice. Eur. J. Immunol. 2003, 33, 798–805. [Google Scholar] [CrossRef]
- Sharifnia, T.; Antoun, J.; Verriere, T.G.C.; Suarez, G.; Wattacheril, J.; Wilson, K.T.; Peek, R.M.; Abumrad, N.N.; Flynn, C.R. Hepatic TLR4 signaling in obese NAFLD. Am. J. Physiol. Liver Physiol. 2015, 309, G270–G278. [Google Scholar] [CrossRef] [Green Version]
- Cox, A.J.; West, N.P.; Cripps, A.W. Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol. 2015, 3, 207–215. [Google Scholar] [CrossRef]
- Damms-Machado, A.; Louis, S.; Schnitzer, A.; Volynets, V.; Rings, A.; Basrai, M.; Bischoff, S.C. Gut permeability is related to body weight, fatty liver disease, and insulin resistance in obese individuals undergoing weight reduction. Am. J. Clin. Nutr. 2016, 105, 127–135. [Google Scholar] [CrossRef] [Green Version]
- Mele, L.; Bidault, G.; Mena, P.; Crozier, A.; Brighenti, F.; Vidal-Puig, A.; Del Rio, D. Dietary (Poly)phenols, Brown Adipose Tissue Activation, and Energy Expenditure: A Narrative Review. Adv. Nutr. 2017, 8, 694–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, T.W.; Pereira, Q.C.; Teixeira, L.; Gambero, A.; Villena, J.A.; Ribeiro, M.L. Effects of Polyphenols on Thermogenesis and Mitochondrial Biogenesis. Int. J. Mol. Sci. 2018, 19, 2757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scazzocchio, B.; Varì, R.; Filesi, C.; D’Archivio, M.; Santangelo, C.; Giovannini, C.; Iacovelli, A.; Silecchia, G.; Volti, G.L.; Galvano, F.; et al. Cyanidin-3-O-β-Glucoside and Protocatechuic Acid Exert Insulin-Like Effects by Upregulating PPARγ Activity in Human Omental Adipocytes. Diabetes 2011, 60, 2234–2244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scazzocchio, B.; Varì, R.; Filesi, C.; Del Gaudio, I.; D’Archivio, M.; Santangelo, C.; Iacovelli, A.; Galvano, F.; Pluchinotta, F.R.; Giovannini, C.; et al. Protocatechuic acid activates key components of insulin signaling pathway mimicking insulin activity. Mol. Nutr. Food Res. 2015, 59, 1472–1481. [Google Scholar] [CrossRef]
- Lee, C.W.; Seo, J.Y.; Lee, J.; Choi, J.W.; Cho, S.; Bae, J.Y.; Sohng, J.; Kim, S.O.; Kim, J.; Park, Y.I. 3- O -Glucosylation of quercetin enhances inhibitory effects on the adipocyte differentiation and lipogenesis. Biomed. Pharmacother. 2017, 95, 589–598. [Google Scholar] [CrossRef]
- Varì, R.; Scazzocchio, B.; Santangelo, C.; Filesi, C.; Galvano, F.; D’Archivio, M.; Masella, R.; Giovannini, C. Protocatechuic Acid Prevents oxLDL-Induced Apoptosis by Activating JNK/Nrf2 Survival Signals in Macrophages. Oxidative Med. Cell. Longev. 2015, 2015, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Moriano, C.; Divakar, P.K.; Crespo, A.; Gómez-Serranillos, M.P. In vitro neuroprotective potential of lichen metabolite fumarprotocetraric acid via intracellular redox modulation. Toxicol. Appl. Pharmacol. 2017, 316, 83–94. [Google Scholar] [CrossRef]








| Gen | Sequence 5′-3′ | Sequence Accession Number | Annealing Temperature (°C) |
|---|---|---|---|
| Gapdh | D: CCATCACCATCTTCCAGGAG R: CCTGCTTCACCACCTTCTTG | AK002273.1 | 60 |
| Il-1β | D: TGATGAGAATGACCTCTTCT R: CTTCTTCAAAGATGAAGGAAA | AK225002.1 | 60 |
| Tnf-α | D: AACTAGTGGTGCCAGCCGAT R: CTTCACAGAGCAATGACTCC | AK159989.1 | 60 |
| Il-6 | D: TAGTCCTTCCTACCCCAATTTCC R: TTGGTCCTTAGCCACTCCTTCC | AK152189.1 | 60 |
| Jnk-1 | D: GATTTTGGACTGGCGAGGACT R: TAGCCCATGCCGAGAATGA | BC053027.1 | 60 |
| Jnk-2 | D: TTGTGCTGCTTTTGATACAGTTCTTGGG R: CTGGAAAGAGCTCTTCAAATTTGAT | AK031959.1 | 62 |
| Mcp-1 | D: AGCCAACTCTCACTGAAG R: TCTCCAGCCTACTCATTG | AK150937.1 | 55 |
| Leptin | D: TTCACACACGCAGTCGGTAT R: GCTGGTGAGGACCTGTTGAT | KX255818.1 | 60 |
| Leptin-r | D: GCAGTCCTCAGTGGCACCTC R: CACCGTGGGGCTACTGGAGAG | AK143989.1 | 60 |
| Ampk | D: GACTTCCTTCACAGCCTCATC R:CGCGCGACTATCAAAGACATACG | AK160612.1 | 60 |
| Ppar-α | D: AGGCTGTAAGGGCTTCTTTCG R: GGCATTTGTTCCGGTTCTTC | AK035676.1 | 62 |
| Ppar-β | D: TAGGACTGGTGATCTGTGAG R: TACAAGTGAGTGGGAGAGAG | AK028464.1 | 60 |
| Ppar-γ | D: CAGTACAGCCCCGATGACTC R: GAAAGCTCGTCCACGTCAG | AH013273.2 | 62 |
| Glut-4 | D: GAGAATACAGCTAGGACCAGTG R:TCTTATTGCAGCAGCGCCTGAG | AK137607.1 | 62 |
| Tlr-4 | D: GCCTTTCAGGGAATTAAGCTCC R: AGATCAACCGATGGACGTGTAA | JX878359.1 | 60 |
| Adiponectin | D: GATGGCAGAGATGGCACTCC R: CTTGCCAGTGCTGCCGTCAT | AK003138.1 | 56 |
| Lpl | D: TTCCAGCCAGGATGCAACA R: GGTCCACGTCTCCGAGTCC | AK150328.1 | 60 |
| Muc-1 | D: GCAGTCCTCAGTGGCACCTC R: CACCGTGGGCTACTGGAGAG | BG005441.1 | 60 |
| Muc-2 | D: GCAGTCCTCAGTGGCACCTC R: CACCGTGGGGCTACTGGAGAG | AK008250.1 | 60 |
| Muc-3 | D: CGTGGTCAACTGCGAGAATGG R: CGGCTCTATCTCTACGCTCTCC | AK136468.1 | 60 |
| Occludin | D: ACGGACCCTGACCACTATGA R: TCAGCAGCAGCCATGTACTC | AK019880.1 | 56 |
| Tff-3 | D: CCTGGTTGCTGGGTCCTCTG R:GCCACGGTTGTTACACTGCTC | D38410.1 | 60 |
| Zo-1 | D: GGGGCCTACACTGATCAAGA R: TGGAGATGAGGCTTCTGCTT | AK144506.1 | 56 |
| Cmpd | RT (min) | m/z Exp | Molecular Formula | Tentative Identification | Presence | |||
|---|---|---|---|---|---|---|---|---|
| I | K | F | V | |||||
| Organic Acids | ||||||||
| 2 | 1.9 | 191.0572 | C7H11O6 | Quinic acid | + | + | + | + |
| Benzoic Acids | ||||||||
| 3 | 5.1 | 315.0722 | C13H15O9 | Protocatechuic acid hexoside | + | + | + | + |
| 4 | 5.6 | 459.1122 | C19H23O13 | Parishin E | + | |||
| 26 | 10.5 | 469.1704 | C22H29O11 | Rengyoside D | + | |||
| 28 | 10.8 | 135.0455 | C8H7O2 | Methyl salicyl aldehyde | + | |||
| 36 | 14.5 | 221.1179 | C13H17O3 | Hexyl salicylate | + | + | + | + |
| 52 | 24.3 | 265.1448 | C15H21O4 | Gingerol | + | + | + | + |
| Cinnamic Acids | ||||||||
| 5 | 5.8 | 353.0881 | C16H17O9 | Neochlorogenic acid | + | + | + | + |
| 7 | 6.6 | 353.0865 | C16H17O9 | Chlorogenic acid | + | + | + | + |
| 8 | 6.7 | 353.0876 | C16H17O9 | Cryptochlorogenic acid | + | + | + | + |
| 9 | 6.9 | 359.0758 | C18H15O8 | Rosemarinic acid | + | + | + | + |
| 11 | 7.2 | 353.085 | C16H17O9 | Chlorogenic acid isomer | + | + | + | + |
| 22 | 9.3 | 515.1186 | C25H23O12 | Caffeoylquinic acid hexoside | + | |||
| 31 | 12.1 | 207.0659 | C11H11O4 | Dimethoxy-cinnamic acid | + | + | + | + |
| 32 | 12.6 | 163.0391 | C9H7O3 | Coumaric acid | + | + | + | |
| Flavonoids | ||||||||
| 6 | 6.2 | 625.1420 | C27H29O17 | Quercetin dihexoside | + | + | + | + |
| 12 | 7.3 | 755.2031 | C33H39O20 | Kaempferol rutinoside hexoside | + | |||
| 14 | 7.7 | 609.1469 | C27H29O16 | Rutin isomer 1 | + | + | ||
| 15 | 8.0 | 609.1467 | C27H29O16 | Rutin isomer 2 | + | + | + | + |
| 17 | 8.4 | 463.0879 | C21H19O12 | Quercetin-3-glucoside | + | + | + | + |
| 18 | 8.6 | 593.1510 | C27H29O15 | Kaempferol-3-o-rutinoside | + | + | + | + |
| 19 | 8.7 | 505.0990 | C23H21O13 | Quercetin-3-O-(6-acetylglucoside) isomer 1 | + | + | + | |
| 20 | 9.0 | 447.0908 | C21H19O11 | Kaempferol 3-o-glucoside | + | + | + | + |
| 21 | 9.2 | 505.0997 | C23H21O13 | Quercetin-3-O-(6-acetylglucoside) isomer 2 | + | + | + | |
| 23 | 9.4 | 489.1040 | C23H21O12 | Kaempferol-3-o-6″-o-acetyl-β-d glucopyranoside isomer 1 | + | + | + | |
| 24 | 9.7 | 505.0997 | C23H21O13 | Quercetin-3-O-(6-acetylglucoside) isomer 3 | + | + | + | |
| 25 | 10.0 | 489.1040 | C23H21O12 | Kaempferol-3-o-6″-o-acetyl-β-d glucopyranoside isomer 2 | + | + | + | |
| 30 | 11.5 | 301.0357 | C15H9O7 | Quercetin | + | + | + | + |
| Fatty Acids | ||||||||
| 33 | 13.1 | 183.1398 | C11H19O2 | Undecenoic acid | + | + | + | + |
| 34 | 13.7 | 309.2057 | C18H29O4 | Linoleic acid hydroperoxide isomer 1 | + | + | + | + |
| 35 | 13.9 | 309.2057 | C18H29O4 | Linoleic acid hydroperoxide isomer 2 | + | + | + | + |
| 37 | 15.7 | 309.2074 | C18H29O4 | Linoleic acid hydroperoxide isomer 3 | + | + | + | + |
| 44 | 18.6 | 337.2356 | C20H33O4 | Dihydroxy eicosatrienoic acid | + | |||
| 46 | 19.2 | 293.2161 | C18H29O3 | Hydroxy octadecatrienoic acid isomer 1 | + | + | + | |
| 47 | 19.5 | 293.2161 | C18H29O3 | Hydroxy octadecatrienoic acid isomer 2 | + | + | + | |
| 48 | 20.6 | 291.1949 | C18H27O3 | Hydroxyperoxy octadecatrienoic acid | + | + | + | |
| 49 | 20.9 | 561.3249 | C28H49O11 | Dioxo-penta-oxatritriacontanedioic acid | + | |||
| 50 | 21.2 | 295.2252 | C18H31O3 | Hydroxy octadecadienoic acid | + | + | + | |
| 51 | 21.5 | 221.1530 | C14H21O2 | Myristic acid | + | + | + | + |
| 55 | 29.4 | 277.2155 | C18H29O2 | Linolenic acid | + | + | + | + |
| 56 | 33.0 | 279.2315 | C18H31O2 | Linoleic acid | + | + | + | + |
| Others | ||||||||
| 1 | 1.9 | 341.1100 | C12H21O11 | Sugar | + | + | + | + |
| 13 | 7.6 | 481.1332 | C22H25O12 | Fumaroprotocetraric acid isomer 1 | + | + | + | + |
| 16 | 8.3 | 481.1338 | C22H25O12 | Fumaroprotocetraric acid isomer 2 | + | + | ||
| 42 | 17.7 | 593.2674 | C30H41O12 | Aceroside VIII | + | + | + | |
| 54 | 27.1 | 555.2819 | C28H43O11 | Picrasinoside F | + | |||
| Unknowns | ||||||||
| 10 | 7.1 | 431.1905 | C20H31O10 | Unk 1 | + | |||
| 27 | 10.5 | 459.2232 | C22H35O10 | Unk 2 | + | |||
| 29 | 11.2 | 357.0613 | C18H13O8 | Unk 3 | + | |||
| 38 | 16.0 | 721.3611 | C34H57O16 | Unk 4 | + | + | + | + |
| 39 | 16.4 | 721.3620 | C34H57O16 | Unk 5 | + | + | + | + |
| 40 | 17.0 | 562.3133 | C26H36 N13O2 | Unk 6 | + | + | + | + |
| 41 | 17.4 | 552.2667 | C24H42NO13 | Unk 7 | + | |||
| 43 | 18.5 | 559.3124 | C28H47O11 | Unk 8 | + | + | + | + |
| 45 | 18.9 | 559.3105 | C28H47O11 | Unk 9 | + | + | + | + |
| 53 | 26.5 | 481.2502 | C25H37O9 | Unk 10 | + | |||
| Compound | Italia | Filipina | Kokuso | Valenciana Temprana |
|---|---|---|---|---|
| Quinic acid | 3711 ± 49 b | 9341 ± 259 a | 10,994 ± 236 a | 1469 ± 55 c |
| Protocatechuic acid glucoside | 4241 ± 380 a | 1809 ± 126 b | 1368 ± 138 b,c | 905 ± 27 c |
| Neochlorogenic acid | 4430 ± 154 c | 5753 ± 43 b | 10,091 ± 737 a | 2260 ± 72 d |
| Chlorogenic acid | 5807 ± 101 b | 9089 ± 774 a | 5394 ± 274 b | 2345 ± 131 c |
| Cryptochlorogenic acid | 10,537 ± 733 b | 12,478 ± 82 a | 16,514 ± 32 a | 4602 ± 289 c |
| Rosmarinic acid | 1611 ± 87 b | 2981 ± 253 a | 1625 ± 85 b | 1343 ± 71 b |
| Chlorogenic acid isomer | 1162 ± 27 a | 1280 ± 19 a | 1288 ± 120 a | 99 ± 11 b |
| Caffeoylquinic acid hexoside | ND | ND | ND | 396 ± 13 a |
| Dimethoxy cinnamic acid | 194 ± 8 c | 1018 ± 33 a | 639 ± 107 b | 1077 ± 55 a |
| p-coumaric acid | 85 ± 5 b | 221 ± 5 a | 95 ± 9 b | ND |
| Quercetin dihexoside | NQ | 94.9 ± 0.2 a | NQ | NQ |
| Rutin isomer 1 | ND | 66 ± 3 a | ND | 23.1 ± 0.4 b |
| Rutin isomer 2 | 2343 ± 54 c | 2039 ± 46 d | 2625 ± 15 b | 2977 ± 69 a |
| Quercetin-3- glucoside | 274 ± 55 d | 3092 ± 214 a | 1302 ± 70 b | 702 ± 51 c |
| Kaempferol-3-rutinoside | 574 ± 5 b | 166 ± 8 d | 275 ± 8 c | 973 ± 33 a |
| Quercetin acetylglucoside isomer 1 | 604 ± 23 c | 2872 ± 221 b | 3353 ± 108 a | ND |
| Kaempferol-3-glucoside | 141 ± 6 c | 675 ± 10 a | 232.7 ± 0.8 b | NQ |
| Quercetin acetylglucoside isomer 2 | NQ | 1122 ± 9 a | 877 ± 22 b | ND |
| Kaempferol acetylglucoside isomer 1 | 605 ± 27 c | 1133 ± 24 b | 1367 ± 33 a | ND |
| Quercetin acetylglucoside isomer 3 | NQ | NQ | NQ | ND |
| Kaempferol acetylglucoside isomer 2 | 95 ± 5 c | 576 ± 11 a | 435 ± 26 b | ND |
| Quercetin | NQ | NQ | NQ | NQ |
| Total phenolic content | 36,414 ± 1719 b | 55,806 ± 2140 a | 58,474 ± 2021 a | 19,171 ± 877 c |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leyva-Jiménez, F.J.; Ruiz-Malagón, A.J.; Molina-Tijeras, J.A.; Diez-Echave, P.; Vezza, T.; Hidalgo-García, L.; Lozano-Sánchez, J.; Arráez-Román, D.; Cenis, J.L.; Lozano-Pérez, A.A.; et al. Comparative Study of the Antioxidant and Anti-Inflammatory Effects of Leaf Extracts from Four Different Morus alba Genotypes in High Fat Diet-Induced Obesity in Mice. Antioxidants 2020, 9, 733. https://doi.org/10.3390/antiox9080733
Leyva-Jiménez FJ, Ruiz-Malagón AJ, Molina-Tijeras JA, Diez-Echave P, Vezza T, Hidalgo-García L, Lozano-Sánchez J, Arráez-Román D, Cenis JL, Lozano-Pérez AA, et al. Comparative Study of the Antioxidant and Anti-Inflammatory Effects of Leaf Extracts from Four Different Morus alba Genotypes in High Fat Diet-Induced Obesity in Mice. Antioxidants. 2020; 9(8):733. https://doi.org/10.3390/antiox9080733
Chicago/Turabian StyleLeyva-Jiménez, Francisco Javier, Antonio Jesús Ruiz-Malagón, José Alberto Molina-Tijeras, Patricia Diez-Echave, Teresa Vezza, Laura Hidalgo-García, Jesús Lozano-Sánchez, David Arráez-Román, José Luis Cenis, Antonio Abel Lozano-Pérez, and et al. 2020. "Comparative Study of the Antioxidant and Anti-Inflammatory Effects of Leaf Extracts from Four Different Morus alba Genotypes in High Fat Diet-Induced Obesity in Mice" Antioxidants 9, no. 8: 733. https://doi.org/10.3390/antiox9080733