Control of Gene Expression by Exosome-Derived Non-Coding RNAs in Cancer Angiogenesis and Lymphangiogenesis
Abstract
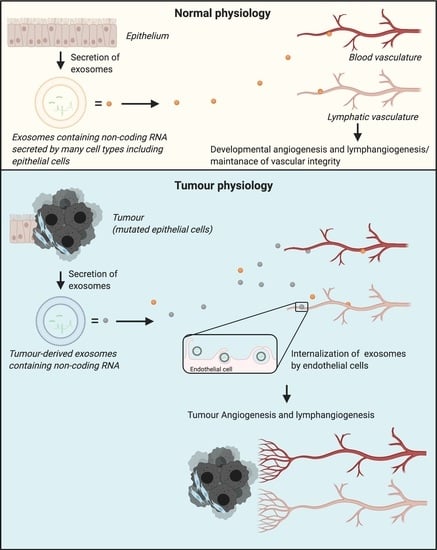
:1. Introduction
2. Tumour Angiogenesis and Lymphangiogenesis
2.1. Tumour Angiogenesis
2.2. Tumour Lymphangiogenesis
2.3. Molecular Regulation of Angiogenesis and Lymphangiogenesis in Cancer
2.3.1. VEGF Signalling Pathways in Angiogenesis and Lymphangiogenesis
2.3.2. Other Signalling Pathways
3. Role of Exosome-Derived Non-Coding RNAs in Tumour Lymphangiogenesis and Angiogenesis
3.1. Role of Exosomal MiRNAs
3.2. Role of Exosomal LncRNAs
4. Relevance of Tumour-Derived Exosomal Non-Coding RNAs for Cancer Biomarkers and Therapeutic Interventions
4.1. Non-Coding RNAs as Cancer Biomarkers
4.2. Non-Coding RNAs and Therapeutics
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Baeriswyl, V.; Christofori, G. The angiogenic switch in carcinogenesis. Semin. Cancer Biol. 2009, 19, 329–337. [Google Scholar] [CrossRef]
- Stacker, S.A.; Williams, S.P.; Karnezis, T.; Shayan, R.; Fox, S.B.; Achen, M.G. Lymphangiogenesis and lymphatic vessel remodelling in cancer. Nat. Rev. Cancer 2014, 14, 159–172. [Google Scholar] [CrossRef]
- Ferrara, N.; Hillan, K.J.; Gerber, H.-P.; Novotny, W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat. Rev. Drug Discov. 2004, 3, 391–400. [Google Scholar] [CrossRef]
- Ferrara, N. Role of vascular endothelial growth factor in the regulation of angiogenesis. Kidney Int. 1999, 56, 794–814. [Google Scholar] [CrossRef] [Green Version]
- Zampetaki, A.; Albrecht, A.; Steinhofel, K. Long Non-coding RNA Structure and Function: Is There a Link? Front. Physiol. 2018, 9, 1201. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlmann, S.; Mannsperger, H.; Zhang, J.D.; Horvat, E.-Á.; Schmidt, C.; Küblbeck, M.; Henjes, F.; Ward, A.; Tschulena, U.; Zweig, K.; et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol. Syst. Biol. 2012, 8, 570. [Google Scholar] [CrossRef] [PubMed]
- Man, H.S.J.; Sukumar, A.N.; Lam, G.C.; Turgeon, P.J.; Yan, M.S.; Ku, K.H.; Dubinsky, M.K.; Ho, J.J.D.; Wang, J.J.; Das, S.; et al. Angiogenic patterning by STEEL, an endothelial-enriched long noncoding RNA. Proc. Natl. Acad. Sci. USA 2018, 115, 2401–2406. [Google Scholar] [CrossRef] [Green Version]
- Fu, W.-M.; Lu, Y.-F.; Hu, B.-G.; Liang, W.-C.; Zhu, X.; Yang, H.-D.; Li, G.; Zhang, J.-F. Long noncoding RNA hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget 2016, 7, 4712–4723. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-X.; Xu, X.-X.; Tan, B.-Z.; Zhang, Z.; Zhou, X.-D. MicroRNA-29b Inhibits Angiogenesis by Targeting VEGFA through the MAPK/ERK and PI3K/Akt Signaling Pathways in Endometrial Carcinoma. Cell. Physiol. Biochem. 2017, 41, 933–946. [Google Scholar] [CrossRef]
- Li, Y.; Cai, B.; Shen, L.; Dong, Y.; Lu, Q.; Sun, S.; Liu, S.; Ma, S.; Ma, P.X.; Chen, J.-H. MiRNA-29b suppresses tumor growth through simultaneously inhibiting angiogenesis and tumorigenesis by targeting Akt3. Cancer Lett. 2017, 397, 111–119. [Google Scholar] [CrossRef]
- Tripathi, V.; Shen, Z.; Chakraborty, A.; Giri, S.; Freier, S.M.; Wu, X.; Zhang, Y.; Gorospe, M.; Prasanth, S.G.; Lal, A.; et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013, 9, e1003368. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-D.; Zhou, D.-M.; Sun, L.-L.; Xiao, L.; Liu, Z.; Zhou, M.; Wang, W.-B.; Li, X.-Q. LncRNA WTAPP1 Promotes Migration and Angiogenesis of Endothelial Progenitor Cells via MMP1 Through MicroRNA 3120 and Akt/PI3K/Autophagy Pathways. Stem Cells 2018, 36, 1863–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Blum, Y.; Verma, A.; Liu, Z.; Pramanik, K.; Leigh, N.R.; Chun, C.Z.; Samant, G.V.; Zhao, B.; Garnaas, M.K.; et al. A noncoding antisense RNA in tie-1 locus regulates tie-1 function in vivo. Blood 2010, 115, 133–139. [Google Scholar] [CrossRef] [Green Version]
- JJia, P.; Cai, H.; Liu, X.; Chen, J.; Ma, J.; Wang, P.; Liu, Y.; Zheng, J.; Xue, Y. Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 2016, 381, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Melo, C.A.; Drost, J.; Wijchers, P.J.; Van De Werken, H.J.; De Wit, E.; Vrielink, J.A.O.; Elkon, R.; Melo, S.A.; Léveillé, N.; Kalluri, R.; et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell 2013, 49, 524–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, M.T.Y.; Cho, H.; Lesch, H.P.; Gosselin, D.; Heinz, S.; Tanaka-Oishi, Y.; Benner, C.; Kaikkonen, M.U.; Kim, A.S.; Kosaka, M.; et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 2013, 498, 511–515. [Google Scholar] [CrossRef]
- Creamer, K.M.; Lawrence, J.B. XIST RNA: A window into the broader role of RNA in nuclear chromosome architecture. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160360. [Google Scholar] [CrossRef] [Green Version]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Kopp, F.; Chang, T.-C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Zhong, M.-E.; Chen, Y.; Zhang, G.; Xu, L.; Ge, W.; Wu, B. LncRNA H19 regulates PI3K-Akt signal pathway by functioning as a ceRNA and predicts poor prognosis in colorectal cancer: Integrative analysis of dysregulated ncRNA-associated ceRNA network. Cancer Cell Int. 2019, 19, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, S.; Lu, Z.; Zheng, S.; Zhang, H.; Zhang, G.; Wang, F.; Huang, J.; Lei, Y.; Wang, X.; Liu, C.; et al. Exosomal miR-141 promotes tumor angiogenesis via KLF12 in small cell lung cancer. J. Exp. Clin. Cancer Res. 2020, 39, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, X.; Yang, Y.; Chen, W.; Zhang, K.; Teng, B.; Huang, C.; Zhao, Q.; Qiu, Z. Hypoxic Tumor-Derived Exosomal Long Noncoding RNA UCA1 Promotes Angiogenesis via miR-96-5p/AMOTL2 in Pancreatic Cancer. Mol. Ther. Nucleic Acids 2020, 22, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Folkman, J. Tumor Angiogenesis: Therapeutic Implications. N. Engl. J. Med. 1971, 285, 1182–1186. [Google Scholar] [CrossRef]
- Tammela, T.; Zarkada, G.; Wallgard, E.; Murtomäki, A.; Suchting, S.; Wirzenius, M.; Waltari, M.; Hellström, M.; Schomber, T.; Peltonen, R.; et al. Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature 2008, 454, 656–660. [Google Scholar] [CrossRef]
- Greenfield, J.P.; Cobb, W.S.; Lyden, D. Resisting arrest: A switch from angiogenesis to vasculogenesis in recurrent malignant gliomas. J. Clin. Investig. 2010, 120, 663–667. [Google Scholar] [CrossRef]
- Baluk, P.; Hashizume, H.; McDonald, D.M. Cellular abnormalities of blood vessels as targets in cancer. Curr. Opin. Genet. Dev. 2005, 15, 102–111. [Google Scholar] [CrossRef]
- Skobe, M.; Hawighorst, T.; Jackson, D.G.; Prevo, R.; Janes, L.; Velasco, P.; Riccardi, L.; Alitalo, K.; Claffey, K.P.; Detmar, M. Induction of tumor lymphangiogenesis by VEGF-C promotes breast cancer metastasis. Nat. Med. 2001, 7, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Mandriota, S.J.; Jussila, L.; Jeltsch, M.; Compagni, A.; Baetens, D.; Prevo, R.; Banerji, S.; Huarte, J.; Montesano, R.; Jackson, D.G.; et al. Vascular endothelial growth factor-C-mediated lymphangiogenesis promotes tumour metastasis. EMBO J 2001, 20, 672–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stacker, S.A.; Caesar, C.; Baldwin, M.E.; Thornton, G.E.; Williams, R.A.; Prevo, R.; Jackson, D.G.; Nishikawa, S.-I.; Kubo, H.; Achen, M.G. VEGF-D promotes the metastatic spread of tumor cells via the lymphatics. Nat. Med. 2001, 7, 186–191. [Google Scholar] [CrossRef]
- Karnezis, T.; Shayan, R.; Caesar, C.; Roufail, S.; Harris, N.C.; Ardipradja, K.; Zhang, Y.F.; Williams, S.P.; Farnsworth, R.H.; Chai, M.G.; et al. VEGF-D promotes tumor metastasis by regulating prostaglandins produced by the collecting lymphatic endothelium. Cancer Cell 2012, 21, 181–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshida, T.; Isaka, N.; Hagendoorn, J.; Di Tomaso, E.; Chen, Y.-L.; Pytowski, B.; Fukumura, D.; Padera, T.P.; Jain, R.K. Imaging steps of lymphatic metastasis reveals that vascular endothelial growth factor-C increases metastasis by increasing delivery of cancer cells to lymph nodes: Therapeutic implications. Cancer Res. 2006, 66, 8065–8075. [Google Scholar] [CrossRef] [Green Version]
- Gogineni, A.; Caunt, M.; Crow, A.; Lee, C.V.; Fuh, G.; Van Bruggen, N.; Ye, W.; Weimer, R.M. Inhibition of VEGF-C modulates distal lymphatic remodeling and secondary metastasis. PLoS ONE 2013, 8, e68755. [Google Scholar] [CrossRef] [PubMed]
- Holmes, D.I.R.; Zachary, I. The vascular endothelial growth factor (VEGF) family- angiogenic factors in health and disease. Genome Biol. 2005, 6, 209. [Google Scholar] [CrossRef] [Green Version]
- Hiratsuka, S.; Minowa, O.; Kuno, J.; Noda, T.; Shibuya, M. Flt-1 lacking the tyrosine kinase domain is sufficient for normal development and angiogenesis in mice. Proc. Natl. Acad. Sci. USA 1998, 95, 9349–9354. [Google Scholar] [CrossRef] [Green Version]
- De Vries, C.; Escobedo, J.A.; Ueno, H.; Houck, K.; Ferrara, N.; Williams, L.T. The fms-Like Tyrosine Kinase, a Receptor for Vascular Endothelial Growth Factor. Science 1992, 255, 989–991. [Google Scholar] [CrossRef]
- Terman, B.I.; Dougher-Vermazen, M.; Carrion, M.E.; Dimitrov, D.; Armellino, D.C.; Gospodarowicz, D.; Böhlen, P. Identification of the KDR tyrosine kinase as a receptor for vascular endothelial cell growth factor. Biochem. Biophys. Res. Commun. 1992, 187, 1579–1586. [Google Scholar] [CrossRef]
- Joukov, V.; Sorsa, T.; Kumar, V.; Jeltsch, M.; Claesson-Welsh, L.; Cao, Y.; Saksela, O.; Kalkkinen, N.; Alitalo, K. Proteolytic processing regulates receptor specificity and activity of VEGF-C. EMBO J. 1997, 16, 3898–3911. [Google Scholar] [CrossRef] [Green Version]
- Achen, M.G.; Jeltsch, M.; Kukk, E.; Mäkinen, T.; Vitali, A.; Wilks, A.F.; Alitalo, K.; Stacker, S.A. Vascular endothelial growth factor D (VEGF-D) is a ligand for the tyrosine kinases VEGF receptor 2 (Flk1) and VEGF receptor 3 (Flt4). Proc. Natl. Acad. Sci. USA 1998, 95, 548–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, N.R.; Baker, D.; James, N.H.; Ratcliffe, K.; Jenkins, M.; Ashton, S.E.; Sproat, G.; Swann, R.; Gray, N.; Ryan, A.; et al. Vascular endothelial growth factor receptors VEGFR-2 and VEGFR-3 are localized primarily to the vasculature in human primary solid cancers. Clin. Cancer Res. 2010, 16, 3548–3561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerber, M.; Reiss, Y.; Wickersheim, A.; Jugold, M.; Kiessling, F.; Heil, M.; Tchaikovski, V.; Waltenberger, J.; Shibuya, M.; Plate, K.H.; et al. Flt-1 signaling in macrophages promotes glioma growth in vivo. Cancer Res. 2008, 68, 7342–7351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuchrist, C.; Erovic, B.M.; Handisurya, A.; Fischer, M.B.; Steiner, G.E.; Hollemann, D.; Gedlicka, C.; Saaristo, A.; Burian, M. Vascular endothelial growth factor C and vascular endothelial growth factor receptor 3 expression in squamous cell carcinomas of the head and neck. Head Neck 2003, 25, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Schoppmann, S.F.; Fenzl, A.; Nagy, K.; Unger, S.; Bayer, G.; Geleff, S.; Gnant, M.; Horvat, R.; Jakesz, R.; Birner, P. VEGF-C expressing tumor-associated macrophages in lymph node positive breast cancer: Impact on lymphangiogenesis and survival. Surgery 2006, 139, 839–846. [Google Scholar] [CrossRef]
- Mäkinen, T.; Veikkola, T.; Mustjoki, S.; Karpanen, T.; Catimel, B.; Nice, E.C.; Wise, L.; Mercer, A.; Kowalski, H.; Kerjaschki, D.; et al. Isolated lymphatic endothelial cells transduce growth, survival and migratory signals via the VEGF-C/D receptor VEGFR-3. EMBO J. 2001, 20, 4762–4773. [Google Scholar] [CrossRef] [Green Version]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef] [Green Version]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 1–13. [Google Scholar] [CrossRef]
- Fagiani, E.; Lorentz, P.; Kopfstein, L.; Christofori, G. Angiopoietin-1 and -2 exert antagonistic functions in tumor angiogenesis, yet both induce lymphangiogenesis. Cancer Res. 2011, 71, 5717–5727. [Google Scholar] [CrossRef] [Green Version]
- Matsuo, M.; Yamada, S.; Koizumi, K.; Sakurai, H.; Saiki, I. Tumour-derived fibroblast growth factor-2 exerts lymphangiogenic effects through Akt/mTOR/p70S6kinase pathway in rat lymphatic endothelial cells. Eur. J. Cancer 2007, 43, 1748–1754. [Google Scholar] [CrossRef]
- Cao, R.; Björndahl, M.A.; Religa, P.; Clasper, S.; Garvin, S.; Galter, D.; Meister, B.; Ikomi, F.; Tritsaris, K.; Dissing, S.; et al. PDGF-BB induces intratumoral lymphangiogenesis and promotes lymphatic metastasis. Cancer Cell 2004, 6, 333–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibot, L.; Galbraith, T.; Kloos, B.; Das, S.; Lacroix, D.A.; Auger, F.A.; Skobe, M. Cell-based approach for 3D reconstruction of lymphatic capillaries in vitro reveals distinct functions of HGF and VEGF-C in lymphangiogenesis. Biomaterials 2016, 78, 129–139. [Google Scholar] [CrossRef] [Green Version]
- Björndahl, M.A.; Cao, R.; Burton, J.B.; Brakenhielm, E.; Religa, P.; Galter, D.; Wu, L.; Cao, Y. Vascular endothelial growth factor-a promotes peritumoral lymphangiogenesis and lymphatic metastasis. Cancer Res. 2005, 65, 9261–9268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marino, D.; Angehrn, Y.; Klein, S.; Riccardi, S.; Baenziger-Tobler, N.; Otto, V.I.; Pittelkow, M.; Detmar, M. Activation of the epidermal growth factor receptor promotes lymphangiogenesis in the skin. J. Dermatol. Sci. 2013, 71, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Oka, M.; Iwata, C.; Suzuki, H.I.; Kiyono, K.; Morishita, Y.; Watabe, T.; Komuro, A.; Kano, M.R.; Miyazono, K. Inhibition of endogenous TGF- signaling enhances lymphangiogenesis. Blood 2008, 111, 4571–4579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noren, N.K.; Lu, M.; Freeman, A.L.; Koolpe, M.; Pasquale, E.B. Interplay between EphB4 on tumor cells and vascular ephrin-B2 regulates tumor growth. Proc. Natl. Acad. Sci. USA 2004, 101, 5583–5588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaahtomeri, K.; Karaman, S.; Mäkinen, T.; Alitalo, K. Lymphangiogenesis guidance by paracrine and pericellular factors. Genes Dev. 2017, 31, 1615–1634. [Google Scholar] [CrossRef]
- Kahlert, C.; Kalluri, R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 2013, 91, 431–437. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Dong, J.; Li, S.; Wang, C.; Ding, H.; Li, H.; Su, X.; Ge, X.; Sun, L.; Bai, C.; et al. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int. J. Biol. Sci. 2015, 11, 961–969. [Google Scholar] [CrossRef]
- Lin, X.-J.; Fang, J.-H.; Yang, X.-J.; Zhang, C.; Yuan, Y.; Zheng, L.; Zhuang, S. Hepatocellular Carcinoma Cell-Secreted Exosomal MicroRNA-210 Promotes Angiogenesis In Vitro and In Vivo. Mol. Ther. Nucleic Acids 2018, 11, 243–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Liu, Y.; Zhang, C.; Duan, C. Multiple Roles of Exosomal Long Noncoding RNAs in Cancers. BioMed Res. Int. 2019, 2019, 1–12. [Google Scholar] [CrossRef]
- Masoumi-Dehghi, S.; Babashah, S.; Sadeghizadeh, M. microRNA-141-3p-containing small extracellular vesicles derived from epithelial ovarian cancer cells promote endothelial cell angiogenesis through activating the JAK/STAT3 and NF-kappaB signaling pathways. J. Cell Commun. Signal. 2020, 14, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Park, S.; Kim, H.; Choi, Y.J.; Kim, S.Y.; Sung, K.J.; Sung, Y.H.; Choi, C.-M.; Yun, M.; Yi, Y.-S.; et al. Tumor-derived exosomal miR-619-5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1.4. Cancer Lett. 2020, 475, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Duan, B.; Shi, S.; Yue, H.; You, B.; Shan, Y.; Zhu, Z.; Bao, L.; You, Y. Exosomal miR-17-5p promotes angiogenesis in nasopharyngeal carcinoma via targeting BAMBI. J. Cancer 2019, 10, 6681–6692. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, H.; Xu, H.; Zhao, H.; Xiong, N. Hypoxic Cancer-Secreted Exosomal miR-182-5p Promotes Glioblastoma Angiogenesis by Targeting Kruppel-like Factor 2 and 4. Mol. Cancer Res. 2020, 18, 1218–1231. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhang, H.; Deng, T.; Ning, T.; Liu, R.; Liu, D.; Bai, M.; Ying, G.; Ba, Y. Exosomes Carrying MicroRNA-155 Target Forkhead Box O3 of Endothelial Cells and Promote Angiogenesis in Gastric Cancer. Mol. Ther. Oncolytics 2019, 15, 223–233. [Google Scholar] [CrossRef] [Green Version]
- Deng, T.; Zhang, H.; Yang, H.; Wang, H.; Bai, M.; Sun, W.; Wang, X.; Si, Y.; Ning, T.; Zhang, L.; et al. Exosome miR-155 Derived from Gastric Carcinoma Promotes Angiogenesis by Targeting the c-MYB/VEGF Axis of Endothelial Cells. Mol. Ther. Nucleic Acids 2020, 19, 1449–1459. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, H.; Ge, S.; Ning, T.; Bai, M.; Li, J.; Li, S.; Sun, W.; Deng, T.; Zhang, L.; et al. Exosome-Derived miR-130a Activates Angiogenesis in Gastric Cancer by Targeting C-MYB in Vascular Endothelial Cells. Mol. Ther. 2018, 26, 2466–2475. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, H.; Cao, J.; Ye, C. Exosomes from Adipose-Derived Stem Cells Promotes VEGF-C-Dependent Lymphangiogenesis by Regulating miRNA-132/TGF-beta Pathway. Cell. Physiol. Biochem. 2018, 49, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.-F.; Ma, J.; Huang, L.; Yi, H.-Y.; Zhang, Y.-M.; Wu, X.-G.; Yan, R.-M.; Liang, L.; Zhong, M.; Yu, Y.-H.; et al. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1. Oncogene 2019, 38, 1256–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Yang, X.; Xue, X.; Sun, D.; Cai, P.; Song, Q.; Zhang, B.; Qin, L. HANR promotes lymphangiogenesis of hepatocellular carcinoma via secreting miR-296 exosome and regulating EAG1/VEGFA signaling in HDLEC cells. J. Cell. Biochem. 2019, 120, 17699–17708. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kgamma to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018, 78, 4586–4598. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Zhang, Y.; Yan, R.; Huang, L.; Mellor, A.L.; Yang, Y.; Chen, X.; Wei, W.; Wu, X.; Yu, L.; et al. Exosome-derived miR-142-5p remodels lymphatic vessels and induces IDO to promote immune privilege in the tumour microenvironment. Cell Death Differ. 2020, 1–15. [Google Scholar] [CrossRef]
- Fan, J.; Xu, G.; Chang, Z.; Zhu, L.; Yao, J. miR-210 transferred by lung cancer cell-derived exosomes may act as proangiogenic factor in cancer-associated fibroblasts by modulating JAK2/STAT3 pathway. Clin. Sci. 2020, 134, 807–825. [Google Scholar] [CrossRef]
- Hsieh, C.-H.; Tai, S.-K.; Yang, M.-H. Snail-overexpressing Cancer Cells Promote M2-Like Polarization of Tumor-Associated Macrophages by Delivering MiR-21-Abundant Exosomes. Neoplasia 2018, 20, 775–788. [Google Scholar] [CrossRef]
- Corliss, B.A.; Azimi, M.S.; Munson, J.M.; Peirce, S.M.; Murfee, W.L. Macrophages: An Inflammatory Link Between Angiogenesis and Lymphangiogenesis. Microcirculation 2016, 23, 95–121. [Google Scholar] [CrossRef]
- Pakravan, K.; Babashah, S.; Sadeghizadeh, M.; Mowla, S.J.; Mossahebi-Mohammadi, M.; Ataei, F.; Dana, N.; Javan, M. MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes suppresses in vitro angiogenesis through modulating the mTOR/HIF-1alpha/VEGF signaling axis in breast cancer cells. Cell. Oncol. 2017, 40, 457–470. [Google Scholar] [CrossRef]
- Diaz-Montero, C.M.; Salem, M.L.; Nishimura, M.I.; Garrett-Mayer, E.; Cole, D.J.; Montero, A.J. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol. Immunother. 2008, 58, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Feng, S.; Cheng, X.; Zhang, L.; Lu, X.; Chaudhary, S.; Teng, R.; Frederickson, C.; Champion, M.M.; Zhao, R.; Cheng, L.; et al. Myeloid-derived suppressor cells inhibit T cell activation through nitrating LCK in mouse cancers. Proc. Natl. Acad. Sci. USA 2018, 115, 10094–10099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Z.; Rong, Y.; Teng, Y.; Zhuang, X.; Samykutty, A.; Mu, J.; Zhang, L.; Cao, P.; Yan, J.; Miller, D.; et al. Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis. Oncogene 2017, 36, 639–651. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Wang, S. Angio-LncRs: LncRNAs that regulate angiogenesis and vascular disease. Theranostics 2018, 8, 3654–3675. [Google Scholar] [CrossRef]
- Ma, X.; Li, Z.; Li, T.; Zhu, L.; Li, Z.; Tian, N. Long non-coding RNA HOTAIR enhances angiogenesis by induction of VEGFA expression in glioma cells and transmission to endothelial cells via glioma cell derived-extracellular vesicles. Am. J. Transl. Res. 2017, 9, 5012–5021. [Google Scholar]
- Lang, H.-L.; Hu, G.-W.; Chen, Y.; Liu, Y.; Tu, W.; Lu, Y.-M.; Wu, L.; Xu, G.-H. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 959–972. [Google Scholar] [PubMed]
- Wang, Y.; Li, Z.; Xu, P.; Huang, L.; Tong, J.; Huang, H.; Meng, A. Angiomotin-like2 gene (amotl2) is required for migration and proliferation of endothelial cells during angiogenesis. J. Biol. Chem. 2011, 286, 41095–41104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Luo, Y.; Cao, J.; Wang, X.; Miao, Z.; Shao, G. Exosomal lncRNA FAM225A accelerates esophageal squamous cell carcinoma progression and angiogenesis via sponging miR-206 to upregulate NETO2 and FOXP1 expression. Cancer Med. 2020, 9, 8600–8611. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Zhang, Z.; Cheng, F.; Shao, Z. Exosomal lncRNA RAMP2-AS1 Derived from Chondrosarcoma Cells Promotes Angiogenesis Through miR-2355-5p/VEGFR2 Axis. OncoTargets Ther. 2020, 13, 3291–3301. [Google Scholar] [CrossRef] [Green Version]
- Lang, H.-L.; Hu, G.-W.; Zhang, B.; Kuang, W.; Chen, Y.; Wu, L.; Xu, G.-H. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol. Rep. 2017, 38, 785–798. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Zhao, Y.; Yu, Z.-Y.; Zhang, R.-D.; Li, S.-A.; Zhang, P.; Shan, T.-K.; Liu, X.-Y.; Wang, Z.-M.; Zhao, P.-C.; et al. Glioma exosomal microRNA-148a-3p promotes tumor angiogenesis through activating the EGFR/MAPK signaling pathway via inhibiting ERRFI1. Cancer Cell Int. 2020, 20, 1–16. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Zhou, X.; Luo, X.; Liu, K.; Jiang, E.; Chen, Y.; Shao, Z.; Shang, Z. OSCC Exosomes Regulate miR-210-3p Targeting EFNA3 to Promote Oral Cancer Angiogenesis through the PI3K/AKT Pathway. BioMed Res. Int. 2020, 2020, 1–13. [Google Scholar] [CrossRef]
- Monaco, F.; Gaetani, S.; Alessandrini, F.; Tagliabracci, A.; Bracci, M.; Valentino, M.; Neuzil, J.; Amati, M.; Bovenzi, M.; Tomasetti, M.; et al. Exosomal transfer of miR-126 promotes the anti-tumour response in malignant mesothelioma: Role of miR-126 in cancer-stroma communication. Cancer Lett. 2019, 463, 27–36. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhu, W.; Chen, Q.; Yuan, Y.; Wang, Y.; Wang, J.; Wu, X. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics 2019, 9, 8206–8220. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Li, J.; Yang, H.; Zhang, H.; Zhou, Z.; Deng, T.; Zhu, K.; Ning, T.; Fan, Q.; Ying, G.; et al. miR-135b Delivered by Gastric Tumor Exosomes Inhibits FOXO1 Expression in Endothelial Cells and Promotes Angiogenesis. Mol. Ther. 2019, 27, 1772–1783. [Google Scholar] [CrossRef]
- Du, L.; Li, G.; Yang, Y.; Yang, G.; Wan, J.; Ma, Z.; Hou, Y. Exosomes from microRNA-199-3p-modified adipose-derived stem cells promote proliferation and migration of endothelial tip cells by downregulation of semaphorin 3A. Int. J. Clin. Exp. Pathol. 2018, 11, 4879–4888. [Google Scholar]
- An, Y.; Zhao, J.; Nie, F.; Qin, Z.; Xue, H.; Wang, G.; Li, D. Exosomes from Adipose-Derived Stem Cells (ADSCs) Overexpressing miR-21 Promote Vascularization of Endothelial Cells. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raimondi, L.; De Luca, A.; Gallo, A.; Costa, V.; Russelli, G.; Cuscino, N.; Manno, M.; Raccosta, S.; Carina, V.; Bellavia, D.; et al. Osteosarcoma cell-derived exosomes affect tumor microenvironment by specific packaging of microRNAs. Carcinogenesis 2020, 41, 666–677. [Google Scholar] [CrossRef]
- Wang, Z.-F.; Liao, F.; Wu, H.; Dai, J. Glioma stem cells-derived exosomal miR-26a promotes angiogenesis of microvessel endothelial cells in glioma. J. Exp. Clin. Cancer Res. 2019, 38, 201. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Ma, X.; Wang, J.; Zhao, Y.; Wang, Y.; Bihl, J.C.; Chen, Y.; Jiang, C. Glioma stem cells-derived exosomes promote the angiogenic ability of endothelial cells through miR-21:VEGF signal. Oncotarget 2017, 8, 36137–36148. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.-G.; Zhou, C.-F.; Zhang, Y.-M.; Yan, R.-M.; Wei, W.-F.; Chen, X.-J.; Yi, H.-Y.; Liang, L.-J.; Fan, L.-S.; Liang, L.; et al. Cancer-derived exosomal miR-221-3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis 2019, 22, 397–410. [Google Scholar] [CrossRef]
- Chen, X.; Yang, F.; Zhang, T.; Wang, W.; Xi, W.; Li, Y.; Zhang, D.; Huo, Y.; Zhang, J.; Yang, A.; et al. MiR-9 promotes tumorigenesis and angiogenesis and is activated by MYC and OCT4 in human glioma. J. Exp. Clin. Cancer Res. 2019, 38, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Bao, L.; You, B.; Shi, S.; Shan, Y.; Zhang, Q.; Yue, H.; Zhang, J.; Zhang, W.; Shi, Y.; Liu, Y.; et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene 2018, 37, 2873–2889. [Google Scholar] [CrossRef]
- Hsu, Y.-L.; Hung, J.-Y.; Chang, W.-A.; Lin, Y.-S.; Pan, Y.-C.; Tsai, P.-H.; Wu, C.-Y.; Kuo, P.-L. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 2017, 36, 4929–4942. [Google Scholar] [CrossRef]
- Liu, J.Y.; Jiang, L.; He, T.; Liu, J.J.; Fan, J.Y.; Xu, X.H.; Tang, B.; Shi, Y.; Zhao, Y.L.; Qian, F.; et al. NETO2 promotes invasion and metastasis of gastric cancer cells via activation of PI3K/Akt/NF-kappaB/Snail axis and predicts outcome of the patients. Cell Death Dis. 2019, 10, 162. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Ridinger, J.; Rupp, A.-K.; Janssen, J.W.G.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.; Siu, L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chang, S.; Li, G.; Sun, Y. Application of liquid biopsy in precision medicine: Opportunities and challenges. Front. Med. 2017, 11, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Tellez-Gabriel, M.; Heymann, D. Exosomal lncRNAs: The newest promising liquid biopsy. Cancer Drug Resist. 2019. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, R.; So, J.B.; Zhu, F.; Too, H.-P.; Yeoh, K.-G.; Yoong, J.S.-Y. Evaluating the Use of microRNA Blood Tests for Gastric Cancer Screening in a Stratified Population-Level Screening Program: An Early Model-Based Cost-Effectiveness Analysis. Value Health 2020, 23, 1171–1179. [Google Scholar] [CrossRef]
- He, C.; Yang, W.; Yang, J.; Ding, J.; Li, S.; Wu, H.; Zhou, F.; Jiang, Y.; Teng, L.; Yang, J. Long Noncoding RNA MEG3 Negatively Regulates Proliferation and Angiogenesis in Vascular Endothelial Cells. DNA Cell Biol. 2017, 36, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Besnier, M.; Shantikumar, S.; Anwar, M.; Dixit, P.; Chamorro-Jorganes, A.; Sweaad, W.; Sala-Newby, G.; Madeddu, P.; Thomas, A.C.; Howard, L.; et al. miR-15a/-16 Inhibit Angiogenesis by Targeting the Tie2 Coding Sequence: Therapeutic Potential of a miR-15a/16 Decoy System in Limb Ischemia. Mol. Ther. Nucleic Acids 2019, 17, 49–62. [Google Scholar] [CrossRef] [Green Version]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Sun, J.; Zhao, Z.; Lei, W.; Chen, Y.; Wang, X.; Yang, J.; Shen, Z. A brief review: Adipose-derived stem cells and their therapeutic potential in cardiovascular diseases. Stem Cell Res. Ther. 2017, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Jin, X.; Hu, C.-F.; Li, R.; Zhou, Z.; Shen, C.-X. Exosomes Derived from Mesenchymal Stem Cells Rescue Myocardial Ischaemia/Reperfusion Injury by Inducing Cardiomyocyte Autophagy Via AMPK and Akt Pathways. Cell. Physiol. Biochem. 2017, 43, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Cells Producing Exosomes | miRNA in Exosomes | Target Gene | Role of MiRNA in Promoting Lymphangiogenesis | Recipient Cells | Reference |
|---|---|---|---|---|---|
| Adipose-derived stem cells | miR-132 | Smad-7 | Regulating TGF-β pathway | LECs | [71] |
| Cervical squamous cell carcinoma cells | miR-221-3p | Vasohibin-1 | Regulating an inhibitor of lymphangiogenesis implicated in microtubule dynamics | LECs | [72] |
| Hepatocellular carcinoma cells | miR-296 | EAG1 | Inhibiting VEGFA expression | LECs | [73] |
| Hypoxic pancreatic cancer cells | miR-301a-3p | PTEN | Promoting M2 polarization of macrophages by activation of the PTEN/PI3Kγ pathway | Macrophages | [74] |
| Cervical squamous cell carcinoma cells | miR-142-5p | ARID2 | Inducing IDO expression via ARID2–DNMT1–IFN-γ signalling to suppress CD8+ T cells | LECs | [72,75] |
| Cells Producing Exosomes | ncRNA in Exosomes | mRNA/miRNA Target | Role of ncRNA in Promoting Angiogenesis | Recipient Cells | Reference |
|---|---|---|---|---|---|
| Hypoxic pancreatic cancer cells | UCA1 lncRNA | miR-96-5p | Sponging miR-96-5p thus derepressing its target AMOTL2 thereby activating ERK1/ERK2 axis | HUVECs | [25] |
| Oesophageal squamous cell carcinoma cells | FAM225A lncRNA | miR-206 | Sponging miR-206 thus derepressing its targets NETO2 and FOXP1 thereby activating PI3K/Akt/NF-κB/Snail axis | HUVECs | [87] |
| Chondrosarcoma cells | RAMP2-AS1 lncRNA | miR-2355-5p | Sponging miR-2355-5p thus derepressing its target VEGFR2 thereby increasing angiogenic cell surface receptors | HUVECs | [88] |
| Glioma cells | POU3F3 lncRNA | bFGF, VEGFA, bFGFR | Increasing the expression of bFGF, VEGFA and bFGFR in endothelial cells | HBMECs | [85] |
| Glioma cells | HOTAIR lncRNA | VEGFA | Increasing the expression of VEGFA in endothelial cells | HBMECs | [84] |
| Glioma cells | CCAT2 lncRNA | VEGFA, TGF-β, Bcl-2, and Bax | Increasing the expression of VEGFA and other angiogenic signalling in endothelial cells plus decreasing apoptosis | HUVECs | [89] |
| Small-cell lung cancer cells | miR-141 | KLF12 | Repressing an anti-angiogenic transcriptional factor | HUVECs | [24] |
| Glioma cells | miR-148a-3p | ERRFI1 | Repressing an anti-angiogenic cell surface receptor | HUVECs | [90] |
| Oesophageal squamous cell carcinoma cells | miR-210-3p | EphrinA3 | Repressing ephrinA3 and therefore activating PI3K/AKT signalling | HUVECs | [91] |
| Glioblastoma multiforme cells | miR-182-5p | KLF2 and KLF4 | Repressing anti-angiogenic transcription factors | HUVECs | [67] |
| Lung cancer cells | miR-210 | TET2 | Reprogramming normal fibroblasts into CAFs | Fibroblasts | [76] |
| Epithelial ovarian cancer cells | miR-141-3p | SOCS5 | Repressing an inhibitor of the JAK/STAT3 and NF-κB signalling pathways | HUVECs | [64] |
| Non-small cell lung cancer cells | miR-619-5p | RCAN1.4 | Repressing an inhibitor of the calcineurin/NFAT pathway | HUVECs | [65] |
| Gastric carcinoma cells | miR-130a | c-MYB | Repressing an inhibitor of the expression of VEGFA | HUVECs | [70] |
| Gastric carcinoma cells | miR-155 | c-MYB | Repressing an inhibitor of the expression of VEGFA | HUVECs | [69] |
| Gastric carcinoma cells | miR-155 | FOXO3a | Repressing anti-angiogenic transcription factors | HUVECs | [68] |
| Nasopharyngeal carcinoma cells | miR-17-5p | BAMBI | Repressing an inhibitor of the VEGFA/AKT axis | HUVECs | [66] |
| HUVECs | miR-126 | IRS1, VEGFA and EGFL7 | Targeting crucial factors involved in this process | Malignant mesothelioma cells | [92] |
| Ovarian cancer cells | miR-205 | PTEN | Repressing a phosphatase that negatively regulates AKT | HUVECs | [93] |
| Gastric cancer cells | miR-135b | FOXO1 | Repressing anti-angiogenic transcription factors | HUVECs | [94] |
| Adipose-derived stem cells | miR-199-3p | Sema3A | Driving the migration of endothelial tip cells and their sprouting | HUVECs | [95] |
| Adipose-derived stem cells | miR-21 | PTEN | Repressing a phosphatase that negatively regulates AKT | HUVECs | [96] |
| Osterosarcoma cells | miR-21 | PTEN | Repressing a phosphatase that negatively regulates AKT | HUVECs | [97] |
| Glioma stem cells | miR-26a | PTEN | Repressing a phosphatase that negatively regulates AKT | HUVECs | [98] |
| Glioma stem cells | miR-21 | Not identified | Upregulating VEGFA expression in endothelial cells | HUVECs | [99] |
| Cervical squamous cell carcinoma cells | miR-221-3p | THBS2 | Repressing a potent endogenous inhibitor of angiogenesis | HUVECs | [100] |
| Glioma cells | miR-9 | COL18A1, THBS2, PTCH1 and PHD3 | Repressing endogenous inhibitors of angiogenesis and initiating HIF-1α/VEGF signal transduction | HUVECs | [101] |
| Nasopharyngeal carcinoma cells | miR-23a | TSGA10 | Repressing TSGA10, a novel inhibitor of angiogenesis | HUVECs | [102] |
| Mesenchymal stem cells | miR-100 | mTOR | Inducing expression of VEGFA in tumour cells | Breast cancer cells | [79] |
| Hypoxic lung cancer cells | miR-23a | PHD1, PHD2 and ZO-1 | Initiating HIF-1α/VEGFA signal transduction, and promoting vascular permeability by destabilising cellular junctions | HUVECs | [103] |
| Hepatocellular carcinoma cells | miR-210 | SMAD4 and STAT6 | Repressing anti-angiogenic transcription factors and signal transducers | HUVECs | [62] |
| Head and neck cancer cells | miR-21 | PTEN, PDCD4 and IGFBP3 | Stimulating M2 polarization of tumour-associated macrophages | Macrophages | [77] |
| Myeloid derived suppressor cells | miR-126a | Not identified | Promoting an inflammatory milieu that leads to metastasis | CD4+ T-helper cell type-2 | [82] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arcucci, V.; Stacker, S.A.; Achen, M.G. Control of Gene Expression by Exosome-Derived Non-Coding RNAs in Cancer Angiogenesis and Lymphangiogenesis. Biomolecules 2021, 11, 249. https://doi.org/10.3390/biom11020249
Arcucci V, Stacker SA, Achen MG. Control of Gene Expression by Exosome-Derived Non-Coding RNAs in Cancer Angiogenesis and Lymphangiogenesis. Biomolecules. 2021; 11(2):249. https://doi.org/10.3390/biom11020249
Chicago/Turabian StyleArcucci, Valeria, Steven A. Stacker, and Marc G. Achen. 2021. "Control of Gene Expression by Exosome-Derived Non-Coding RNAs in Cancer Angiogenesis and Lymphangiogenesis" Biomolecules 11, no. 2: 249. https://doi.org/10.3390/biom11020249