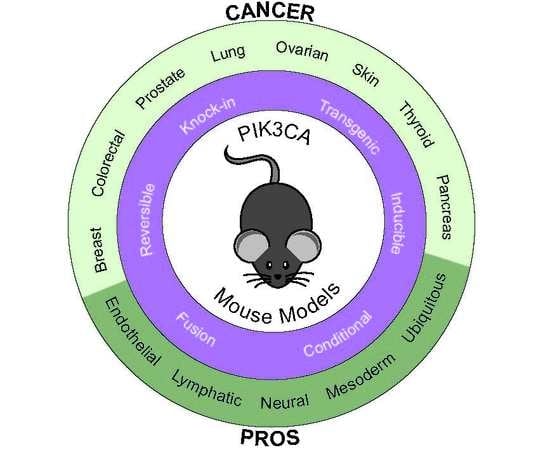
Mouse Models for Exploring the Biological Consequences and Clinical Significance of PIK3CA Mutations
Abstract
:1. Introduction
2. Modelling PIK3CA-Induced Cancers with Genetically Engineered Mouse Models
2.1. Fusion Models
2.2. Mutation-Specific Transgenic Models
2.3. Reversible Models
2.4. Conditional Knock-In Models
3. Mouse Models of Non-Malignant PIK3CA-Related Conditions
4. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Vanhaesebroeck, B.; Guillermet-Guibert, J.; Graupera, M.; Bilanges, B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell. Biol. 2010, 11, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Vogt, P.K. Class I PI3K in oncogenic cellular transformation. Oncogene 2008, 27, 5486–5496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, L.M.; Yuzugullu, H.; Zhao, J.J. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2015, 15, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Marone, R.; Cmiljanovic, V.; Giese, B.; Wymann, M.P. Targeting phosphoinositide 3-kinase—moving towards therapy. Biochim. Biophys. Acta Proteins Proteom. 2008, 1784, 159–185. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Stephens, L.; Hawkins, P. PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell. Biol. 2012, 13, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Campbell, I.G.; Russell, S.E.; Choong, D.Y.; Montgomery, K.G.; Ciavarella, M.L.; Hooi, C.S.; Cristiano, B.E.; Pearson, R.B.; Phillips, W.A. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004, 64, 7678–7681. [Google Scholar] [CrossRef]
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef]
- Millis, S.Z.; Ikeda, S.; Reddy, S.; Gatalica, Z.; Kurzrock, R. Landscape of phosphatidylinositol-3-kinase pathway alterations across 19 784 diverse solid tumors. JAMA Oncol. 2016, 2, 1565–1573. [Google Scholar] [CrossRef]
- Zhang, Y.; Kwok-Shing Ng, P.; Kucherlapati, M.; Chen, F.; Liu, Y.; Tsang, Y.H.; de Velasco, G.; Jeong, K.J.; Akbani, R.; Hadjipanayis, A.; et al. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell 2017, 31, 820–832.e3. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Xing, M.; Mambo, E.; Huang, X.; Liu, J.; Guo, Z.; Chatterjee, A.; Goldenberg, D.; Gollin, S.M.; Sukumar, S.; et al. Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 2005, 7, R609. [Google Scholar] [CrossRef]
- Levine, D.A.; Bogomolniy, F.; Yee, C.J.; Lash, A.; Barakat, R.R.; Borgen, P.I.; Boyd, J. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin. Cancer Res. 2005, 11, 2875–2878. [Google Scholar] [CrossRef]
- Dogruluk, T.; Tsang, Y.H.; Espitia, M.; Chen, F.; Chen, T.; Chong, Z.; Appadurai, V.; Dogruluk, A.; Eterovic, A.K.; Bonnen, P.E.; et al. Identification of variant-specific functions of PIK3CA by rapid phenotyping of rare mutations. Cancer Res. 2015, 75, 1–14. [Google Scholar] [CrossRef]
- Kurek, K.C.; Luks, V.L.; Ayturk, U.M.; Alomari, A.I.; Fishman, S.J.; Spencer, S.A.; Mulliken, J.B.; Bowen, M.E.; Yamamoto, G.L.; Kozakewich, H.P.W.; et al. Somatic Mosaic Activating Mutations in PIK3CA Cause CLOVES Syndrome. Am. J. Hum. Genet. 2012, 90, 1108–1115. [Google Scholar] [CrossRef] [Green Version]
- Lindhurst, M.J.; Parker, V.E.; Payne, F.; Sapp, J.C.; Rudge, S.; Harris, J.; Witkowski, A.M.; Zhang, Q.; Groeneveld, M.P.; Scott, C.E.; et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat. Genet. 2012, 44, 928–933. [Google Scholar] [CrossRef]
- Luks, V.L.; Kamitaki, N.; Vivero, M.P.; Uller, W.; Rab, R.; Bovée, J.V.; Rialon, K.L.; Guevara, C.J.; Alomari, A.I.; Greene, A.K.; et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J. Pediatr. 2015, 166, 1048–1054.e5. [Google Scholar] [CrossRef]
- Lee, J.H.; Huynh, M.; Silhavy, J.L.; Kim, S.; Dixon-Salazar, T.; Heiberg, A.; Scott, E.; Bafna, V.; Hill, K.J.; Collazo, A.; et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat. Genet. 2012, 44, 941–945. [Google Scholar] [CrossRef] [Green Version]
- Keppler-Noreuil, K.M.; Sapp, J.C.; Lindhurst, M.J.; Parker, V.E.; Blumhorst, C.; Darling, T.; Tosi, L.L.; Huson, S.M.; Whitehouse, R.W.; Jakkula, E.; et al. Clinical delineation and natural history of the PIK3CA-related overgrowth spectrum. Am. J. Med. Genet. A 2014, 164, 1713–1733. [Google Scholar] [CrossRef]
- Castillo, S.D.; Vanhaesebroeck, B.; Sebire, N.J. Phosphoinositide 3-kinase: a new kid on the block in vascular anomalies. J. Pathol. 2016, 240, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Leiter, S.M.; Parker, V.E.; Welters, A.; Knox, R.; Rocha, N.; Clark, G.; Payne, F.; Lotta, L.; Harris, J.; Guerrero-Fernández, J.; et al. Hypoinsulinaemic, hypoketotic hypoglycaemia due to mosaic genetic activation of PI3-kinase. Eur. J. Endocrinol. 2017, 177, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Mirzaa, G.; Timms, A.E.; Conti, V.; Boyle, E.A.; Girisha, K.M.; Martin, B.; Kircher, M.; Olds, C.; Juusola, J.; Collins, S.; et al. PIK3CA-associated developmental disorders exhibit distinct classes of mutations with variable expression and tissue distribution. JCI insight 2016, 1, e87623. [Google Scholar] [CrossRef]
- Castel, P.; Carmona, F.J.; Grego-Bessa, J.; Berger, M.F.; Viale, A.; Anderson, K.V.; Bague, S.; Scaltriti, M.; Antonescu, C.R.; Baselga, E.; et al. Somatic PIK3CA mutations as a driver of sporadic venous malformations. Sci. Transl. Med. 2016, 8, 332ra42. [Google Scholar] [CrossRef]
- Castillo, S.D.; Tzouanacou, E.; Zaw-Thin, M.; Berenjeno, I.M.; Parker, V.E.R.; Chivite, I.; Milà-Guasch, M.; Pearce, W.; Solomon, I.; Angulo-Urarte, A.; et al. Somatic activating mutations in Pik3ca cause sporadic venous malformations in mice and humans. Sci. Transl. Med. 2016, 8, 332ra43. [Google Scholar] [CrossRef]
- Mirzaa, G.M.; Conway, R.L.; Gripp, K.W.; Lerman-Sagie, T.; Siegel, D.H.; deVries, L.S.; Lev, D.; Kramer, N.; Hopkins, E.; Graham, J.M., Jr.; et al. Megalencephaly-capillary malformation (MCAP) and megalencephaly-polydactyly-polymicrogyria-hydrocephalus (MPPH) syndromes: two closely related disorders of brain overgrowth and abnormal brain and body morphogenesis. Am. J. Med. Genet. A 2012, 158a, 269–291. [Google Scholar] [CrossRef]
- Roy, A.; Skibo, J.; Kalume, F.; Ni, J.; Rankin, S.; Lu, Y.; Dobyns, W.B.; Mills, G.B.; Zhao, J.J.; Baker, S.J.; et al. Mouse models of human PIK3CA-related brain overgrowth have acutely treatable epilepsy. Elife 2015, 4, e12703. [Google Scholar] [CrossRef]
- Smith, H.W.; Muller, W.J. Transgenic mouse models—A seminal breakthrough in oncogene research. Cold Spring Harb. Protoc. 2013, 2013, 1099–1108. [Google Scholar] [CrossRef]
- Adams, J.M.; Cory, S. Transgenic models of tumor development. Science 1991, 254, 1161–1167. [Google Scholar] [CrossRef]
- Renner, O.; Fominaya, J.; Alonso, S.; Blanco-Aparicio, C.; Leal, J.F.; Carnero, A. Mst1, RanBP2 and eIF4G are new markers for in vivo PI3K activation in murine and human prostate. Carcinogenesis 2007, 28, 1418–1425. [Google Scholar] [CrossRef] [Green Version]
- Renner, O.; Blanco-Aparicio, C.; Grassow, M.; Cañamero, M.; Leal, J.F.M.; Carnero, A. Activation of Phosphatidylinositol 3-Kinase by Membrane Localization of p110α Predisposes Mammary Glands to Neoplastic Transformation. Cancer Res. 2008, 68, 9643–9653. [Google Scholar] [CrossRef]
- Shioi, T.; Kang, P.M.; Douglas, P.S.; Hampe, J.; Yballe, C.M.; Lawitts, J.; Cantley, L.C.; Izumo, S. The conserved phosphoinositide 3-kinase pathway determines heart size in mice. EMBO J. 2000, 19, 2537–2548. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, L.; Sasaki, Y.; Calado, D.P.; Zhang, B.; Paik, J.H.; DePinho, R.A.; Kutok, J.L.; Kearney, J.F.; Otipoby, K.L.; Rajewsky, K. PI3 Kinase Signals BCR-Dependent Mature B Cell Survival. Cell 2009, 139, 573–586. [Google Scholar] [CrossRef]
- Leystra, A.A.; Deming, D.A.; Zahm, C.D.; Farhoud, M.; Olson, T.J.P.; Hadac, J.N.; Nettekoven, L.A.; Albrecht, D.M.; Clipson, L.; Sullivan, R.; et al. Mice expressing activated PI3K develop advanced colon cancer. Cancer Res. 2012, 72, 2931–2936. [Google Scholar] [CrossRef]
- Deming, D.A.; Leystra, A.A.; Nettekoven, L.; Sievers, C.; Miller, D.; Middlebrooks, M.; Clipson, L.; Albrecht, D.; Bacher, J.; Washington, M.K.; et al. PIK3CA and APC mutations are synergistic in the development of intestinal cancers. Oncogene 2013, 33, 2245–2254. [Google Scholar] [CrossRef] [Green Version]
- Venot, Q.; Blanc, T.; Rabia, S.H.; Berteloot, L.; Ladraa, S.; Duong, J.-P.; Blanc, E.; Johnson, S.C.; Hoguin, C.; Boccara, O.; et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature 2018, 558, 540–546. [Google Scholar] [CrossRef]
- Sheen, M.R.; Warner, S.L.; Fields, J.L.; Conejo-Garcia, J.R.; Fiering, S. Myristoylated p110α causes embryonic death due to developmental and vascular defects. Open Life Sci. 2015, 10, 461–478. [Google Scholar] [CrossRef]
- Sheen, M.R.; Marotti, J.D.; Allegrezza, M.J.; Rutkowski, M.; Conejo-Garcia, J.R.; Fiering, S. Constitutively activated PI3K accelerates tumor initiation and modifies histopathology of breast cancer. Oncogenesis 2016, 5, e267. [Google Scholar] [CrossRef]
- Engelman, J.A.; Chen, L.; Tan, X.; Crosby, K.; Guimaraes, A.R.; Upadhyay, R.; Maira, M.; McNamara, K.; Perera, S.A.; Song, Y.; et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008, 14, 1351–1356. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.R.; Xu, K.; Liu, J.C.; Agamez, N.M.R.; Loch, A.J.; Wong, R.G.; Wang, W.; Wright, K.L.; Lane, T.F.; Zacksenhaus, E.; et al. Cooperation between Pik3ca and p53 mutations in mouse mammary tumor formation. Cancer Res. 2011, 71, 1–12. [Google Scholar] [CrossRef]
- Liu, P.; Cheng, H.; Santiago, S.; Raeder, M.; Zhang, F.; Isabella, A.; Yang, J.; Semaan, D.J.; Chen, C.; Fox, E.A.; et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway–dependent and PI3K pathway–independent mechanisms. Nat. Med. 2011, 17, 1116–1121. [Google Scholar] [CrossRef]
- di Blasio, L.; Puliafito, A.; Gagliardi, P.A.; Comunanza, V.; Somale, D.; Chiaverina, G.; Bussolino, F.; Primo, L. PI3K/mTOR inhibition promotes the regression of experimental vascular malformations driven by PIK3CA-activating mutations. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Hanker, A.B.; Pfefferle, A.D.; Balko, J.M.; Kuba, M.G.; Young, C.D.; Sánchez, V.; Sutton, C.R.; Cheng, H.; Perou, C.M.; Zhao, J.J.; et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc. Natl. Acad. Sci. 2013, 110, 14372–14377. [Google Scholar] [CrossRef]
- Meyer, D.S.; Brinkhaus, H.; Müller, U.; Müller, M.; Cardiff, R.D.; Bentires-Alj, M. Luminal Expression of PIK3CA Mutant H1047R in the Mammary Gland Induces Heterogeneous Tumors. Cancer Res. 2011, 71, 4344–4351. [Google Scholar] [CrossRef]
- Koren, S.; Reavie, L.; Couto, J.P.; De Silva, D.; Stadler, M.B.; Roloff, T.; Britschgi, A.; Eichlisberger, T.; Kohler, H.; Aina, O.; et al. PIK3CAH1047R induces multipotency and multi-lineage mammary tumours. Nature 2015, 525, 114–118. [Google Scholar] [CrossRef]
- Kinross, K.M.; Montgomery, K.G.; Kleinschmidt, M.; Waring, P.; Ivetac, I.; Tikoo, A.; Saad, M.; Hare, L.; Roh, V.; Mantamadiotis, T.; et al. An activating Pik3ca mutation coupled with Pten loss is sufficient to initiate ovarian tumorigenesis in mice. J. Clin. Invest. 2012, 122, 553–557. [Google Scholar] [CrossRef] [Green Version]
- Tikoo, A.; Roh, V.; Montgomery, K.G.; Ivetac, I.; Waring, P.; Pelzer, R.; Hare, L.; Shackleton, M.; Humbert, P.; Phillips, W.A. Physiological Levels of Pik3caH1047R Mutation in the Mouse Mammary Gland Results in Ductal Hyperplasia and Formation of ERα-Positive Tumors. PLoS One 2012, 7, e36924. [Google Scholar] [CrossRef]
- Van Keymeulen, A.; Lee, M.Y.; Ousset, M.; Brohée, S.; Rorive, S.; Giraddi, R.R.; Wuidart, A.; Bouvencourt, G.; Dubois, C.; Salmon, I.; et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 2015, 525, 119–123. [Google Scholar] [CrossRef]
- Green, S.; Trejo, C.L.; McMahon, M. PIK3CAH1047R accelerates and enhances KRASG12D-driven lung tumorigenesis. Cancer Res. 2015, 75, 5378–5391. [Google Scholar] [CrossRef] [Green Version]
- Trejo, C.L.; Green, S.; Marsh, V.; Collisson, E.A.; Iezza, G.; Phillips, W.A.; McMahon, M. Mutationally Activated PIK3CAH1047R Cooperates with BRAFV600E to Promote Lung Cancer Progression. Cancer Res. 2013, 73, 1–14. [Google Scholar] [CrossRef]
- Deuker, M.M.; Marsh Durban, V.; Phillips, W.A.; McMahon, M. PI3′-Kinase Inhibition Forestalls the Onset of MEK1/2 Inhibitor Resistance in BRAF-Mutated Melanoma. Cancer Discov. 2015, 5, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Durban, V.M.; Deuker, M.M.; Bosenberg, M.W.; Phillips, W.; McMahon, M. Differential AKT dependency displayed by mouse models of BRAF V600E-Initiated melanoma. J. Clin. Invest. 2013, 123, 5104–5118. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Trejo, C.L.; Silva, J.M.; Gu, S.; Korkola, J.E.; Heiser, L.M.; Charles, R.-P.; Rabinovich, B.A.; Hann, B.; Dankort, D.; et al. A Central Role for RAF→MEK→ERK Signaling in the Genesis of Pancreatic Ductal Adenocarcinoma. Cancer Discov. 2012, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Charles, R.-P.; Silva, J.; Iezza, G.; Phillips, W.A.; McMahon, M. Activating BRAF and PIK3CA Mutations Cooperate to Promote Anaplastic Thyroid Carcinogenesis. Mol. Cancer Res. 2014, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- ElMokh, O.; Ruffieux-Daidié, D.; Roelli, M.A.; Stooss, A.; Phillips, W.A.; Gertsch, J.; Dettmer, M.S.; Charles, R.-P. Combined MEK and Pi3’-kinase inhibition reveals synergy in targeting thyroid cancer in vitro and in vivo. Oncotarget 2017, 8, 24604–24620. [Google Scholar] [CrossRef] [PubMed]
- Roelli, M.A.; Ruffieux-Daidié, D.; Stooss, A.; ElMokh, O.; Phillips, W.A.; Dettmer, M.S.; Charles, R.-P. PIK3CA(H1047R)-induced paradoxical ERK activation results in resistance to BRAF(V600E) specific inhibitors in BRAF(V600E) PIK3CA(H1047R) double mutant thyroid tumors. Oncotarget 2017, 8, 103207–103222. [Google Scholar] [CrossRef]
- Ying, Z.; Sandoval, M.; Beronja, S. Oncogenic activation of PI3K induces progenitor cell differentiation to suppress epidermal growth. Nat. Cell Biol. 2018, 20, 1256–1266. [Google Scholar] [CrossRef]
- Hare, L.M.; Phesse, T.J.; Waring, P.M.; Montgomery, K.G.; Kinross, K.M.; Mills, K.; Roh, V.; Heath, J.K.; Ramsay, R.G.; Ernst, M.; et al. Physiological expression of the PI3K-activating mutation Pik3caH1047R combines with Apc loss to promote development of invasive intestinal adenocarcinomas in mice. Biochem. J. 2014, 458, 251–258. [Google Scholar] [CrossRef]
- Pearson, H.B.; Li, J.; Meniel, V.S.; Fennell, C.M.; Waring, P.; Montgomery, K.G.; Rebello, R.J.; Macpherson, A.A.; Koushyar, S.; Furic, L.; et al. Identification of Pik3ca Mutation as a Genetic Driver of Prostate Cancer That Cooperates with Pten Loss to Accelerate Progression and Castration-Resistant Growth. Cancer Discov. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Daniel, P.M.; Filiz, G.; Brown, D.V.; Christie, M.; Waring, P.M.; Zhang, Y.; Haynes, J.M.; Pouton, C.; Flanagan, D.; Vincan, E.; et al. PI3K activation in neural stem cells drives tumorigenesis which can be ameliorated by targeting the cAMP response element binding protein. Neuro Oncol. 2018, 20, 1344–1355. [Google Scholar] [CrossRef]
- Kinross, K.M.; Montgomery, K.G.; Mangiafico, S.P.; Hare, L.M.; Kleinschmidt, M.; Bywater, M.J.; Poulton, I.J.; Vrahnas, C.; Henneicke, H.; Malaterre, J.; et al. Ubiquitous expression of the Pik3caH1047R mutation promotes hypoglycemia, hypoinsulinemia, and organomegaly. FASEB J. 2015, 29, 1426–1434. [Google Scholar] [CrossRef] [Green Version]
- Hare, L.M.; Schwarz, Q.; Wiszniak, S.; Gurung, R.; Montgomery, K.G.; Mitchell, C.A.; Phillips, W.A. Heterozygous expression of the oncogenic Pik3ca H1047R mutation during murine development results in fatal embryonic and extraembryonic defects. Dev. Biol. 2015, 404, 14–26. [Google Scholar] [CrossRef]
- Robinson, G.; Parker, M.; Kranenburg, T.A.; Lu, C.; Chen, X.; Ding, L.; Phoenix, T.N.; Hedlund, E.; Wei, L.; Zhu, X.; et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012, 488, 43–48. [Google Scholar] [CrossRef] [Green Version]
- Meyer, D.S.; Koren, S.; Leroy, C.; Brinkhaus, H.; Müller, U.; Klebba, I.; Müller, M.; Cardiff, R.D.; Bentires-Alj, M. Expression of PIK3CA mutant E545K in the mammary gland induces heterogeneous tumors but is less potent than mutant H1047R. Oncogenesis 2013, 2, e74. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Stawiski, E.; Janakiraman, V.; Chan, E.; Durinck, S.; Edgar, K.; Kljavin, N.; Rivers, C.; Gnad, F.; Roose-Girma, M.; et al. Conditional activation of Pik3ca H1047R in a knock-in mouse model promotes mammary tumorigenesis and emergence of mutations. Oncogene 2013, 32, 318–326. [Google Scholar] [CrossRef]
- Berenjeno, I.M.; Piñeiro, R.; Castillo, S.D.; Pearce, W.; McGranahan, N.; Dewhurst, S.M.; Meniel, V.; Birkbak, N.J.; Lau, E.; Sansregret, L.; et al. Oncogenic PIK3CA induces centrosome amplification and tolerance to genome doubling. Nat. Commun. 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Stratikopoulos, E.E.; Dendy, M.; Szabolcs, M.; Khaykin, A.J.; Lefebvre, C.; Zhou, M.-M.; Parsons, R. Kinase and BET Inhibitors Together Clamp Inhibition of PI3K Signaling and Overcome Resistance to Therapy. Cancer Cell 2015, 27, 837–851. [Google Scholar] [CrossRef] [Green Version]
- Stratikopoulos, E.E.; Kiess, N.; Szabolcs, M.; Pegno, S.; Kakit, C.; Wu, X.; Poulikakos, P.I.; Cheung, P.; Schmidt, H.; Parsons, R. Mouse ER+/PIK3CAH1047R breast cancers caused by exogenous estrogen are heterogeneously dependent on estrogen and undergo BIM-dependent apoptosis with BH3 and PI3K agents. Oncogene 2019, 38, 47–59. [Google Scholar] [CrossRef]
- Rollini, P.; Billotte, J.; Kolb, E.; Diggelmann, H. Expression pattern of mouse mammary tumor virus in transgenic mice carrying exogenous proviruses of different origins. J. Virol. 1992, 66, 4580–4586. [Google Scholar] [PubMed]
- Coelho, P.A.; Bury, L.; Shahbazi, M.N.; Liakath-Ali, K.; Tate, P.H.; Wormald, S.; Hindley, C.J.; Huch, M.; Archer, J.; Skarnes, W.C.; et al. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 2015, 5, 150209. [Google Scholar] [CrossRef] [Green Version]
- Serçin, Ö.; Larsimont, J.-C.; Karambelas, A.E.; Marthiens, V.; Moers, V.; Boeckx, B.; Le Mercier, M.; Lambrechts, D.; Basto, R.; Blanpain, C. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat. Cell Biol. 2015, 18, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Rivière, J.-B.; Mirzaa, G.M.; O’Roak, B.J.; Beddaoui, M.; Alcantara, D.; Conway, R.L.; St-Onge, J.; Schwartzentruber, J.A.; Gripp, K.W.; Nikkel, S.M.; et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat. Genet. 2012, 44, 934–940. [Google Scholar] [CrossRef] [Green Version]
- D’gama, A.M.; Geng, Y.; Couto, J.A.; Martin, B.; Boyle, E.A.; LaCoursiere, C.M.; Hossain, A.; Hatem, N.E.; Barry, B.J.; Kwiatkowski, D.J.; et al. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann. Neurol. 2015, 77, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Jansen, L.A.; Mirzaa, G.M.; Ishak, G.E.; O’roak, B.J.; Hiatt, J.B.; Roden, W.H.; Gunter, S.A.; Christian, S.L.; Collins, S.; Adams, C.; et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain 2015, 138, 1613–1628. [Google Scholar] [CrossRef] [Green Version]



| Original Ref | Mouse Model | Genetic Approach | Inducible? | Target Tissue (Promoter) |
|---|---|---|---|---|
| Shioi et al., 2000 [32] | iSH2p110α* | Transgenic | Non-inducible | Cardiac myocytes (αMyHC) [32] |
| Srinivasan et al., 2009 [33] | iSH2p110α* | Transgenic | Non-inducible | Mature B cells (CD21) [33] |
| Distal small bowel epithelial cells (Fatty acid binding protein) [34,35] | ||||
| All cells (CAGG) [36] | ||||
| Renner et al., 2007 [30] | MYR-p110α | Transgenic | Non-inducible | Prostate epithelial cells (MMTV) [30] |
| Mammary epithelial cells (MMTV) [31] | ||||
| All cells, early embryos (CMV) [37] | ||||
| Mammary duct (adenoviral) [38] | ||||
| Engleman et al., 2008 [39] | rtTA-Tet-op-PIK3CAH1047R | Transgenic | Tetracycline inducible (doxycycline) | Type II alveolar epithelial cells (CCSP) [39] |
| Adams et al., 2011 [40] | CreNLST; Pik3caH1047R | Transgenic | Non-inducible | Mammary epithelial cells (MMTV) [40] |
| Luminal and glandular uterine epithelial cells (Sprr2f); All cells (UBC); Blood vessels (Tie2) [24] | ||||
| Liu et al., 2011 [41] | rtTA TetO-Pik3caH1047R | Transgenic | Tetracycline inducible (doxycycline) | Mammary epithelial cells (MMTV) [41] |
| Blood vessels (Tie2); VE-cadherin (Cdh5) [42] | ||||
| Mammary epithelial cells (MMTV) [43] | ||||
| Meyer et al., 2011 [44] | Cre; Pik3caH1047R | Transgenic | Non-inducible | Mammary epithelial cells (MMTV); Alveolar progenitor cells (WAPi) [44] |
| Neural progenitor cells (hGFAP) [27] | ||||
| Basal and luminal mammary epithelium cells (Lgr5, K8) [45] | ||||
| Kinross et al., 2012 [46] | Cre; Pik3caH1047R | Knock-in | Oestrogen Receptor inducible (Tamoxifen) | Ovarian bursal cells (adenoviral) [46] |
| Mammary epithelial cells (MMTV, K5, K8) [47,48] | ||||
| Lung (adenoviral) [49,50] | ||||
| Melanocytes (Tyrosinase) [51,52] | ||||
| Pancreatic cells (p48, Pdx1) [53] | ||||
| Thyroid (thyroglobulin) [54,55,56] | ||||
| Epidermis (lentivirus) [57] | ||||
| Intestinal epithelial cells (Gpa33) [58] | ||||
| Prostate cancer (probasin) [59] | ||||
| Neural stem/progenitor cells (nestin) [60] | ||||
| Embryonic mesoderm (T gene); Endothelial cells (Pdgfb) [25] | ||||
| All cells (UBC) [61] | ||||
| Endothelial cells (Tie2) [62] | ||||
| Robinson et al., 2012 [63] | Cre; Pik3caE545K | Knock-in | Inducible | Lower rhombic lip progenitor cells (Blbp) [63] |
| Neural progenitor cells (hGFAP, Nestin) [27] | ||||
| Meyer et al., 2013 [64] | Cre; Pik3caE545K | Transgenic | Non-inducible | Alveolar progenitor cells (WAPi) [64] |
| Yuan et al., 2013 [65] | Cre; Pik3cae20H1047R | Knock-in | Inducible | Mammary epithelial cells (MMTV) [65] |
| Berenjeno et al., 2017 [66] | Flpe-ERT2; Pik3caH1047R | Knock-in | Tamoxifen Inducible | All cells (CAG) [66] |
| Stratikopoulos et al., 2015 [67] | Myc; Pik3caE545K Myc; Pik3caH1047R | Knock-in | Non-inducible | Mammary epithelial cells (MMTV, WAP) [67,68] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitchell, C.B.; Phillips, W.A. Mouse Models for Exploring the Biological Consequences and Clinical Significance of PIK3CA Mutations. Biomolecules 2019, 9, 158. https://doi.org/10.3390/biom9040158
Mitchell CB, Phillips WA. Mouse Models for Exploring the Biological Consequences and Clinical Significance of PIK3CA Mutations. Biomolecules. 2019; 9(4):158. https://doi.org/10.3390/biom9040158
Chicago/Turabian StyleMitchell, Camilla B., and Wayne A. Phillips. 2019. "Mouse Models for Exploring the Biological Consequences and Clinical Significance of PIK3CA Mutations" Biomolecules 9, no. 4: 158. https://doi.org/10.3390/biom9040158