Proteomic Studies of Psoriasis
Abstract
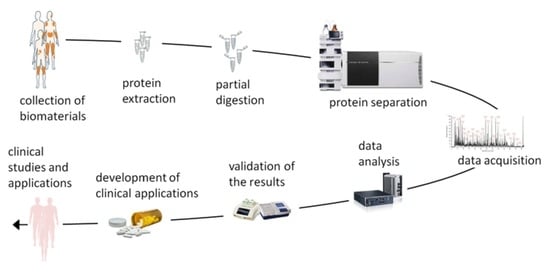
:1. Introduction
2. The Analysis of Differentially Expressed Proteins in the Skin
3. The Studies of Patients’ Blood
4. Clinical Applications
4.1. Monitoring the Therapeutic Response
4.2. Drug Evaluation
4.3. Discovering Risk Factors of Comorbidities and Their Analysis
4.4. Assessment of Adverse Effects
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Schön, M.P.; Boehncke, W.H. Psoriasis. N. Engl. J. Med. 2005, 352, 1899–1912. [Google Scholar] [CrossRef] [PubMed]
- Mogulevtseva, J.A.; Mezentsev, A.V.; Bruskin, S.A. The Role of Matrix Metalloproteinases in the Pathogenesis of Psoriasis. In A Closer Look at Metalloproteinases; Nova Science Publishers: Hauppauge, NY, USA, 2019; pp. 97–130. [Google Scholar]
- Yadav, K.; Singh, D.; Singh, M.R. Protein biomarker for psoriasis: A systematic review on their role in the pathomechanism, diagnosis, potential targets and treatment of psoriasis. Int. J. Biol. Macromol. 2018, 118, 1796–1810. [Google Scholar] [CrossRef] [PubMed]
- Samuelsson, L.; Enlund, F.; Torinsson, A.; Yhr, M.; Inerot, A.; Enerbäck, C.; Wahlström, J.; Swanbeck, G.; Martinsson, T. A genome-wide search for genes predisposing to familial psoriasis by using a stratification approach. Hum. Genet. 1999, 105, 523–529. [Google Scholar] [CrossRef]
- O’Rielly, D.D.; Rahman, P. Genetic, Epigenetic and Pharmacogenetic Aspects of Psoriasis and Psoriatic Arthritis. Rheum. Dis. Clin. N. Am. 2015, 41, 623–642. [Google Scholar] [CrossRef] [PubMed]
- Jianu, R.; Yu, K.; Cao, L.; Nguyen, V.; Salomon, A.R.; Laidlaw, D.H. Visual integration of quantitative proteomic data, pathways, and protein interactions. IEEE Trans. Vis. Comput. Graph. 2010, 16, 609–620. [Google Scholar] [CrossRef] [Green Version]
- Wasinger, V.C.; Cordwell, S.J.; Cerpa-Poljak, A.; Yan, J.X.; Gooley, A.A.; Wilkins, M.R.; Duncan, M.W.; Harris, R.; Williams, K.L.; Humphery-Smith, I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 1995, 16, 1090–1094. [Google Scholar] [CrossRef] [PubMed]
- Chandramouli, K.; Qian, P.Y. Proteomics: Challenges, techniques and possibilities to overcome biological sample complexity. Hum. Genom. Proteom. HGP 2009, 2009, 239204. [Google Scholar] [CrossRef] [Green Version]
- Chularojanamontri, L.; Charoenpipatsin, N.; Silpa-Archa, N.; Wongpraparut, C.; Thongboonkerd, V. Proteomics in Psoriasis. Int. J. Mol. Sci. 2019, 20, 1141. [Google Scholar] [CrossRef] [Green Version]
- Carlén, L.M.; Sánchez, F.; Bergman, A.C.; Becker, S.; Hirschberg, D.; Franzén, B.; Coffey, J.; Jörnvall, H.; Auer, G.; Alaiya, A.A.; et al. Proteome analysis of skin distinguishes acute guttate from chronic plaque psoriasis. J. Investig. Dermatol. 2005, 124, 63–69. [Google Scholar] [CrossRef] [Green Version]
- Bonnekoh, B.; Pommer, A.J.; Böckelmann, R.; Hofmeister, H.; Philipsen, L.; Gollnick, H. Topo-proteomic in situ analysis of psoriatic plaque under efalizumab treatment. Ski. Pharmacol. Physiol. 2007, 20, 237–252. [Google Scholar] [CrossRef]
- Plavina, T.; Wakshull, E.; Hancock, W.S.; Hincapie, M. Combination of abundant protein depletion and multi-lectin affinity chromatography (M-LAC) for plasma protein biomarker discovery. J. Proteome Res. 2007, 6, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Plavina, T.; Hincapie, M.; Wakshull, E.; Subramanyam, M.; Hancock, W.S. Increased plasma concentrations of cytoskeletal and Ca2+-binding proteins and their peptides in psoriasis patients. Clin. Chem. 2008, 54, 1805–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, J.; Park, S.G.; Park, B.C.; Choe, M.; Lee, K.S.; Cho, J.W. Proteomic analysis of psoriatic skin tissue for identification of differentially expressed proteins: Up-regulation of GSTP1, SFN and PRDX2 in psoriatic skin. Int. J. Mol. Med. 2011, 28, 785–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piruzian, E.; Bruskin, S.; Ishkin, A.; Abdeev, R.; Moshkovskii, S.; Melnik, S.; Nikolsky, Y.; Nikolskaya, T. Integrated network analysis of transcriptomic and proteomic data in psoriasis. BMC Syst. Biol. 2010, 4, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schonthaler, H.B.; Guinea-Viniegra, J.; Wculek, S.K.; Ruppen, I.; Ximénez-Embún, P.; Guío-Carrión, A.; Navarro, R.; Hogg, N.; Ashman, K.; Wagner, E.F. S100A8–S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity 2013, 39, 1171–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Swelm, R.P.; Laarakkers, C.M.; Kooijmans-Otero, M.; de Jong, E.M.; Masereeuw, R.; Russel, F.G. Biomarkers for methotrexate-induced liver injury: Urinary protein profiling of psoriasis patients. Toxicol. Lett. 2013, 221, 219–224. [Google Scholar] [CrossRef]
- Williamson, J.C.; Scheipers, P.; Schwämmle, V.; Zibert, J.R.; Beck, H.C.; Jensen, O.N. A proteomics approach to the identification of biomarkers for psoriasis utilising keratome biopsy. J. Proteom. 2013, 94, 176–185. [Google Scholar] [CrossRef]
- Fattahi, S.; Kazemipour, N.; Hashemi, M.; Sepehrimanesh, M. Alpha-1 antitrypsin, retinol binding protein and keratin 10 alterations in patients with psoriasis vulgaris, a proteomic approach. Iran. J. Basic Med. Sci. 2014, 17, 651–655. [Google Scholar]
- Cretu, D.; Liang, K.; Saraon, P.; Batruch, I.; Diamandis, E.P.; Chandran, V. Quantitative tandem mass-spectrometry of skin tissue reveals putative psoriatic arthritis biomarkers. Clin. Proteom. 2015, 12, 1. [Google Scholar] [CrossRef] [Green Version]
- Swindell, W.R.; Remmer, H.A.; Sarkar, M.K.; Xing, X.; Barnes, D.H.; Wolterink, L.; Voorhees, J.J.; Nair, R.P.; Johnston, A.; Elder, J.T.; et al. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med. 2015, 7, 86. [Google Scholar] [CrossRef] [Green Version]
- Reindl, J.; Pesek, J.; Krüger, T.; Wendler, S.; Nemitz, S.; Muckova, P.; Büchler, R.; Opitz, S.; Krieg, N.; Norgauer, J.; et al. Proteomic biomarkers for psoriasis and psoriasis arthritis. J. Proteom. 2016, 140, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Brunner, P.M.; Suárez-Fariñas, M.; He, H.; Malik, K.; Wen, H.C.; Gonzalez, J.; Chan, T.C.; Estrada, Y.; Zheng, X.; Khattri, S.; et al. The atopic dermatitis blood signature is characterized by increases in inflammatory and cardiovascular risk proteins. Sci. Rep. 2017, 7, 8707. [Google Scholar] [CrossRef] [PubMed]
- Kolbinger, F.; Loesche, C.; Valentin, M.A.; Jiang, X.; Cheng, Y.; Jarvis, P.; Peters, T.; Calonder, C.; Bruin, G.; Polus, F.; et al. β-Defensin 2 is a responsive biomarker of IL-17A-driven skin pathology in patients with psoriasis. J. Allergy Clin. Immunol. 2017, 139, 923–932.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuura, T.; Sato, M.; Nagai, K.; Sato, T.; Arito, M.; Omoteyama, K.; Suematsu, N.; Okamoto, K.; Kato, T.; Soma, Y.; et al. Serum peptides as putative modulators of inflammation in psoriasis. J. Dermatol. Sci. 2017, 87, 36–49. [Google Scholar] [CrossRef] [Green Version]
- Méhul, B.; Laffet, G.; Séraïdaris, A.; Russo, L.; Fogel, P.; Carlavan, I.; Pernin, C.; Andres, P.; Queille-Roussel, C.; Voegel, J.J. Noninvasive proteome analysis of psoriatic stratum corneum reflects pathophysiological pathways and is useful for drug profiling. Br. J. Dermatol. 2017, 177, 470–488. [Google Scholar] [CrossRef]
- Méhul, B.; Ménigot, C.; Fogel, P.; Seraidaris, A.; Genette, A.; Pascual, T.; Duvic, M.; Voegel, J.J. Proteomic analysis of stratum corneum in Cutaneous T-Cell Lymphomas and psoriasis. Exp. Dermatol. 2019, 28, 317–321. [Google Scholar] [CrossRef]
- Wang, J.; Suárez-Fariñas, M.; Estrada, Y.; Parker, M.L.; Greenlees, L.; Stephens, G.; Krueger, J.; Guttman-Yassky, E.; Howell, M.D. Identification of unique proteomic signatures in allergic and non-allergic skin disease. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2017, 47, 1456–1467. [Google Scholar] [CrossRef] [Green Version]
- Gęgotek, A.; Domingues, P.; Wroński, A.; Wójcik, P.; Skrzydlewska, E. Proteomic plasma profile of psoriatic patients. J. Pharm. Biomed. Anal. 2018, 155, 185–193. [Google Scholar] [CrossRef]
- Kim, J.; Tomalin, L.; Lee, J.; Fitz, L.J.; Berstein, G.; Correa-da Rosa, J.; Garcet, S.; Lowes, M.A.; Valdez, H.; Wolk, R.; et al. Reduction of Inflammatory and Cardiovascular Proteins in the Blood of Patients with Psoriasis: Differential Responses between Tofacitinib and Etanercept after 4 Weeks of Treatment. J. Investig. Dermatol. 2018, 138, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Wang, Y.; Li, Y.; Yang, H.; Wang, H. Quantitative Analysis of the Global Proteome in Peripheral Blood Mononuclear Cells from Patients with New-Onset Psoriasis. Proteomics 2018, 18, e1800003. [Google Scholar] [CrossRef]
- Foulkes, A.C.; Watson, D.S.; Carr, D.F.; Kenny, J.G.; Slidel, T.; Parslew, R.; Pirmohamed, M.; Anders, S.; Reynolds, N.J.; Griffiths, C.E.M.; et al. A Framework for Multi-Omic Prediction of Treatment Response to Biologic Therapy for Psoriasis. J. Investig. Dermatol. 2019, 139, 100–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garshick, M.S.; Barrett, T.J.; Wechter, T.; Azarchi, S.; Scher, J.U.; Neimann, A.; Katz, S.; Fuentes-Duculan, J.; Cannizzaro, M.V.; Jelic, S.; et al. Inflammasome Signaling and Impaired Vascular Health in Psoriasis. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Gęgotek, A.; Domingues, P.; Wroński, A.; Ambrożewicz, E.; Skrzydlewska, E. The Proteomic Profile of Keratinocytes and Lymphocytes in Psoriatic Patients. Proteom. Clin. Appl. 2019, 13, e1800119. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Li, B.; Chen, W.; Xu, Q.; Chen, S.; Zhang, H.; Wu, J.; Zhen, Q.; Li, Y.; Yong, L.; et al. Differential occurrence of lysine 2-hydroxyisobutyrylation in psoriasis skin lesions. J. Proteom. 2019, 205, 103420. [Google Scholar] [CrossRef]
- Szél, E.; Bozó, R.; Hunyadi-Gulyás, É.; Manczinger, M.; Szabó, K.; Kemény, L.; Bata-Csörgő, Z.; Groma, G. Comprehensive Proteomic Analysis Reveals Intermediate Stage of Non-Lesional Psoriatic Skin and Points out the Importance of Proteins Outside this Trend. Sci. Rep. 2019, 9, 11382. [Google Scholar] [CrossRef]
- Xu, M.; Deng, J.; Xu, K.; Zhu, T.; Han, L.; Yan, Y.; Yao, D.; Deng, H.; Wang, D.; Sun, Y.; et al. In-depth serum proteomics reveals biomarkers of psoriasis severity and response to traditional Chinese medicine. Theranostics 2019, 9, 2475–2488. [Google Scholar] [CrossRef]
- Medvedeva, I.V.; Stokes, M.E.; Eisinger, D.; LaBrie, S.T.; Ai, J.; Trotter, M.W.B.; Schafer, P.; Yang, R. Large-scale Analyses of Disease Biomarkers and Apremilast Pharmacodynamic Effects. Sci. Rep. 2020, 10, 605. [Google Scholar] [CrossRef] [Green Version]
- Gęgotek, A.; Domingues, P.; Wroński, A.; Skrzydlewska, E. Changes in Proteome of Fibroblasts Isolated from Psoriatic Skin Lesions. Int. J. Mol. Sci. 2020, 21, 5363. [Google Scholar] [CrossRef]
- Li, Y.; Lin, P.; Wang, S.; Li, S.; Wang, R.; Yang, L.; Wang, H. Quantitative analysis of differentially expressed proteins in psoriasis vulgaris using tandem mass tags and parallel reaction monitoring. Clin. Proteom. 2020, 17, 30. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, P.; Yan, B.X.; Chen, X.Y.; Landeck, L.; Wang, Z.Y.; Li, X.X.; Zhang, J.; Zheng, M.; Man, X.Y. Quantitative Proteomic Profile of Psoriatic Epidermis Identifies OAS2 as a Novel Biomarker for Disease Activity. Front. Immunol. 2020, 11, 1432. [Google Scholar] [CrossRef]
- Elnabawi, Y.A.; Garshick, M.S.; Tawil, M.; Barrett, T.J.; Fisher, E.A.; Lo Sicco, K.; Neimann, A.L.; Scher, J.U.; Krueger, J.; Berger, J.S. CCL20 in psoriasis: A potential biomarker of disease severity, inflammation, and impaired vascular health. J. Am. Acad. Dermatol. 2021, 84, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Glickman, J.W.; Dubin, C.; Renert-Yuval, Y.; Dahabreh, D.; Kimmel, G.W.; Auyeung, K.; Estrada, Y.D.; Singer, G.; Krueger, J.G.; Pavel, A.B.; et al. Cross-sectional study of blood biomarkers of patients with moderate to severe alopecia areata reveals systemic immune and cardiovascular biomarker dysregulation. J. Am. Acad. Dermatol. 2021, 84, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, H.; Wang, X.; Kvist-Hansen, A.; Krakauer, M.; Gørtz, P.M.; McCauley, B.D.; Skov, L.; Becker, C.; Hansen, P.R. Biomarkers of subclinical atherosclerosis in patients with psoriasis. Sci. Rep. 2021, 11, 21438. [Google Scholar] [CrossRef] [PubMed]
- Leijten, E.; Tao, W.; Pouw, J.; van Kempen, T.; Olde Nordkamp, M.; Balak, D.; Tekstra, J.; Muñoz-Elías, E.; DePrimo, S.; Drylewicz, J.; et al. Broad proteomic screen reveals shared serum proteomic signature in patients with psoriatic arthritis and psoriasis without arthritis. Rheumatology 2021, 60, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Navrazhina, K.; Renert-Yuval, Y.; Frew, J.W.; Grand, D.; Gonzalez, J.; Williams, S.C.; Garcet, S.; Krueger, J.G. Large-scale serum analysis identifies unique systemic biomarkers in psoriasis and hidradenitis suppurativa. Br. J. Dermatol. 2021. [Google Scholar] [CrossRef]
- Sobolev, V.V.; Mezentsev, A.V.; Ziganshin, R.H.; Soboleva, A.G.; Denieva, M.; Korsunskaya, I.M.; Svitich, O.A. LC-MS/MS analysis of lesional and normally looking psoriatic skin reveals significant changes in protein metabolism and RNA processing. PLoS ONE 2021, 16, e0240956. [Google Scholar] [CrossRef]
- Sobolev, V.; Soboleva, A.; Denisova, E.; Denieva, M.; Dvoryankova, E.; Suleymanov, E.; Zhukova, O.V.; Potekaev, N.; Korsunskaya, I.; Mezentsev, A. Differential Expression of Estrogen-Responsive Genes in Women with Psoriasis. J. Pers. Med. 2021, 11, 925. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Q.; Li, B.; Li, H.; Shen, S.; Wu, J.; Ge, H.; Zhang, H.; Chen, S.; Chen, W.; et al. Proteomic analysis of psoriatic skin lesions in a Chinese population. J. Proteom. 2021, 240, 104207. [Google Scholar] [CrossRef]
- Zhu, J.; Han, L.; Liu, R.; Zhang, Z.; Huang, Q.; Fang, X.; Yang, K.; Huang, G.; Zheng, Z.; Yawalkar, N.; et al. Identification of proteins associated with development of psoriatic arthritis in peripheral blood mononuclear cells: A quantitative iTRAQ-based proteomics study. J. Transl. Med. 2021, 19, 331. [Google Scholar] [CrossRef]
- Navrazhina, K.; Garcet, S.; Frew, J.W.; Zheng, X.; Coats, I.; Guttman-Yassky, E.; Krueger, J.G. The inflammatory proteome of hidradenitis suppurativa skin is more expansive than that of psoriasis vulgaris. J. Am. Acad. Dermatol. 2022, 86, 322–330. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Hinchliffe, T.E.; Wu, T. Biomarkers of An Autoimmune Skin Disease—Psoriasis. Genom. Proteom. Bioinform. 2015, 13, 224–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boersema, P.J.; Raijmakers, R.; Lemeer, S.; Mohammed, S.; Heck, A.J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009, 4, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Vélez-Bermúdez, I.C.; Wen, T.N.; Lan, P.; Schmidt, W. Isobaric Tag for Relative and Absolute Quantitation (iTRAQ)-Based Protein Profiling in Plants. Methods Mol. Biol. 2016, 1450, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Ritchlin, C.T.; Pennington, S.R.; Reynolds, N.J.; FitzGerald, O. Moving toward Precision Medicine in Psoriasis and Psoriatic Arthritis. J. Rheumatol. Suppl. 2020, 96, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Zhang, W.J.; McMillen, T.S.; Leboeuf, R.C.; Frei, B. Copper chelation by tetrathiomolybdate inhibits vascular inflammation and atherosclerotic lesion development in apolipoprotein E-deficient mice. Atherosclerosis 2012, 223, 306–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ioannou, M.; Sourli, F.; Mylonis, I.; Barbanis, S.; Papamichali, R.; Kouvaras, E.; Zafiriou, E.; Siomou, P.; Klimi, E.; Simos, G.; et al. Increased HIF-1 alpha immunostaining in psoriasis compared to psoriasiform dermatitides. J. Cutan. Pathol. 2009, 36, 1255–1261. [Google Scholar] [CrossRef]
- De Saedeleer, C.J.; Copetti, T.; Porporato, P.E.; Verrax, J.; Feron, O.; Sonveaux, P. Lactate activates HIF-1 in oxidative but not in Warburg-phenotype human tumor cells. PLoS ONE 2012, 7, e46571. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Tang, D.; Dai, Y. Metabolic Functions of Lysine 2-Hydroxyisobutyrylation. Cureus 2020, 12, e9651. [Google Scholar] [CrossRef]
- Huang, H.; Tang, S.; Ji, M.; Tang, Z.; Shimada, M.; Liu, X.; Qi, S.; Locasale, J.W.; Roeder, R.G.; Zhao, Y.; et al. p300-Mediated Lysine 2-Hydroxyisobutyrylation Regulates Glycolysis. Mol. Cell 2018, 70, 984. [Google Scholar] [CrossRef] [Green Version]
- Scigelova, M.; Hornshaw, M.; Giannakopulos, A.; Makarov, A. Fourier transform mass spectrometry. Mol. Cell. Proteom. MCP 2011, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gudjonsson, J.E.; Ding, J.; Li, X.; Nair, R.P.; Tejasvi, T.; Qin, Z.S.; Ghosh, D.; Aphale, A.; Gumucio, D.L.; Voorhees, J.J.; et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J. Investig. Dermatol. 2009, 129, 2795–2804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suárez-Fariñas, M.; Li, K.; Fuentes-Duculan, J.; Hayden, K.; Brodmerkel, C.; Krueger, J.G. Expanding the psoriasis disease profile: Interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J. Investig. Dermatol. 2012, 132, 2552–2564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Tsoi, L.C.; Swindell, W.R.; Gudjonsson, J.E.; Tejasvi, T.; Johnston, A.; Ding, J.; Stuart, P.E.; Xing, X.; Kochkodan, J.J.; et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J. Investig. Dermatol. 2014, 134, 1828–1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiricozzi, A.; Guttman-Yassky, E.; Suárez-Fariñas, M.; Nograles, K.E.; Tian, S.; Cardinale, I.; Chimenti, S.; Krueger, J.G. Integrative responses to IL-17 and TNF-α in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J. Investig. Dermatol. 2011, 131, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Batycka-Baran, A.; Hattinger, E.; Zwicker, S.; Summer, B.; Zack Howard, O.M.; Thomas, P.; Szepietowski, J.C.; Ruzicka, T.; Prinz, J.C.; Wolf, R. Leukocyte-derived koebnerisin (S100A15) and psoriasin (S100A7) are systemic mediators of inflammation in psoriasis. J. Dermatol. Sci. 2015, 79, 214–221. [Google Scholar] [CrossRef]
- Mezentsev, A.; Bezsonov, E.; Kashirskikh, D.; Baig, M.S.; Eid, A.H.; Orekhov, A. Proatherogenic Sialidases and Desialylated Lipoproteins: 35 Years of Research and Current State from Bench to Bedside. Biomedicines 2021, 9, 600. [Google Scholar] [CrossRef]
- Łuczaj, W.; Gęgotek, A.; Skrzydlewska, E. Analytical approaches to assess metabolic changes in psoriasis. J. Pharm. Biomed. Anal. 2021, 205, 114359. [Google Scholar] [CrossRef]
- Iskandar, I.Y.K.; Warren, R.B.; Lunt, M.; Mason, K.J.; Evans, I.; McElhone, K.; Smith, C.H.; Reynolds, N.J.; Ashcroft, D.M.; Griffiths, C.E.M. Differential Drug Survival of Second-Line Biologic Therapies in Patients with Psoriasis: Observational Cohort Study from the British Association of Dermatologists Biologic Interventions Register (BADBIR). J. Investig. Dermatol. 2018, 138, 775–784. [Google Scholar] [CrossRef] [Green Version]
- Johnston, A.; Guzman, A.M.; Swindell, W.R.; Wang, F.; Kang, S.; Gudjonsson, J.E. Early tissue responses in psoriasis to the antitumour necrosis factor-α biologic etanercept suggest reduced interleukin-17 receptor expression and signalling. Br. J. Dermatol. 2014, 171, 97–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correa da Rosa, J.; Kim, J.; Tian, S.; Tomalin, L.E.; Krueger, J.G.; Suárez-Fariñas, M. Shrinking the Psoriasis Assessment Gap: Early Gene-Expression Profiling Accurately Predicts Response to Long-Term Treatment. J. Investig. Dermatol. 2017, 137, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorgensen, A.L.; Williamson, P.R. Methodological quality of pharmacogenetic studies: Issues of concern. Stat. Med. 2008, 27, 6547–6569. [Google Scholar] [CrossRef] [PubMed]
- Menter, M.A.; Armstrong, A.W.; Gordon, K.B.; Wu, J.J. Common and Not-So-Common Comorbidities of Psoriasis. Semin. Cutan. Med. Surg. 2018, 37, S48–S51. [Google Scholar] [CrossRef]
- Masson, W.; Lobo, M.; Molinero, G. Psoriasis and Cardiovascular Risk: A Comprehensive Review. Adv. Ther. 2020, 37, 2017–2033. [Google Scholar] [CrossRef] [Green Version]
- Bachelez, H.; van de Kerkhof, P.C.; Strohal, R.; Kubanov, A.; Valenzuela, F.; Lee, J.H.; Yakusevich, V.; Chimenti, S.; Papacharalambous, J.; Proulx, J.; et al. Tofacitinib versus etanercept or placebo in moderate-to-severe chronic plaque psoriasis: A phase 3 randomised non-inferiority trial. Lancet 2015, 386, 552–561. [Google Scholar] [CrossRef]
- Chima, M.; Lebwohl, M. TNF inhibitors for psoriasis. Semin. Cutan. Med. Surg. 2018, 37, 134–142. [Google Scholar] [CrossRef]
- Azevedo, A.; Torres, T. Tofacitinib: A New Oral Therapy for Psoriasis. Clin. Drug Investig. 2018, 38, 101–112. [Google Scholar] [CrossRef]
- Ciechanowicz, P.; Rakowska, A.; Sikora, M.; Rudnicka, L. JAK-inhibitors in dermatology: Current evidence and future applications. J. Dermatol. Treat. 2019, 30, 648–658. [Google Scholar] [CrossRef]
- Ritchlin, C.T.; Colbert, R.A.; Gladman, D.D. Psoriatic Arthritis. N. Engl. J. Med. 2017, 376, 957–970. [Google Scholar] [CrossRef] [Green Version]
- Roubille, C.; Richer, V.; Starnino, T.; McCourt, C.; McFarlane, A.; Fleming, P.; Siu, S.; Kraft, J.; Lynde, C.; Pope, J.; et al. The effects of tumour necrosis factor inhibitors, methotrexate, non-steroidal anti-inflammatory drugs and corticosteroids on cardiovascular events in rheumatoid arthritis, psoriasis and psoriatic arthritis: A systematic review and meta-analysis. Ann. Rheum. Dis. 2015, 74, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Boehncke, W.H. Systemic Inflammation and Cardiovascular Comorbidity in Psoriasis Patients: Causes and Consequences. Front. Immunol. 2018, 9, 579. [Google Scholar] [CrossRef] [PubMed]
- Calvayrac, O.; Rodríguez-Calvo, R.; Alonso, J.; Orbe, J.; Martín-Ventura, J.L.; Guadall, A.; Gentile, M.; Juan-Babot, O.; Egido, J.; Beloqui, O.; et al. CCL20 is increased in hypercholesterolemic subjects and is upregulated by LDL in vascular smooth muscle cells: Role of NF-κB. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2733–2741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsay, K.; Fraser, A.D.; Layton, A.; Goodfield, M.; Gruss, H.; Gough, A. Liver fibrosis in patients with psoriasis and psoriatic arthritis on long-term, high cumulative dose methotrexate therapy. Rheumatology 2009, 48, 569–572. [Google Scholar] [CrossRef] [Green Version]
- Visser, K.; van der Heijde, D.M. Risk and management of liver toxicity during methotrexate treatment in rheumatoid and psoriatic arthritis: A systematic review of the literature. Clin. Exp. Rheumatol. 2009, 27, 1017–1025. [Google Scholar]
- Barker, J.; Horn, E.J.; Lebwohl, M.; Warren, R.B.; Nast, A.; Rosenberg, W.; Smith, C. Assessment and management of methotrexate hepatotoxicity in psoriasis patients: Report from a consensus conference to evaluate current practice and identify key questions toward optimizing methotrexate use in the clinic. J. Eur. Acad. Dermatol. Venereol. JEADV 2011, 25, 758–764. [Google Scholar] [CrossRef]
- Andersen, V.; Holmskov, U.; Sørensen, S.B.; Jawhara, M.; Andersen, K.W.; Bygum, A.; Hvid, L.; Grauslund, J.; Wied, J.; Glerup, H.; et al. A Proposal for a Study on Treatment Selection and Lifestyle Recommendations in Chronic Inflammatory Diseases: A Danish Multidisciplinary Collaboration on Prognostic Factors and Personalised Medicine. Nutrients 2017, 9, 499. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, H.; Kvist-Hansen, A.; Becker, C.; Wang, X.; McCauley, B.D.; Krakauer, M.; Gørtz, P.M.; Henningsen, K.M.A.; Zachariae, C.; Skov, L.; et al. Multiscale Biology of Cardiovascular Risk in Psoriasis: Protocol for a Case-Control Study. JMIR Res. Protoc. 2021, 10, e28669. [Google Scholar] [CrossRef]






| Year | Author/Reference | Proteomics Methodology | Samples | Patients | Key Findings |
|---|---|---|---|---|---|
| 2005 | Carlén et al. [10] | 2D-electrophoresis, MALDI-TOF MS, Q-TOF-MS/MS | Skin | Psoriasis patients: lesional skin—7 samples; normal-looking skin—3 samples); patients with acute glutate psoriasis: lesional skin—6 samples; normal-looking skin—5 samples; healthy individuals—4 samples. | The first known proteomic study of lesional psoriatic skin; the first comparative analysis of patients with plaque psoriasis and acute guttate psoriasis. |
| 2007 | Bonnekoh et al. [11] | Multi-epitope ligand cartography (MELC) robot technology | Skin | Psoriasis patients: 6 samples of lesional skin; healthy volunteers 6 samples of healthy skin. | The authors showed a significant diversity in location of inflammatory epitopes after immunotherapy with efalizumab. They proposed CD138 and TRF1/CD71 as prognostic biomarkers of treatment outcome with efalizumab. |
| 2007 | Plavina et al. [12] | M-LAC coupled with LC-MS/MS | Serum | Psoriasis patients—20; healthy volunteers—20. | Depletion of immunoglobulins and albumin revealed upregulation of cytoskeletal and actin-binding proteins in plasma of psoriasis undetectable in regular serum. |
| 2008 | Plavina et al. [13] | M-LAC coupled with LC-MS/MS; ultracentrifugation followed by bynano LC-MS/MS | Serum | Psoriasis patients—20; healthy volunteers—20. | The authors identified 21 DEPs previously associated with other autoimmune disorders (e.g., thymosin β4, talin 1, γ-actin, filamin, profilin, S100A8, and S100A9) in serum of psoriasis patients. These DEPs were previously undetectable in the serum due to a higher abundance of immunoglobulins and serum albumin. |
| 2011 | Ryu et al. [14] | 2D-electrophoresis, nanoLC-MS/MS | Two pools of skin samples (n = 8 and 28) | Psoriasis patients, 36 paired samples of lesional and normal-looking skin. | One of the first comparative analyses of lesional and normal-looking skin; the first known ontology analysis of DEPs in psoriatic skin. |
| 2010 | Piruzian et al. [15] | 2D-electrophoresis, nanoLC-MS/MS | Pooled skin samples | Psoriasis patients, 3 paired samples of lesional and normal-looking skin. | The authors reported of the 10 most upregulated proteins in lesional skin. |
| 2013 | Schonthaler et al. [16] | iTRAQ-2DLC-MS/MS | Epidermis | Psoriasis patients, 19 paired samples of lesional and normal-looking skin. | The authors identified S100A8, S100A9, and complement C3 as the three most upregulated proteins in lesional skin: they also showed that knocking S100A9 out in JunB-Jun double knockout mice attenuated psoriasis-like skin disorder. |
| 2013 | van Swelm et al. [17] | MALDI-TOF MS | Urine | Psoriasis patients—60, with and without liver fibrosis | The authors proposed ITIH4 and CDH2 as candidate biomarkers of methotrexate-induced hepatic fibrosis in psoriasis patients. |
| 2013 | Williamson et al. [18] | Dimethyl labelling, LTQ-Orbitrapnano LC-MS/MS | Skin and serum | Psoriasis patients—4 paired samples of lesional and normal-looking skin; 4 samples of blood plasma; commercial blood plasma of healthy donors (n = 2). | The authors identified several dozen DEPs comparing lesional vs. normal-looking skin of psoriasis patients: they also proposed profilin as a biomarker of psoriasis. |
| 2014 | Fattahi et al. [19] | MALDI/TOF-TOF | Serum | Psoriasis patients—20; and 16 healthy volunteers. | The authors found a lower abundance of retinol-binding protein RBP4, a higher abundance of KRT10 and the unique expression pattern of α1 antitrypsin isoforms in sera of psoriasis patients. |
| 2015 | Cretu et al. [20] | LC-MS/MS | Pooled skin samples (n = 5) | Psoriasis patients with and without psoriatic arthritis (10 patients in each group), samples of lesional and normal-looking skin. | The authors proposed ITGB5 as a potential biomarker of psoriatic arthritis in psoriasis patients. |
| 2015 | Swindell et al. [21] | Label-free LC-MS/MS, LTQ-Orbitrap nanoLC-MS/MS | Skin | Psoriasis patients—14 paired samples of lesional and normal-looking skin. | The first known “bi-omic study” of psoriatic skin. The authors identified 748 DEPs in lesional and normal-looking psoriatic skin. They also discovered a modest correlation between protein and gene expression in psoriasis patients and characterized the role of IL-17A in disease-associated gene expression. |
| 2016 | Reindl et al. [22] | LTQ-Orbitrap nanoLC-MS/MS | Serum | Psoriasis patients—6; healthy volunteers—6. | The authors proposed AZGP1, complement C3, polymeric immunoglobulin receptor PIGR, and plasma kallikrein KLKB1 as disease-associated biomarkers. They discovered a moderate correlation between disease severity and the expression of DSP, complement C3, PIGR, and KRT17. |
| 2017 | Brunner [23] | Proximity extension assay | Serum | Patients with atopic dermatitis—59; psoriasis patients—22. | The authors found that inflammatory potential in patients with atopic dermatitis is higher than in psoriasis patients. They also showed a higher risk of cardiovascular disorders in both groups of patients. |
| 2017 | Kolbinger et al. [24] | Proximity extension assay | Serum, skin and dermis | Psoriasis patients—8 paired samples of lesional and normally-looking skin; healthy volunteers 8 skin samples; blood serum of the same individuals. | The authors showed how increased expression of antimicrobial peptides, proinflammatory cytokines and neutrophil chemoattractants normalizes in psoriasis patients after their treatment with secukinumab. They also proposed DEFB4 as a biomarker of the therapeutic response. |
| 2017 | Matsuura et al. [25] | MALDI-TOF MS, TripleTOF-MS/MS | Serum | Psoriasis patients with and without psoriatic arthritis (n = 10 and 24, respectively); 14 patients with atopic dermatitis; 23 healthy volunteers. | The authors identified several psoriasis/psoriatic arthritis associated DEPt originated from FGA, FLG, TMSB4X, and FLJ55606 in the sera of psoriasis patients. |
| 2017 | Méhul et al. [26] | qTOF-MS/MS and protein array | Stratum corneum | 40 paired samples stripped from lesional and normal-looking skin of psoriasis patients. | The first comparative study of stratum corneum of psoriasis patients; the authors proposed 21 candidate biomarkers of lesional psoriatic stratum corneum. |
| 2017 | Méhul et al. [27] | qTOF-MS/MS and protein array | Stratum corneum | Patients with CTCL—10; psoriasis patients—24 (paired samples of stratum corneum stripped from lesional and normal-looking skin). | The first comparative proteomic study of patients with psoriasis and CTCL. The authors established a molecular signature of 112 DEPs to distinguish the samples of psoriasis patients and patients with CTCL. |
| 2017 | Wang et al., [28] | SomaScan | Serum | Patients with atopic dermatitis—20; patients with contact dermatitis—10; patients with atopic and contact dermatitis—10; psoriasis patients—12. | The authors reported 4 DEPs, namely KYNU, LG3BP, TPSB2, and CA6 associated with psoriasis and proposed KYNU as a disease-associated biomarker. |
| 2018 | Gęgotek et al. [29] | GeLC-MS/MS, LTQ Orbitrap-nanoLC-MS/MS | Serum | Psoriasis patients—6; healthy volunteers—6. | The authors detected a higher level of adducts in plasma of psoriasis patients. They also found a decreased level of vitamin D and proteins involved in lipid metabolism. In addition, they demonstrated higher abundance of proteins involved in immune response and signal transduction. |
| 2018 | Kim et al. [30] | Proximity extension assay | Serum | Psoriasis patients—266. | The authors discovered the strongest correlation between PASI and the expression levels of IL17A and IL17C, IL20, and CCL20 among the reponders to tofacitinib. |
| 2018 | Li et al. [31] | TMT labeling, LC-MS/MS | PBMC | New onset psoriasis patients (n = 31) and healthy volunteers (n = 32). | The authors identified new disease-associated proteins, namely ATM, SLFN5, ZNF512, SPATA13, DOCK2, ARSB, VIRMA, and NRGN. |
| 2019 | Foulkes et al. [32] | SomaScan | Serum | Psoriasis patients—10. | The authors reported increased expression of TNF- and interferon-dependent proteins. |
| 2019 | Garshick et al. [33] | Proximity extension assay | Serum and endothelial cells of brachial vein | Psoriasis patients—20. | The authors established a molecular signature of 8 DEPs, namely IL1β, CXCL10, VCAM-1, IL-8, CXCL1, LTB, ICAM-1, and CCL3, that characterizes the risk of atherosclerosis in psoriasis patients. They also discovered a correlation of the named biomarkers and PASI. In addition, they proposed an existence of a mechanism that damages different tissues in psoriasis. |
| 2019 | Gęgotek et al. [34] | GeLC-MS/MS, LTQ Orbitrap-nanoLC-MS/MS | Keratino-cytes and lympho-cytes | Psoriasis patients—6; healthy volunteers—6. | The authors discovered a higher level of adducts in plasma, skin and primary cells of psoriasis patients, lower expression of TXNRD1, higher expression of the glycolytic isoenzymes, namely PGAM1 and -2. |
| 2019 | Ge et al. [35] | UPLC-MS/MS with Q-Exactive Plus Hybrid Quadrupole-Orbitrap | Pooled skin samples (n = 15) | Psoriasis patients—45 paired samples of lesional and normal-looking skin. | The first study presenting a comprehensive analysis of 2-hydroxyisobutyrylation in lesional and normal-looking skin of psoriasis patients. |
| 2019 | Szél, E. et al. [36] | 2D-electrophoresis, LC-MS/MS | Skin | Psoriasis patients—3 paired samples of lesional and normal-looking skin; healthy volunteers—3 skin samples, | The authors reported ~30 DEPs previously not associated with the disease. They also proposed PRKDC and MYBBP1A as potential key regulators of hyperproliferation and altered differentiation of skin cells, stress, and immune response in psoriasis, |
| 2019 | Xu et al. [37] | Custome-made array of specific antibodies directed to disease associated biomarkers, DIA-MS | Serum | Psoriasis patients—16; healthy volunteers—23, | The authors designed and tested a custom-made array of antibodies specific to 112 previously discovered disease-associated biomarkers. They also found a moderate correlation of PASI and the expression of PI3, CCL22, and IL12B. In addition, they proposed three predictive biomarkers, namely FCN2, MIF, and MMP1, to identify the responders to the traditional Chinese medicine YinXieLing. |
| 2020 | Medvedeva et al. [38] | Proximity extension assay | Serum | Psoriasis patients—150. | The authors proposed IL17A and KLK7 as biomarkers of disease severity and they also established the molecular signature of 4 DEPs, namely KLK7, PEDF, MDC, and ANGPTL4, to predict the outcome of the therapy to apremilast. |
| 2020 | Gęgotek et al. [39] | GeLC-MS/MS, LTQ Orbitrap-nanoLC-MS/MS | Fibroblasts | Psoriasis patients—5 samples of lesional skin; healthy volunteers—6 skin samples. | The authors discovered a higher abundance of TXNRD1 and a lower abundance of several glycolytic enzymes (PK, PGK2, ALDOL, and GAPDH) in dermal fibroblasts. |
| 2020 | Li et al. [40] | TMT labeling, LC-MS/MS | Skin | Psoriasis patients—11 samples of lesional skin; healthy volunteers—11 skin samples. | The authors identified 9 DEPs previously not associated with psoriasis: MPO, TYMP, IMPDH2, GSTM4, and ALDH3A1 that were upregulated and CES1, MAOB, MGST1, and GSTT1—that were downregulated in lesional skin. |
| 2020 | Zhou et al. [41] | iTRAQ-Labeling, LC-MS/MS | Skin and serum | Psoriasis patients—16 paired samples of lesional and normal-looking skin; 32 blood samples; healthy volunteers—15 skin samples and 24 blood samples. | The authors identified 4 new proteins, namely OAS2, IFIT3, IRF3, and MeCP2, previously not associated with psoriasis, and proposed OAS2 as a disease-associated biomarker to analyze both skin and sera samples. They also presented an optimized protocol for the obtaining of skin samples and their processing. |
| 2021 | Elnabawi et al. [42] | Proximity extension assay | Endothe-lial cells of brachial vein and serum | Psoriasis patients—23; healthy volunteers—10. | The authors found that the expression of CCL20 and IL6 correlates with LDL-cholesterol, endothelial inflammation score and PASI. They proposed CCL6 as a biomarker of impaired vascular health in psoriasis patients. |
| 2021 | Glickman et al. [43] | Proximity extension assay | Serum | Patients with moderate-to-severe alopecia areata (n = 35), atopic dermatitis (n = 49), moderate-to-severe psoriasis (n = 19) and healthy volunteers (n = 36). | The authors found that patients with the advanced forms of alopecia areata exhibit the highest systemic inflammatory tone and higher expression of cardiovascular risk biomarkers compared to the other groups of patients and their fellow groupmates without total involvement. They presented evidence that alopecia areata is a systemic disorder. |
| 2021 | Kaiser et al. [44] | Proximity extension assay | Serum | Psoriasis patients with and without the signs of atherosclerosis—85. | The authors discovered a negative correlation of GDF15 and vascular inflammation in the ascending aorta and entire aorta. They found that the expression of GDF15 positively correlates with carotid intima-media thickness and coronary artery calcium score in psoriasis patients without cardiovascular disease and statin treatment. |
| 2021 | Leijten et al. [45] | Proximity extension assay | Serum | Psoriasis patients with and without psoriatic arthritis (n = 20 and 18, respectively); healthy volunteers (n = 19). | The authors discovered strong correlations of joint swollenness and the expression levels of of ICAM-1 and CCL18. They also found a strong correlation of PASI and the expression of PI3 and IL17RA. |
| 2021 | Navrazhina et al. [46] | Proximity extension assay | Serum | Patients with moderate-to-severe hidradenitis suppurativa (n = 11), patients with psoriasis (n = 10) and healthy volunteers (n = 10). | The authors discovered that patients with hidradenitis suppurativa exhibited a significantly more intense inflammatory burden and an increase in cardiovascular/atherosclerosis-related biomarkers than psoriasis patients. They also proposed a computer model to distinguish sera samples of patients with hidradenitis suppurativa and psoriasis. |
| 2021 | Sobolev et al. [47] | LC-MS/MS | Skin | Psoriasis patients—5 paired samples of lesional and normal-looking skin; healthy volunteers—5 skin samples. | The authors proposed an existence of two adaptive mechanisms in normal-looking skin aimed to modulate there the development of the inflammatory response and accelerate the protein metabolism in the diseased cells, respectively. They reported a suppression of kallikrein-kinin system in normal-looking skin. |
| 2021 | Sobolev et al. [48] | LC-MS/MS | Skin | Psoriasis patients—5 paired samples of lesional and normal-looking skin; healthy volunteers—5 skin samples. | The authors discovered a set of 6 estrogen-dependent DEPs that modulate psoriasis in female skin. They proposed an existence of adaptive mechanism in female patients that facilitates the disease flow. |
| 2021 | Wang et al. [49] | TMT labeling, LC-MS/MS | pooled skin samples (n = 15) | Psoriasis patients—30 paired samples of lesional and normal-looking skin; healthy volunteers—30 skin samples. | The study compared psoriasis patients of Chinese and Caucasian descent pointing to the differences in protein expression in both populations. They identified GSTP1, SFN, KRT77, FLG2, and TREX2 as DEPs differentially expressed in Caucasians and SIRT1—as differentially expressed in Chinese patients. |
| 2021 | Zue et al. [50] | iTRAQ-Labeling, LC-MS/MS | PBMC | Two groups of 4 psoriasis patients with and without psoriatic arthritis. | The authors proposed SIRT2 as a potential biomarker of psoriatic arthritis in psoriasis patients. |
| 2022 | Navrazhina et al. [51] | Proximity extension assay | Skin | Patients with hidradenitis suppurativa—13 paired samples of lesional and normal-looking skin; psoriasis patients—11 paired samples of lesional and normal-looking skin; healthy individuals—11 skin samples. | The authors found that skin inflammation in the patients with hidradenitis suppurativa extends far beyond the skin lesions and sustains on a comparable level. They provided evidence that hidradenitis suppurativa is a systemic disorder. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobolev, V.V.; Soboleva, A.G.; Denisova, E.V.; Pechatnikova, E.A.; Dvoryankova, E.; Korsunskaya, I.M.; Mezentsev, A. Proteomic Studies of Psoriasis. Biomedicines 2022, 10, 619. https://doi.org/10.3390/biomedicines10030619
Sobolev VV, Soboleva AG, Denisova EV, Pechatnikova EA, Dvoryankova E, Korsunskaya IM, Mezentsev A. Proteomic Studies of Psoriasis. Biomedicines. 2022; 10(3):619. https://doi.org/10.3390/biomedicines10030619
Chicago/Turabian StyleSobolev, Vladimir V., Anna G. Soboleva, Elena V. Denisova, Eva A. Pechatnikova, Eugenia Dvoryankova, Irina M. Korsunskaya, and Alexandre Mezentsev. 2022. "Proteomic Studies of Psoriasis" Biomedicines 10, no. 3: 619. https://doi.org/10.3390/biomedicines10030619