Functional Therapeutic Target Validation Using Pediatric Zebrafish Xenograft Models
Abstract
:Simple Summary
Abstract
1. Introduction
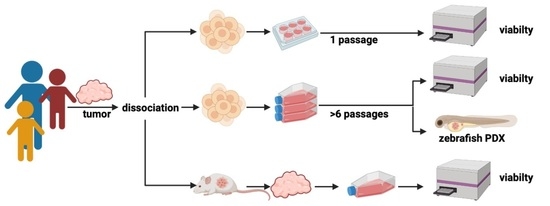
2. Materials and Methods
2.1. Primary Cell Isolation from Fresh Tumor Tissue
2.2. Tumor Cell Isolation from Mouse PDX
2.3. Molecular Diagnosis
2.4. Drug Screening and Metabolic Activity Assays
2.5. Zebrafish Lines
2.6. Zebrafish Embryo Toxicity Assays
2.7. Cell Preparation for Zebrafish Embryo Xenotransplantation
2.8. Drug Treatment of Zebrafish Embryos Bearing Human Tumor Cells
2.9. Imaging and Analysis of Zebrafish Embryos Bearing Human Tumor Cells
2.10. Immunohistochemistry (IHC)
2.11. Reagents for the Treatment of Zebrafish Embryos
2.12. Ethical Approval
3. Results
3.1. Characterization and Comparison of the Three Patient-Derived Samples
3.2. Comparison of Drug Sensitivities of Short-Term, Long-Term and Mouse-Xenograft Derived-Cultures Obtained from the Same Original Tumor
3.3. Drug Hit Selection and Zebrafish Embryo Toxicity Test
3.4. Drug Hit Validation with zPDX
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. (Eds.) SEER Cancer Statistics Review, 1975–2018; National Cancer Institute: Bethesda, MD, USA, 2021. Available online: https://seer.cancer.gov/csr/1975_2018/ (accessed on 15 April 2021).
- Blattner-Johnson, M.; Jones, D.T.W.; Pfaff, E. Precision medicine in pediatric solid cancers. Semin. Cancer Biol. 2021. in print. [Google Scholar] [CrossRef]
- Worst, B.C.; van Tilburg, C.M.; Balasubramanian, G.P.; Fiesel, P.; Witt, R.; Freitag, A.; Boudalil, M.; Previti, C.; Wolf, S.; Schmidt, S.; et al. Next-generation personalised medicine for high-risk paediatric cancer patients—The INFORM pilot study. Eur. J. Cancer 2016, 65, 91–101. [Google Scholar] [CrossRef] [Green Version]
- Van Tilburg, C.M.; Pfaff, E.; Pajtler, K.W.; Langenberg, K.P.S.; Fiesel, P.; Jones, B.C.; Balasubramanian, G.P.; Stark, S.; Johann, P.D.; Blattner-Johnson, M.; et al. The pediatric precision oncology INFORM registry: Clinical outcome and benefit for patients with very high-evidence targets. Cancer Discov. 2021. [Google Scholar] [CrossRef] [PubMed]
- Letai, A. Functional precision cancer medicine-moving beyond pure genomics. Nat. Med. 2017, 23, 1028–1035. [Google Scholar] [CrossRef] [PubMed]
- Bingel, C.; Koeneke, E.; Ridinger, J.; Bittmann, A.; Sill, M.; Peterziel, H.; Wrobel, J.K.; Rettig, I.; Milde, T.; Fernekorn, U.; et al. Three-dimensional tumor cell growth stimulates autophagic flux and recapitulates chemotherapy resistance. Cell Death Dis. 2017, 8, e3013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levinger, I.; Ventura, Y.; Vago, R. Life is three dimensional-as in vitro cancer cultures should be. Adv. Cancer Res. 2014, 121, 383–414. [Google Scholar] [CrossRef] [PubMed]
- Pompili, L.; Porru, M.; Caruso, C.; Biroccio, A.; Leonetti, C. Patient-derived xenografts: A relevant preclinical model for drug development. J. Exp. Clin. Cancer Res. CR 2016, 35, 189. [Google Scholar] [CrossRef] [Green Version]
- Barriuso, J.; Nagaraju, R.; Hurlstone, A. Zebrafish: A new companion for translational research in oncology. Clin. Cancer Res. 2015, 21, 969–975. [Google Scholar] [CrossRef] [Green Version]
- Griffin, A.; Hamling, K.R.; Knupp, K.; Hong, S.; Lee, L.P.; Baraban, S.C. Clemizole and modulators of serotonin signalling suppress seizures in Dravet syndrome. Brain 2017, 140, 669–683. [Google Scholar] [CrossRef]
- Asimaki, A.; Kapoor, S.; Plovie, E.; Karin Arndt, A.; Adams, E.; Liu, Z.; James, C.A.; Judge, D.P.; Calkins, H.; Churko, J.; et al. Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Sci. Transl. Med. 2014, 6, 240ra74. [Google Scholar] [CrossRef] [Green Version]
- Yu, P.B.; Hong, C.C.; Sachidanandan, C.; Babitt, J.L.; Deng, D.Y.; Hoyng, S.A.; Lin, H.Y.; Bloch, K.D.; Peterson, R.T. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 2008, 4, 33–41. [Google Scholar] [CrossRef]
- Leung, A.W.Y.; Veinotte, C.J.; Melong, N.; Oh, M.H.; Chen, K.; Enfield, K.S.S.; Backstrom, I.; Warburton, C.; Yapp, D.; Berman, J.N.; et al. In Vivo Validation of PAPSS1 (3’-phosphoadenosine 5’-phosphosulfate synthase 1) as a Cisplatin-sensitizing Therapeutic Target. Clin. Cancer Res. 2017, 23, 6555–6566. [Google Scholar] [CrossRef] [Green Version]
- Ibhazehiebo, K.; Gavrilovici, C.; de la Hoz, C.L.; Ma, S.C.; Rehak, R.; Kaushik, G.; Meza Santoscoy, P.L.; Scott, L.; Nath, N.; Kim, D.Y.; et al. A novel metabolism-based phenotypic drug discovery platform in zebrafish uncovers HDACs 1 and 3 as a potential combined anti-seizure drug target. Brain 2018, 141, 744–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandelbaum, J.; Shestopalov, I.A.; Henderson, R.E.; Chau, N.G.; Knoechel, B.; Wick, M.J.; Zon, L.I. Zebrafish blastomere screen identifies retinoic acid suppression of MYB in adenoid cystic carcinoma. J. Exp. Med. 2018, 215, 2673–2685. [Google Scholar] [CrossRef]
- North, T.E.; Goessling, W.; Walkley, C.R.; Lengerke, C.; Kopani, K.R.; Lord, A.M.; Weber, G.J.; Bowman, T.V.; Jang, I.H.; Grosser, T.; et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 2007, 447, 1007–1011. [Google Scholar] [CrossRef]
- Wertman, J.; Veinotte, C.J.; Dellaire, G.; Berman, J.N. The Zebrafish Xenograft Platform: Evolution of a Novel Cancer Model and Preclinical Screening Tool. Adv Exp. Med. Biol. 2016, 916, 289–314. [Google Scholar] [CrossRef]
- Veinotte, C.J.; Dellaire, G.; Berman, J.N. Hooking the big one: The potential of zebrafish xenotransplantation to reform cancer drug screening in the genomic era. Dis. Models Mech. 2014, 7, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Kirchberger, S.; Sturtzel, C.; Pascoal, S.; Distel, M. Quo natas, Danio?-Recent Progress in Modeling Cancer in Zebrafish. Front. Oncol. 2017, 7, 186. [Google Scholar] [CrossRef]
- Stewart, E.; Federico, S.M.; Chen, X.; Shelat, A.A.; Bradley, C.; Gordon, B.; Karlstrom, A.; Twarog, N.R.; Clay, M.R.; Bahrami, A.; et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 2017, 549, 96–100. [Google Scholar] [CrossRef]
- Müller, M.; Rösch, L.; Najafi, S.; Gatzweiler, C.; Ridinger, J.; Gerloff, X.F.; Jones, D.T.W.; Baßler, J.; Kreth, S.; Stainczyk, S.; et al. Combining APR-246 and HDAC-Inhibitors: A Novel Targeted Treatment Option for Neuroblastoma. Cancers 2021, 13, 4476. [Google Scholar] [CrossRef]
- Schmitt, M.; Pawlita, M. High-throughput detection and multiplex identification of cell contaminations. Nucleic Acids Res. 2009, 37, e119. [Google Scholar] [CrossRef]
- Koelsche, C.; Schrimpf, D.; Stichel, D.; Sill, M.; Sahm, F.; Reuss, D.E.; Blattner, M.; Worst, B.; Heilig, C.E.; Beck, K.; et al. Sarcoma classification by DNA methylation profiling. Nat. Commun. 2021, 12, 498. [Google Scholar] [CrossRef]
- Henrich, K.O.; Bender, S.; Saadati, M.; Dreidax, D.; Gartlgruber, M.; Shao, C.; Herrmann, C.; Wiesenfarth, M.; Parzonka, M.; Wehrmann, L.; et al. Integrative Genome-Scale Analysis Identifies Epigenetic Mechanisms of Transcriptional Deregulation in Unfavorable Neuroblastomas. Cancer Res. 2016, 76, 5523–5537. [Google Scholar] [CrossRef] [Green Version]
- ElHarouni, D.; Berker, Y.; Peterziel, H.; Gopisetty, A.; Turunen, L.; Kreth, S.; Stainczyk, S.A.; Oehme, I.; Pietiainen, V.; Jager, N.; et al. iTReX: Interactive exploration of mono- and combination therapy dose response profiling data. Pharmacol. Res. 2021, 175, 105996. [Google Scholar] [CrossRef]
- Wrobel, J.K.; Najafi, S.; Ayhan, S.; Gatzweiler, C.; Krunic, D.; Ridinger, J.; Milde, T.; Westermann, F.; Peterziel, H.; Meder, B.; et al. Rapid In Vivo Validation of HDAC Inhibitor-Based Treatments in Neuroblastoma Zebrafish Xenografts. Pharmaceuticals 2020, 13, 345. [Google Scholar] [CrossRef]
- Fior, R.; Povoa, V.; Mendes, R.V.; Carvalho, T.; Gomes, A.; Figueiredo, N.; Ferreira, M.G. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc. Natl. Acad. Sci. USA 2017, 114, E8234–E8243. [Google Scholar] [CrossRef] [Green Version]
- Yadav, B.; Pemovska, T.; Szwajda, A.; Kulesskiy, E.; Kontro, M.; Karjalainen, R.; Majumder, M.M.; Malani, D.; Murumagi, A.; Knowles, J.; et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci. Rep. 2014, 4, 5193. [Google Scholar] [CrossRef] [Green Version]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Hee, E.; Wong, M.K.; Tan, S.H.; Choo, Z.; Kuick, C.H.; Ling, S.; Yong, M.H.; Jain, S.; Lian, D.W.Q.; Ng, E.H.Q.; et al. Neuroblastoma patient-derived cultures are enriched for a mesenchymal gene signature and reflect individual drug response. Cancer Sci. 2020, 111, 3780–3792. [Google Scholar] [CrossRef]
- Vaira, V.; Fedele, G.; Pyne, S.; Fasoli, E.; Zadra, G.; Bailey, D.; Snyder, E.; Faversani, A.; Coggi, G.; Flavin, R.; et al. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc. Natl. Acad. Sci. USA 2010, 107, 8352–8356. [Google Scholar] [CrossRef] [Green Version]
- Sidarovich, V.; De Mariano, M.; Aveic, S.; Pancher, M.; Adami, V.; Gatto, P.; Pizzini, S.; Pasini, L.; Croce, M.; Parodi, F.; et al. A High-Content Screening of Anticancer Compounds Suggests the Multiple Tyrosine Kinase Inhibitor Ponatinib for Repurposing in Neuroblastoma Therapy. Mol. Cancer Ther. 2018, 17, 1405–1415. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.K.; Liu, Z.; Sa, J.K.; Shin, S.; Wang, J.; Bordyuh, M.; Cho, H.J.; Elliott, O.; Chu, T.; Choi, S.W.; et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat. Genet. 2018, 50, 1399–1411. [Google Scholar] [CrossRef]
- Sa, J.K.; Hwang, J.R.; Cho, Y.J.; Ryu, J.Y.; Choi, J.J.; Jeong, S.Y.; Kim, J.; Kim, M.S.; Paik, E.S.; Lee, Y.Y.; et al. Pharmacogenomic analysis of patient-derived tumor cells in gynecologic cancers. Genome Biol. 2019, 20, 253. [Google Scholar] [CrossRef]
- Chia, S.; Low, J.L.; Zhang, X.; Kwang, X.L.; Chong, F.T.; Sharma, A.; Bertrand, D.; Toh, S.Y.; Leong, H.S.; Thangavelu, M.T.; et al. Phenotype-driven precision oncology as a guide for clinical decisions one patient at a time. Nat. Commun. 2017, 8, 435. [Google Scholar] [CrossRef]
- Pemovska, T.; Kontro, M.; Yadav, B.; Edgren, H.; Eldfors, S.; Szwajda, A.; Almusa, H.; Bespalov, M.M.; Ellonen, P.; Elonen, E.; et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov. 2013, 3, 1416–1429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermsen, S.A.; van den Brandhof, E.J.; van der Ven, L.T.; Piersma, A.H. Relative embryotoxicity of two classes of chemicals in a modified zebrafish embryotoxicity test and comparison with their in vivo potencies. Toxicol. In Vitro 2011, 25, 745–753. [Google Scholar] [CrossRef] [Green Version]
- Chahardehi, A.M.; Arsad, H.; Lim, V. Zebrafish as a Successful Animal Model for Screening Toxicity of Medicinal Plants. Plants 2020, 9, 345. [Google Scholar] [CrossRef]
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Reviews Clin. Oncol. 2012, 9, 338–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Franco, G.; Usai, A.; Funel, N.; Palmeri, M.; Montesanti, I.E.R.; Bianchini, M.; Gianardi, D.; Furbetta, N.; Guadagni, S.; Vasile, E.; et al. Use of zebrafish embryos as avatar of patients with pancreatic cancer: A new xenotransplantation model towards personalized medicine. World J. Gastroenterol. 2020, 26, 2792–2809. [Google Scholar] [CrossRef] [PubMed]
- Versteege, I.; Sevenet, N.; Lange, J.; Rousseau-Merck, M.F.; Ambros, P.; Handgretinger, R.; Aurias, A.; Delattre, O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998, 394, 203–206. [Google Scholar] [CrossRef]
- Biegel, J.A.; Zhou, J.Y.; Rorke, L.B.; Stenstrom, C.; Wainwright, L.M.; Fogelgren, B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999, 59, 74–79. [Google Scholar] [PubMed]
- Hoy, S.M. Tazemetostat: First Approval. Drugs 2020, 80, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Rare Tumors in Kids May Respond to Tazemetostat. Cancer Discov. 2018, 8, OF5. [CrossRef] [Green Version]
- Massaro, F.; Molica, M.; Breccia, M. Ponatinib: A Review of Efficacy and Safety. Curr. Cancer Drug Targets 2018, 18, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.P.; Todd, J.R.; Finetti, M.A.; McCarthy, F.; Broncel, M.; Vyse, S.; Luczynski, M.T.; Crosier, S.; Ryall, K.A.; Holmes, K.; et al. Dual Targeting of PDGFRalpha and FGFR1 Displays Synergistic Efficacy in Malignant Rhabdoid Tumors. Cell Rep. 2016, 17, 1265–1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauvin, C.; Leruste, A.; Tauziede-Espariat, A.; Andrianteranagna, M.; Surdez, D.; Lescure, A.; Han, Z.Y.; Anthony, E.; Richer, W.; Baulande, S.; et al. High-Throughput Drug Screening Identifies Pazopanib and Clofilium Tosylate as Promising Treatments for Malignant Rhabdoid Tumors. Cell Rep. 2017, 21, 1737–1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerl, K.; Ries, D.; Unland, R.; Borchert, C.; Moreno, N.; Hasselblatt, M.; Jurgens, H.; Kool, M.; Gorlich, D.; Eveslage, M.; et al. The histone deacetylase inhibitor SAHA acts in synergism with fenretinide and doxorubicin to control growth of rhabdoid tumor cells. BMC Cancer 2013, 13, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harttrampf, A.C.; da Costa, M.E.M.; Renoult, A.; Daudigeos-Dubus, E.; Geoerger, B. Histone deacetylase inhibitor panobinostat induces antitumor activity in epithelioid sarcoma and rhabdoid tumor by growth factor receptor modulation. BMC Cancer 2021, 21, 833. [Google Scholar] [CrossRef]
- Hou, H.; Sun, D.; Zhang, X. The role of MDM2 amplification and overexpression in therapeutic resistance of malignant tumors. Cancer Cell Int. 2019, 19, 216. [Google Scholar] [CrossRef] [Green Version]
- Taylor, A.C.; Shu, L.; Danks, M.K.; Poquette, C.A.; Shetty, S.; Thayer, M.J.; Houghton, P.J.; Harris, L.C. P53 mutation and MDM2 amplification frequency in pediatric rhabdomyosarcoma tumors and cell lines. Med. Pediatric Oncol. 2000, 35, 96–103. [Google Scholar] [CrossRef]
- Zafar, A.; Wang, W.; Liu, G.; Wang, X.; Xian, W.; McKeon, F.; Foster, J.; Zhou, J.; Zhang, R. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med. Res. Rev. 2021, 41, 961–1021. [Google Scholar] [CrossRef]
- Passoni, L.; Longo, L.; Collini, P.; Coluccia, A.M.; Bozzi, F.; Podda, M.; Gregorio, A.; Gambini, C.; Garaventa, A.; Pistoia, V.; et al. Mutation-independent anaplastic lymphoma kinase overexpression in poor prognosis neuroblastoma patients. Cancer Res. 2009, 69, 7338–7346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, L.M.; Seftor, E.A.; Bonde, G.; Cornell, R.A.; Hendrix, M.J. The fate of human malignant melanoma cells transplanted into zebrafish embryos: Assessment of migration and cell division in the absence of tumor formation. Dev. Dyn. 2005, 233, 1560–1570. [Google Scholar] [CrossRef]
- Konantz, M.; Balci, T.B.; Hartwig, U.F.; Dellaire, G.; Andre, M.C.; Berman, J.N.; Lengerke, C. Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann. N. Y. Acad. Sci. 2012, 1266, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Fazio, M.; Ablain, J.; Chuan, Y.; Langenau, D.M.; Zon, L.I. Zebrafish patient avatars in cancer biology and precision cancer therapy. Nat. Rev. Cancer 2020, 20, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.W.; Oh, E.S.; Park, S.H.; Chang, Y.T.; Kim, C.H.; Choi, S.Y.; Williams, D.R. A novel zebrafish human tumor xenograft model validated for anti-cancer drug screening. Mol. Biosyst. 2012, 8, 1930–1939. [Google Scholar] [CrossRef] [PubMed]
- Usai, A.; Di Franco, G.; Colucci, P.; Pollina, L.E.; Vasile, E.; Funel, N.; Palmeri, M.; Dente, L.; Falcone, A.; Morelli, L.; et al. A Model of a Zebrafish Avatar for Co-Clinical Trials. Cancers 2020, 12, 677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usai, A.; Di Franco, G.; Piccardi, M.; Cateni, P.; Pollina, L.E.; Vivaldi, C.; Vasile, E.; Funel, N.; Palmeri, M.; Dente, L.; et al. Zebrafish Patient-Derived Xenografts Identify Chemo-Response in Pancreatic Ductal Adenocarcinoma Patients. Cancers 2021, 13, 4131. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Sainz, P.; Guerra-Varela, J.; Carreira, M.J.; Mariscal, J.; Roel, M.; Rubiolo, J.A.; Sciara, A.A.; Abal, M.; Botana, L.M.; Lopez, R.; et al. Improving zebrafish embryo xenotransplantation conditions by increasing incubation temperature and establishing a proliferation index with ZFtool. BMC Cancer 2018, 18, 3. [Google Scholar] [CrossRef] [PubMed]
- Almeida, V.M.; Marana, S.R. Optimum temperature may be a misleading parameter in enzyme characterization and application. PLoS ONE 2019, 14, e0212977. [Google Scholar] [CrossRef] [Green Version]
- Peterson, M.E.; Daniel, R.M.; Danson, M.J.; Eisenthal, R. The dependence of enzyme activity on temperature: Determination and validation of parameters. Biochem. J. 2007, 402, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Avci, M.E.; Keskus, A.G.; Targen, S.; Isilak, M.E.; Ozturk, M.; Atalay, R.C.; Adams, M.M.; Konu, O. Development of a novel zebrafish xenograft model in ache mutants using liver cancer cell lines. Sci. Rep. 2018, 8, 1570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabezas-Sainz, P.; Coppel, C.; Pensado-Lopez, A.; Fernandez, P.; Muinelo-Romay, L.; Lopez-Lopez, R.; Rubiolo, J.A.; Sanchez, L. Morphological Abnormalities and Gene Expression Changes Caused by High Incubation Temperatures in Zebrafish Xenografts with Human Cancer Cells. Genes 2021, 12, 113. [Google Scholar] [CrossRef] [PubMed]
- Cabezas-Sainz, P.; Pensado-Lopez, A.; Sainz, B., Jr.; Sanchez, L. Modeling Cancer Using Zebrafish Xenografts: Drawbacks for Mimicking the Human Microenvironment. Cells 2020, 9, 1978. [Google Scholar] [CrossRef]
- Pascoal, S.; Salzer, B.; Scheuringer, E.; Wenninger-Weinzierl, A.; Sturtzel, C.; Holter, W.; Taschner-Mandl, S.; Lehner, M.; Distel, M. A Preclinical Embryonic Zebrafish Xenograft Model to Investigate CAR T Cells In Vivo. Cancers 2020, 12, 567. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Culture | Cells/Well | Pre-Culture Time 1/Passage |
|---|---|---|
| FTC_INF_R_1288_r1 | 908 | 5 d after fresh tissue dissociation |
| LTC_INF_R_1288_r1 | 1000 | passage 22 |
| PDX-C_INF_R_1288_r1 | 1000 | 2d after PDX tumor dissociation |
| FTC_INF_R_1467_r1 | 1000 | 3 d after fresh tissue dissociation |
| LTC_INF_R_1467_r1 | 1000 | passage 15 |
| PDX-C_INF_R_1467_r1 | 1000 | 6 days after PDX tumor dissociation |
| FTC_ INF_R_359_r3 | 1000 | 3 d after fresh tissue dissociation |
| LTC_ INF_R_359_r3 | 1000 | passage 10 |
| PDX-C_ INF_R_359_r3 | 1000 | 14 d after PDX tumor dissociation |
| Reagent | Stock Concentration/Solvent | Supplier (Name, City, State) |
|---|---|---|
| A-1151852 | 100 mM/DMSO | ChemieTek, Indianapolis, IN, USA |
| alpelisib | 10 mM/DMSO | MedChemExpress, Monmouth Junction, NJ, USA |
| AMG-232 | 10 mM/DMSO | MedChemExpress, Monmouth Junction, NJ, USA |
| crizotinib | 50 mM/DMSO | Selleckchem, Houston, TX, USA |
| dactinomycin | 10 mM/DMSO | MedChemExpress, Monmouth Junction, NJ, USA |
| entrectinib | 10 mM/DMSO | MedChemExpress, Monmouth Junction, NJ, USA |
| everolimus | 10 mM/DMSO | LC Laboratories, Woburn, MA, USA |
| gemcitabine | 10 mM/DMSO | MedChemExpress, Monmouth Junction, NJ, USA |
| idasanutlin | 100 mM/DMSO | MedChemExpress, Monmouth Junction, NJ, USA |
| methotrexate | 50 mM/DMSO | Sigma-Aldrich Chemie, Taufkirchen, Germany |
| navitoclax | 100 mM/DMSO | MedChemExpress, Monmouth Junction, NJ, USA |
| olaparib | 100 mM/DMSO | LC Laboratories, Woburn, MA, USA |
| panobinostat | 10 mM/DMSO | LC Laboratories, Woburn, MA, USA |
| ponatinib | 10 mM/DMSO | Selleckchem, Houston, TX, USA |
| selinexor | 100 mM/DMSO | ChemieTek, Indianapolis, IN, USA |
| tazemetostat | 100 mM/DMSO | ChemieTek, Indianapolis, IN, USA |
| trametinib | 25 mM/DMSO | ChemieTek, Indianapolis, IN, USA |
| Patient Sample | Entity 1 | Molecular Aberrations 2 |
|---|---|---|
| INF_R_1288_r1 | rhabdoid tumor | SMARCB1 del; PRKCDmut; DDR2mut |
| INF_R_1467_r1 | embryonal RMS (eRMS) | MDM2amp; CDKN2Amut&LOH |
| INF_R_359_r3 | neuroblastoma | MYCNamp; PIK3CAmut |
| Basis for Drug Selection | INF_R_1288_r1 (Rhabdoid Tumor) | INF_R_1467_r1 (Embryonal RMS) | INF_R_359_r3 (Neuroblastoma) |
|---|---|---|---|
| NGS based molecular targets 1 and matching drugs | tazemetostat (EZH2i; SMARCB1del); ponatinib (RTKi; DDR2mut) | idasanutlin (MDM2i; MDM2amp); olaparib (PARPi; genomic instability) | alpelisib (PI3Ki; PIK3CAmut); everolimus (mTORi; PIK3CAmut) |
| DSP-derived hits (TOP25 hit for all three models) | AMG-232 crizotinib dactinomycin idasanutlin navitoclax panobinostat ponatinib selinexor trametinib | A-1151852 AMG-232 dactinomycin idasanutlin navitoclax olaparib selinexor | ceritinib gemcitabine idasanutlin selinexor panobinostat |
| Negative controls (solvent and DSP noneffective drug) | DMSO methotrexate | DMSO entrectinib | DMSO tazemetostat |
| Drug | Concentration Range | MTD 1 | LD 2 |
|---|---|---|---|
| A-1151852 | 1–500 µM | 10 µM | not detected |
| alpelisib | 0.1–50 µM | 50 µM | not detected |
| AMG-232 | 0.1–50 µM | 10 µM | 50 µM |
| crizotinib | 0.05–10 µM | 10 µM | not detected |
| dactinomycin | 0.001–20 µM | 20 µM | not detected |
| entrectinib | 0.02–20 µM | 2 µM | not detected |
| everolimus | 0.04–10 µM | 1 µM | 4 µM |
| gemcitabine | 0.1–10 µM | 10 µM | not detected |
| idasanutlin | 0.01–50 µM | 10 µM | 50 µM |
| methotrexate | 1.5–150 µM | 150 µM | not detected |
| navitoclax | 1–50 µM | 10 µM | 50 µM 3 |
| olaparib | 0.1–30 µM | 30 µM | not detected |
| panobinostat | 0.1–20 µM | 1 µM | 10 µM 4 |
| ponatinib | 0.01–2 µM | 1 µM | 2 µM 5 |
| selinexor | 0.1–20 µM | 1 µM | 10 µM |
| tazemetostat | 0.1–30 µM | 30 µM | not detected |
| trametinib | 0.1–50 µM | 0.1 µM | 1 µM 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gatzweiler, C.; Ridinger, J.; Herter, S.; Gerloff, X.F.; ElHarouni, D.; Berker, Y.; Imle, R.; Schmitt, L.; Kreth, S.; Stainczyk, S.; et al. Functional Therapeutic Target Validation Using Pediatric Zebrafish Xenograft Models. Cancers 2022, 14, 849. https://doi.org/10.3390/cancers14030849
Gatzweiler C, Ridinger J, Herter S, Gerloff XF, ElHarouni D, Berker Y, Imle R, Schmitt L, Kreth S, Stainczyk S, et al. Functional Therapeutic Target Validation Using Pediatric Zebrafish Xenograft Models. Cancers. 2022; 14(3):849. https://doi.org/10.3390/cancers14030849
Chicago/Turabian StyleGatzweiler, Charlotte, Johannes Ridinger, Sonja Herter, Xenia F. Gerloff, Dina ElHarouni, Yannick Berker, Roland Imle, Lukas Schmitt, Sina Kreth, Sabine Stainczyk, and et al. 2022. "Functional Therapeutic Target Validation Using Pediatric Zebrafish Xenograft Models" Cancers 14, no. 3: 849. https://doi.org/10.3390/cancers14030849