Large-Scale Analysis of X Inactivation Variations between Primed and Naïve Human Embryonic Stem Cells
Abstract
:1. Introduction
2. Results
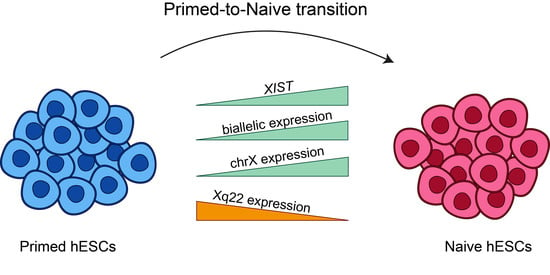
2.1. XIST Expression in Primed-To-Naïve Transition
2.2. Chromosome X Genes and Allelic Expression in Naïve Samples
2.3. Geographical Pattern of Expression of X-Linked Genes and Their Allelic Representation
3. Discussion
4. Experimental Procedure
4.1. Sample Collection
4.2. Bulk RNA Gene and Allelic Expression Analysis
4.3. Gene Expression TPM Calculation and Normalization
4.4. Pseudo-Bulk RNA Analysis of Human Epiblasts
4.5. Quantifying Naïve: Primed Moving Average Plots
4.6. Quantifying X: Autosomes Allelic Ratio
4.7. Statistical Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Disteche, C.M. Dosage Compensation of the Sex Chromosomes. Annu. Rev. Genet. 2012, 46, 537–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ercan, S.; Lieb, J.D.C. Elegans Dosage Compensation: A Window into Mechanisms of Domain-Scale Gene Regulation. Chromosome Res. 2009, 17, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Hiatt, J.B.; Nguyen, D.K.; Ercan, S.; Sturgill, D.; Hillier, L.W.; Schlesinger, F.; Davis, C.A.; Reinke, V.J.; Gingeras, T.R.; et al. Evidence for Compensatory Upregulation of Expressed X-Linked Genes in Mammals, Caenorhabditis Elegans and Drosophila Melanogaster. Nat. Genet. 2011, 43, 1179–1185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nora, E.P.; Heard, E. X Chromosome Inactivation: When Dosage Counts. Cell 2009, 139, 865–867. [Google Scholar] [CrossRef] [Green Version]
- Heard, E.; Disteche, C.M. Dosage Compensation in Mammals: Fine-Tuning the Expression of the X Chromosome. Genes Dev. 2006, 20, 1848–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, C.J.; Hendrich, B.D.; Rupert, J.L.; Lafrenière, R.G.; Xing, Y.; Lawrence, J.; Willard, H.F. The Human XIST Gene: Analysis of a 17 Kb Inactive X-Specific RNA That Contains Conserved Repeats and Is Highly Localized within the Nucleus. Cell 1992, 71, 527–542. [Google Scholar] [CrossRef]
- De Los Angeles, A.; Ferrari, F.; Xi, R.; Fujiwara, Y.; Benvenisty, N.; Deng, H.; Hochedlinger, K.; Jaenisch, R.; Lee, S.; Leitch, H.G.; et al. Hallmarks of Pluripotency. Nature 2015, 525, 469–478. [Google Scholar] [CrossRef]
- Hoffman, L.M.; Hall, L.; Batten, J.L.; Young, H.; Pardasani, D.; Baetge, E.E.; Lawrence, J.; Carpenter, M.K. X-Inactivation Status Varies in Human Embryonic Stem Cell Lines. Stem Cells 2005, 23, 1468–1478. [Google Scholar] [CrossRef]
- Xie, P.; Ouyang, Q.; Leng, L.; Hu, L.; Cheng, D.; Tan, Y.; Lu, G.; Lin, G. The Dynamic Changes of X Chromosome Inactivation during Early Culture of Human Embryonic Stem Cells. Stem Cell Res. 2016, 17, 84–92. [Google Scholar] [CrossRef] [Green Version]
- Dvash, T.; Lavon, N.; Fan, G. Variations of X Chromosome Inactivation Occur in Early Passages of Female Human Embryonic Stem Cells. PLoS ONE 2010, 5, e11330. [Google Scholar] [CrossRef]
- Vallot, C.; Ouimette, J.F.; Makhlouf, M.; Féraud, O.; Pontis, J.; Côme, J.; Martinat, C.; Bennaceur-Griscelli, A.; Lalande, M.; Rougeulle, C. Erosion of X Chromosome Inactivation in Human Pluripotent Cells Initiates with XACT Coating and Depends on a Specific Heterochromatin Landscape. Cell Stem Cell 2015, 16, 533–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, L.L.; Byron, M.; Butler, J.; Becker, K.A.; Nelson, A.; Amit, M.; Itskovitz-Eldor, J.; Stein, J.; Stein, G.; Ware, C.; et al. X-Inactivation Reveals Epigenetic Anomalies in Most HESC but Identifies Sublines That Initiate as Expected. J. Cell. Physiol. 2008, 216, 445–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Matsuno, Y.; Fouse, S.D.; Rao, N.; Root, S.; Xu, R.; Pellegrini, M.; Riggs, A.D.; Fan, G. X-Inactivation in Female Human Embryonic Stem Cells Is in a Nonrandom Pattern and Prone to Epigenetic Alterations. Proc. Natl. Acad. Sci. USA 2008, 105, 4709–4714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, S.S.; Rowntree, R.K.; Mekhoubad, S.; Lee, J.T. X-Chromosome Inactivation and Epigenetic Fluidity in Human Embryonic Stem Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 4820–4825. [Google Scholar] [CrossRef] [Green Version]
- Vallot, C.; Ouimette, J.F.; Rougeulle, C. Establishment of X Chromosome Inactivation and Epigenomic Features of the Inactive X Depend on Cellular Contexts. BioEssays 2016, 38, 869–880. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.; Bonora, G.; Sahakyan, A.; Kim, R.; Chronis, C.; Langerman, J.; Fitz-Gibbon, S.; Rubbi, L.; Skelton, R.J.P.; Ardehali, R.; et al. Human Embryonic Stem Cells Do Not Change Their X Inactivation Status during Differentiation. Cell Rep. 2017, 18, 54–67. [Google Scholar] [CrossRef]
- Bar, S.; Seaton, L.R.; Weissbein, U.; Eldar-Geva, T.; Benvenisty, N. Global Characterization of X Chromosome Inactivation in Human Pluripotent Stem Cells. Cell Rep. 2019, 27, 20–29.e3. [Google Scholar] [CrossRef] [Green Version]
- Bruck, T.; Benvenisty, N. Meta-Analysis of the Heterogeneity of X Chromosome Inactivation in Human Pluripotent Stem Cells. Stem Cell Res. 2011, 6, 187–193. [Google Scholar] [CrossRef] [Green Version]
- Takashima, Y.; Guo, G.; Loos, R.; Nichols, J.; Ficz, G.; Krueger, F.; Oxley, D.; Santos, F.; Clarke, J.; Mansfield, W.; et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell 2014, 158, 1254–1269. [Google Scholar] [CrossRef] [Green Version]
- Di Stefano, B.; Ueda, M.; Sabri, S.; Brumbaugh, J.; Huebner, A.J.; Sahakyan, A.; Clement, K.; Clowers, K.J.; Erickson, A.R.; Shioda, K.; et al. Reduced MEK Inhibition Preserves Genomic Stability in Naive Human Embryonic Stem Cells. Nat. Methods 2018, 15, 732–740. [Google Scholar] [CrossRef]
- Theunissen, T.W.; Friedli, M.; He, Y.; Planet, E.; O’Neil, R.C.; Markoulaki, S.; Pontis, J.; Wang, H.; Iouranova, A.; Imbeault, M.; et al. Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 2016, 19, 502–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, T.S.; Zimmerlin, L.; Evans-Moses, R.; Thomas, J.; Huo, J.S.; Kanherkar, R.; He, A.; Ruzgar, N.; Grebe, R.; Bhutto, I.; et al. Vascular Progenitors Generated from Tankyrase Inhibitor-Regulated Naïve Diabetic Human IPSC Potentiate Efficient Revascularization of Ischemic Retina. Nat. Commun. 2020, 11, 1195. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Li, H.; Jiang, H.; Ren, Y.; Yu, X.; Qiu, J.; Stablewski, A.B.; Zhang, B.; Buck, M.J.; Feng, J. Transient Inhibition of MTOR in Human Pluripotent Stem Cells Enables Robust Formation of Mouse-Human Chimeric Embryos. Sci. Adv. 2020, 6, eaaz0298. [Google Scholar] [CrossRef] [PubMed]
- An, C.; Feng, G.; Zhang, J.; Cao, S.; Wang, Y.; Wang, N.; Lu, F.; Zhou, Q.; Wang, H. Overcoming Autocrine FGF Signaling-Induced Heterogeneity in Naive Human ESCs Enables Modeling of Random X Chromosome Inactivation. Cell Stem Cell 2020, 27, 482–497.e4. [Google Scholar] [CrossRef] [PubMed]
- Gafni, O.; Weinberger, L.; Mansour, A.A.; Manor, Y.S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of Novel Human Ground State Naive Pluripotent Stem Cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, A.; Benvenisty, N. Defining Human Pluripotency. Cell Stem Cell 2019, 25, 9–22. [Google Scholar] [CrossRef]
- Chan, Y.S.; Göke, J.; Ng, J.H.; Lu, X.; Gonzales, K.A.U.; Tan, C.P.; Tng, W.Q.; Hong, Z.Z.; Lim, Y.S.; Ng, H.H. Induction of a Human Pluripotent State with Distinct Regulatory Circuitry That Resembles Preimplantation Epiblast. Cell Stem Cell 2013, 13, 663–675. [Google Scholar] [CrossRef] [Green Version]
- Sperber, H.; Mathieu, J.; Wang, Y.; Ferreccio, A.; Hesson, J.; Xu, Z.; Fischer, K.A.; Devi, A.; Detraux, D.; Gu, H.; et al. The Metabolome Regulates the Epigenetic Landscape during Naive-to-Primed Human Embryonic Stem Cell Transition. Nat. Cell Biol. 2015, 17, 1523–1535. [Google Scholar] [CrossRef]
- Ji, X.; Dadon, D.B.; Powell, B.E.; Fan, Z.P.; Borges-Rivera, D.; Shachar, S.; Weintraub, A.S.; Hnisz, D.; Pegoraro, G.; Lee, T.I.; et al. 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell 2016, 18, 262–275. [Google Scholar] [CrossRef] [Green Version]
- Guo, G.; von Meyenn, F.; Rostovskaya, M.; Clarke, J.; Dietmann, S.; Baker, D.; Sahakyan, A.; Myers, S.; Bertone, P.; Reik, W.; et al. Epigenetic Resetting of Human Pluripotency. Development 2017, 144, 2748–2763. [Google Scholar] [CrossRef] [Green Version]
- Sahakyan, A.; Kim, R.; Chronis, C.; Sabri, S.; Bonora, G.; Theunissen, T.W.; Kuoy, E.; Langerman, J.; Clark, A.T.; Jaenisch, R.; et al. Human Naïve Pluripotent Stem Cells Model X-Chromosome Dampening and X-Inactivation. Cell Stem Cell 2017, 20, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collier, A.J.; Panula, S.P.; Schell, J.P.; Chovanec, P.; Plaza Reyes, A.; Petropoulos, S.; Corcoran, A.E.; Walker, R.; Douagi, I.; Lanner, F.; et al. Comprehensive Cell Surface Protein Profiling Identifies Specific Markers of Human Naive and Primed Pluripotent States. Cell Stem Cell 2017, 20, 874–890.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Nefzger, C.M.; Rossello, F.J.; Chen, J.; Knaupp, A.S.; Firas, J.; Ford, E.; Pflueger, J.; Paynter, J.M.; Chy, H.S.; et al. Comprehensive Characterization of Distinct States of Human Naive Pluripotency Generated by Reprogramming. Nat. Methods 2017, 14, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Sagi, I.; Benvenisty, N. Stem Cells: Aspiring to Naivety. Nature 2016, 540, 211–212. [Google Scholar] [CrossRef] [PubMed]
- Petropoulos, S.; Edsgärd, D.; Reinius, B.; Deng, Q.; Panula, S.P.; Codeluppi, S.; Plaza Reyes, A.; Linnarsson, S.; Sandberg, R.; Lanner, F. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 2016, 165, 1012–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stirparo, G.G.; Boroviak, T.; Guo, G.; Nichols, J.; Smith, A.; Bertone, P. Integrated Analysis of Single-Cell Embryo Data Yields a Unified Transcriptome Signature for the Human Preimplantation Epiblast. Development 2018, 7, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krawchuk, D.; Honma-Yamanaka, N.; Anani, S.; Yamanaka, Y. FGF4 Is a Limiting Factor Controlling the Proportions of Primitive Endoderm and Epiblast in the ICM of the Mouse Blastocyst. Dev. Biol. 2013, 384, 65–71. [Google Scholar] [CrossRef] [Green Version]
- Ying, Q.L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The Ground State of Embryonic Stem Cell Self-Renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.; Maruyama, T.; Fan, G. The Naive State of Human Pluripotent Stem Cells: A Synthesis of Stem Cell and Preimplantation Embryo Transcriptome Analyses. Cell Stem Cell 2014, 15, 410–415. [Google Scholar] [CrossRef] [Green Version]
- Pastor, W.A.; Chen, D.; Liu, W.; Kim, R.; Sahakyan, A.; Lukianchikov, A.; Plath, K.; Jacobsen, S.E.; Clark, A.T. Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell 2016, 18, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Lyon, M.F. The Lyon and the LINE Hypothesis. Semin. Cell Dev. Biol. 2003, 14, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.A.; Carrel, L.; Chakravarti, A.; Eichler, E.E. Molecular Evidence for a Relationship between LINE-1 Elements and X Chromosome Inactivation: The Lyon Repeat Hypothesis. Proc. Natl. Acad. Sci. USA 2000, 97, 6634–6639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Song, F.; Zhu, J.; Zhang, S.; Yang, Y.; Chen, T.; Tang, B.; Dong, L.; Ding, N.; Zhang, Q.; et al. GSA: Genome Sequence Archive. Genom. Proteom. Bioinform. 2017, 15, 14–18. [Google Scholar] [CrossRef]
- Wan, H.; Fu, R.; Tong, M.; Wang, Y.; Wang, L.; Wang, S.; Zhang, Y.; Li, W.; Wang, X.J.; Feng, G. Influence of Feeder Cells on Transcriptomic Analysis of Pluripotent Stem Cells. Cell Prolif. 2022, 55, e13189. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Kluin, R.J.C.; Kemper, K.; Kuilman, T.; de Ruiter, J.R.; Iyer, V.; Forment, J.V.; Cornelissen-Steijger, P.; de Rink, I.; ter Brugge, P.; Song, J.Y.; et al. XenofilteR: Computational Deconvolution of Mouse and Human Reads in Tumor Xenograft Sequence Data. BMC Bioinform. 2018, 19, 366. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Lezmi, E.; Benvenisty, N. Identification of Cancer-Related Mutations in Human Pluripotent Stem Cells Using RNA-Seq Analysis. Nat. Protoc. 2021, 16, 4522–4537. [Google Scholar] [CrossRef]
- Keshet, G.; Benvenisty, N. Large-Scale Analysis of Imprinting in Naive Human Pluripotent Stem Cells Reveals Recurrent Aberrations and a Potential Link to FGF Signaling. Stem Cell Rep. 2021, 16, 2520–2533. [Google Scholar] [CrossRef]
- Weissbein, U.; Schachter, M.; Egli, D.; Benvenisty, N. Analysis of Chromosomal Aberrations and Recombination by Allelic Bias in RNA-Seq. Nat. Commun. 2016, 7, 12144. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarel-Gallily, R.; Benvenisty, N. Large-Scale Analysis of X Inactivation Variations between Primed and Naïve Human Embryonic Stem Cells. Cells 2022, 11, 1729. https://doi.org/10.3390/cells11111729
Sarel-Gallily R, Benvenisty N. Large-Scale Analysis of X Inactivation Variations between Primed and Naïve Human Embryonic Stem Cells. Cells. 2022; 11(11):1729. https://doi.org/10.3390/cells11111729
Chicago/Turabian StyleSarel-Gallily, Roni, and Nissim Benvenisty. 2022. "Large-Scale Analysis of X Inactivation Variations between Primed and Naïve Human Embryonic Stem Cells" Cells 11, no. 11: 1729. https://doi.org/10.3390/cells11111729