Technologies Enabling Single-Molecule Super-Resolution Imaging of mRNA
Abstract
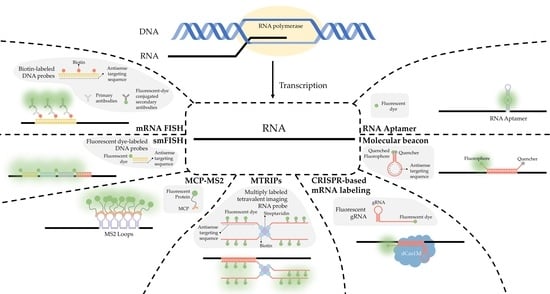
:1. Introduction
1.1. Why Image RNA?
1.2. RNA Localization and Imaging: Seeing Is Believing
1.3. Difficulties Associated with Imaging mRNA
1.4. Single-Molecule Super-Resolution Imaging
1.5. Utilizing Single-Molecule Super-Resolution Imaging for mRNA
2. RNA Imaging Methods
2.1. FISH

2.2. smFISH
2.3. seqFISH

2.4. MERFISH
2.5. Single-Molecule Localization Microscopy (SMLM)
3. RNA Labeling Strategies
3.1. MCP-MS2 Loop System
3.2. Antibody Labeling
3.2.1. RNA:DNA Hybrids and S9.6 Antibody Labeling
3.2.2. New Probes for RNA:DNA Hybrid Labeling
3.3. Multiply-Labeled Tetravalent RNA Imaging Probes (MTRIPS)
3.4. CRISPR-Based Labeling Strategies
3.5. RNA Molecular Beacons
3.6. RNA Aptamer
4. Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Noller, H.F. Ribosomal RNA and translation. Annu. Rev. Biochem. 1991, 60, 191–227. [Google Scholar] [CrossRef] [PubMed]
- Elela, S.A.; Nazar, R.N. Role of the 5.8 S rRNA in ribosome translocation. Nucleic Acids Res. 1997, 25, 1788–1794. [Google Scholar] [CrossRef] [PubMed]
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mehmandoost, N.; Moazzen, F.; Mazraeh, A.; Marmari, V. Molecular mechanisms and biological functions of siRNA. Int. J. Biomed. Sci. IJBS 2017, 13, 48. [Google Scholar] [PubMed]
- Moore, C.B.; Guthrie, E.H.; Huang, M.T.; Taxman, D.J. Short hairpin RNA (shRNA): Design, delivery, and assessment of gene knockdown. Methods Mol. Biol. 2010, 629, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Sheng, P.; Flood, K.A.; Xie, M. Short hairpin RNAs for strand-specific small interfering RNA production. Front. Bioeng. Biotechnol. 2020, 8, 940. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Becker, S.; Feldmann, J.; Wiedemann, S.; Okamura, H.; Schneider, C.; Iwan, K.; Crisp, A.; Rossa, M.; Amatov, T.; Carell, T. Unified prebiotically plausible synthesis of pyrimidine and purine RNA ribonucleotides. Science 2019, 366, 76–82. [Google Scholar] [CrossRef]
- Müller, F.; Escobar, L.; Xu, F.; Węgrzyn, E.; Nainytė, M.; Amatov, T.; Chan, C.Y.; Pichler, A.; Carell, T. A prebiotically plausible scenario of an RNA–peptide world. Nature 2022, 605, 279–284. [Google Scholar] [CrossRef]
- Pearce, B.K.; Pudritz, R.E.; Semenov, D.A.; Henning, T.K. Origin of the RNA world: The fate of nucleobases in warm little ponds. Proc. Natl. Acad. Sci. USA 2017, 114, 11327–11332. [Google Scholar] [CrossRef] [Green Version]
- Lincoln, T.A.; Joyce, G.F. Self-sustained replication of an RNA enzyme. Science 2009, 323, 1229–1232. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, C. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Thomas, S.J.; Moreira Jr, E.D.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Polack, F.P.; Zerbini, C. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine through 6 months. N. Engl. J. Med. 2021, 385, 1761–1773. [Google Scholar] [CrossRef]
- Setten, R.L.; Rossi, J.J.; Han, S.-p. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef]
- Byron, S.A.; Keuren-Jensen, V.; Kendall, R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef]
- Kloc, M.; Zearfoss, N.R.; Etkin, L.D. Mechanisms of subcellular mRNA localization. Cell 2002, 108, 533–544. [Google Scholar] [CrossRef]
- Shestakova, E.A.; Singer, R.H.; Condeelis, J. The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc. Natl. Acad. Sci. USA 2001, 98, 7045–7050. [Google Scholar] [CrossRef]
- Trovisco, V.; Belaya, K.; Nashchekin, D.; Irion, U.; Sirinakis, G.; Butler, R.; Lee, J.J.; Gavis, E.R.; St Johnston, D. bicoid mRNA localises to the Drosophila oocyte anterior by random Dynein-mediated transport and anchoring. eLife 2016, 5, e17537. [Google Scholar] [CrossRef]
- Delanoue, R.; Herpers, B.; Soetaert, J.; Davis, I.; Rabouille, C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev. Cell 2007, 13, 523–538. [Google Scholar] [CrossRef]
- Herpers, B.; Rabouille, C. mRNA localization and ER-based protein sorting mechanisms dictate the use of transitional endoplasmic reticulum-golgi units involved in gurken transport in Drosophila oocytes. Mol. Biol. Cell 2004, 15, 5306–5317. [Google Scholar] [CrossRef] [Green Version]
- Galej, W.P.; Toor, N.; Newman, A.J.; Nagai, K. Molecular mechanism and evolution of nuclear pre-mRNA and group II intron splicing: Insights from cryo-electron microscopy structures. Chem. Rev. 2018, 118, 4156–4176. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.E.; Lin, P.-C.; Plaschka, C.; Nagai, K. Cryo-EM studies of pre-mRNA splicing: From sample preparation to model visualization. Annu. Rev. Biophys. 2018, 47, 175–199. [Google Scholar] [CrossRef] [PubMed]
- Schnell, U.; Dijk, F.; Sjollema, K.A.; Giepmans, B.N. Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods 2012, 9, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Weil, T.T.; Parton, R.M.; Davis, I. Making the message clear: Visualizing mRNA localization. Trends Cell Biol. 2010, 20, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, D.; Su, N.; Bao, B.; Xie, X.; Zuo, F.; Yang, L.; Wang, H.; Jiang, L.; Lin, Q. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat. Biotechnol. 2019, 37, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Weber, G.; Teale, F. Determination of the absolute quantum yield of fluorescent solutions. Trans. Faraday Soc. 1957, 53, 646–655. [Google Scholar] [CrossRef]
- Song, L.; Hennink, E.; Young, I.T.; Tanke, H.J. Photobleaching kinetics of fluorescein in quantitative fluorescence microscopy. Biophys. J. 1995, 68, 2588–2600. [Google Scholar] [CrossRef]
- Liu, Y.; Le, P.; Lim, S.J.; Ma, L.; Sarkar, S.; Han, Z.; Murphy, S.J.; Kosari, F.; Vasmatzis, G.; Cheville, J.C. Enhanced mRNA FISH with compact quantum dots. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Ma, Y.; Mao, G.; Huang, W.; Wu, G.; Yin, W.; Ji, X.; Deng, Z.; Cai, Z.; Zhang, X.-E.; He, Z. Quantum dot nanobeacons for single RNA labeling and imaging. J. Am. Chem. Soc. 2019, 141, 13454–13458. [Google Scholar] [CrossRef]
- Zimyanin, V.L.; Belaya, K.; Pecreaux, J.; Gilchrist, M.J.; Clark, A.; Davis, I.; St Johnston, D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 2008, 134, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Schermelleh, L.; Ferrand, A.; Huser, T.; Eggeling, C.; Sauer, M.; Biehlmaier, O.; Drummen, G.P. Super-resolution microscopy demystified. Nat. Cell Biol. 2019, 21, 72–84. [Google Scholar] [CrossRef]
- Huang, B.; Bates, M.; Zhuang, X. Super resolution fluorescence microscopy. Annu. Rev. Biochem. 2009, 78, 993. [Google Scholar] [CrossRef]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3, 793–796. [Google Scholar] [CrossRef]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef]
- Hell, S.W.; Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994, 19, 780–782. [Google Scholar] [CrossRef]
- Gustafsson, M.G. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 2000, 198, 82–87. [Google Scholar] [CrossRef]
- Sigal, Y.M.; Zhou, R.; Zhuang, X. Visualizing and discovering cellular structures with super-resolution microscopy. Science 2018, 361, 880–887. [Google Scholar] [CrossRef]
- Somasekharan, S.P.; Saxena, N.; Zhang, F.; Beraldi, E.; Huang, J.N.; Gentle, C.; Fazli, L.; Thi, M.; Sorensen, P.H.; Gleave, M. Regulation of AR mRNA translation in response to acute AR pathway inhibition. Nucleic Acids Res. 2022, 50, 1069–1091. [Google Scholar] [CrossRef]
- Harbauer, A.B.; Hees, J.T.; Wanderoy, S.; Segura, I.; Gibbs, W.; Cheng, Y.; Ordonez, M.; Cai, Z.; Cartoni, R.; Ashrafi, G. Neuronal mitochondria transport Pink1 mRNA via Synaptojanin 2 to support local mitophagy. Neuron 2022, 110, 1516–1531.e1519. [Google Scholar] [CrossRef]
- Zhang, W.I.; Röhse, H.; Rizzoli, S.O.; Opazo, F. Fluorescent in situ hybridization of synaptic proteins imaged with super-resolution STED microscopy. Microsc. Res. Tech. 2014, 77, 517–527. [Google Scholar] [CrossRef]
- Li, Y.; Aksenova, V.; Tingey, M.; Yu, J.; Ma, P.; Arnaoutov, A.; Chen, S.; Dasso, M.; Yang, W. Distinct roles of nuclear basket proteins in directing the passage of mRNA through the nuclear pore. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Paramasivam, P.; Franke, C.; Stöter, M.; Höijer, A.; Bartesaghi, S.; Sabirsh, A.; Lindfors, L.; Arteta, M.Y.; Dahlén, A.; Bak, A. Endosomal escape of delivered mRNA from endosomal recycling tubules visualized at the nanoscale. J. Cell Biol. 2021, 221, e202110137. [Google Scholar] [CrossRef]
- Lelek, M.; Gyparaki, M.T.; Beliu, G.; Schueder, F.; Griffié, J.; Manley, S.; Jungmann, R.; Sauer, M.; Lakadamyali, M.; Zimmer, C. Single-molecule localization microscopy. Nat. Rev. Methods Primers 2021, 1, 1–27. [Google Scholar] [CrossRef]
- Vangindertael, J.; Camacho, R.; Sempels, W.; Mizuno, H.; Dedecker, P.; Janssen, K. An introduction to optical super-resolution microscopy for the adventurous biologist. Methods Appl. Fluoresc. 2018, 6, 022003. [Google Scholar] [CrossRef]
- Sung, M.H.; McNally, J.G. Live cell imaging and systems biology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 167–182. [Google Scholar] [CrossRef]
- von Diezmann, L.; Shechtman, Y.; Moerner, W. Three-dimensional localization of single molecules for super-resolution imaging and single-particle tracking. Chem. Rev. 2017, 117, 7244–7275. [Google Scholar] [CrossRef]
- Eliscovich, C.; Shenoy, S.M.; Singer, R.H. Imaging mRNA and protein interactions within neurons. Proc. Natl. Acad. Sci. USA 2017, 114, E1875–E1884. [Google Scholar] [CrossRef]
- Ma, J.; Liu, Z.; Michelotti, N.; Pitchiaya, S.; Veerapaneni, R.; Androsavich, J.R.; Walter, N.G.; Yang, W. High-resolution three-dimensional mapping of mRNA export through the nuclear pore. Nat. Commun. 2013, 4, 1–9. [Google Scholar] [CrossRef]
- Li, Y.; Tingey, M.; Ruba, A.; Yang, W. High-speed super-resolution imaging of rotationally symmetric structures using SPEED microscopy and 2D-to-3D transformation. Nat. Protoc. 2021, 16, 532–560. [Google Scholar] [CrossRef]
- Abrahamsson, S.; Chen, J.; Hajj, B.; Stallinga, S.; Katsov, A.Y.; Wisniewski, J.; Mizuguchi, G.; Soule, P.; Mueller, F.; Darzacq, C.D. Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat. Methods 2013, 10, 60–63. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.S.; Preibisch, S.; Joseph, A.; Abrahamsson, S.; Rieger, B.; Myers, E.; Singer, R.H.; Grunwald, D. Nuclear accessibility of β-actin mRNA is measured by 3D single-molecule real-time tracking. J. Cell Biol. 2015, 209, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Bauman, J.; Wiegant, J.; Borst, P.; Van Duijn, P. A new method for fluorescence microscopical localization of specific DNA sequences by in situ hybridization of fluorochrome-labelled RNA. Exp. Cell Res. 1980, 128, 485–490. [Google Scholar] [CrossRef]
- Rudkin, G.T.; Stollar, B. High resolution detection of DNA–RNA hybrids in situ by indirect immunofluorescence. Nature 1977, 265, 472–473. [Google Scholar] [CrossRef] [PubMed]
- Singer, R.H.; Ward, D.C. Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc. Natl. Acad. Sci. USA 1982, 79, 7331–7335. [Google Scholar] [CrossRef]
- Young, A.P.; Jackson, D.J.; Wyeth, R.C. A technical review and guide to RNA fluorescence in situ hybridization. PeerJ 2020, 8, e8806. [Google Scholar] [CrossRef]
- Kearney, L. Multiplex-FISH (M-FISH): Technique, developments and applications. Cytogenet. Genome Res. 2006, 114, 189–198. [Google Scholar] [CrossRef]
- Speicher, M.R.; Ballard, S.G.; Ward, D.C. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat. Genet. 1996, 12, 368–375. [Google Scholar] [CrossRef]
- Raj, A.; Van Den Bogaard, P.; Rifkin, S.A.; Van Oudenaarden, A.; Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 2008, 5, 877–879. [Google Scholar] [CrossRef]
- Femino, A.M.; Fay, F.S.; Fogarty, K.; Singer, R.H. Visualization of single RNA transcripts in situ. Science 1998, 280, 585–590. [Google Scholar] [CrossRef]
- Randolph, J.B.; Waggoner, A.S. Stability, specificity and fluorescence brightness of multiply-labeled fluorescent DNA probes. Nucleic Acids Res. 1997, 25, 2923–2929. [Google Scholar] [CrossRef]
- Tingey, M.; Mudumbi, K.C.; Schirmer, E.C.; Yang, W. Casting a Wider Net: Differentiating between Inner Nuclear Envelope and Outer Nuclear Envelope Transmembrane Proteins. Int. J. Mol. Sci. 2019, 20, 5248. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 2009, 37, e45. [Google Scholar] [CrossRef]
- Conn, V.; Conn, S.J. SplintQuant: A method for accurately quantifying circular RNA transcript abundance without reverse transcription bias. RNA 2019, 25, 1202–1210. [Google Scholar] [CrossRef]
- Ma, J.; Yang, W. Three-dimensional distribution of transient interactions in the nuclear pore complex obtained from single-molecule snapshots. Proc. Natl. Acad. Sci. USA 2010, 107, 7305–7310. [Google Scholar] [CrossRef]
- Ruba, A.; Kelich, J.; Ma, J.; Yang, W. Reply to ‘Deconstructing transport-distribution reconstruction in the nuclear-pore complex’. Nat. Struct. Mol. Biol. 2018, 25, 1062–1064. [Google Scholar] [CrossRef]
- Schnell, S.J.; Tingey, M.; Yang, W. Speed Microscopy: High-Speed Single Molecule Tracking and Mapping of Nucleocytoplasmic Transport. In The Nuclear Pore Complex: Methods and Protocols; Goldberg, M.W., Ed.; Springer: New York, NY, USA, 2022; pp. 353–371. [Google Scholar] [CrossRef]
- Schnell, S.J.; Ma, J.; Yang, W. Three-dimensional mapping of mRNA export through the nuclear pore complex. Genes 2014, 5, 1032–1049. [Google Scholar] [CrossRef]
- Bates, M.; Dempsey, G.T.; Chen, K.H.; Zhuang, X. Multicolor super-resolution fluorescence imaging via multi-parameter fluorophore detection. ChemPhysChem 2012, 13, 99–107. [Google Scholar] [CrossRef]
- Westphal, V.; Hell, S.W. Nanoscale resolution in the focal plane of an optical microscope. Phys. Rev. Lett. 2005, 94, 143903. [Google Scholar] [CrossRef]
- Miklyaev, Y.V.; Asselborn, S.; Zaytsev, K.; Darscht, M.Y. Superresolution microscopy in far-field by near-field optical random mapping nanoscopy. Appl. Phys. Lett. 2014, 105, 113103. [Google Scholar] [CrossRef]
- Lubeck, E.; Cai, L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat. Methods 2012, 9, 743–748. [Google Scholar] [CrossRef]
- Shah, S.; Lubeck, E.; Zhou, W.; Cai, L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 2016, 92, 342–357. [Google Scholar] [CrossRef]
- Lubeck, E.; Coskun, A.F.; Zhiyentayev, T.; Ahmad, M.; Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 2014, 11, 360–361. [Google Scholar] [CrossRef]
- Eng, C.-H.L.; Shah, S.; Thomassie, J.; Cai, L. Profiling the transcriptome with RNA SPOTs. Nat. Methods 2017, 14, 1153–1155. [Google Scholar] [CrossRef]
- Chen, K.H.; Boettiger, A.N.; Moffitt, J.R.; Wang, S.; Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348, aaa6090. [Google Scholar] [CrossRef]
- D’Alessio, L.; Liu, L.; Duffy, K.; Eldar, Y.C.; Médard, M.; Babadi, M. A coding theory perspective on multiplexed molecular profiling of biological tissues. In Proceedings of the 2020 International Symposium on Information Theory and Its Applications (ISITA), Kapolei, HI, USA, 24–27 October 2020; pp. 309–313. [Google Scholar]
- Beliveau, B.J.; Joyce, E.F.; Apostolopoulos, N.; Yilmaz, F.; Fonseka, C.Y.; McCole, R.B.; Chang, Y.; Li, J.B.; Senaratne, T.N.; Williams, B.R. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. USA 2012, 109, 21301–21306. [Google Scholar] [CrossRef]
- Moffitt, J.R.; Zhuang, X. RNA Imaging with Multiplexed Error-Robust Fluorescence In Situ Hybridization (MERFISH). Methods Enzymol 2016, 572, 1–49. [Google Scholar] [CrossRef]
- Heinrich, S.; Derrer, C.P.; Lari, A.; Weis, K.; Montpetit, B. Temporal and spatial regulation of mRNA export: Single particle RNA-imaging provides new tools and insights. Bioessays 2017, 39. [Google Scholar] [CrossRef]
- Li, Y.; Junod, S.L.; Ruba, A.; Kelich, J.M.; Yang, W. Nuclear export of mRNA molecules studied by SPEED microscopy. Methods 2019, 153, 46–62. [Google Scholar] [CrossRef]
- Huang, B.; Wang, W.; Bates, M.; Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 2008, 319, 810–813. [Google Scholar] [CrossRef] [Green Version]
- Baddeley, D.; Cannell, M.B.; Soeller, C. Three-dimensional sub-100 nm super-resolution imaging of biological samples using a phase ramp in the objective pupil. Nano Res. 2011, 4, 589–598. [Google Scholar] [CrossRef]
- Pavani, S.R.P.; Thompson, M.A.; Biteen, J.S.; Lord, S.J.; Liu, N.; Twieg, R.J.; Piestun, R.; Moerner, W.E. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl. Acad. Sci. USA 2009, 106, 2995–2999. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Vaughan, J.C.; Zhuang, X. Isotropic three-dimensional super-resolution imaging with a self-bending point spread function. Nat. Photonics 2014, 8, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Lew, M.D.; Lee, S.F.; Badieirostami, M.; Moerner, W. Corkscrew point spread function for far-field three-dimensional nanoscale localization of pointlike objects. Opt. Lett. 2011, 36, 202–204. [Google Scholar] [CrossRef] [PubMed]
- Shechtman, Y.; Sahl, S.J.; Backer, A.S.; Moerner, W.E. Optimal point spread function design for 3D imaging. Phys. Rev. Lett. 2014, 113, 133902. [Google Scholar] [CrossRef]
- Ruba, A.; Luo, W.; Kelich, J.; Tingey, M.; Yang, W. 3D Tracking-Free Approach for Obtaining 3D Super-Resolution Information in Rotationally Symmetric Biostructures. J. Phys. Chem. B 2019, 123, 5107–5120. [Google Scholar] [CrossRef]
- Tingey, M.; Li, Y.; Yu, W.; Young, A.; Yang, W. Spelling out the roles of individual nucleoporins in nuclear export of mRNA. Nucleus 2022, 13, 170–193. [Google Scholar] [CrossRef]
- Kelich, J.M.; Yang, W. High-Resolution Imaging Reveals New Features of Nuclear Export of mRNA through the Nuclear Pore Complexes. Int. J. Mol. Sci. 2014, 15, 14492–14504. [Google Scholar] [CrossRef]
- Bertrand, E.; Chartrand, P.; Schaefer, M.; Shenoy, S.M.; Singer, R.H.; Long, R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 1998, 2, 437–445. [Google Scholar] [CrossRef]
- Grünwald, D.; Singer, R.H. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 2010, 467, 604–607. [Google Scholar] [CrossRef] [Green Version]
- Larson, D.R.; Zenklusen, D.; Wu, B.; Chao, J.A.; Singer, R.H. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 2011, 332, 475–478. [Google Scholar] [CrossRef]
- Wu, B.; Eliscovich, C.; Yoon, Y.J.; Singer, R.H. Translation dynamics of single mRNAs in live cells and neurons. Science 2016, 352, 1430–1435. [Google Scholar] [CrossRef]
- Halstead, J.M.; Lionnet, T.; Wilbertz, J.H.; Wippich, F.; Ephrussi, A.; Singer, R.H.; Chao, J.A. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science 2015, 347, 1367–1671. [Google Scholar] [CrossRef]
- Morisaki, T.; Lyon, K.; DeLuca, K.F.; DeLuca, J.G.; English, B.P.; Zhang, Z.; Lavis, L.D.; Grimm, J.B.; Viswanathan, S.; Looger, L.L. Real-time quantification of single RNA translation dynamics in living cells. Science 2016, 352, 1425–1429. [Google Scholar] [CrossRef]
- Lim, B.; Levine, M.; Yamazaki, Y. Transcriptional pre-patterning of Drosophila gastrulation. Curr. Biol. 2017, 27, 286–290. [Google Scholar] [CrossRef]
- Tantale, K.; Mueller, F.; Kozulic-Pirher, A.; Lesne, A.; Victor, J.-M.; Robert, M.-C.; Capozi, S.; Chouaib, R.; Bäcker, V.; Mateos-Langerak, J. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Paranchych, W. Attachment, ejection and penetration stages of the RNA phage infectious process. RNA Phages 1975, 85–111. [Google Scholar]
- Fiers, W.; Contreras, R.; Duerinck, F.; Haegeman, G.; Iserentant, D.; Merregaert, J.; Min Jou, W.; Molemans, F.; Raeymaekers, A.; Van den Berghe, A. Complete nucleotide sequence of bacteriophage MS2 RNA: Primary and secondary structure of the replicase gene. Nature 1976, 260, 500–507. [Google Scholar] [CrossRef]
- Zinder, N.D. Portraits of Viruses: RNA Phage. Intervirology 1980, 13, 257–270. [Google Scholar] [CrossRef]
- Gorzelnik, K.V.; Zhang, J. Cryo-EM reveals infection steps of single-stranded RNA bacteriophages. Prog. Biophys Mol. Biol. 2021, 160, 79–86. [Google Scholar] [CrossRef]
- Rolfsson, Ó.; Middleton, S.; Manfield, I.W.; White, S.J.; Fan, B.; Vaughan, R.; Ranson, N.A.; Dykeman, E.; Twarock, R.; Ford, J. Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J. Mol. Biol. 2016, 428, 431–448. [Google Scholar] [CrossRef] [Green Version]
- Mor, A.; Suliman, S.; Ben-Yishay, R.; Yunger, S.; Brody, Y.; Shav-Tal, Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 2010, 12, 543–552. [Google Scholar] [CrossRef]
- Garcia, J.F.; Parker, R. MS2 coat proteins bound to yeast mRNAs block 5′ to 3′ degradation and trap mRNA decay products: Implications for the localization of mRNAs by MS2-MCP system. RNA 2015, 21, 1393–1395. [Google Scholar] [CrossRef]
- Garcia, J.F.; Parker, R. Ubiquitous accumulation of 3′ mRNA decay fragments in Saccharomyces cerevisiae mRNAs with chromosomally integrated MS2 arrays. RNA 2016, 22, 657–659. [Google Scholar] [CrossRef]
- Haimovich, G.; Zabezhinsky, D.; Haas, B.; Slobodin, B.; Purushothaman, P.; Fan, L.; Levin, J.Z.; Nusbaum, C.; Gerst, J.E. Use of the MS2 aptamer and coat protein for RNA localization in yeast: A response to “MS2 coat proteins bound to yeast mRNAs block 5′ to 3′ degradation and trap mRNA decay products: Implications for the localization of mRNAs by MS2-MCP system”. RNA 2016, 22, 660–666. [Google Scholar] [CrossRef]
- Heinrich, S.; Sidler, C.L.; Azzalin, C.M.; Weis, K. Stem–loop RNA labeling can affect nuclear and cytoplasmic mRNA processing. RNA 2017, 23, 134–141. [Google Scholar] [CrossRef]
- Vera, M.; Tutucci, E.; Singer, R.H. Imaging Single mRNA Molecules in Mammalian Cells Using an Optimized MS2-MCP System. In Imaging Gene Expression: Methods and Protocols; Shav-Tal, Y., Ed.; Springer New York: New York, NY, USA, 2019; pp. 3–20. [Google Scholar] [CrossRef]
- Tutucci, E.; Vera, M.; Biswas, J.; Garcia, J.; Parker, R.; Singer, R.H. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat. Methods 2018, 15, 81–89. [Google Scholar] [CrossRef]
- Chedin, F.; Benham, C.J. Emerging roles for R-loop structures in the management of topological stress. J. Biol. Chem. 2020, 295, 4684–4695. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, C.; Pizzul, P.; Longhese, M.P.; Bonetti, D. Sensing R-Loop-Associated DNA Damage to Safeguard Genome Stability. Front. Cell Dev. Biol. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Boguslawski, S.J.; Smith, D.E.; Michalak, M.A.; Mickelson, K.E.; Yehle, C.O.; Patterson, W.L.; Carrico, R.J. Characterization of monoclonal antibody to DNA · RNA and its application to immunodetection of hybrids. J. Immunol. Methods 1986, 89, 123–130. [Google Scholar] [CrossRef]
- El Hage, A.; Webb, S.; Kerr, A.; Tollervey, D. Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria. PLOS Genet. 2014, 10, e1004716. [Google Scholar] [CrossRef] [PubMed]
- Bayona-Feliu, A.; Casas-Lamesa, A.; Reina, O.; Bernués, J.; Azorín, F. Linker histone H1 prevents R-loop accumulation and genome instability in heterochromatin. Nat. Commun. 2017, 8, 283. [Google Scholar] [CrossRef]
- Smolka, J.A.; Sanz, L.A.; Hartono, S.R.; Chédin, F. Recognition of RNA by the S9.6 antibody creates pervasive artifacts when imaging RNA:DNA hybrids. J. Cell Biol. 2021, 220. [Google Scholar] [CrossRef]
- Hartono, S.R.; Malapert, A.; Legros, P.; Bernard, P.; Chédin, F.; Vanoosthuyse, V. The Affinity of the S9.6 Antibody for Double-Stranded RNAs Impacts the Accurate Mapping of R-Loops in Fission Yeast. J. Mol. Biol. 2018, 430, 272–284. [Google Scholar] [CrossRef]
- Phillips, D.D.; Garboczi, D.N.; Singh, K.; Hu, Z.; Leppla, S.H.; Leysath, C.E. The sub-nanomolar binding of DNA–RNA hybrids by the single-chain Fv fragment of antibody S9.6. J. Mol. Recognit. 2013, 26, 376–381. [Google Scholar] [CrossRef]
- Bates, A.; Power, C.A. David vs. Goliath: The Structure, Function, and Clinical Prospects of Antibody Fragments. Antibodies 2019, 8, 28. [Google Scholar] [CrossRef]
- Kinman, A.W.L.; Pompano, R.R. Optimization of Enzymatic Antibody Fragmentation for Yield, Efficiency, and Binding Affinity. Bioconjugate Chem. 2019, 30, 800–807. [Google Scholar] [CrossRef]
- Nowotny, M.; Cerritelli, S.M.; Ghirlando, R.; Gaidamakov, S.A.; Crouch, R.J.; Yang, W. Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD. EMBO J. 2008, 27, 1172–1181. [Google Scholar] [CrossRef]
- Nowotny, M.; Gaidamakov, S.A.; Ghirlando, R.; Cerritelli, S.M.; Crouch, R.J.; Yang, W. Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription. Mol. Cell 2007, 28, 264–276. [Google Scholar] [CrossRef]
- Bhatia, V.; Barroso, S.I.; García-Rubio, M.L.; Tumini, E.; Herrera-Moyano, E.; Aguilera, A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 2014, 511, 362–365. [Google Scholar] [CrossRef]
- Santangelo, P.J.; Lifland, A.W.; Curt, P.; Sasaki, Y.; Bassell, G.J.; Lindquist, M.E.; Crowe, J.E. Single molecule–sensitive probes for imaging RNA in live cells. Nat. Methods 2009, 6, 347–349. [Google Scholar] [CrossRef]
- Jung, J.; Lifland, A.W.; Zurla, C.; Alonas, E.J.; Santangelo, P.J. Quantifying RNA–protein interactions in situ using modified-MTRIPs and proximity ligation. Nucleic Acids Res. 2012, 41, e12. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. eLife 2013, 2, e00471. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.L.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.B.; Jiang, W.Y.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Spille, J.H.; Hecht, M.; Grube, V.; Cho, W.K.; Lee, C.; Cissé, I.I. A CRISPR/Cas9 platform for MS2-labelling of single mRNA in live stem cells. Methods 2019, 153, 35–45. [Google Scholar] [CrossRef]
- Wang, M.; Chen, K.; Wu, Q.; Peng, R.; Zhang, R.; Li, J. RCasFISH: CRISPR/dCas9-Mediated in Situ Imaging of mRNA Transcripts in Fixed Cells and Tissues. Anal. Chem. 2020, 92, 2468–2475. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Liu, J.; Chang, J.; Jiang, Y.; Meng, X.; Sun, T.; Mao, L.; Xu, Q.; Wang, M. Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles. Adv. Mater. 2019, 31, e1902575. [Google Scholar] [CrossRef]
- Hashimoto, M.; Takemoto, T. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci. Rep. 2015, 5, 11315. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Perez-Leal, O.; Nixon-Abell, J.; Barrero, C.A.; Gordon, J.C.; Oesterling, J.; Rico, M.C. Multiplex Gene Tagging with CRISPR-Cas9 for Live-Cell Microscopy and Application to Study the Role of SARS-CoV-2 Proteins in Autophagy, Mitochondrial Dynamics, and Cell Growth. CRISPR J. 2021, 4, 854–871. [Google Scholar] [CrossRef] [PubMed]
- Nelles, D.A.; Fang, M.Y.; O’Connell, M.R.; Xu, J.L.; Markmiller, S.J.; Doudna, J.A.; Yeo, G.W. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell 2016, 165, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Tanenbaum, M.E.; Gilbert, L.A.; Qi, L.S.; Weissman, J.S.; Vale, R.D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014, 159, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.H.; Chen, D.Y.; Ye, L.P.; Sheng, G.; Gong, J.J.; Chen, B.H.; Lu, Y.M.; Han, F. CRISPR-Sunspot: Imaging of endogenous low-abundance RNA at the single-molecule level in live cells. Theranostics 2020, 10, 10993–11012. [Google Scholar] [CrossRef] [PubMed]
- Nelles, D.A.; Fang, M.Y.; Aigner, S.; Yeo, G.W. Applications of Cas9 as an RNA-programmed RNA-binding protein. Bioessays 2015, 37, 732–739. [Google Scholar] [CrossRef] [PubMed]
- Balderston, S.; Clouse, G.; Ripoll, J.J.; Pratt, G.K.; Gasiunas, G.; Bock, J.O.; Bennett, E.P.; Aran, K. Diversification of the CRISPR Toolbox: Applications of CRISPR-Cas Systems Beyond Genome Editing. CRISPR J. 2021, 4, 400–415. [Google Scholar] [CrossRef] [PubMed]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Shmakov, S.; Abudayyeh, O.O.; Makarova, K.S.; Wolf, Y.I.; Gootenberg, J.S.; Semenova, E.; Minakhin, L.; Joung, J.; Konermann, S.; Severinov, K.; et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 2015, 60, 385–397. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Zhou, Y.; Xiao, Q.; He, B.; Geng, G.; Wang, Z.; Cao, B.; Dong, X.; Bai, W.; Wang, Y.; et al. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat. Methods 2021, 18, 499–506. [Google Scholar] [CrossRef]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630.e617. [Google Scholar] [CrossRef]
- Yang, L.Z.; Wang, Y.; Li, S.Q.; Yao, R.W.; Luan, P.F.; Wu, H.; Carmichael, G.G.; Chen, L.L. Dynamic Imaging of RNA in Living Cells by CRISPR-Cas13 Systems. Mol. Cell 2019, 76, 981–997.e987. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, L.Z.; Chen, L.L. Protocol for Dynamic Imaging of RNA in Living Cells by CRISPR-Cas13 System. STAR Protoc. 2020, 1, 100037. [Google Scholar] [CrossRef]
- Davis, B.J.; O’Connell, M.R. Put on Your Para-spectacles: The Development of Optimized CRISPR-Cas13-Based Approaches to Image RNA Dynamics in Real Time. Mol. Cell 2020, 77, 207–209. [Google Scholar] [CrossRef]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef]
- George, L.; Indig, F.E.; Abdelmohsen, K.; Gorospe, M. Intracellular RNA-tracking methods. Open Biol. 2018, 8. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, X.; Ye, D. Molecular beacons: An optimal multifunctional biological probe. Biochem. Biophys. Res. Commun. 2008, 373, 457–461. [Google Scholar] [CrossRef]
- Monroy-Contreras, R.; Vaca, L. Molecular Beacons: Powerful Tools for Imaging RNA in Living Cells. J. Nucleic Acids 2011, 2011, 741723. [Google Scholar] [CrossRef]
- Chen, A.K.; Rhee, W.J.; Bao, G.; Tsourkas, A. Delivery of molecular beacons for live-cell imaging and analysis of RNA. Methods Mol Biol 2011, 714, 159–174. [Google Scholar] [CrossRef]
- Sherrill-Mix, S.; Hwang, Y.; Roche, A.M.; Glascock, A.; Weiss, S.R.; Li, Y.; Haddad, L.; Deraska, P.; Monahan, C.; Kromer, A.; et al. Detection of SARS-CoV-2 RNA using RT-LAMP and molecular beacons. Genome Biol. 2021, 22, 169. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Trachman, R.J.; Ferré-D’Amaré, A.R. Tracking RNA with light: Selection, structure, and design of fluorescence turn-on RNA aptamers. Q. Rev. Biophys. 2019, 52, e8. [Google Scholar] [CrossRef]
- Sato, H.; Das, S.; Singer, R.H.; Vera, M. Imaging of DNA and RNA in Living Eukaryotic Cells to Reveal Spatiotemporal Dynamics of Gene Expression. Annu. Rev. Biochem. 2020, 89, 159–187. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.; Yin, F.; Liu, Y.; Xu, S.; Jiang, W.; Wang, R.; Xue, Q. In situ generation spatial confinement fluorescence RNA for sensitive and stable imaging of telomerase RNA in cells. Sens. Actuators B Chem. 2021, 345, 130414. [Google Scholar] [CrossRef]
- Laprade, H.; Lalonde, M.; Guérit, D.; Chartrand, P. Live-cell imaging of budding yeast telomerase RNA and TERRA. Methods 2017, 114, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Tingey, M. Yeast Npl3 Regulates TERRA Production and Prevents Faster Senescence in Telomerase-Null Cells. Master’s Thesis, Saint Joseph’s University, Francis A. Drexel Library, Philadelphia, PA, USA, 2017. [Google Scholar]
- Zappulla, D.C. Yeast Telomerase RNA Flexibly Scaffolds Protein Subunits: Results and Repercussions. Molecules 2020, 25, 2750. [Google Scholar] [CrossRef]
- Bettin, N.; Oss Pegorar, C.; Cusanelli, E. The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability. Cells 2019, 8, 246. [Google Scholar] [CrossRef]
- Dolgosheina, E.V.; Jeng, S.C.; Panchapakesan, S.S.; Cojocaru, R.; Chen, P.S.; Wilson, P.D.; Hawkins, N.; Wiggins, P.A.; Unrau, P.J. RNA mango aptamer-fluorophore: A bright, high-affinity complex for RNA labeling and tracking. ACS Chem. Biol. 2014, 9, 2412–2420. [Google Scholar] [CrossRef]
- Wirth, R.; Gao, P.; Nienhaus, G.U.; Sunbul, M.; Jaschke, A. SiRA: A Silicon Rhodamine-Binding Aptamer for Live-Cell Super-Resolution RNA Imaging. J. Am. Chem. Soc. 2019, 141, 7562–7571. [Google Scholar] [CrossRef]
- Park, S.Y.; Moon, H.C.; Park, H.Y. Live-cell imaging of single mRNA dynamics using split superfolder green fluorescent proteins with minimal background. RNA 2020, 26, 101–109. [Google Scholar] [CrossRef]
- Padrón, A.; Iwasaki, S.; Ingolia, N.T. Proximity RNA Labeling by APEX-Seq Reveals the Organization of Translation Initiation Complexes and Repressive RNA Granules. Mol. Cell 2019, 75, 875–887.e875. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Y.; Wang, Y.; Zhang, L.; Brinkman, E.K.; Adam, S.A.; Goldman, R.; Van Steensel, B.; Ma, J.; Belmont, A.S. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J. Cell Biol. 2018, 217, 4025–4048. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Bi, X.; Pan, Y.; Gao, B.; Wu, J.; Yin, Y.; Liu, Z.; Peng, M.; Zhang, W.; Jiang, X.; et al. Phase separation of RNA-binding protein promotes polymerase binding and transcription. Nat. Chem. Biol. 2022, 18, 70–80. [Google Scholar] [CrossRef]




| Methodology | Single-Molecule Precision | Live-Cell Imaging | Signal to Noise Ratio | Challenging Technique | Quantitative Measurement | Other Advantages | Other Limitations |
|---|---|---|---|---|---|---|---|
| FISH | No | No | Poor Potential off-target interactions and high background noise | No Established technique that is well characterized and optimized | No Provides only an ensemble average of detected fluorescence | Able to multiplex and label up to 24 different targets with different fluorophores using M-FISH. | Severely limited by photostability of the fluorophores used |
| smFISH | Yes When combined with an appropriate technique | No | Good Multiple probes binding a single target provides higher signal-to-noise ratio | Yes Technically challenging | Yes Combing this technique with qRT-PCR provides quantification of RNA. | Provides spatial localization at the cellular level. | Cannot localize within subcellular compartments. |
| seqFISH | Yes | No | N/A Requires optimization to visualize SPOTS. | Yes Time consuming | Yes Capable of relative quantification of RNA. | Can differentiate a theoretically unlimited number of mRNA species | Repeated rounds of imaging may cause photodamage. |
| merFISH | Yes | No | N/A Requires optimization to localize probes. | Yes Requires a degree of coding knowledge as well as optimization. | Yes Capable of relative quantification | Capable of imaging the full transcriptome using a 16-bit coding approach. | Multiple rounds of photobleaching may cause photodamage. |
| RNA Aptamer | Yes When combined with an appropriate technique | Yes | Good Fluorescent Dyes only emit fluorescence when bound to Aptamer. | No Widely used and well characterized | Yes Provides relative quantification of RNA. | A wide variety of aptamer probes are available that cater to specific requirements. | Different aptamers have different viability. Many suffer from poor folding, poor quantum yield, and rapid photobleaching. |
| MCP-MS2 loop System | Yes When combined with an appropriate technique | Yes | Poor High background Noise | No Widely used and well characterized | No Provides only an ensemble average of detected fluorescence | Widespread use allows for many readily available plasmids utilizing this system. | Can interfere with mammalian cellular processes. |
| MTRIPS | Yes When combined with an appropriate technique | Yes | Good Multiple probes binding a single target provides higher signal-to-noise ratio | Yes Requires significant post-imaging processing. | Yes Provides relative quantification of RNA and associated proteins. | Capable of use proximity ligation assays. | Requires membrane permeabilization to introduce the probe to the cell. |
| CRISPR based labeling strategies | Yes When combined with an appropriate technique | Yes | Poor High background Noise | Yes Technically challenging. | No Provides only an ensemble average of detected fluorescence | Highly robust and versatile system. Can either engineer a tag into RNA or utilize fluorescent gRNA to label RNA. | Can potentially interfere with mammalian cellular processes. |
| Molecular Beacons | Yes When combined with an appropriate technique | Yes | Good Low background noise | No Straightforward and streamlined technique | Yes Provides relative quantification of RNA. | Highly specific to the target sequence. | May interfere with cellular machinery. Requires membrane permeabilization to introduce into the cell. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tingey, M.; Schnell, S.J.; Yu, W.; Saredy, J.; Junod, S.; Patel, D.; Alkurdi, A.A.; Yang, W. Technologies Enabling Single-Molecule Super-Resolution Imaging of mRNA. Cells 2022, 11, 3079. https://doi.org/10.3390/cells11193079
Tingey M, Schnell SJ, Yu W, Saredy J, Junod S, Patel D, Alkurdi AA, Yang W. Technologies Enabling Single-Molecule Super-Resolution Imaging of mRNA. Cells. 2022; 11(19):3079. https://doi.org/10.3390/cells11193079
Chicago/Turabian StyleTingey, Mark, Steven J. Schnell, Wenlan Yu, Jason Saredy, Samuel Junod, Dhrumil Patel, Abdullah A. Alkurdi, and Weidong Yang. 2022. "Technologies Enabling Single-Molecule Super-Resolution Imaging of mRNA" Cells 11, no. 19: 3079. https://doi.org/10.3390/cells11193079