Towards Accurate Skin Lesion Classification across All Skin Categories Using a PCNN Fusion-Based Data Augmentation Approach
Abstract
:1. Introduction
2. Literature Review
2.1. Data Augmentation Methods
2.2. Images Fusion Methods
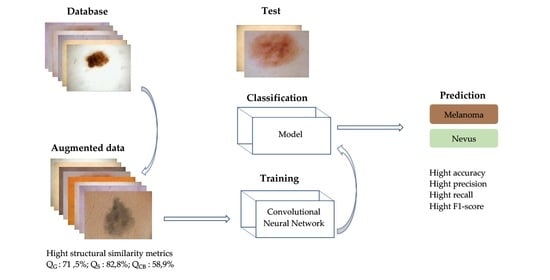
3. Proposed Method
3.1. Non-Subsampled Shearlet Transform (NSST)
3.2. Pulse Coupled Neural Network (PCNN)
3.3. Detailed Algorithm
| Algorithm 1. Proposed fusion algorithm |
| Input: Dermoscopic images in dataset A and unaffected darker tones images in dataset B Output: Fused image Step 1: Image decomposition with NSST |
| Randomly select source images and decompose each source image into five levels with NSST, to obtain (,) and (,). and are the low-frequency coefficients of A and B. and represent the l-th high-frequency sub-band coefficients in the kth decomposition layer of A and B. |
| Step 2: Fusion strategy |
| The low-frequency sub-band contains texture structure and background of source images. The fused low-frequency coefficients are obtained as follows: |
| High-frequency sub-bands contain information about details in images. The fused high-frequency sub-bands are obtained by computing the following operations on each pixel of each high-frequency sub-bands. |
| 1. Normalize between 0 and 1 high-frequency coefficients; |
| 2. Initialize , , and to 0 and to 1 to accelerate neurons activation; |
| 3. Stimulate the PCNN respectively with the normalized coefficients; |
| 4. Compute , , , and until the neuron is activated (equal to 1); |
| 5. Fuse high-frequency sub-bands coefficients as follows: |
| Step 3: Image reconstruction with inverse NSST |
4. Experimental Results and Discussion
4.1. Dataset
4.2. Experimental Setup
4.3. Results and Discussion
4.3.1. Visual and Qualitative Evaluation
4.3.2. Impact of Data Augmentation in Skin Lesion Classification on All Existing Tones
5. Conclusions and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Li, X.; Wang, L.; He, J.; Li, D.; Zhou, J. How Does the Data set Affect CNN-based Image Classification Performance? In Proceedings of the 2018 5th International Conference on Systems and Informatics (ICSAI), Nanjing, China, 10–12 November 2018; pp. 361–366. [Google Scholar] [CrossRef]
- Cunniff, C.; Byrne, J.L.; Hudgins, L.M.; Moeschler, J.B.; Olney, A.H.; Pauli, R.M.; Seaver, L.H.; Stevens, C.A.; Figone, C. Informed consent for medical photographs. Genet. Med. 2000, 2, 353–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simard, P.Y.; Steinkraus, D.; Platt, J.C. Best practices for convolutional neural networks applied to visual document analysis. In Proceedings of the 7th International Conference on Document Analysis and Recognition, Edinburgh, UK, 3–6 August 2003; pp. 958–963. [Google Scholar] [CrossRef]
- Li, S.; Kang, X.; Fang, L.; Hu, J.; Yin, H. Pixel-level image fusion: A survey of the state of the art. Inf. Fusion 2017, 33, 100–112. [Google Scholar] [CrossRef]
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the International Interdisciplinary PhD Workshop (IIPhDW), Swinoujscie, Poland, 9–12 May 2018; pp. 117–122. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, B.; Wang, L.; Zu, C.; Lalush, D.S.; Lin, W.; Wu, X.; Zhou, J.; Shen, D.; Zhou, L. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. NeuroImage 2018, 174, 550–562. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, X.-J. DenseFuse: A Fusion Approach to Infrared and Visible Images. IEEE Trans. Image Process. 2019, 28, 2614–2623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, Y.; Sun, P.; Yan, H.; Zhao, X.; Zhang, L. IFCNN: A general image fusion framework based on convolutional neural network. Inf. Fusion 2020, 54, 99–118. [Google Scholar] [CrossRef]
- Parvathy, V.S.; Pothiraj, S.; Sampson, J. A novel approach in multimodality medical image fusion using optimal shearlet and deep learning. Int. J. Imaging Syst. Technol. 2020, 30, 847–859. [Google Scholar] [CrossRef]
- Bowles, C.; Chen, L.; Guerrero, R.; Bentley, P.; Gunn, R.; Hammers, A.; Dickie, D.A.; Hernández, M.V.; Wardlaw, J.; Rueckert, D. GAN Augmentation: Augmenting Training Data using Generative Adversarial Networks. arXiv 2018, arXiv:1810.10863. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Karras, T.; Aila, T.; Laine, S.; Lehtinen, J. Progressive Growing of Gans for Improved Quality, Stability, and Variation. arXiv 2018, arXiv:1710.10196. [Google Scholar]
- Wolterink, J.M.; Leiner, T.; Viergever, M.A.; Isgum, I. Generative Adversarial Networks for Noise Reduction in Low-Dose CT. IEEE Trans. Med. Imaging 2017, 36, 2536–2545. [Google Scholar] [CrossRef] [PubMed]
- Shitrit, O.; Raviv, T.R. Accelerated Magnetic Resonance Imaging by Adversarial Neural Network. In Lecture Notes in Computer Science; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2017; pp. 30–38. [Google Scholar]
- Mahapatra, D.; Bozorgtabar, B. Retinal Vasculature Segmentation Using Local Saliency Maps and Generative Adversarial Networks For Image Super Resolution. arXiv 2018, arXiv:1710.04783. [Google Scholar]
- Madani, A.; Moradi, M.; Karargyris, A.; Syeda-Mahmood, T. Chest x-ray generation and data augmentation for cardiovascular abnormality classification. In Medical Imaging 2018: Image Processing; SPIE Medical Imaging: Houston, TX, US, 2018; p. 105741M. [Google Scholar] [CrossRef]
- Lu, C.-Y.; Rustia, D.J.A.; Lin, T.-T. Generative Adversarial Network Based Image Augmentation for Insect Pest Classification Enhancement. IFAC-PapersOnLine 2019, 52, 1–5. [Google Scholar] [CrossRef]
- Chuquicusma, M.J.M.; Hussein, S.; Burt, J.; Bagci, U. How to fool radiologists with generative adversarial networks? A visual turing test for lung cancer diagnosis. arXiv 2018, arXiv:1710.09762. [Google Scholar]
- Calimeri, F.; Marzullo, A.; Stamile, C.; Terracina, G. Biomedical Data Augmentation Using Generative Adversarial Neural Networks. In Artificial Neural Networks and Machine Learning—ICANN 2017, Proceedings of the 26th International Conference on Artificial Neural Networks, Alghero, Italy, 11–14 September 2017; Springer: Cham, Switzerland, 2017; pp. 626–634. [Google Scholar] [CrossRef]
- Plassard, A.J.; Davis, L.T.; Newton, A.T.; Resnick, S.M.; Landman, B.A.; Bermudez, C. Learning implicit brain MRI manifolds with deep learning. In Medical Imaging 2018: Image Processing; International Society for Optics and Photonics: Bellingham, WA, USA, 2018; Volume 10574, p. 105741L. [Google Scholar] [CrossRef]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.; Zheng, J.; Liu, Z.; Zheng, Y.; Chen, Y.; Xu, X.; Lu, J.; Xie, J. High-resolution dermoscopy image synthesis with conditional generative adversarial networks. Biomed. Signal Process. Control 2021, 64, 102224. [Google Scholar] [CrossRef]
- Bissoto, A.; Perez, F.; Valle, E.; Avila, S. Skin lesion synthesis with Generative Adversarial Networks. In OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11041, pp. 294–302. [Google Scholar] [CrossRef] [Green Version]
- Qin, Z.; Liu, Z.; Zhu, P.; Xue, Y. A GAN-based image synthesis method for skin lesion classification. Comput. Methods Programs Biomed. 2020, 195, 105568. [Google Scholar] [CrossRef]
- Venu, S.K.; Ravula, S. Evaluation of Deep Convolutional Generative Adversarial Networks for Data Augmentation of Chest X-ray Images. Future Internet 2020, 13, 8. [Google Scholar] [CrossRef]
- Goodfellow, I. NIPS 2016 Tutorial: Generative Adversarial Networks. arXiv 2016, arXiv:1701.00160. [Google Scholar]
- Wang, K.; Gou, C.; Duan, Y.; Lin, Y.; Zheng, X.; Wang, F.-Y. Generative adversarial networks: Introduction and outlook. IEEE/CAA J. Autom. Sin. 2017, 4, 588–598. [Google Scholar] [CrossRef]
- Wang, N.; Wang, W. An image fusion method based on wavelet and dual-channel pulse coupled neural network. In Proceedings of the 2015 IEEE International Conference on Progress in Informatics and Computing (PIC), Nanjing, China, 18–20 December 2015; pp. 270–274. [Google Scholar]
- Li, Y.; Sun, Y.; Huang, X.; Qi, G.; Zheng, M.; Zhu, Z. An Image Fusion Method Based on Sparse Representation and Sum Modified-Laplacian in NSCT Domain. Entropy 2018, 20, 522. [Google Scholar] [CrossRef] [Green Version]
- Biswas, B.; Sen, B.K. Color PET-MRI Medical Image Fusion Combining Matching Regional Spectrum in Shearlet Domain. Int. J. Image Graph. 2019, 19, 1950004. [Google Scholar] [CrossRef]
- Li, B.; Peng, H.; Luo, X.; Wang, J.; Song, X.; Pérez-Jiménez, M.J.; Riscos-Núñez, A. Medical Image Fusion Method Based on Coupled Neural P Systems in Nonsubsampled Shearlet Transform Domain. Int. J. Neural Syst. 2021, 31, 2050050. [Google Scholar] [CrossRef]
- Li, L.; Ma, H. Pulse Coupled Neural Network-Based Multimodal Medical Image Fusion via Guided Filtering and WSEML in NSCT Domain. Entropy 2021, 23, 591. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, H.; Jia, Z.; Si, Y. A novel multiscale transform decomposition based multi-focus image fusion framework. Multimed. Tools Appl. 2021, 80, 12389–12409. [Google Scholar] [CrossRef]
- Li, X.; Zhou, F.; Tan, H. Joint image fusion and denoising via three-layer decomposition and sparse representation. Knowl.-Based Syst. 2021, 224, 107087. [Google Scholar] [CrossRef]
- Shehanaz, S.; Daniel, E.; Guntur, S.R.; Satrasupalli, S. Optimum weighted multimodal medical image fusion using particle swarm optimization. Optik 2021, 231, 166413. [Google Scholar] [CrossRef]
- Maqsood, S.; Javed, U. Multi-modal Medical Image Fusion based on Two-scale Image Decomposition and Sparse Representation. Biomed. Signal Process. Control 2020, 57, 101810. [Google Scholar] [CrossRef]
- Qi, G.; Hu, G.; Mazur, N.; Liang, H.; Haner, M. A Novel Multi-Modality Image Simultaneous Denoising and Fusion Method Based on Sparse Representation. Computers 2021, 10, 129. [Google Scholar] [CrossRef]
- Wang, M.; Tian, Z.; Gui, W.; Zhang, X.; Wang, W. Low-Light Image Enhancement Based on Nonsubsampled Shearlet Transform. IEEE Access 2020, 8, 63162–63174. [Google Scholar] [CrossRef]
- Johnson, J.L.; Ritter, D. Observation of periodic waves in a pulse-coupled neural network. Opt. Lett. 1993, 18, 1253–1255. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhang, L.; Lei, Y. Novel fusion method for visible light and infrared images based on NSST–SF–PCNN. Infrared Phys. Technol. 2014, 65, 103–112. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.C.F.; Gutman, D.; Emre Celebi, M.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H. Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2017, arXiv:1710.05006. [Google Scholar]
- Combalia, M.; Codella, N.C.F.; Rotemberg, V.; Helba, B.; Vilaplana, V.; Reiter, O.; Carrera, C.; Barreiro, A.; Halpern, A.C.; Puig, S.; et al. BCN20000: Dermoscopic Lesions in the Wild. arXiv 2019, arXiv:1908.02288. [Google Scholar]
- Kinyanjui, N.M.; Odonga, T.; Cintas, C.; Codella, N.C.F.; Panda, R.; Sattigeri, P.; Varshney, K.R. Fairness of Classifiers Across Skin Tones in Dermatology. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2017; Springer: Cham, Switzerland, 2020; pp. 320–329. [Google Scholar]
- Groh, M.; Harris, C.; Soenksen, L.; Lau, F.; Han, R.; Kim, A.; Koochek, A.; Badri, O. Evaluating Deep Neural Networks Trained on Clinical Images in Dermatology with the Fitzpatrick 17k Dataset. arXiv 2021, arXiv:2104.09957. [Google Scholar]
- Xiao, Y.; Decenciere, E.; Velasco-Forero, S.; Burdin, H.; Bornschlogl, T.; Bernerd, F.; Warrick, E.; Baldeweck, T. A New Color Augmentation Method for Deep Learning Segmentation of Histological Images. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 886–890. [Google Scholar]
- Liu, Z.; Blasch, E.; Xue, Z.; Zhao, J.; Laganiere, R.; Wu, W. Objective Assessment of Multiresolution Image Fusion Algorithms for Context Enhancement in Night Vision: A Comparative Study. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 94–109. [Google Scholar] [CrossRef]
- Adjobo, E.C.; Mahama, A.T.S.; Gouton, P.; Tossa, J. Proposition of Convolutional Neural Network Based System for Skin Cancer Detection. In Proceedings of the 2019 15th International Conference on Signal-Image Technology & Internet-Based Systems (SITIS), Sorrento, Italy, 26–29 November 2019; pp. 35–39. [Google Scholar]
- Lallas, A.; Reggiani, C.; Argenziano, G.; Kyrgidis, A.; Bakos, R.; Masiero, N.; Scheibe, A.; Cabo, H.; Ozdemir, F.; Sortino-Rachou, A.; et al. Dermoscopic nevus patterns in skin of colour: A prospective, cross-sectional, morphological study in individuals with skin type V and VI. J. Eur. Acad. Dermatol. Venereol. 2014, 28, 1469–1474. [Google Scholar] [CrossRef]







| Categories | Techniques, Reference and Year | Strengths and Limatations |
|---|---|---|
| Fusion methods | - DWT and dual-channel PCNN [29]-(2015) - NSCT and sparse representation [30]-(2018) - Shearlet transforms and matching regional spectrum [31]-(2019) - NSST and coupled neural P systems [32]- (2020) - Two-scale decomposition and sparse representation [10]-(2020) - NSCT and PCNN guided filtering and WSEML [33]-(2021) - NSCT and successive integrating average filter and median filter [34]-(2021) - NSCT, interval-gradient-filtering and sparse representation [35]-(2021) - DWT_PSO [36]-(2021) | Strengths - Decomposition includes texture and edge features - Low information loss - Shift-invariant: NSST and NSCT - Easy to implement - No training - Fast computation Limitations - Depend on decomposition level - Some decomposition methods are noise sensitive - Subjective fusion strategy - Low generalization |
| Deep-learning-based methods | - GAN [24]-(2018) - Densefuse [8]-(2019) - IFCNN [9]-(2020) - GAN [25]-(2020) - Optimal shearlet and deep learning [10]-(2020) - DCGAN [26]-(2021) | Strengths - Generates realistic images - Good generalization ability for different tasks - Low information loss - Low noise sensitivity Limitations - Requires training data - Requires training - High power computation - Mode collapse - Time-consuming |
| ITA Range | Skin Tone Category |
|---|---|
| >55° | Very Light |
| >41° and ≤55° | Light |
| >28° and ≤41° | Intermediate |
| >10° and ≤28° | Tanned |
| >−30° and ≤10° | Brown |
| ≤−30 | Dark |
| Obtained Images | Methods | QG | QS | QC | QY | QCB |
|---|---|---|---|---|---|---|
| b1 | CT | 0.7390 | 0.8080 | 0.7910 | 0.7820 | 0.5470 |
| c1 | DWT_SR | 0.4990 | 0.6750 | 0.8330 | 0.5630 | 0.5530 |
| d1 | DWT_CT | 0.6600 | 0.7700 | 0.8400 | 0.7170 | 0.5050 |
| e1 | NSCT_SR_SML | 0.4440 | 0.5780 | 0.6410 | 0.4190 | 0.4450 |
| f1 | NSST_PCNN | 0.8060 | 0.8390 | 0.8810 | 0.8240 | 0.6190 |
| b2 | CT | 0.7730 | 0.9430 | 0.8900 | 0.9140 | 0.6310 |
| c2 | DWT_SR | 0.6660 | 0.7560 | 0.8170 | 0.7900 | 0.6280 |
| d2 | DWT_CT | 0.7540 | 0.7280 | 0.9320 | 0.9050 | 0.6130 |
| e2 | NSCT_SR_SML | 0.4920 | 0.6200 | 0.8640 | 0.5050 | 0.4690 |
| f2 | NSST_PCNN | 0.8030 | 0.8800 | 0.9680 | 0.9750 | 0.6580 |
| b3 | CT | 0.4500 | 0.8380 | 0.7530 | 0.7610 | 0.4840 |
| c3 | DWT_SR | 0.5930 | 0.8090 | 0.8020 | 0.7320 | 0.4590 |
| d3 | DWT_CT | 0.6900 | 0.7930 | 0.7430 | 0.7570 | 0.5000 |
| e3 | NSCT_SR_SML | 0.4880 | 0.7390 | 0.4280 | 0.5330 | 0.4800 |
| f3 | NSST_PCNN | 0.7690 | 0.8470 | 0.8090 | 0.8450 | 0.5740 |
| All images | CT | 0.5760 | 0.8350 | 0.7640 | 0.7810 | 0.5260 |
| DWT_SR | 0.5080 | 0.7190 | 0.7910 | 0.8440 | 0.5190 | |
| DWT_CT | 0.6360 | 0.7360 | 0.8380 | 0.6570 | 0.5120 | |
| NSCT_SR_SML | 0.3970 | 0.6180 | 0.5970 | 0.4480 | 0.4370 | |
| NSST_PCNN | 0.7150 | 0.8280 | 0.7700 | 0.7550 | 0.5890 |
| Methods | CT | DWT_SR | DWT_CT | NSCT_SR_SML | NSST_PCNN |
|---|---|---|---|---|---|
| Times/s | 0.6685 | 13.1643 | 6.9492 | 24.2362 | 23.9312 |
| Metrics | Very Light | Light | Intermediate | Tanned | Brown |
|---|---|---|---|---|---|
| Accuracy | 0.9434 | 0.9480 | 0.9333 | 0.7255 | 0.7692 |
| Precision | 0.9091 | 0.9297 | 0.8900 | 0.7826 | 0.7500 |
| Recall | 0.9662 | 0.9557 | 0.9082 | 0.6667 | 1.0000 |
| F1-score | 0.9368 | 0.9425 | 0.8990 | 0.7200 | 0.8571 |
| Metrics | Very Light | Light | Intermediate | Tanned | Brown |
|---|---|---|---|---|---|
| Accuracy | 0.9518 | 0.9553 | 0.9500 | 0.8824 | 0.9231 |
| Precision | 0.9227 | 0.9365 | 0.9300 | 0.9130 | 0.9167 |
| Recall | 0.9713 | 0.9650 | 0.9208 | 0.8400 | 1.0000 |
| F1-score | 0.9464 | 0.9505 | 0.9254 | 0.8750 | 0.9565 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chabi Adjobo, E.; Sanda Mahama, A.T.; Gouton, P.; Tossa, J. Towards Accurate Skin Lesion Classification across All Skin Categories Using a PCNN Fusion-Based Data Augmentation Approach. Computers 2022, 11, 44. https://doi.org/10.3390/computers11030044
Chabi Adjobo E, Sanda Mahama AT, Gouton P, Tossa J. Towards Accurate Skin Lesion Classification across All Skin Categories Using a PCNN Fusion-Based Data Augmentation Approach. Computers. 2022; 11(3):44. https://doi.org/10.3390/computers11030044
Chicago/Turabian StyleChabi Adjobo, Esther, Amadou Tidjani Sanda Mahama, Pierre Gouton, and Joël Tossa. 2022. "Towards Accurate Skin Lesion Classification across All Skin Categories Using a PCNN Fusion-Based Data Augmentation Approach" Computers 11, no. 3: 44. https://doi.org/10.3390/computers11030044