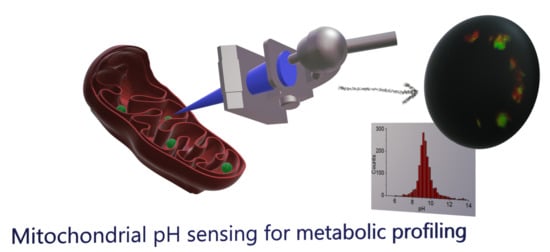
Mitochondrial pH Nanosensors for Metabolic Profiling of Breast Cancer Cell Lines
Abstract
:1. Introduction
2. Results
2.1. QD-SS-MPA Nanosensor Preparation, Characterization, and Calibration
2.2. QD-SS20-MPA Mitochondrial Localization
2.3. Intramitochondrial pHm Estimation through FLIM Imaging
2.4. Correlation with Different Metabolic Features
3. Discussion
4. Materials and Methods
4.1. Synthesis of SS Peptide-MPA-Capped CdSe/ZnS Nanoparticles
4.2. Mitochondrial Localization and FLIM Imaging of QD-SS-MPA pH Nanosensors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| FLIM | Fluorescence lifetime imaging microscopy |
| QD | Quantum Dots |
| SS | Szeto-Schiller peptide |
| MPA | Mercaptopropionic acid |
| PL | Photoluminescence |
| pHm | Intramitochondrial pH |
| MT | MitoTracker Deep Red |
| PIE | Pulsed interleaved excitation |
References
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menendez, J.A.; Joven, J.; Cufí, S.; Corominas-Faja, B.; Oliveras-Ferraros, C.; Cuyàs, E.; Martin-Castillo, B.; López-Bonet, E.; Alarcón, T.; Vazquez-Martin, A. The Warburg effect version 2.0: Metabolic reprogramming of cancer stem cells. Cell Cycle 2013, 12, 1166–1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramaniam, S.R.; Chesselet, M.F. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease. Prog. Neurobiol. 2013, 106–107, 17–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruhoy, I.S.; Saneto, R.P. The genetics of Leigh syndrome and its implications for clinical practice and risk management. Appl. Clinical Genet. 2014, 7, 221–234. [Google Scholar]
- Bonnen, P.E.; Yarham, J.W.; Besse, A.; Wu, P.; Faqeih, E.A.; Al-Asmari, A.M.; Saleh, M.A.; Eyaid, W.; Hadeel, A.; He, L.; et al. Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am. J. Hum. Genet. 2013, 93, 471–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorman, G.S.; Chinnery, P.F.; DiMauro, S.; Hirano, M.; Koga, Y.; McFarland, R.; Suomalainen, A.; Thorburn, D.R.; Zeviani, M.; Turnbull, D.M. Mitochondrial diseases. Nat. Rev. Dis. Primers 2016, 2, 16080. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Kang, J.M.; Kim, D.-J.; Park, S.H.; Jeong, H.Y.; Lee, Y.H.; Kim, Y.G.; Yang, D.H.; Lee, S.H. PGC1α Activators Mitigate Diabetic Tubulopathy by Improving Mitochondrial Dynamics and Quality Control. J. Diabetes Res. 2017, 2017, 6483572. [Google Scholar]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [Green Version]
- Boland, M.L.; Chourasia, A.H.; Macleod, K.F. Mitochondrial dysfunction in cancer. Front. Oncol. 2013, 3, 292. [Google Scholar] [CrossRef] [Green Version]
- Wallace, D.C. Mitochondria and cancer. Nat. Rev. Cancer 2012, 12, 685–698. [Google Scholar] [CrossRef] [Green Version]
- Berezin, M.Y.; Achilefu, S. Fluorescence Lifetime Measurements and Biological Imaging. Chem. Rev. 2010, 110, 2641–2684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruedas-Rama, M.; Alvarez-Pez, J.; Crovetto, L.; Paredes, J.; Orte, A. FLIM Strategies for Intracellular Sensing. In Advanced Photon Counting; Kapusta, P., Wahl, M., Erdmann, R., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 15, pp. 191–223. [Google Scholar]
- Martin, K.J.; McGhee, E.J.; Schwarz, J.P.; Drysdale, M.; Brachmann, S.M.; Stucke, V.; Sansom, O.J.; Anderson, K.I. Accepting from the best donor; analysis of long-lifetime donor fluorescent protein pairings to optimise dynamic FLIM-based FRET experiments. PLoS ONE 2018, 13, e0183585. [Google Scholar] [CrossRef] [PubMed]
- Esposito, A.; Gralle, M.; Dani, M.A.C.; Lange, D.; Wouters, F.S. pHlameleons: A Family of FRET-Based Protein Sensors for Quantitative pH Imaging. Biochemistry 2008, 47, 13115–13126. [Google Scholar] [CrossRef] [PubMed]
- Burgstaller, S.; Bischof, H.; Gensch, T.; Stryeck, S.; Gottschalk, B.; Ramadani-Muja, J.; Eroglu, E.; Rost, R.; Balfanz, S.; Baumann, A.; et al. pH-Lemon, a Fluorescent Protein-Based pH Reporter for Acidic Compartments. ACS Sens. 2019, 4, 883–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, A.F.; Yasuda, R. An Improved Ras Sensor for Highly Sensitive and Quantitative FRET-FLIM Imaging. PLoS ONE 2013, 8, e52874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hum, M.J.; Siegel, P.A.; Pavalko, M.F.; Day, N.R. Monitoring Biosensor Activity in Living Cells with Fluorescence Lifetime Imaging Microscopy. Int. J. Mol. Sci. 2012, 13, 14385–14400. [Google Scholar] [CrossRef]
- Yang, Z.; He, Y.; Lee, J.-H.; Park, N.; Suh, M.; Chae, W.-S.; Cao, J.; Peng, X.; Jung, H.; Kang, C.; et al. A Self-Calibrating Bipartite Viscosity Sensor for Mitochondria. J. Am. Chem. Soc. 2013, 135, 9181–9185. [Google Scholar] [CrossRef]
- Sherin, P.S.; Lopez-Duarte, I.; Dent, M.R.; Kubankova, M.; Vysniauskas, A.; Bull, J.A.; Reshetnikova, E.S.; Klymchenko, A.S.; Tsentalovich, Y.P.; Kuimova, M.K. Visualising the membrane viscosity of porcine eye lens cells using molecular rotors. Chem. Sci. 2017, 8, 3523–3528. [Google Scholar] [CrossRef] [Green Version]
- Kuimova, M.K.; Yahioglu, G.; Levitt, J.A.; Suhling, K. Molecular Rotor Measures Viscosity of Live Cells via Fluorescence Lifetime Imaging. J. Am. Chem. Soc. 2008, 130, 6672–6673. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Purkayastha, P. Modulating Thiazole Orange Aggregation in Giant Lipid Vesicles: Photophysical Study Associated with FLIM and FCS. ACS Omega 2017, 2, 5036–5043. [Google Scholar] [CrossRef]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Ruedas-Rama, M.J.; Orte, A.; Hall, E.A.H.; Alvarez-Pez, J.M.; Talavera, E.M. A chloride ion nanosensor for time-resolved fluorimetry and fluorescence lifetime imaging. Analyst 2012, 137, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Ripoll, C.; Martin, M.; Roldan, M.; Talavera, E.M.; Orte, A.; Ruedas-Rama, M.J. Intracellular Zn2+ detection with quantum dot-based FLIM nanosensors. Chem. Commun. 2015, 51, 16964–16967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orte, A.; Alvarez-Pez, J.M.; Ruedas-Rama, M.J. Fluorescence Lifetime Imaging Microscopy for the Detection of Intracellular pH with Quantum Dot Nanosensors. ACS Nano 2013, 7, 6387–6395. [Google Scholar] [CrossRef]
- Ruedas-Rama, M.J.; Orte, A.; Hall, E.A.H.; Alvarez-Pez, J.M.; Talavera, E.M. Quantum dot photoluminescence lifetime-based pH nanosensor. Chem. Commun. 2011, 47, 2898–2900. [Google Scholar] [CrossRef]
- Huang, C.-P.; Li, Y.-K.; Chen, T.-M. A highly sensitive system for urea detection by using CdSe/ZnS core-shell quantum dots. Biosens. Bioelectron. 2007, 22, 1835–1838. [Google Scholar] [CrossRef]
- Deng, Z.; Zhang, Y.; Yue, J.; Tang, F.; Wei, Q. Green and Orange CdTe Quantum Dots as Effective pH-Sensitive Fluorescent Probes for Dual Simultaneous and Independent Detection of Viruses. J. Phys. Chem. B 2007, 111, 12024–12031. [Google Scholar] [CrossRef]
- Ruedas-Rama, M.J.; Hall, E.A.H. Analytical Nanosphere Sensors Using Quantum Dot-Enzyme Conjugates for Urea and Creatinine. Anal. Chem. 2010, 82, 9043–9049. [Google Scholar] [CrossRef]
- Gao, X.; Chan, W.C.W.; Nie, S. Quantum-dot nanocrystals for ultrasensitive biological labeling and multicolor optical encoding. J. Biomed. Opt. 2002, 7, 532–537. [Google Scholar] [CrossRef]
- Szeto, H.H. Development of mitochondria-targeted aromatic-cationic peptides for neurodegenerative diseases. Ann. N. Y. Acad. Sci. 2008, 1147, 112–121. [Google Scholar] [CrossRef]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castello, F.; Paredes, J.M.; Ruedas-Rama, M.J.; Martin, M.; Roldan, M.; Casares, S.; Orte, A. Two-Step Amyloid Aggregation: Sequential Lag Phase Intermediates. Sci. Rep. 2017, 7, 40065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azarias, G.; Chatton, J.-Y. Selective Ion Changes during Spontaneous Mitochondrial Transients in Intact Astrocytes. PLoS ONE 2011, 6, e28505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poburko, D.; Santo-Domingo, J.; Demaurex, N. Dynamic Regulation of the Mitochondrial Proton Gradient during Cytosolic Calcium Elevations. J. Biol. Chem. 2011, 286, 11672–11684. [Google Scholar] [CrossRef] [Green Version]
- Cao, L.; Zhao, Z.; Zhang, T.; Guo, X.; Wang, S.; Li, S.; Li, Y.; Yang, G. In vivo observation of the pH alternation in mitochondria for various external stimuli. Chem. Commun. 2015, 51, 17324–17327. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, C.; Cen, J.; Bai, Y.; He, W.; Guo, Z. Ratiometric detection of pH fluctuation in mitochondria with a new fluorescein/cyanine hybrid sensor. Chem. Sci. 2015, 6, 3187–3194. [Google Scholar] [CrossRef] [Green Version]
- Llopis, J.; McCaffery, J.M.; Miyawaki, A.; Farquhar, M.G.; Tsien, R.Y. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 6803–6808. [Google Scholar] [CrossRef] [Green Version]
- Balut, C.; vandeVen, M.; Despa, S.; Lambrichts, I.; Ameloot, M.; Steels, P.; Smets, I. Measurement of cytosolic and mitochondrial pH in living cells during reversible metabolic inhibition. Kidney Int. 2008, 73, 226–232. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, S.E.; Chandel, N.S. Targeting mitochondria metabolism for cancer therapy. Nature Chem. Biol. 2014, 11, 9–15. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Huang, J.; Xie, C.; Li, X.; Liu, L.; He, J.; Pan, H.; Huang, L.; Fan, X.; Yao, X.; et al. Combined use of PI3K and MEK inhibitors synergistically inhibits lung cancer with EGFR and KRAS mutations. Oncol. Rep. 2016, 36, 365–375. [Google Scholar] [CrossRef]
- Szeto, H.H.; Liu, S. Cardiolipin-targeted peptides rejuvenate mitochondrial function, remodel mitochondria, and promote tissue regeneration during aging. Archiv. Biochem. Biophys. 2018, 660, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, W.; Ng, E.A.; Tamucci, J.D.; Boyd, K.J.; Sathappa, M.; Coscia, A.; Pan, M.; Han, X.; Eddy, N.A.; May, E.R.; et al. The mitochondria-targeted peptide SS-31 binds lipid bilayers and modulates surface electrostatics as a key component of its mechanism of action. J. Biol. Chem. 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szeto, H.H.; Schiller, P.W. Novel Therapies Targeting Inner Mitochondrial Membrane—From Discovery to Clinical Development. Pharm. Res. 2011, 28, 2669–2679. [Google Scholar] [CrossRef] [PubMed]
- Amelia, M.; Avellini, T.; Monaco, S.; Impellizzeri, S.; Yildiz, I.; Raymo, F.M.; Credi, A. Redox properties of CdSe and CdSe–ZnS quantum dots in solution. Pure Appl. Chem. 2011, 83, 1–8. [Google Scholar] [CrossRef]
- Davies, K.M.; Strauss, M.; Daum, B.; Kief, J.H.; Osiewacz, H.D.; Rycovska, A.; Zickermann, V.; Kühlbrandt, W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl. Acad. Sci. USA 2011, 108, 14121–14126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, K.L.; Stewart, K.M.; Fonseca, S.B.; Guo, Q.; Kelley, S.O. Mitochondria-Penetrating Peptides. Chem. Biol. 2008, 15, 375–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santo-Domingo, J.; Demaurex, N. The renaissance of mitochondrial pH. J. Gen. Physiol. 2012, 139, 415–423. [Google Scholar] [CrossRef] [Green Version]
- Zimmer, M. Green Fluorescent Protein (GFP): Applications, Structure, and Related Photophysical Behavior. Chem. Rev. 2002, 102, 759–782. [Google Scholar] [CrossRef]
- Lee, M.H.; Park, N.; Yi, C.; Han, J.H.; Hong, J.H.; Kim, K.P.; Kang, D.H.; Sessler, J.L.; Kang, C.; Kim, J.S. Mitochondria-Immobilized pH-Sensitive Off–On Fluorescent Probe. J. Am. Chem. Soc. 2014, 136, 14136–14142. [Google Scholar] [CrossRef]
- Datta, R.; Heaster, T.M.; Sharick, J.T.; Gillette, A.A.; Skala, M.C. Fluorescence lifetime imaging microscopy: Fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt. 2020, 25, 1–43. [Google Scholar] [CrossRef]
- Liu, X.; Lin, D.; Becker, W.; Niu, J.; Yu, B.; Liu, L.; Qu, J. Fast fluorescence lifetime imaging techniques: A review on challenge and development. J. Innov. Opt. Health Sci. 2019, 12, 1930003. [Google Scholar] [CrossRef] [Green Version]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ripoll, C.; Roldan, M.; Contreras-Montoya, R.; Diaz-Mochon, J.J.; Martin, M.; Ruedas-Rama, M.J.; Orte, A. Mitochondrial pH Nanosensors for Metabolic Profiling of Breast Cancer Cell Lines. Int. J. Mol. Sci. 2020, 21, 3731. https://doi.org/10.3390/ijms21103731
Ripoll C, Roldan M, Contreras-Montoya R, Diaz-Mochon JJ, Martin M, Ruedas-Rama MJ, Orte A. Mitochondrial pH Nanosensors for Metabolic Profiling of Breast Cancer Cell Lines. International Journal of Molecular Sciences. 2020; 21(10):3731. https://doi.org/10.3390/ijms21103731
Chicago/Turabian StyleRipoll, Consuelo, Mar Roldan, Rafael Contreras-Montoya, Juan J. Diaz-Mochon, Miguel Martin, Maria J. Ruedas-Rama, and Angel Orte. 2020. "Mitochondrial pH Nanosensors for Metabolic Profiling of Breast Cancer Cell Lines" International Journal of Molecular Sciences 21, no. 10: 3731. https://doi.org/10.3390/ijms21103731