Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription
Abstract
:1. The PIKK Family and TRRAP
2. TRRAP and HAT in Transcription
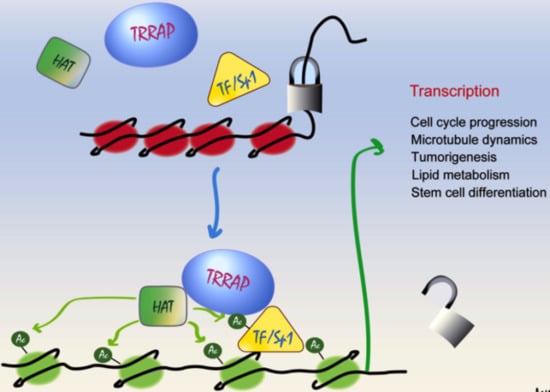
2.1. TRRAP as an Adaptor Protein of HAT
2.2. The Role of TRRAP in Transcription
2.3. The Biological Function of TRRAP in Different Cellular and Animal Models
3. Sp1 Is a Ubiquitous TF
3.1. Overview
3.2. Sp1 Structure
3.3. Transcription Initiation and Transactivation by Sp1
3.4. Regulation of Sp1 Activity
3.5. Post-Translational Modification (PTM) on Sp1
3.6. Novel Functions of Sp1 in the Nervous System and Diseases
4. Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Elías-Villalobos, A.; Fort, P.; Helmlinger, D. New insights into the evolutionary conservation of the sole PIKK pseudokinase Tra1/TRRAP. Biochem. Soc. Trans. 2019, 47, 1597–1608. [Google Scholar] [CrossRef]
- Thiruvenkatam, V. Structural and strategic landscape of PIKK protein family and their inhibitors an overview. Front. Biosci. 2020, 25, 1538–1567. [Google Scholar] [CrossRef] [PubMed]
- Magaway, C.; Kim, E.; Jacinto, E. Targeting mTOR and Metabolism in Cancer: Lessons and Innovations. Cells 2019, 8, 1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasif, S.; Contu, L.; Mühlemann, O. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol. 2018, 75, 78–87. [Google Scholar] [CrossRef]
- Lempiäinen, H.; Halazonetis, T.D. Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J. 2009, 28, 3067–3073. [Google Scholar] [CrossRef] [Green Version]
- Kwon, A.; Scott, S.; Taujale, R.; Yeung, W.; Kochut, K.J.; Eyers, P.A.; Kannan, N. Tracing the origin and evolution of pseudokinases across the tree of life. Sci. Signal. 2019, 12, eaav3810. [Google Scholar] [CrossRef]
- McMahon, S.B.; Van Buskirk, H.A.; Dugan, K.A.; Copeland, T.D.; Cole, M.D. The Novel ATM-Related Protein TRRAP is an Essential Cofactor for the c-Myc and E2F Oncoproteins. Cell 1998, 94, 363–374. [Google Scholar] [CrossRef] [Green Version]
- Helmlinger, D.; Marguerat, S.; Villen, J.; Swaney, D.L.; Gygi, S.P.; Bahler, J.; Winston, F. Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. EMBO J. 2011, 30, 2843–2852. [Google Scholar] [CrossRef]
- Ryu, H.-Y.; Hochstrasser, M. Histone sumoylation and chromatin dynamics. Nucleic Acids Res. 2021, 49, 6043–6052. [Google Scholar] [CrossRef] [PubMed]
- Soffers, J.H.; Workman, J.L. The SAGA chromatin-modifying complex: The sum of its parts is greater than the whole. Genes Dev. 2020, 34, 1287–1303. [Google Scholar] [CrossRef]
- Murr, R.; Vaissière, T.; Sawan, C.; Shukla, V.; Herceg, Z. Orchestration of chromatin-based processes: Mind the TRRAP. Oncogene 2007, 26, 5358–5372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, C.E.; Howe, L.; Sousa, K.; Alley, S.C.; Carrozza, M.J.; Tan, S.; Workman, J.L. Recruitment of HAT Complexes by Direct Activator Interactions with the ATM-Related Tra1 Subunit. Science 2001, 292, 2333–2337. [Google Scholar] [CrossRef] [PubMed]
- Ard, P.G.; Chatterjee, C.; Kunjibettu, S.; Adside, L.R.; Gralinski, L.E.; McMahon, S.B. Transcriptional Regulation of the mdm2 Oncogene by p53 Requires TRRAP Acetyltransferase Complexes. Mol. Cell. Biol. 2002, 22, 5650–5661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, A.S.; Jin, V.X.; Fan, M.; Smith, L.T.; Liyanarachchi, S.; Yan, P.S.; Leu, Y.-W.; Chan, M.W.; Plass, C.; Nephew, K.P.; et al. Combinatorial Analysis of Transcription Factor Partners Reveals Recruitment of c-MYC to Estrogen Receptor-α Responsive Promoters. Mol. Cell 2006, 21, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Sierra, J.; Yoshida, T.; Joazeiro, C.A.; Jones, K.A. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006, 20, 586–600. [Google Scholar] [CrossRef] [Green Version]
- Unno, A.; Takada, I.; Takezawa, S.; Oishi, H.; Baba, A.; Shimizu, T.; Tokita, A.; Yanagisawa, J.; Kato, S. TRRAP as a hepatic coactivator of LXR and FXR function. Biochem. Biophys. Res. Commun. 2005, 327, 933–938. [Google Scholar] [CrossRef]
- De la Cruz, X.; Lois, S.; Sánchez-Molina, S.; Martinez-Balbás, M. Do protein motifs read the histone code? BioEssays 2005, 27, 164–175. [Google Scholar] [CrossRef] [Green Version]
- Mavros, C.F.; Brownstein, C.A.; Thyagrajan, R.; Genetti, C.A.; Tembulkar, S.; Graber, K.; Murphy, Q.; Cabral, K.; Vannoy, G.E.; Bainbridge, M.; et al. De novo variant of TRRAP in a patient with very early onset psychosis in the context of non-verbal learning disability and obsessive-compulsive disorder: A case report. BMC Med. Genet. 2018, 19, 197. [Google Scholar] [CrossRef]
- Brownell, J.E.; Zhou, J.; Ranalli, T.; Kobayashi, R.; Edmondson, D.G.; Roth, S.Y.; Allis, C. Tetrahymena Histone Acetyltransferase A: A Homolog to Yeast Gcn5p Linking Histone Acetylation to Gene Activation. Cell 1996, 84, 843–851. [Google Scholar] [CrossRef] [Green Version]
- Vignali, M.; Steger, D.J.; Neely, K.E.; Workman, J.L. Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 2000, 19, 2629–2640. [Google Scholar] [CrossRef] [Green Version]
- Abbey, D.; Conlon, D.; Rainville, C.; Elwyn, S.; Quiroz-Figueroa, K.; Billheimer, J.; Schultz, D.C.; Hand, N.J.; Cherry, S.; Rader, D.J. Lipid droplet screen in human hepatocytes identifies TRRAP as a regulator of cellular triglyceride metabolism. Clin. Transl. Sci. 2021, 14, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Tapias, A.; Zhou, Z.; Shi, Y.; Chong, Z.; Wang, P.; Groth, M.; Platzer, M.; Huttner, W.; Herceg, Z.; Yang, Y.-G.; et al. Trrap-Dependent Histone Acetylation Specifically Regulates Cell-Cycle Gene Transcription to Control Neural Progenitor Fate Decisions. Cell Stem Cell 2014, 14, 632–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loizou, J.I.; Oser, G.; Shukla, V.; Sawan, C.; Murr, R.; Wang, Z.-Q.; Trumpp, A.; Herceg, Z. Histone Acetyltransferase Cofactor Trrap Is Essential for Maintaining the Hematopoietic Stem/Progenitor Cell Pool. J. Immunol. 2009, 183, 6422–6431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeRan, M.; Pulvino, M.; Greene, E.; Su, C.; Zhao, J. Transcriptional Activation of Histone Genes Requires NPAT-Dependent Recruitment of TRRAP-Tip60 Complex to Histone Promoters during the G1/S Phase Transition. Mol. Cell. Biol. 2008, 28, 435–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Cuenin, C.; Murr, R.; Wang, Z.-Q.; Herceg, Z. HAT cofactor Trrap regulates the mitotic checkpoint by modulation of Mad1 and Mad2 expression. EMBO J. 2004, 23, 4824–4834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Plasschaert, L.W.; Aryal, S.; Renaud, N.A.; Yang, Z.; Choo-Wing, R.; Pessotti, A.D.; Kirkpatrick, N.D.; Cochran, N.R.; Carbone, W.; et al. TRRAP is a central regulator of human multiciliated cell formation. J. Cell Biol. 2018, 217, 1941–1955. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.T.; Kwon, Y.W.; Kim, D.K.; Lee, S.I.; Kim, K.-H.; Suh, D.-S.; Kim, J.H. TRRAP stimulates the tumorigenic potential of ovarian cancer stem cells. BMB Rep. 2018, 51, 514–519. [Google Scholar] [CrossRef]
- Sawan, C.; Hernandez-Vargas, H.; Murr, R.; Lopez, F.; Vaissière, T.; Ghantous, A.Y.; Cuenin, C.; Imbert, J.; Wang, Z.-Q.; Ren, B.; et al. Histone Acetyltransferase Cofactor Trrap Maintains Self-Renewal and Restricts Differentiation of Embryonic Stem Cells. Stem Cells 2013, 31, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Tapias, A.; Lázaro, D.; Yin, B.-K.; Rasa, S.M.M.; Krepelova, A.; Sacramento, E.K.; Grigaravicius, P.; Koch, P.; Kirkpatrick, J.; Ori, A.; et al. HAT cofactor TRRAP modulates microtubule dynamics via SP1 signaling to prevent neurodegeneration. eLife 2021, 10, e61531. [Google Scholar] [CrossRef]
- Kwan, S.; Sheel, A.; Song, C.; Zhang, X.-O.; Jiang, T.; Dang, H.; Cao, Y.; Ozata, D.M.; Mou, H.; Yin, H.; et al. Depletion of TRRAP Induces p53-Independent Senescence in Liver Cancer by Down-Regulating Mitotic Genes. Hepatology 2020, 71, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Herceg, Z.; Hulla, W.; Gell, D.; Cuenin, C.; Lleonart, M.E.; Jackson, S.P.; Wang, Z.-Q. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat. Genet. 2001, 29, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Cogné, B.; Ehresmann, S.; Beauregard-Lacroix, É.; Rousseau, J.; Besnard, T.; Garcia, T.; Petrovski, S.; Avni, S.; McWalter, K.; Blackburn, P.R.; et al. Missense Variants in the Histone Acetyltransferase Complex Component Gene TRRAP Cause Autism and Syndromic Intellectual Disability. Am. J. Hum. Genet. 2019, 104, 530–541. [Google Scholar] [CrossRef] [Green Version]
- Creppe, C.; Malinouskaya, L.; Volvert, M.-L.; Gillard, M.; Close, P.; Malaise, O.; Laguesse, S.; Cornez, I.; Rahmouni, S.; Ormenese, S.; et al. Elongator Controls the Migration and Differentiation of Cortical Neurons through Acetylation of α-Tubulin. Cell 2009, 136, 551–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montgomery, R.L.; Hsieh, J.; Barbosa, A.C.; Richardson, J.A.; Olson, E.N. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci. USA 2009, 106, 7876–7881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connor, L.; Gilmour, J.; Bonifer, C. The Role of the Ubiquitously Expressed Transcription Factor Sp1 in Tissue-specific Transcriptional Regulation and in Disease. Yale J. Biol. Med. 2016, 89, 513–525. [Google Scholar] [PubMed]
- Philipsen, S.; Suske, G. A tale of three fingers: The family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 1999, 27, 2991–3000. [Google Scholar] [CrossRef] [Green Version]
- Kadonaga, J.T.; Carner, K.R.; Masiarz, F.R.; Tjian, R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 1987, 51, 1079–1090. [Google Scholar] [CrossRef]
- Li, L.; Davie, J. The role of Sp1 and Sp3 in normal and cancer cell biology. Ann. Anat.-Anat. Anz. 2010, 192, 275–283. [Google Scholar] [CrossRef]
- Beishline, K.; Azizkhan-Clifford, J. Sp1 and the ‘hallmarks of cancer’. FEBS J. 2015, 282, 224–258. [Google Scholar] [CrossRef]
- Pascall, J.C.; Brown, K.D. Identification of a minimal promoter element of the mouse epidermal growth factor gene. Biochem. J. 1997, 324, 869–875. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.; Heckman, C.A.; Boxer, L.M. Histone Deacetylase Inhibitors Down-Regulate bcl-2 Expression and Induce Apoptosis in t(14;18) Lymphomas. Mol. Cell. Biol. 2005, 25, 1608–1619. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, M.; Ono, M.; Uchiumi, T.; Ueno, H.; Kohno, K.; Sugimachi, K.; Kuwano, M. Up-regulation of thrombospondin-1 gene by epidermal growth factor and transforming growth factor β in human cancer cells—Transcriptional activation and messenger RNA stabilization. Biochim. Et Biophys. Acta (BBA)-Gene Struct. Expr. 2002, 1574, 24–34. [Google Scholar] [CrossRef]
- Hung, W.-C.; Tseng, W.-L.; Shiea, J.; Chang, H.-C. Skp2 overexpression increases the expression of MMP-2 and MMP-9 and invasion of lung cancer cells. Cancer Lett. 2010, 288, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Maor, S.; Papa, M.Z.; Yarden, R.I.; Friedman, E.; Lerenthal, Y.; Lee, S.W.; Mayer, D.; Werner, H. Insulin-like Growth Factor-I Controls BRCA1 Gene Expression through Activation of Transcription Factor Sp1. Horm. Metab. Res. 2007, 39, 179–185. [Google Scholar] [CrossRef]
- Blom-Dahl, D.; Córdoba, S.; Gabilondo, H.; Carr-Baena, P.; Díaz-Benjumea, F.J.; Estella, C. In vivo analysis of the evolutionary conserved BTD-box domain of Sp1 and Btd during Drosophila development. Dev. Biol. 2020, 466, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Courey, A.J.; Holtzman, D.A.; Jackson, S.P.; Tjian, R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell 1989, 59, 827–836. [Google Scholar] [CrossRef]
- Elia, A.E.; Boardman, A.P.; Wang, D.C.; Huttlin, E.; Everley, R.A.; Dephoure, N.; Zhou, C.; Koren, I.; Gygi, S.P.; Elledge, S.J. Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA Damage Response. Mol. Cell 2015, 59, 867–881. [Google Scholar] [CrossRef] [Green Version]
- Rajagopalan, D.; Pandey, A.K.; Xiuzhen, M.C.; Lee, K.K.; Hora, S.; Zhang, Y.; Chua, B.H.; Kwok, H.S.; Bhatia, S.S.; Deng, L.W.; et al. TIP60 represses telomerase expression by inhibiting Sp1 binding to the TERT promoter. PLoS Pathog. 2017, 13, e1006681. [Google Scholar] [CrossRef] [Green Version]
- Saluja, D.; Vassallo, M.F.; Tanese, N. Distinct Subdomains of Human TAF II 130 Are Required for Interactions with Glutamine-Rich Transcriptional Activators. Mol. Cell. Biol. 1998, 18, 5734–5743. [Google Scholar] [CrossRef] [Green Version]
- Gill, G.; Pascal, E.; Tseng, Z.H.; Tjian, R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl. Acad. Sci. USA 1994, 91, 192–196. [Google Scholar] [CrossRef] [Green Version]
- Wierstra, I. Sp1: Emerging roles—Beyond constitutive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 2008, 372, 1–13. [Google Scholar] [CrossRef]
- Blais, A.; Monté, D.; Pouliot, F.; Labrie, C. Regulation of the Human Cyclin-dependent Kinase Inhibitor p18 by the Transcription Factors E2F1 and Sp1. J. Biol. Chem. 2002, 277, 31679–31693. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, Y.; Shibahara, S.; Takahashi, K. Synergistic activation of the human adrenomedullin gene promoter by Sp1 and AP-2α. Peptides 2008, 29, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Ström, A.-C.; Forsberg, M.; Lillhager, P.; Westin, G. The transcription factors Sp1 and Oct-1 interact physically to regulate human U2 snRNA gene expression. Nucleic Acids Res. 1996, 24, 1981–1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascal, E.; Tjian, R. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev. 1991, 5, 1646–1656. [Google Scholar] [CrossRef] [Green Version]
- Mastrangelo, I.A.; Courey, A.J.; Wall, J.S.; Jackson, S.P.; Hough, P.V. DNA looping and Sp1 multimer links: A mechanism for transcriptional synergism and enhancement. Proc. Natl. Acad. Sci. USA 1991, 88, 5670–5674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Kimura, A.; Nagai, R.; Horikoshi, M. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells 2000, 5, 29–41. [Google Scholar] [CrossRef]
- Davie, J.R.; He, S.; Li, L.; Sekhavat, A.; Espino, P.; Drobic, B.; Dunn, K.L.; Sun, J.-M.; Chen, H.Y.; Yu, J.; et al. Nuclear organization and chromatin dynamics–Sp1, Sp3 and histone deacetylases. Adv. Enzym. Regul. 2008, 48, 189–208. [Google Scholar] [CrossRef]
- Billon, N.; Carlisi, D.; Datto, M.B.; van Grunsven, L.; Watt, A.; Wang, X.-F.; Rudkin, B.B. Cooperation of Sp1 and p300 in the induction of the CDK inhibitor p21WAF1/CIP1 during NGF-mediated neuronal differentiation. Oncogene 1999, 18, 2872–2882. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, A.; Mahata, B.; Dhir, A.; Mandal, T.K.; Biswas, K. Elevated histone H3 acetylation and loss of the Sp1–HDAC1 complex de-repress the GM2-synthase gene in renal cell carcinoma. J. Biol. Chem. 2019, 294, 1005–1018. [Google Scholar] [CrossRef] [Green Version]
- Hung, J.-J.; Wang, Y.-T.; Chang, W.-C. Sp1 Deacetylation Induced by Phorbol Ester Recruits p300 To Activate 12(S)-Lipoxygenase Gene Transcription. Mol. Cell. Biol. 2006, 26, 1770–1785. [Google Scholar] [CrossRef] [Green Version]
- Samson, S.L.A.; Wong, N.C.W. Role of Sp1 in insulin regulation of gene expression. J. Mol. Endocrinol. 2002, 29, 265–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roos, M.D.; Su, K.; Baker, J.R.; Kudlow, J.E. O glycosylation of an Sp1-derived peptide blocks known Sp1 protein interactions. Mol. Cell. Biol. 1997, 17, 6472–6480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spengler, M.L.; Guo, L.-W.; Brattain, M.G. Phosphorylation mediates Sp1 coupled activities of proteolytic processing, desumoylation and degradation. Cell Cycle 2008, 7, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Spengler, M.L.; Brattain, M.G. Sumoylation Inhibits Cleavage of Sp1 N-terminal Negative Regulatory Domain and Inhibits Sp1-dependent Transcription. J. Biol. Chem. 2006, 281, 5567–5574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaniolo, K.; Desnoyers, S.; Leclerc, S.; Guérin, S.L. Regulation of poly(ADP-ribose) polymerase-1 (PARP-1) gene expression through the post-translational modification of Sp1: A nuclear target protein of PARP-1. BMC Mol. Biol. 2007, 8, 96. [Google Scholar] [CrossRef] [Green Version]
- Bonello, M.R.; Khachigian, L. Fibroblast Growth Factor-2 Represses Platelet-derived Growth Factor Receptor-α (PDGFR-α) Transcription via ERK1/2-dependent Sp1 Phosphorylation and an Atypical cis-Acting Element in the Proximal PDGFR-α Promoter. J. Biol. Chem. 2004, 279, 2377–2382. [Google Scholar] [CrossRef] [Green Version]
- D’Addario, M.; Arora, P.D.; Ellen, R.P.; McCulloch, C.A. Interaction of p38 and Sp1 in a Mechanical Force-induced, β1 Integrin-mediated Transcriptional Circuit That Regulates the Actin-binding Protein Filamin-A. J. Biol. Chem. 2002, 277, 47541–47550. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-T.; Yang, W.-B.; Chang, W.-C.; Hung, J.-J. Interplay of Posttranslational Modifications in Sp1 Mediates Sp1 Stability during Cell Cycle Progression. J. Mol. Biol. 2011, 414, 1–14. [Google Scholar] [CrossRef]
- Ramanan, V.K.; Saykin, A.J. Pathways to neurodegeneration: Mechanistic insights from GWAS in Alzheimer’s disease, Parkinson’s disease, and related disorders. Am. J. Neurodegener. Dis. 2013, 2, 145–175. [Google Scholar]
- Citron, B.A.; Dennis, J.S.; Zeitlin, R.S.; Echeverria, V. Transcription factor Sp1 dysregulation in Alzheimer’s disease. J. Neurosci. Res. 2008, 86, 2499–2504. [Google Scholar] [CrossRef]
- Dunah, A.W.; Jeong, H.; Griffin, A.; Kim, Y.-M.; Standaert, D.G.; Hersch, S.M.; Mouradian, M.M.; Young, A.B.; Tanese, N.; Krainc, D. Sp1 and TAFII130 Transcriptional Activity Disrupted in Early Huntington’s Disease. Science 2002, 296, 2238–2243. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Li, L.; Yang, S.; Wang, W.; Ye, C.; Li, H. Disruption of zinc transporter ZnT3 transcriptional activity and synaptic vesicular zinc in the brain of Huntington’s disease transgenic mouse. Cell Biosci. 2020, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Norflus, F.; Singh, B.; Swindell, M.K.; Buzescu, R.; Bejarano, M.; Chopra, R.; Zucker, B.; Benn, C.L.; DiRocco, D.P.; et al. Sp1 Is Up-regulated in Cellular and Transgenic Models of Huntington Disease, and Its Reduction Is Neuroprotective. J. Biol. Chem. 2006, 281, 16672–16680. [Google Scholar] [CrossRef] [Green Version]
- Saunders, J.; Wisidagama, D.R.; Morford, T.; Malone, C.S. Maximal Expression of the Evolutionarily Conserved Slit2 Gene Promoter Requires Sp1. Cell. Mol. Neurobiol. 2015, 36, 955–964. [Google Scholar] [CrossRef] [Green Version]
- García-Huerta, P.; Diaz-Hernandez, M.; Delicado, E.G.; Pimentel-Santillana, M.; Miras-Portugal, M.T.; Gomez-Villafuertes, R. The Specificity Protein Factor Sp1 Mediates Transcriptional Regulation of P2X7 Receptors in the Nervous System. J. Biol. Chem. 2012, 287, 44628–44644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Kundakovic, M.; Agís-Balboa, R.C.; Pinna, G.; Grayson, D.R. Induction of the reelin promoter by retinoic acid is mediated by Sp1. J. Neurochem. 2007, 103, 650–665. [Google Scholar] [CrossRef]
- Penazzi, L.; Bakota, L.; Brandt, R. Microtubule Dynamics in Neuronal Development, Plasticity, and Neurodegeneration. Int. Rev. Cell Mol. Biol. 2016, 321, 89–169. [Google Scholar] [CrossRef]
- Majello, B.; De Luca, P.; Suske, G.; Lania, L. Differential transcriptional regulation of c-myc promoter through the same DNA binding sites targeted by Sp1-like proteins. Oncogene 1995, 10, 1841–1848. [Google Scholar]
- Karlseder, J.; Rotheneder, H.; Wintersberger, E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol. 1996, 16, 1659–1667. [Google Scholar] [CrossRef] [Green Version]
- Boyault, C.; Khochbin, S. Regulatory cross-talk between lysine acetylation and ubiquitination: Role in the control of protein stability. BioEssays 2005, 27, 408–415. [Google Scholar] [CrossRef]


| TRRAP Target Genes | Cell Type | Cellular Process and Reference |
|---|---|---|
| ABCA1, ABCG1, SCD, HBP | Hepatic cell lines | Lipid metabolism [16] |
| APOC3 | Hepatocytes | Triglyceride metabolism [21] |
| Cdc25A, CycA2, TopA2 | Neural progenitors | Stem cell differentiation [22] |
| CyclinD2, ID2, MCM7 | Hematopoietic stem cells | Maintenance of the hematopoietic stem cell pool [23] |
| H2B, H4 | HEK293T cells | G1/S-phase transition [24] |
| Mad1, Mad2 | Embryonic fibroblasts | Cell cycle progression [25] |
| MCIDAS, CCNO, MYB | Airway epithelial cells | Multiciliated cell formation [26] |
| NANOG | Ovarian cancer cells | Tumorigenic potential of ovarian cancer stem cells [27] |
| Nanog, Oct4, Sox2 | Embryonic stem cells | Maintenance of cell stemness [28] |
| STMN3, STMN4 | Postmitotic neurons | Microtubule dynamics [29] |
| TOP2A | Hepatocellular carcinoma cells | Proliferation of tumor cells [27] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, B.-K.; Wang, Z.-Q. Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription. Int. J. Mol. Sci. 2021, 22, 12445. https://doi.org/10.3390/ijms222212445
Yin B-K, Wang Z-Q. Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription. International Journal of Molecular Sciences. 2021; 22(22):12445. https://doi.org/10.3390/ijms222212445
Chicago/Turabian StyleYin, Bo-Kun, and Zhao-Qi Wang. 2021. "Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription" International Journal of Molecular Sciences 22, no. 22: 12445. https://doi.org/10.3390/ijms222212445