BRM Complex in Arabidopsis Adopts ncBAF-like Composition and Requires BRD Subunits for Assembly and Stability
Abstract
:1. Introduction
2. Results
2.1. Isolation of the SWI/SNF Complexes Associated with BRM Identifies BCL7-like Proteins
2.2. Bdh Mutants Display Similar Phenotypic Traits to Other Mutants in SWI/SNF Subunits
2.3. BRM-Associated SWI/SNF Complex Is Homologous to Mammalian ncBAF
2.4. Mutations in BRD Genes Differentially Affect BRM Protein Levels
2.5. BRD Subunits Are Necessary for the Integrity of the SWI/SNF Complex
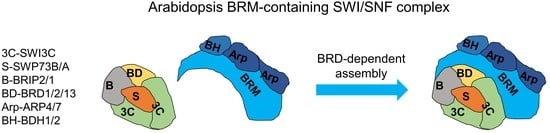
2.6. BRD Subunits Are Required for the Assembly of the Complete BRM Complex
3. Discussion
4. Materials and Methods
4.1. Plant Lines
4.2. Growth Conditions
4.3. Protein Extraction and Immunoprecipitation
4.4. LC-MS/MS Analysis
4.5. MS Data Analysis
4.6. Western Blot
4.7. RNA Isolation and RT-qPCR
4.8. Localization Analysis
4.9. Confocal Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clapier, C.R.; Cairns, B.R. The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Flaus, A.; Martin, D.M.A.; Barton, G.J.; Owen-Hughes, T. Identification of Multiple Distinct Snf2 Subfamilies with Conserved Structural Motifs. Nucleic Acids Res. 2006, 34, 2887–2905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knizewski, L.; Ginalski, K.; Jerzmanowski, A. Snf2 Proteins in Plants: Gene Silencing and Beyond. Trends Plant Sci. 2008, 13, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Hurtado, L.; Farrona, S.; Reyes, J.C. The Putative SWI/SNF Complex Subunit BRAHMA Activates Flower Homeotic Genes in Arabidopsis Thaliana. Plant Mol. Biol. 2006, 62, 291–304. [Google Scholar] [CrossRef]
- Wagner, D.; Meyerowitz, E.M. SPLAYED, a Novel SWI/SNF ATPase Homolog, Controls Reproductive Development in Arabidopsis. Curr. Biol. 2002, 12, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Sang, Y.; Silva-Ortega, C.O.; Wu, S.; Yamaguchi, N.; Wu, M.-F.; Pfluger, J.; Gillmor, C.S.; Gallagher, K.L.; Wagner, D. Mutations in Two Non-Canonical Arabidopsis SWI2/SNF2 Chromatin Remodeling ATPases Cause Embryogenesis and Stem Cell Maintenance Defects. Plant J. 2012, 72, 1000–1014. [Google Scholar] [CrossRef] [Green Version]
- Sarnowski, T.J.; Ríos, G.; Jásik, J.; Swiezewski, S.; Kaczanowski, S.; Li, Y.; Kwiatkowska, A.; Pawlikowska, K.; Koźbiał, M.; Koźbiał, P.; et al. SWI3 Subunits of Putative SWI/SNF Chromatin-Remodeling Complexes Play Distinct Roles during Arabidopsis Development. Plant Cell 2005, 17, 2454–2472. [Google Scholar] [CrossRef] [Green Version]
- Sacharowski, S.P.; Gratkowska, D.M.; Sarnowska, E.A.; Kondrak, P.; Jancewicz, I.; Porri, A.; Bucior, E.; Rolicka, A.T.; Franzen, R.; Kowalczyk, J.; et al. SWP73 Subunits of Arabidopsis SWI/SNF Chromatin Remodeling Complexes Play Distinct Roles in Leaf and Flower Development. Plant Cell 2015, 27, 1889–1906. [Google Scholar] [CrossRef]
- Jégu, T.; Latrasse, D.; Delarue, M.; Hirt, H.; Domenichini, S.; Ariel, F.; Crespi, M.; Bergounioux, C.; Raynaud, C.; Benhamed, M. The BAF60 Subunit of the SWI/SNF Chromatin-Remodeling Complex Directly Controls the Formation of a Gene Loop at FLOWERING LOCUS C in Arabidopsis. Plant Cell 2014, 26, 538–551. [Google Scholar] [CrossRef] [Green Version]
- Buszewicz, D.; Archacki, R.; Palusiński, A.; Kotliński, M.; Fogtman, A.; Iwanicka-Nowicka, R.; Sosnowska, K.; Kuciński, J.; Pupel, P.; Olędzki, J.; et al. HD2C Histone Deacetylase and a SWI/SNF Chromatin Remodelling Complex Interact and Both Are Involved in Mediating the Heat Stress Response in Arabidopsis. Plant Cell Environ. 2016, 39, 2108–2122. [Google Scholar] [CrossRef]
- Jarończyk, K.; Sosnowska, K.; Zaborowski, A.; Pupel, P.; Bucholc, M.; Małecka, E.; Siwirykow, N.; Stachula, P.; Iwanicka-Nowicka, R.; Koblowska, M.; et al. Bromodomain-Containing Subunits BRD1, BRD2, and BRD13 Are Required for Proper Functioning of SWI/SNF Complexes in Arabidopsis. Plant Commun. 2021, 2, 100174. [Google Scholar] [CrossRef]
- Li, C.; Gu, L.; Gao, L.; Chen, C.; Wei, C.-Q.; Qiu, Q.; Chien, C.-W.; Wang, S.; Jiang, L.; Ai, L.-F.; et al. Concerted Genomic Targeting of H3K27 Demethylase REF6 and Chromatin-Remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016, 48, 687–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vercruyssen, L.; Verkest, A.; Gonzalez, N.; Heyndrickx, K.S.; Eeckhout, D.; Han, S.-K.; Jégu, T.; Archacki, R.; Van Leene, J.; Andriankaja, M.; et al. ANGUSTIFOLIA3 Binds to SWI/SNF Chromatin Remodeling Complexes to Regulate Transcription during Arabidopsis Leaf Development. Plant Cell 2014, 26, 210–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Liang, Z.; Song, X.; Fu, W.; Xu, J.; Lei, Y.; Yuan, L.; Ruan, J.; Chen, C.; Fu, W.; et al. BRAHMA-Interacting Proteins BRIP1 and BRIP2 Are Core Subunits of Arabidopsis SWI/SNF Complexes. Nat. Plants 2020, 6, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-K.; Wu, M.-F.; Cui, S.; Wagner, D. Roles and Activities of Chromatin Remodeling ATPases in Plants. Plant J. 2015, 83, 62–77. [Google Scholar] [CrossRef] [Green Version]
- Ojolo, S.P.; Cao, S.; Priyadarshani, S.V.G.N.; Li, W.; Yan, M.; Aslam, M.; Zhao, H.; Qin, Y. Regulation of Plant Growth and Development: A Review from a Chromatin Remodeling Perspective. Front. Plant Sci. 2018, 9, 1232. [Google Scholar] [CrossRef] [Green Version]
- Reyes, J.C. The Many Faces of Plant SWI/SNF Complex. Mol. Plant 2014, 7, 454–458. [Google Scholar] [CrossRef] [Green Version]
- Sarnowska, E.; Gratkowska, D.M.; Sacharowski, S.P.; Cwiek, P.; Tohge, T.; Fernie, A.R.; Siedlecki, J.A.; Koncz, C.; Sarnowski, T.J. The Role of SWI/SNF Chromatin Remodeling Complexes in Hormone Crosstalk. Trends Plant Sci. 2016, 21, 594–608. [Google Scholar] [CrossRef]
- Yu, Y.; Fu, W.; Xu, J.; Lei, Y.; Song, X.; Liang, Z.; Zhu, T.; Liang, Y.; Hao, Y.; Yuan, L.; et al. Bromodomain-Containing Proteins BRD1, BRD2, and BRD13 Are Core Subunits of SWI/SNF Complexes and Vital for Their Genomic Targeting in Arabidopsis. Mol. Plant 2021, 14, 888–904. [Google Scholar] [CrossRef]
- Hernández-García, J.; Diego-Martin, B.; Kuo, P.H.; Jami-Alahmadi, Y.; Vashisht, A.A.; Wohlschlegel, J.; Jacobsen, S.E.; Blázquez, M.A.; Gallego-Bartolomé, J. Comprehensive Identification of SWI/SNF Complex Subunits Underpins Deep Eukaryotic Ancestry and Reveals New Plant Components. Commun. Biol. 2022, 5, 549. [Google Scholar] [CrossRef]
- Archacki, R.; Buszewicz, D.; Sarnowski, T.J.; Sarnowska, E.; Rolicka, A.T.; Tohge, T.; Fernie, A.R.; Jikumaru, Y.; Kotlinski, M.; Iwanicka-Nowicka, R.; et al. BRAHMA ATPase of the SWI/SNF Chromatin Remodeling Complex Acts as a Positive Regulator of Gibberellin-Mediated Responses in Arabidopsis. PLoS ONE 2013, 8, e58588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.-K.; Sang, Y.; Rodrigues, A.; BIOL425 F2010; Wu, M.-F.; Rodriguez, P.L.; Wagner, D. The SWI2/SNF2 Chromatin Remodeling ATPase BRAHMA Represses Abscisic Acid Responses in the Absence of the Stress Stimulus in Arabidopsis. Plant Cell 2012, 24, 4892–4906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarnowska, E.A.; Rolicka, A.T.; Bucior, E.; Cwiek, P.; Tohge, T.; Fernie, A.R.; Jikumaru, Y.; Kamiya, Y.; Franzen, R.; Schmelzer, E.; et al. DELLA-Interacting SWI3C Core Subunit of Switch/Sucrose Nonfermenting Chromatin Remodeling Complex Modulates Gibberellin Responses and Hormonal Cross Talk in Arabidopsis. Plant Physiol. 2013, 163, 305–317. [Google Scholar] [CrossRef] [Green Version]
- Brzeski, J.; Podstolski, W.; Olczak, K.; Jerzmanowski, A. Identification and Analysis of the Arabidopsis Thaliana BSH Gene, a Member of the SNF5 Gene Family. Nucleic Acids Res. 1999, 27, 2393–2399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Yuan, C.; Zhu, B.; Yuan, T.; Li, X.; Yuan, S.; Cui, S.; Zhao, H. LFR Physically and Genetically Interacts With SWI/SNF Component SWI3B to Regulate Leaf Blade Development in Arabidopsis. Front. Plant Sci. 2021, 12, 717649. [Google Scholar] [CrossRef]
- Alpsoy, A.; Dykhuizen, E.C. Glioma Tumor Suppressor Candidate Region Gene 1 (GLTSCR1) and Its Paralog GLTSCR1-like Form SWI/SNF Chromatin Remodeling Subcomplexes. J. Biol. Chem. 2018, 293, 3892–3903. [Google Scholar] [CrossRef] [Green Version]
- Mashtalir, N.; D’Avino, A.R.; Michel, B.C.; Luo, J.; Pan, J.; Otto, J.E.; Zullow, H.J.; McKenzie, Z.M.; Kubiak, R.L.; St Pierre, R.; et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 2018, 175, 1272–1288.e20. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, T.; Tsujimoto-Inui, Y.; Sotta, N.; Hirakawa, T.; Matsunaga, T.M.; Fukao, Y.; Matsunaga, S.; Fujiwara, T. Proteasomal Degradation of BRAHMA Promotes Boron Tolerance in Arabidopsis. Nat. Commun. 2018, 9, 5285. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Cai, G.; Li, Y.-Q.; Zhang, Y.-X.; Su, Y.-N.; Yuan, D.-Y.; Zhang, Z.-C.; Liu, Z.-Z.; Cai, X.-W.; Guo, J.; et al. Comprehensive Characterization of Three Classes of Arabidopsis SWI/SNF Chromatin Remodelling Complexes. Nat. Plants 2022, 8, 1423–1439. [Google Scholar] [CrossRef]
- Archacki, R.; Sarnowski, T.J.; Halibart-Puzio, J.; Brzeska, K.; Buszewicz, D.; Prymakowska-Bosak, M.; Koncz, C.; Jerzmanowski, A. Genetic Analysis of Functional Redundancy of BRM ATPase and ATSWI3C Subunits of Arabidopsis SWI/SNF Chromatin Remodelling Complexes. Planta 2009, 229, 1281–1292. [Google Scholar] [CrossRef]
- Dutta, A.; Sardiu, M.; Gogol, M.; Gilmore, J.; Zhang, D.; Florens, L.; Abmayr, S.M.; Washburn, M.P.; Workman, J.L. Composition and Function of Mutant Swi/Snf Complexes. Cell Rep. 2017, 18, 2124–2134. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.H.; Muiño, J.M.; Blanvillain, R.; Busscher, M.; Busscher-Lange, J.; Dinh, Q.D.P.; Liu, S.; Westphal, A.H.; Boeren, S.; et al. Characterization of MADS-Domain Transcription Factor Complexes in Arabidopsis Flower Development. Proc. Natl. Acad. Sci. USA 2012, 109, 1560–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, S.; Mano, S.; Tanaka, Y.; Ohnishi, M.; Nakamori, C.; Araki, M.; Niwa, T.; Nishimura, M.; Kaminaka, H.; Nakagawa, T.; et al. Gateway Binary Vectors with the Bialaphos Resistance Gene, Bar, as a Selection Marker for Plant Transformation. Biosci. Biotechnol. Biochem. 2010, 74, 1315–1319. [Google Scholar] [CrossRef] [Green Version]
- Archacki, R.; Yatusevich, R.; Buszewicz, D.; Krzyczmonik, K.; Patryn, J.; Iwanicka-Nowicka, R.; Biecek, P.; Wilczynski, B.; Koblowska, M.; Jerzmanowski, A.; et al. Arabidopsis SWI/SNF Chromatin Remodeling Complex Binds Both Promoters and Terminators to Regulate Gene Expression. Nucleic Acids Res. 2017, 45, 3116–3129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-Wide Identification and Testing of Superior Reference Genes for Transcript Normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]







| SWI/SNF Subunit | BRM-GFP/brm-1 | BRM-GFP/brm-1 brdx3 | BRM-GFP/brm-1 brd1/2 | BRM-GFP/brm-1 brd1/13 | BRM-GFP/brm-1 brd2/13 |
|---|---|---|---|---|---|
| BRM | 8/8 | 6/6 | 4/4 | 4/4 | 3/3 |
| ARP4 | 8/8 | 4/6 | 4/4 | 4/4 | 3/3 |
| ARP7 | 8/8 | 6/6 | 3/4 | 4/4 | 3/3 |
| SWP73B | 8/8 | 0/6 | 3/4 | 4/4 | 3/3 |
| SWI3C | 8/8 | 0/6 | 3/4 | 4/4 | 3/3 |
| BRIP2 | 8/8 | 0/6 | 0/4 | 4/4 | 3/3 |
| BRD1 | 8/8 | 0/6 | 0/4 | 0/4 | 3/3 |
| BRD2 | 4/8 | 0/6 | 0/4 | 4/4 | 0/3 |
| BRD13 | 8/8 | 0/6 | ¾ | 0/4 | 0/3 |
| SWP73A | 2/8 | 0/6 | 0/4 | 0/4 | 0/3 |
| BRIP1 | 2/8 | 0/6 | 0/4 | 0/4 | 2/3 |
| BDH2 | 4/8 | 2/6 | 1/4 | 1/4 | 3/3 |
| BDH1 | 1/8 | 0/6 | 1/4 | 0/4 | 0/3 |
| GIF1 | 0/8 | 0/6 | 0/4 | 0/4 | 0/3 |
| GIF2 | 1/8 | 0/6 | 0/4 | 0/4 | 1/3 |
| GIF3 | 1/8 | 0/6 | 0/4 | 0/4 | 0/3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stachula, P.; Kapela, K.; Malecka, E.; Jaronczyk, K.; Patryn, J.; Siwirykow, N.; Bucholc, M.; Marczak, M.; Kotlinski, M.; Archacki, R. BRM Complex in Arabidopsis Adopts ncBAF-like Composition and Requires BRD Subunits for Assembly and Stability. Int. J. Mol. Sci. 2023, 24, 3917. https://doi.org/10.3390/ijms24043917
Stachula P, Kapela K, Malecka E, Jaronczyk K, Patryn J, Siwirykow N, Bucholc M, Marczak M, Kotlinski M, Archacki R. BRM Complex in Arabidopsis Adopts ncBAF-like Composition and Requires BRD Subunits for Assembly and Stability. International Journal of Molecular Sciences. 2023; 24(4):3917. https://doi.org/10.3390/ijms24043917
Chicago/Turabian StyleStachula, Paulina, Katarzyna Kapela, Ewelina Malecka, Kamila Jaronczyk, Jacek Patryn, Nina Siwirykow, Maria Bucholc, Malgorzata Marczak, Maciej Kotlinski, and Rafal Archacki. 2023. "BRM Complex in Arabidopsis Adopts ncBAF-like Composition and Requires BRD Subunits for Assembly and Stability" International Journal of Molecular Sciences 24, no. 4: 3917. https://doi.org/10.3390/ijms24043917